Abstract
It is well established that transcription and alternative splicing events are functionally coupled during gene expression. Here, we report that protein arginine N-methyltransferase 6 (PRMT6) may play a key role in this coupling process by functioning as a transcriptional coactivator that can also regulate alternative splicing. PRMT6 coactivates the progesterone, glucocorticoid and oestrogen receptors in luciferase reporter assays in a hormone-dependent manner. In addition, small interfering RNA (siRNA) oligonucleotide duplex knockdown of PRMT6 disrupts oestrogen-stimulated transcription of endogenous GREB1 and progesterone receptor in MCF-7 breast cancer cells, demonstrating the importance of PRMT6 in hormone-dependent transcription. In contrast, the regulation of alternative splicing by PRMT6 is hormone independent. siRNA knockdown of PRMT6 increases the exon inclusion:skipping ratio of alternatively spliced exons in endogenous vascular endothelial growth factor and spleen tyrosine kinase RNA transcripts in both the presence and absence of oestrogen. These results demonstrate that PRMT6 has a dual role in regulating gene expression and that these two functions can occur independently of each other.
INTRODUCTION
Steroid hormone receptors (SHRs) and nuclear receptors (NRs) are members of a large superfamily of ligand-inducible transcription factors (1,2). SHRs/NRs activate transcription in a ligand-dependent manner by binding to hormone response elements in promoters and enhancer regions of target genes. DNA-bound SHRs/NRs recruit coactivators, which are often recruited to gene promoters in multi-protein complexes where they help to reorganize chromatin into a transcriptionally active state by remodelling nucleosomes or modifying histone tails (3,4). The p160/steroid receptor coactivator (SRC) family of SHR/NR coactivators is comprised of three related proteins: SRC-1, SRC-2/GRIP1/TIF2 and SRC-3/pCIP/ACTR/AIB1/RAC3/TRAM1. The p160/SRC coactivators bind directly to the carboxyl terminal activation function-2 domain of SHRs/NRs via three NR boxes, each consisting of an LXXLL motif (where L is a leucine and X is any amino acid), and recruit secondary coactivators via their activation domains (ADs) (5). The basic-helix–loop–helix/Per-Arnt-Sim (bHLH/PAS) domain of p160/SRC proteins binds to the secondary coactivators CoCoA, GAC63, hMMS19 and Fli-I (6–9); the AD1 binds to the acetyltransferases CREB-binding protein (CBP) and p300 (10,11); and the AD2 recruits the protein N-arginine methyltransferases (PRMTs) PRMT1 and coactivator associated arginine methyltransferase 1 (CARM1/PRMT4) (12,13).
PRMT1 and CARM1 both function as secondary coactivators for the oestrogen receptor-α (ERα), thyroid hormone receptor (TR) and androgen receptor (AR) (12,13). They require their enzymatic activity for this process, which results in the transfer of either one or two methyl groups from the methyl donor S-adenosyl-l-methionine to guanidino nitrogen atoms in arginine residues. (14,15). Methylation of arginine residues on histone tails is believed to play a major role in transcriptional activation by PRMTs (16–18). In addition, PRMTs have been demonstrated to methylate an increasing number of non-histone proteins involved in transcription (19,20). For example, methylation of various sites of CBP, p300 and RAC3 by CARM1 regulates the ability of these proteins to bind to other coactivators, and thus may play a role in controlling the assembly and disassembly of coactivator complexes on an active promoter (21–23). This regulation of coactivator complex formation means that CARM1 activity enhances the ability of other coactivators to promote transcription. Indeed, CARM1 acts synergistically with PRMT1, Fli-I, CoCoA, TIF1α, CCAR1, SRCAP, GAC63 or any of the three acetyltransferases p300, CBP or p/CAF to stimulate steroid hormone-dependent transcription (6,8,9,13,24–28). This demonstrates that PRMTs are an integral component of the steroid-hormone-signalling pathway and are required to fully activate steroid-hormone-regulated genes.
Transcriptional coactivators, including CoAA, p72, CAPERα and CAPERβ, Sam68, p68, ASC-1, ASC-2 and CARM1, have been found to also regulate pre-mRNA processing (29–34). Indeed, over 20 transcriptional coregulators are structurally or functionally related to known splicing factors (35). CARM1 is the only PRMT previously shown to directly regulate alternative splicing. CARM1 methylates the splicing factors CA150, SAP49, SmB and U1C and promotes skipping of both a splicing reporter and the endogenous CD44 gene in a methylation-dependent manner (31). The exact mechanism behind splicing regulation by CARM1 is unknown. However, arginine methylation can promote the assembly of small nuclear ribonucleoproteins (snRNPs), and regulate the localization of many RNA-binding proteins, such as heterogeneous nuclear ribonucleoproteins (hnRNPs) (36–39).
Protein arginine N-methyltransferase 6 (PRMT6) is a predominantly nuclear, type-I arginine methyltransferase, that has been demonstrated to methylate Histone 3 on arginine 2 (H3R2), H4R3 and H2AR3 (40–42). PRMT6 has also been found to methylate the HIV Tat, nucleocapsid and Rev proteins, which interferes with the ability of these proteins to interact with RNA (43–45). PRMT6 can also methylate the splicing factors RDA288 and hnRNP D (31). As PRMT6 targets are involved in both transcription and RNA processing, we investigated the ability of PRMT6 to regulate these events. Here, we report that PRMT6 coactivates SHR-dependent transcription in a hormone-dependent manner, and regulates alternative splicing by means of a hormone-independent mechanism.
MATERIALS AND METHODS
Plasmids
The pCDNA3.1-PR, pCDNA3.1-ERα, pCDNA3.1-ERβ, pCMX-β-galactosidase, pCMX-VP16-N, pCMX-GAL4-N, pG5E1b-luciferase, pGL3-E1b-luciferase, pGL3-PRE-E1b-luciferase, pGL3-ERE-E1b-luciferase, pSG5 (Stratagene), pCMX and pCM5 expression and reporter plasmids have all been described previously (29,30,46). The RARE-E1b-luciferase was constructed by inserting the sequence 5′-GTACCGGGTAGGGTTCACCGAAAGT TCACTCGACGGGTAGGGTTCACCGAAAGTTCACTCGAG-3′ containing two retinoic acid receptor (RAR) response elements and its corresponding complementary sequence into the Asp718/Nhe1 sites of pGL3-E1b-luciferase. The human SRC-1 complementary DNA (cDNA) was a gift from Dr William W. Chin and was cloned into a pCMX, pCMX-GAL4-N or pSG5-based expression vectors. The pSV-GR and pCMX-RARα plasmids were gifts from Dr David D. Moore. The pSG5-HA-CARM1 was a gift from Dr Michael R. Stallcup (12). The peroxisome proliferator activated receptor-γ (PPARγ) and the PPAR-TK-luciferase vectors were a gift from Dr Jon Whitehead. The cDNAs for human PRMT6 and human PRMT1 were obtained from the I.M.A.G.E. consortium (47). The PRMT6 and PRMT1 cDNA clones were sequenced and the cDNAs were cloned into a pCMX or pCM5-based expression vectors. The PRMT6 mutant containing the open reading frame of PRMT6 with amino acids 86 and 88 changed from valine/aspartic acid to lysine/alanine respectively (V86K/D88A), was produced by low-cycle polymerase chain reaction (PCR) using pfu-turbo DNA polymerase (Stratagene). A wild-type PRMT6 clone containing only the PRMT6 open reading frame was also produced by low-cycle PCR using pfu-turbo DNA polymerase and cloned into pCMX-VP16-N and pCMX or pCM5-based expression vectors. All PCR-generated clones were verified by sequencing and were tested by transfection and western blotting to ensure that they generate similar expression levels as vectors expressing wild-type proteins. The precise cloning steps for the clones used are available on request. The RHCglo minigene was a gift from Dr Thomas A. Cooper (48). To make the RHCglo-SYK-Exon 7 minigene, PCR primers targeting intron 6 (5′-GCGCGTCGACGCTCGGTGAGACAGATCCATA-3′) and intron 7 (5′-GCGCGGCTAGCCTCAGATTACACCTCTTCTTTAC-3′) were used with HeLa cell genomic DNA, PfuUltra II Fusion HS DNA polymerase and PCR to produce a human genomic DNA fragment encompassing 602 bp of intronic sequence upstream of Syk Exon 7, and 532 bp of intronic sequence downstream of Exon 7. This PCR product was digested with Sal1/Nhe1, agarose gel isolated and cloned into the Sal1/Xba1 sites of the RHCglo minigene reporter. The cloned Syk PCR sequence was confirmed by sequencing.
Cell culture and transfection
HeLa and CV-1 cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C and 5% CO2. For luciferase assays, HeLa or CV-1 cells were plated in 24-well plates and cultured for 24 h in DMEM without phenol red, supplemented with 10% charcoal-stripped FBS before transfection. Cells were transiently transfected at 60–70% confluence by using calcium phosphate precipitation with 100 ng reporter, 0.15–15 ng individual SHR/NR (15 ng ERα, 15 ng ERβ, 12 ng PR, 15 ng GR, 2.5 ng RARα or 15 ng PPARγ unless stated otherwise), 50 ng PRMT6, 50 ng PRMT6 V86K/D88A, 50 ng PRMT1, 50 ng CARM1, 100 ng SRC-1 as indicated and 10 ng β-galactosidase expression vectors per well. An appropriate amount of empty expression vector was used in assays without cofactor and the total amount of DNA was brought to 1 µg/well using pGEM4. Post-transfection, cells were washed with phosphate-buffered saline (PBS) and maintained in DMEM without phenol red plus 10% charcoal-stripped serum. The media was supplemented with the appropriate ligand as indicated: 10–8 M progesterone (Pg), 10–9 M estradiol (E2), 10–7 M all-trans retinoic acid (RA), 10–8 M Dexamethasone (Dex), 10–5 M Ciglitazone (Cig) or vehicle alone (ethanol). After 24–36 h incubation, cells were harvested and cell extracts assayed for luciferase and β-galactosidase activity on a Berthold luminometer. Mammalian-2-hybrid assays were performed in HeLa cells cultured in DMEM supplemented with 10% FBS at 37°C and 5% CO2. Mammalian-2-hybrid transfections were carried out using 100 ng pG5E1b-luciferase reporter, 25 ng Gal4 or Gal4-SRC-1, 300 ng VP16 or VP16-PRMT6 and 10 ng β-galactosidase expression vectors per well. After 24–36 h incubation, cells were harvested and cell extracts assayed for luciferase and β-galactosidase activity. The luciferase assay and Tropix β-galactosidase chemiluminescent assay were done according to the manufacturer’s directions (Applied Biosystems) and β-galactosidase activity was used as an internal control to monitor for transfection efficiency. For luciferase assays, data shown are the means and standard deviations (SD) of results from four transfected cultures.
Splicing assays using the RHCglo-Syk-Exon 7 minigene were performed by transiently transfecting HeLa cells grown to 50% confluency in 6-well plates with 400 ng of RHCglo-Syk-Exon 7 reporter, 75 ng of PRMT6, PRMT6 V86K/D88A, CARM1 or empty expression vector, and pGEM4 to bring up total DNA to 1.5 µg/well. Transfections were performed by using Lipofectamine™ 2000 (Invitrogen) as described by manufacturer’s instructions, and the cells were harvested 24 h post-transfection. RNA transcript quantification using γ-32P labelled primers and real-time (RT)-PCR were performed as previously described (29,33), using the RHCglo specific primers RSV5U and RTRHC as previously described (48) and RT-PCR without reverse transcriptase did not produce any detectable PCR products (data not shown). An autoradiograph of the radiolabeled RT-PCR products from a representative experiment is shown. Six replicate wells of cells were used for each experimental condition and all experiments were repeated twice.
MCF-7 cells were maintained in DMEM nutrient mixture F-12 plus 10% FBS. During experiments utilising hormone, cells were plated and maintained in phenol red-free DMEM nutrient mixture F-12 containing 10% charcoal-stripped serum at 37°C and 5% CO2. To knock down PRMT6 expression levels, MCF-7 cells were transfected with small interfering RNA (siRNA) oligonucleotide duplex at a final concentration of 10 nM using RNAiMAX (Invitrogen) according to manufacturer’s instructions. The annealed siRNA duplex sequences used were: PRMT6-siRNA-1, sense 5′-CGGGACCAGCUGUACUACGTT-3′ and PRMT6-siRNA-1, anti-sense 5′-CGUAGUACAGCUGGUCCCGTT-3′. A scrambled control was used as a negative control: Control-siRNA, sense 5′-CAGCGACUAAACACAUCAATT-3′ and Control-siRNA, anti-sense 5′-UUGAUGUGUUUAGUCGCUGTT-3′. After 48 h post-transfection, cells were treated for 12 h with 10–9 M E2 or vehicle control (ethanol) prior to harvesting RNA.
RNA extraction and quantitative RT–PCR
Total RNA was extracted from MCF-7 cells using Trizol Reagent (Invitrogen), according to manufacturer's; protocol. Total RNA for quantitative (Q)-RT-PCR was further purified by using RNeasy RNA mini-purification columns with on column DNase treatment according to manufacturer's; instructions (Qiagen). RNA was normalized using UV spectrophotometry and agarose gel electrophoresis. cDNA was synthesized from 600 ng of total RNA using TaqMan Reverse Transcription Reagents, according to the manufacturer's; instructions (Applied Biosystems). Gene expression levels were determined by Q-RT-PCR on a 7900HT Fast Real-Time PCR System (Applied Biosystems) using Assay on Demand Taqman primer/probes (Applied Biosystems); PRMT6 (Hs00250803_s1), CARM1/PRMT4 (Hs00406354_m1), PRMT1 (Hs01587651_g1), GREB1 (Hs00536409_m1), PR (Hs01556701_m1), ERα (Hs01046818_m1) and normalised to GAPDH (Cat#4326317E) and RPLP0 (Cat#4326314E) expression levels. Target cDNA levels were analyzed by Q-RT-PCR in 10 µl reactions using Taqman PCR master mix, Taqman probe/primer sets and cDNA (5% of the starting 600 ng of RNA). PCR was initiated at 95°C for 10 min to activate Amplitaq Gold DNA polymerase, followed by 45 cycles of 95°C for 15 s and 60°C for a 1-min two-step thermal cycling. Relative changes in gene expression were calculated using the ΔΔCt method. For each experiment, four replicate wells of cells were used for each experimental condition and all experiments were repeated twice.
In order to determine the effects of PRMT6 on splicing of the endogenous vascular endothelial growth factor (VEGF) nascent RNA, a modified version of the protocol described by Wellmann et al. (49) was used. In order to detect levels of total VEGF transcript (VEGFtotal) and three splice products VEGF121, VEGF165 and VEGF189, a common forward primer (5′-CCCTGATGAGATCGAGTACATCTT-3′) and common fluorescent hydrolysation probe (5′-ATCCTGTGTGCCCCTGATGCGATGCGGT-3′) both located on exon 3 were used (Supplementary Figure S6A). Specific amplification of each spliced product was achieved by using reverse primers spanning exon boundaries specific for the relevant product. Reverse primers used were: VEGF121 (5′-GCCTCGGCTTGTCACATTTT-3′), VEGF165 (5′-AGCAAGGCCCACAGGGATTT-3′), VEGF189 (5′-AACGCTCCAGGACTTATACCG-3′) and VEGFtotal (5′-ACCGCCTCGGCTTGTCAC-3′). Expression levels were normalized to GAPDH and RPLP0, and changes in the expression of the spliced variants was normalized to VEGFtotal. Relative changes in gene expression were calculated using the ΔΔCt method. Four replicate wells of cells were used for each experimental condition and all experiments were repeated twice.
Alterations in the splicing of spleen tyrosine kinase (Syk) were detected using a modified protocol described by Wang et al. (50). The primers used to amplify Syk (Syk[L]) and the shorter alternatively spliced isoform (Syk[S]) were Syk-for 5′-AATCGGCACACAGGGAAATG-3′ and Syk-rev 5′-AGCTTTCGGTCCAGGTAAAC-3′, and to amplify β2-microglobulin were β2-for 5′-GATGAGTATGCCTGCCGTGTG-3′ and β2-rev 5′-CAATCCAAATGCGGCATCT-3′. The primers were radiolabelled using 32P-γATP (3000 Ci/mmol) and T4 kinase (Invitrogen). To amplify the Syk and β2-microglobulin cDNA, PCR was conducted for 30 cycles (30 s at 94°C, 45 s at 56°C and 1 min at 68°C) and radioactive PCR products were separated on non-denaturing 5% polyacrylamide gels. Dried gels were subject to autoradiography and phosphorimaging using a Storm 860 phosphorimager and ImageQuant software (Molecular Dynamics). Levels of Syk alternatively spliced products were normalized to β2-microglobulin expression levels. An autoradiograph of the radiolabelled PCR products from a representative experiment is shown. Four replicate wells of cells were used for each experimental condition and all experiments were repeated twice. DNA products from the alternative splicing experiments were sub-cloned into pCR2.1 using the TOPO-TA cloning kit (Invitrogen) and DNA sequences confirmed by DNA sequencing on a 3730xl DNA Analyzer (Applied Biosystems).
Cell proliferation assays
Cell proliferation assays were performed in 96-well plates in phenol red-free DMEM nutrient mixture F-12 containing 10% charcoal-stripped serum. siRNA was transfected into MCF-7 cells (10 000 cells/well) using RNAiMAX (Invitrogen) according to manufacturer’s instructions. Cells were transfected either with 20 nM control siRNA, 10 nM siRNA targeting PRMT6 and/or 10 nM siRNA targeting CARM1; CARM1-siRNA-1, sense 5′-ACCAAAUGAUGUCCCUGCCTT-3′ and CARM1-siRNA-1, anti-sense 5′-GGCAGGGACAUCAUUUGGUTT-3′. When transfecting siRNA targeting either PRMT6 or CARM1, 10 nM control siRNA was added to make a final siRNA concentration of 20 nM. After 24 h, cells were treated with 10–9 M E2 or vehicle control (ethanol). The proliferation assay was conducted by incubating the cells with a final concentration of 10 µCi/ml 3H-thymidine for 19 h. The cells were then lysed and incorporated 3H-thymidine was harvested onto a filter mat (Wallac). The 3H-thymidine levels were counted using a 1450 MicroBeta TriLux scintillation counter (Wallac) according to manufacturer’s instructions.
Western blotting
Cellular extracts (20 µg) were separated by 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted/transferred on to polyvinylidene fluoride membrane (Sigma–Aldrich). Non-specific binding sites were blocked by immersing the membranes in 5% skim milk/TBS buffer (20 mM Tris–HCl, pH 7.6, 137 mM NaCl, 0.5% Tween 20) for 1–2 h at room temperature and the membrane was subsequently incubated with polyclonal PRMT6 (Sigma–Aldrich), CARM1 (Bethyl Laboratories), ERα (Santa Cruz Biotechnology) or β-tubulin (Sigma–Aldrich) antibodies in 1% skim milk/TBS at a concentration of 1 µg/ml O/N at 4°C, then washed three times in TBS containing 0.05% Tween 20. Further steps involving goat anti-rabbit or goat anti-mouse horseradish peroxidase-conjugated secondary antibodies (Invitrogen) as required and the catalysed oxidation of luminal were carried out with Pierce ECL western blotting detection reagents according to the manufacturer's; protocols. Western blot images were obtained and recorded using a CHEMI5000 chemiluminescence documentation system and Chemi-Capt software. Densitrometry was performed using Bio-profil Bio-1D software.
In vitro binding assay and chromatin immunoprecipitation assay
Glutathione S-transferase (GST) and GST-PRMT6 were isolated from Escherichia coli BL21 or JM109 cells following 5 h induction with 0.1 mM isopropyl thio-β-d-galactosidase. Cells were harvested in MTPBS buffer [150 mM NaCl, 16 mM Na2HPO4, 4 mM NaH2PO4 (pH 7.3) plus protease inhibitors (Roche)], sonicated and centrifuged at 3000g to remove cell debris. Five-hundred nanograms of GST-fusion proteins were isolated by incubation with GST beads (Amersham Biosciences) for 3 h with rotation at 4°C. Beads were washed five times in buffer C [20 mM HEPES (pH 7.6), 100 mM KCl, 0.2 mM EDTA, 20% (v/v) glycerol, 1 mM DTT plus protease inhibitors (Roche)] and resuspended in 200 µl HEMG buffer [100 mM KCl, 40 mM HEPES (pH 7.5), 0.2 mM EDTA, 0.1% NP-40 and 10% glycerol] plus 5%w/v glycine and 0.4 mg/ml BSA. 35S-methione labelled proteins were synthesized using TNT Quick Coupled Transcription/Translation System (Promega). In vitro binding assays were performed by incubating 10–20 µl 35S-methione labelled protein with GST-protein bound beads for 16 h at 4°C with rotation. Beads were then washed three times with HEMG buffer plus 5% w/v glycine and 0.4 mg/ml BSA and then twice in HEMG buffer alone. Proteins were eluted from the beads with Laemmli sample buffer and analysed by SDS-PAGE and autoradiography. All experiments were repeated twice. Chromatin immunoprecipitation (ChIP) was performed as previously described (51), using the MCF-7 breast cancer cell line and anti-PRMT6 monoclonal antibody (Sigma–Aldrich). To analyse the levels of recruitment of PRMT6 to the endogenous GREB1 and PR genes, immunoprecipitated chromatin was quantified by SYBR-green RT-PCR using previously described primer sequences (52–54).
Statistical analysis
Statistical analysis was performed using either one- or two-tailed Student’s t-tests as appropriate.
RESULTS
PRMT6 enhances steroid-hormone-dependent transcription
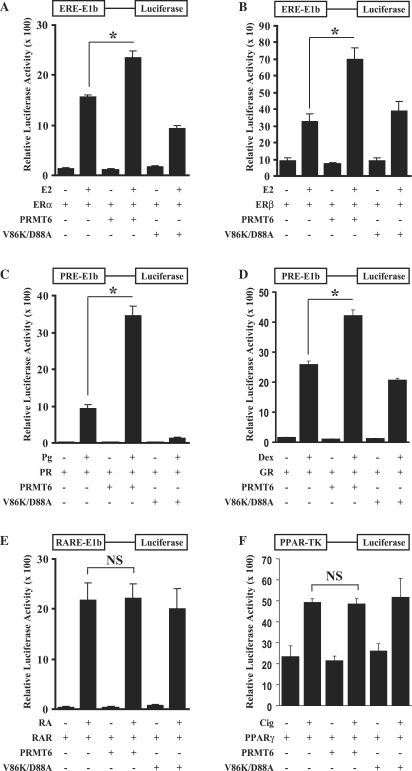
PRMT6 is a nuclear protein that is related to the SHR/NR coactivators PRMT1 and CARM1 (55). We therefore investigated whether PRMT6 can enhance the transcriptional activity of SHRs in transient transfection assays in mammalian cells (Supplementary Figure 1 and Figure 1). HeLa cells were transfected with a SHR/NR expression vector, an appropriate luciferase reporter and an expression vector for PRMT6 as indicated. As shown in Figure 1A, PRMT6 significantly increased the activity of ERα on an ERE-E1b-luciferase reporter in a ligand-dependent manner by ∼1.5-fold. In a similar experiment investigating the activity of ERβ, PRMT6 increased transcriptional activity by ∼2.1-fold (Figure 1B). Furthermore, PRMT6 enhanced the transcriptional activity of the progesterone receptor (PR) by ∼3.7-fold and the glucocorticoid receptor (GR) by ∼1.7-fold (Figures 1C and D). Similar results were also obtained in CV-1 cells (Supplementary Figure S1A–D). All increases in transcriptional activity were ligand-dependent, showing that PRMT6 acts as a coactivator for SHRs rather than a general transcription enhancer. In contrast, PRMT6 was unable to coactivate several non-steroidal NRs tested, including the RARα, PPARγ and TRβ (Figure 1E and F, and data not shown). These NRs were still responsive to other known coactivators, demonstrating that the reporter had not reached its maximal level in the presence of ligand-bound receptor (Supplementary Figure S2). Therefore, transcriptional coactivation stimulated by PRMT6 has specificity in relation to certain SHR/NRs.
Figure 1.
PRMT6 coactivates SHRs in HeLa cells. (A) PRMT6 coactivates ERα transcriptional activity from an oestrogen response element linked to a minimal promoter. HeLa cells were co-transfected with an ERE-E1b-luciferase reporter along with expression vectors for ERα alone, or with PRMT6 or PRMT6 V86K/D88A as indicated. Cells were treated with vehicle (ethanol) or 10−9 M E2 as indicated and tested for luciferase activity (see ‘Materials and Methods’ section). (B) PRMT6 coactivates ERβ transcriptional activity from the ERE-E1b-luciferase reporter. HeLa cells were co-transfected with an ERβ expression plasmid as in (A). (C) PRMT6 enhances the transcriptional activity of PR. HeLa cells were co-transfected with a PRE-E1b-luciferase reporter along with expression vectors for PR alone, or with PRMT6 or PRMT6 V86K/D88A mutant and treated with vehicle (ethanol) or 10−8 M Pg as indicated. (D) PRMT6 enhances the transcriptional activity of GR. HeLa cells were co-transfected with PRE-E1b-luciferase reporter along with expression vectors for GR alone, or PRMT6 or PRMT6 V86K/D88A mutant and treated with vehicle (ethanol) or 10−8 M Dex as indicated. (E) PRMT6 does not coactivate the RARα. HeLa cells were co-transfected with RARE-E1b-luciferase reporter along with expression vectors for RARα alone, or with PRMT6 or PRMT6 V86K/D88A mutant and treated with vehicle (ethanol) or 10–7 M RA as indicated. (F) PRMT6 does not coactivate PPARγ transcriptional activity. HeLa cells were co-transfected with a PPAR-TK-luciferase reporter along with expression vectors for PPARγ alone, or with PRMT6 or PRMT6 V86K/D88A mutant and treated with vehicle (ethanol) or with 10–5 M Cig as indicated. Each data point represents the mean and standard deviation (SD) of results from four transfected cultures. Results shown are from a single experiment, which is representative of three independent experiments. *P < 0.001; NS, no significant change.
PRMT6 requires its enzymatic activity to function as a coactivator
In order to determine the importance of arginine methylation on the ability of PRMT6 to function as a transcriptional coactivator, we engineered a previously described PRMT6 methylation-deficient mutant (VLD/KLA at amino acids 86/88) (43). In HeLa and CV-1 cells, the PRMT6 V86K/D88A mutant was unable to function as a coactivator for SHRs/NRs and did not coactivate ERα, ERβ, PR or GR (Figure 1A–D and Supplementary Figure S1A–D). In both HeLa and CV-1 cells, the PRMT6 V86K/D88A mutant functioned in a dominant-negative fashion by decreasing the transcriptional activity of the PR to lower than observed with the receptor alone. From these experiments, we determined that PRMT6 requires its enzymatic activity to function as a transcriptional coactivator for SHRs.
Mechanism of recruitment of PRMT6 to SHRs
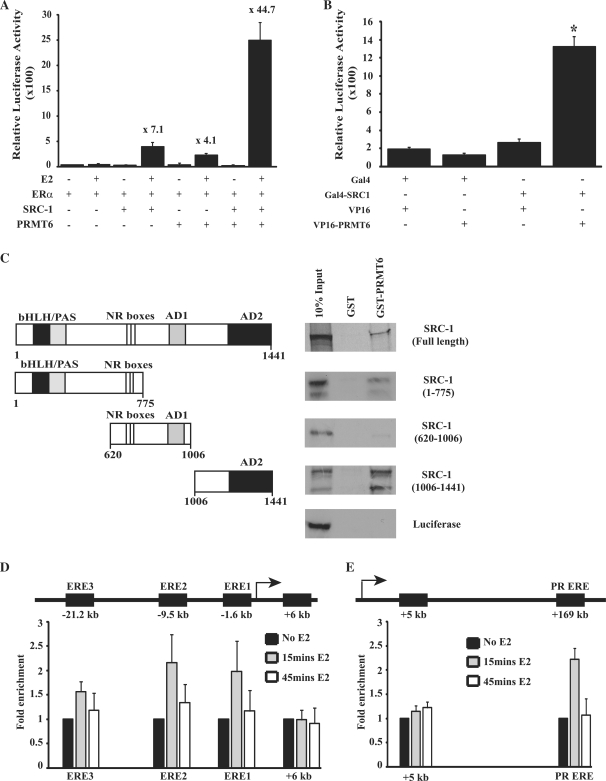
PRMT1 and CARM1 have both been shown to be secondary coactivators that are recruited to SHRs/NRs via the AD2 domains of p160/SRC proteins. It has been demonstrated that this leads to a synergistic response in transcriptional activation when either PRMT1 and CARM1 is co-transfected with a member of the p160/SRC family (12,13). In order to determine whether PRMT6 is also a secondary coactivator, we investigated the transcriptional response of ERα on an ERE-E1b-luciferase reporter in the presence of both PRMT6 and SRC-1 in CV-1 cells (Figure 2A). For this experiment, we reduced the levels of ERα to 5 ng as synergy between multiple PRMTs and p160/SRC proteins has been demonstrated to require low levels of SHR/NRs (13). This lower ERα level reduces the response of the reporter to ERα and E2 alone, while allowing greater levels of coactivation to be observed by exogenous coactivators. At this lower ERα level, SRC-1 increased transcriptional activity by ∼7.1-fold and PRMT6 by ∼4.1-fold compared to the response to ERα alone. However, SRC-1 and PRMT6 together lead to a synergistic enhancement of transcription of ∼44.7-fold. We therefore investigated the binding of PRMT6 to SRC-1 using a mammalian-two-hybrid assay. PRMT6 was cloned into pCMX-VP16-N to produce a VP16 transcriptional activation domain/PRMT6 chimera (VP16-PRMT6) and SRC-1 was cloned into pCMX-GAL4-N to produce a Gal4 DNA-binding domain/SRC-1 chimera (Gal4-SRC-1). Co-transfection of VP16-PRMT6 and Gal4-SRC-1 lead to a significant increase in luciferase activity compared to transfection of either plasmid alone, demonstrating that PRMT6 is able to associate with full-length SRC-1 within the context of mammalian cells (Figure 2B). This interaction was confirmed using GST-pull-down assays with GST-tagged PRMT6 and 35S-labelled SRC-1 (Figure 2C). Luciferase, used as a negative control, showed no interaction with PRMT6 in GST-pull-down assays. In order to determine which regions of SRC-1 interact with PRMT6, we divided the SRC-1 protein into three regions (Figure 2C). An amino-terminal region of SRC-1, containing the bHLH/PAS domain and the NR boxes, weakly interacted with PRMT6. The central region of SRC-1, from amino acids 620–1006, containing the NR boxes and AD1 domain, showed no interaction with PRMT6. However, PRMT6 did bind strongly to the carboxyl-terminal region of SRC-1 composed of amino acids 1006–1441, which contains the AD2 domain.
Figure 2.
PRMT6 synergistically coactivates ERα transcriptional activity in the presence of SRC-1. (A) CV-1 cells were transiently co-transfected with an ERE-E1b-luciferase reporter, 5-ng expression vector for ERα, along with expression vectors for PRMT6 or SRC-1 as indicated. Following treatment with vehicle (ethanol) or 10–9 M E2, cells were assayed for luciferase activity. Numbers above bars show fold increase in luciferase activity compared to transfection with ERα alone and 10–9M E2 stimulation. Each data point represents the mean and SD of results from four transfected cultures. Results shown are from a single experiment, which is representative of three independent experiments. (B) Mammalian-2-hybrid analysis demonstrates that PRMT6 interacts with full-length SRC-1. HeLa cells were co-transfected with pG5-E1b-luciferase reporter plasmid along with expression vectors for the Gal4 DNA-binding domain alone (Gal4), Gal4-SRC-1 chimera, the VP16 transcriptional activation domain alone (VP16) or VP16-PRMT6 chimera as indicated. Cell extracts were tested for luciferase activity. Each data point represents the mean and SD of results from four transfected cultures. Results shown are from a single experiment, which is representative of three independent experiments. *P < 0.001. (C) GST pull-down assays were used to test the ability of GST-PRMT6 to interact with full length or fragments of 35S-radiolabelled SRC-1. GST alone and in vitro translated 35S-radiolabelled luciferase served as negative controls. A schematic representation of the SRC-1 protein fragments used in the assays is shown in the panel, with amino acid positions of the SRC-1 protein or SRC-1 protein fragments indicated. The major functional domains of SRC-1 are indicated, bHLH/PAS, NR Boxes, AD1 and AD2. (D) Recruitment of PRMT6 to oestrogen response elements (EREs) located in promoter regions of the GREB1 gene. Following 0-, 15- and 45-min treatment of MCF-7 cells with 10−9 M E2, recruitment of PRMT6 to EREs in the GREB1 promoter and to a site downstream of the GREB1 transcriptional start site (+6 kb) was determined by chromatin immunoprecipitation as detailed in ‘Materials and Methods’ section. Results show the average and SD of four independent experiments. (E) Recruitment of PRMT6 to an oestrogen-receptor-binding enhancer region of the PR gene. Following 0-, 15- and 45-min treatment of MCF-7 cells with 10−9 M E2, recruitment of PRMT6 to a known oestrogen-receptor-binding enhancer region of the PR gene (PR ERE) and to a site downstream of the PR gene transcriptional start site (+5 kb) was determined by chromatin immunoprecipitation as detailed in ‘Materials and methods’ section. Results show the average and SD of four independent experiments.
We then investigated whether PRMT6 is recruited to an active oestrogen-dependent promoter in response to oestrogen. ERα binds to three EREs to activate transcription of the GREB1 gene, but does not bind at a region 6 kb downstream of the GREB1a transcription start site (52). We, therefore, investigated whether PRMT6 is recruited to these sites in response to oestrogen treatment using ChIP assays. Following 15 min oestrogen stimulation, PRMT6 was recruited to EREs in the GREB1 promoter (Figure 2D). This recruitment was cyclical and returned to baseline levels after 45 min oestrogen treatment. There was no recruitment of PRMT6 to the 6 kb downstream site, which served as a negative control. We then examined PRMT6 recruitment to a second oestrogen-dependent gene, the progesterone receptor. ERα binds to an enhancer located +169 kb downstream of the PR transcriptional start site (53,56). ChIP assays demonstrated that PRMT6 is recruited to this enhancer region following 15 min oestrogen stimulation (Figure 2E). Again, PRMT6 occupancy returned to baseline levels after 45 min oestrogen treatment. A region 5 kb downstream of the PR transcriptional start site, that does not show ERα binding (54), served as a negative control and showed no PRMT6 occupancy.
PRMT6 enhances transcriptional activation with CARM1 in the presence of SRC-1
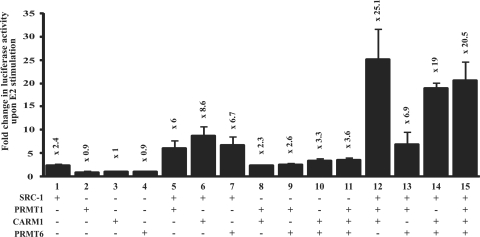
It has been previously demonstrated that transient transfection experiments using very low levels of SHR display a greater requirement for several coactivators (13). Under these conditions, CARM1 and PRMT1 cooperate to synergistically enhance transcription in the presence of a p160/SRC protein. In order to determine whether PRMT6 is also capable of cooperating with other PRMTs, we transfected CV-1 cells with a very low level of ERα (0.15 ng) and various combinations of PRMT1, CARM1, PRMT6 and SRC-1 in luciferase reporter assays (Figure 3). At this low level of SHR, none of the PRMTs alone could stimulate transcription, as observed in previous studies (13). This is due to the requirement of several coactivators to stimulate transcription at this level of SHR. However, co-transfection of PRMTs with SRC-1 led to an increase in ERα transcriptional activity, confirming the roles of PRMTs as secondary coactivators (Figure 3; compare lanes 5 to 2, 6 to 3 and 7 to 4). Co-transfecting combinations of two of the PRMTs together in the absence of SRC-1 had little effect on transcriptional activation. However, PRMT1 and CARM1 cooperated to stimulate ERα activity in the presence of SRC-1 (Figure 3, lane 12). Similar results were obtained when PRMT6 was substituted for PRMT1, indicating that PRMT6 acts cooperatively with CARM1 and SRC-1 to promote ERα-dependent transcription (Figure 3, lane 14). Interestingly, CARM1 was necessary for this cooperative effect, as the same stimulation was not seen when SRC-1, PRMT1 and PRMT6 were co-transfected (Figure 3, lane 13). In addition, transfecting all three PRMTs together gave a similar transcriptional response as transfecting CARM1 with either PRMT1 or PRMT6 (Figure 3, lane 15). Therefore, both PRMT1 and PRMT6 cooperate with CARM1 to stimulate ERα-dependent transcription, but do not cooperate together. Furthermore, this enhanced transcriptional response is dependent on the presence of SRC-1.
Figure 3.
PRMT6 and CARM1 synergistically coactivate ERα transcriptional activity in the presence of SRC-1. CV-1 cells were co-transfected with an ERE-E1b-luciferase reporter, 0.15-ng expression vector for ERα, and combinations of expression vectors for PRMT6, CARM1, PRMT1 or SRC-1 as indicated. Following oestrogen treatment, cells were assayed for luciferase activity. Numbers above bars show fold increase in luciferase activity upon 10−9 M E2 stimulation. Each data point represents the mean and SD of results from four transfected cultures. Results shown are from a single experiment, which is representative of three independent experiments.
PRMT6 is an integral component of the oestrogen-signalling pathway
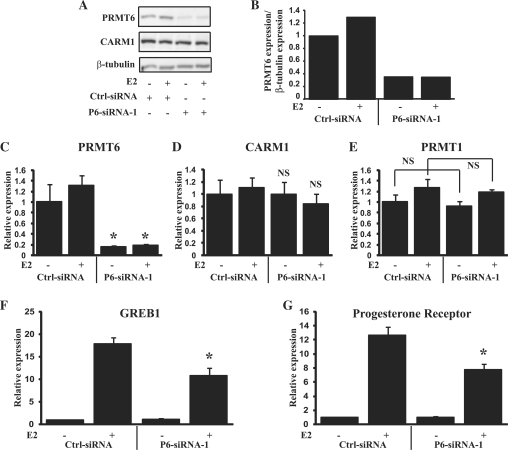
In order to determine the importance of PRMT6 in the transcription of hormone-responsive genes within mammalian cells, we used siRNA interference to knockdown PRMT6 in MCF-7 cells and determined the effects on transcription of endogenous oestrogen-regulated genes. Transfection of PRMT6-siRNA-1 into MCF-7 cells reduced PRMT6 levels by ∼80% at both the RNA and protein levels (Figure 4A–C). Importantly, knockdown by PRMT6-siRNA-1 was specific for PRMT6 and had no effect on the RNA levels of CARM1 and PRMT1, and no effect on the protein levels of CARM1 (Figure 4A, D and E). Knockdown of PRMT6 also had no effect on the levels of ERα at either the RNA or the protein level (Supplementary Figure S3). As PRMT6 is recruited to the promoter and enhancer regions of the GREB1 and PR genes (Figure 2D and E), we determined whether PRMT6 is required for the activation of these genes. Compared to treatment with a control siRNA, knockdown of PRMT6 had no significant effect on the transcription of GREB1 and PR in the absence of oestrogen (Figure 4F and G). However, the oestrogen-activated expression of both GREB1 and PR was significantly reduced by PRMT6 knockdown. This result establishes a role for PRMT6 in the activation of oestrogen-dependent transcription and the expression of endogenous oestrogen-regulated genes. A second PRMT6 siRNA targeting the 3′UTR of PRMT6 (PRMT6-siRNA-2) was also tested and produced similar results (Supplementary Figure S4).
Figure 4.
Knockdown of PRMT6 expression disrupts oestrogen signalling. (A) Western blot showing PRMT6, CARM1 and β-tubulin expression in MCF-7 cells following transfection with control siRNA (Ctrl-siRNA) or siRNA targeting PRMT6 (P6-siRNA-1) and treatment with or without 10−9 M E2 for 12 h as indicated. (B) Graphical representation of PRMT6 expression levels of western blot shown in (A). (C) Q-RT-PCR analysis of PRMT6 RNA levels in MCF-7 cells following transfection by control siRNA or siRNA targeting PRMT6 and treatment with or without 10−9 M E2 for 12 h as indicated. (D) CARM1 expression as detected in (C). (E) PRMT1 expression as detected in (C). (F) GREB1 expression levels following PRMT6 knockdown. RNA was analysed by Q-RT-PCR for expression of GREB1 as in (C). (G) PR expression levels following PRMT6 knockdown. PR levels were examined as in (C). Each data point represents the mean and SD of results from four transfected cultures. Results shown are from a single experiment, which is representative of two independent experiments. *P < 0.005; NS, no significant change compared to treatment with control siRNA.
PRMT6 mediates oestrogen-dependent proliferation
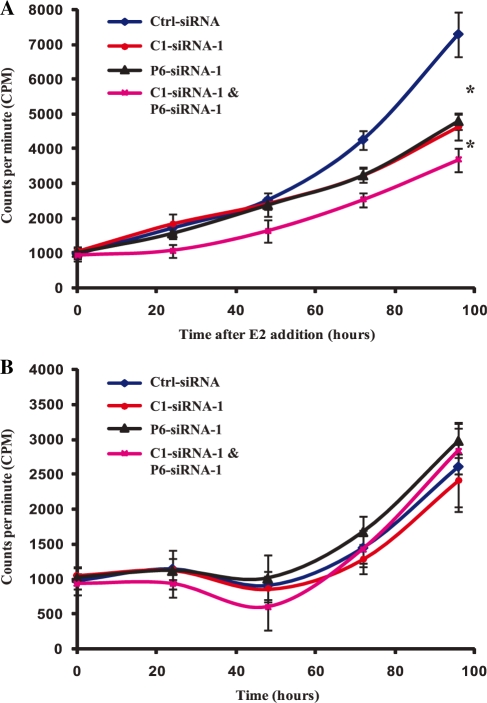
As PRMT6 plays a role in oestrogen signalling, we investigated whether PRMT6 is involved in the oestrogen-stimulated proliferation of ERα-expressing breast cancer cells. We used PRMT6-siRNA-1 to knockdown PRMT6 expression in MCF-7 cells and measured cellular proliferation following oestrogen treatment. As CARM1 is known to play a role in MCF-7 cell proliferation in response to oestrogen (57), we also knocked down CARM1 expression to allow the effects of these two PRMTs to be compared. In addition, as PRMT6 and CARM1 can function synergistically in oestrogen signalling (Figure 3), we also knocked down the two proteins together to see if they function cooperatively in regulating oestrogen-induced proliferation. The siRNA trigger targeting CARM1 reduced CARM1 expression by ∼80% without affecting PRMT6 or PRMT1 expression (Supplementary Figure S5A–C). Knockdown of either PRMT6 or CARM1 levels significantly reduced the proliferation of MCF-7 cells 72 and 96 h after oestrogen treatment compared to treatment with control siRNA. Reducing the levels of either PRMT6 or CARM1 resulted in a similar inhibition of oestrogen-stimulated proliferation (Figure 5A). In addition, treating the cells with siRNAs targeting both PRMT6 and CARM1 together had an additive effect, significantly lowering proliferation when compared to treatment with either siRNA individually. These effects were oestrogen dependent, as knocking down PRMT6 and CARM1 either individually or together had no effect on MCF-7 proliferation in the absence of oestrogen (Figure 5B). Similar results were also obtained using P6-siRNA-2 (Supplementary Figure S5D and E). Therefore, PRMT6 is an integral component of the oestrogen-signalling pathway, and is required to stimulate both oestrogen-dependent transcription and cellular proliferation.
Figure 5.
Knockdown of PRMT6 and CARM1 expression inhibits oestrogen-stimulated proliferation of breast cancer cells. (A) MCF-7 cells were transfected with control siRNA (Ctrl-siRNA) or siRNA targeting PRMT6 (P6-siRNA-1), CARM1 (C1-siRNA-1) or both PRMT6 and CARM1 (P6-siRNA-1 and C1-siRNA-1). Following treatment with 10−9 M E2, cell proliferation was determined by 3H-thymidine incorporation. (B) MCF-7 cells were transfected as in (A) and cell proliferation was determined by 3H-thymidine incorporation in the absence of 10−9 M E2. Each data point represents the mean and SD of results from eight individual cultures. Results are shown from a single experiment, which is representative of two independent experiments. *P < 0.001.
PRMT6 regulates alternative splicing
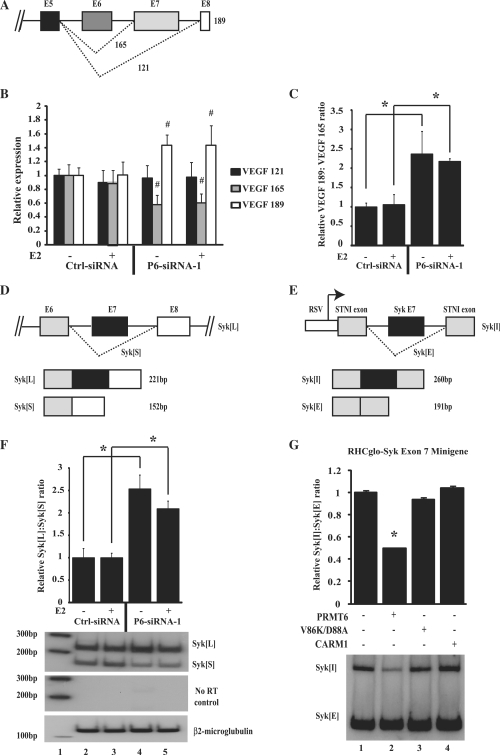
To investigate the ability of PRMT6 to regulate alternative splicing, we examined the splicing of the VEGF gene following knockdown of PRMT6 in MCF-7 cells. Alternative splicing of VEGF produces several major spliced forms of mRNA (58). We used Q-RT-PCR analysis to determine the relative levels of three of these isoforms: full-length VEGF (VEGF189), VEGF following skipping of exon 6 (VEGF165) and VEGF following skipping of exons 6 and 7 (VEGF121) (Figure 6A). MCF-7 cells were transfected with either control siRNA or PRMT6-siRNA-1 and knockdown of PRMT6 was verified by Q-RT-PCR (Supplementary Figure S6B–D). Whilst reduction of PRMT6 transcript had no effect on levels of VEGF121, it did produce a significant increase in the levels of VEGF189 and a decrease in the levels of VEGF165 compared to treatment with control siRNA (Figure 6B). This leads to a >2-fold increase in the VEGF189:VEGF165 ratio (Figure 6C). This change was observed in both the presence and absence of oestrogen, despite the transcription of VEGF being oestrogen-regulated (Supplementary Figure S6H). Therefore, the regulation of alternative splicing of VEGF by PRMT6 does not require steroid hormone stimulation. A similar change in VEGF splicing was observed following PRMT6 knockdown by a different siRNA targeting PRMT6 (PRMT6-siRNA-2) (Supplementary Figure S6E–G and S6I-K). These results suggest that PRMT6 can play a role in the alternative splicing of endogenous genes in MCF-7 cells, and in contrast to its effects on transcription, the regulation of alternative splicing is hormone-independent. As CARM1 can also regulate alternative splicing in a steroid hormone-independent manner, we investigated whether PRMT6 and CARM1 are redundant for splicing of VEGF. Two siRNAs were designed to significantly reduce the levels of CARM1, but not PRMT6 or PRMT1 (Supplementary Figure S7A–C). Similar to PRMT6, knockdown of CARM1 in MCF-7 cells had no effect on VEGF121 and significantly reduced the levels of VEGF165 (Supplementary Figure S7E). However, in contrast to PRMT6, CARM1 knockdown had no effect on the levels of VEGF189 (Supplementary Figure S7E). Therefore, PRMT6 and CARM1 display similar but not redundant effects on the splicing of VEGF.
Figure 6.
PRMT6 regulates alternative splicing of endogenous VEGF and Syk. (A) Schematic representation of the major spliced products of the VEGF gene (not to scale). (B) Effect of PRMT6 knockdown on alternative splicing of VEGF. MCF-7 cells were transfected either with control siRNA (Ctrl-siRNA) or siRNA targeting PRMT6 (P6-siRNA-1) and treated either with or without 10−9 M E2 for 12 h as indicated. RNA was harvested from the cells and the relative expression level of each spliced isoform was determined by Q-RT-PCR analysis as detailed in ‘Materials and Methods’ section. (C) The relative VEGF 189:VEGF 165 ratio was obtained by dividing the relative VEGF 189 cDNA level by the relative VEGF 165 cDNA level for each experimental condition. (D) Schematic representation of the major alternatively spliced products of the endogenous Syk gene (not to scale). (E) Representation of the RHCglo-Syk-Exon 7 minigene construct, not to scale. (F) Effect of PRMT6 knockdown on splicing of endogenous Syk. MCF-7 cells were transfected either with control siRNA (Ctrl-siRNA) or siRNA targeting PRMT6 (P6-siRNA-1) and treated either with or without 10–9 M E2 for 12 h as indicated. RNA was harvested from the cells and Syk splicing was determined by reverse transcriptase-PCR. The relative Syk[L]:Syk[S] ratio was obtained by dividing the relative Syk[L] cDNA level by the relative Syk[S] cDNA level for each experimental condition. An autoradiograph of the radiolabeled Syk and β-2-microglobulin reverse transcriptase-PCR products from a representative experiment is shown. Lane 1; radiolabelled 100-bp DNA marker; lane 2; Ctrl-siRNA without 10−9 M E2; lane 3; Ctrl-siRNA with 10−9 M E2, lane 4; P6-siRNA-1 without 10−9 M E2; lane 5; P6-siRNA-1 with 10−9 M E2. (G) Effect of PRMT6 on the splicing of an RHCglo-Syk-Exon 7 minigene. HeLa cells were transiently co-transfected with a RHCglo-Syk-Exon 7 minigene along with expression vectors for PRMT6, the PRMT6 V86K/D88A mutant or CARM1. RNA was harvested from the cells and Syk splicing determined by reversetranscriptase-PCR. The relative RHCglo-Syk-Exon 7 inclusion (Syk[I]): RHCglo-Syk-Exon 7 exclusion (Syk[E]) ratio was obtained by dividing the relative Syk[I] cDNA level by the relative Syk[E] cDNA level for each experimental condition. RT-PCR without reverse transcriptase did not produce any detectable PCR products (data not shown). A representative autoradiograph of the radiolabelled RHCglo-Syk-Exon 7 minigene products is shown. For all experiments, each data point represents the mean and SD of results from four transfected cultures (or six transfected cultures for the RHCglo-Syk-Exon 7 minigene experiment). Results shown are from a single experiment, which is representative of two independent experiments. *P < 0.005, #P < 0.05.
To further investigate the ability of PRMT6 to regulate RNA processing, we examined the effect of PRMT6 on the alternative splicing of Syk. Syk is expressed in breast tissue and has been implicated to play a role in breast cancer (59). Two alternatively spliced isoforms of Syk have been documented (Figure 6D): full-length Syk (Syk[L]) and a shorter form that lacks a 69-bp sequence comprising exon 7 (Syk[S]) (60–62). In order to investigate the regulation of alternative splicing of the endogenous Syk gene by PRMT6, PRMT6 levels were reduced in MCF-7 cells using PRMT6-siRNA-1. The two different Syk transcripts were then detected by reverse transcriptase-PCR with 32P-radiolabelled primers that span the alternative exon of Syk[L]. PRMT6 knockdown led to a significant increase in the Syk[L]:Syk[S] ratio (Figure 6F and Supplementary Figure S8A). A similar result was seen using a second siRNA directed against PRMT6 (PRMT6-siRNA-2) (Supplementary Figure S8B). Taqman Q-RT-PCR using primers and probes designed to detect the alternative isoforms of Syk gave similar results (Supplementary Figure S8C and D). As with the alternative splicing of VEGF, the effects of PRMT6 on alternative splicing of Syk were hormone independent. In order to investigate the effects of CARM1 on alternative splicing of endogenous Syk, two siRNAs were employed that specifically target CARM1 (Supplementary Figure S7A–C). Following reduction of CARM1 levels in MCF-7 cells, Taqman Q-RT-PCR was used to detect the endogenous levels of the Syk isoforms. CARM1 knockdown did not influence splicing of the endogenous Syk RNA transcripts (Supplementary Figure S8E). Therefore, as with the VEGF gene, PRMT6 and CARM1 are not redundant in the splicing of Syk.
In order to verify the observed effects of PRMT6 and CARM1 on the splicing of Syk, we developed a minigene containing the alternatively spliced exon 7 of the Syk gene (Figure 6E). We transfected this minigene into HeLa cells along with PRMT6, PRMT6 V86K/D88A mutant or CARM1 and determined the levels of the Syk isoforms by reverse transcriptase-PCR with 32P-radiolabelled primers. Over-expression of PRMT6 significantly reduced the Syk Exon 7 Inclusion:Exclusion ratio to ∼50% of control levels (Figure 6G). This is in agreement with our data showing that knocking down PRMT6 leads to an approximate 2-fold increase in the Syk[L]:Syk[S] ratio (Figure 6F). The PRMT6 V86K/D88A mutant had no effect on the splicing of the RHCglo-Syk-Exon 7 minigene, demonstrating that PRMT6 requires its enzymatic activity to regulate RNA processing (Figure 6G). In addition, CARM1 had no affect on the alternative splicing of the RHCglo-Syk-Exon 7 minigene, as observed with the splicing of the endogenous Syk gene (Figure 6G and Supplementary Figure S8E). This validates our findings on the regulation of the endogenous Syk gene, and demonstrates that PRMT6 and CARM1 regulate the alternative splicing of a different subset of genes.
DISCUSSION
Coupling of RNA polymerase-II-mediated transcription and RNA processing is a well-established concept (63). These two processes can be functionally linked by the activity of SHR/NR coactivators. This was first demonstrated by the PPARγ coactivator PGC-1, which can regulate alternative splicing of the fibronectin minigene (64). Further investigations have demonstrated that other coactivators, including CoAA, p72, Sam68, p68, ASC-1 and ASC-2, can play this dual role, whilst the CAPER α and β proteins have been shown to regulate both transcription and alternative splicing in a SHR-dependent manner (29,30,34,65). Recently, CARM1, a PRMT known to function as a SHR/NR coactivator, has been shown to influence alternative splicing (31). Here, we report that another PRMT protein, PRMT6, is a SHR/NR coactivator that can regulate both hormone-dependent transcription and hormone-independent alternative splicing.
We have demonstrated that PRMT6 can function as a coactivator for ERα, ERβ, PR and GR (Figure 1 and Supplementary Figure S1). Similar to PRMT1 and CARM1, we also demonstrate that PRMT6 requires its enzymatic activity to function as a coactivator (Figure 1 and Supplementary Figure S1). We have further characterized the involvement of PRMT6 on the expression of two oestrogen-stimulated genes, GREB1 and PR. PRMT6 is cyclically recruited to the promoter and enhancer regions of these genes in response to oestrogen (Figure 2D and E). The recruitment of PRMT6 was detected at these sites following 15 min oestrogen treatment, but had returned to baseline levels after 45 min. PRMT6 is also required to fully activate these genes, as knockdown of PRMT6 leads to a decrease in their oestrogen-stimulated transcription (Figure 4). This demonstrates that PRMT6 is an integral component of the oestrogen-signalling pathway and is required to fully activate hormone-dependent transcription.
Even though PRMT6 can coactivate SHRs in the absence of other exogenous coactivators, the mode of action of PRMT6 is similar to that of PRMT1 and CARM1, in that it functions as a secondary coactivator to p160/SRC proteins. PRMT6 binds to the AD2 domain of SRC-1, and functions synergistically with SRC-1 to coactivate ERα (Figure 2A–C). Therefore, PRMT6 can be added to the list of SHR coactivators that are able to interact with SRC-1 to enhance transcriptional activation. Two PRMTs previously identified to interact with the p160/SRC proteins, PRMT1 and CARM1, also function synergistically to coactivate SHRs in luciferase reporter experiments (13). In order to determine whether PRMT6 also acts in concert with these PRMTs to stimulate gene expression, we transfected combinations of PRMT6, PRMT1, CARM1 and SRC-1 into CV-1 cells. We found that PRMT6 acts synergistically with CARM1 but not PRMT1, and synergy was dependent on the presence of SRC-1 (Figure 3). PRMT6 and PRMT1 share methylation targets, and so it is possible that methylation of a common target by either PRMT6 or PRMT1 could be required for CARM1 to further activate transcription. However, PRMT6 and PRMT1 are not redundant in stimulating hormone-induced gene activation, as siRNA knockdown of PRMT6 alone leads to a significant decrease in oestrogen-induced GREB1 and PR expression (Figure 4).
Potential methylation targets that may be responsible for PRMT6 coactivation include H4R3, a target that is shared with PRMT1 and is methylated as an early step in oestrogen-induced gene activation (40,66,67). Other targets include the high-mobility group (HMG) A1 proteins HMGA1a and HMGA1b (68–70). These non-histone chromosomal proteins are involved in chromatin structure organization and gene transcription, although the effect of arginine methylation on these proteins is not currently known. Another PRMT6 methylation target is H3R2, which has been reported to be a repressive histone mark (40–42); however, in certain conditions both of the known coactivators PRMT1 and CARM1 can methylate this histone residue (16,17,41,71). H3R2 methylation has been shown to prevent transcription primarily by preventing the mixed lineage leukaemia (MLL) K4 methyltransferase unit binding to and methylating H3K4 (40,42). However, this mechanism only represses a defined subset of genes that are regulated by the MLL complex (40). It is known that transcription of oestrogen-regulated genes can occur independently of MLL1, as siRNA targeting MLL1 has little effect on transcription of the prototypical oestrogen-regulated gene pS2 (72). Further evidence that H3K4 methylation is not required for steroid hormone-activated transcription is provided by the fact that the H3K4 demethylase LSD1 is recruited to the GREB1 enhancer site and is required for oestrogen-dependent GREB1 transcription (72). Indeed, 58% of ERα-enriched promoters examined by Garcia-Bassets et al. (72) showed recruitment of LSD1, and both glucocorticoid and DHT stimulation lead to a decrease in dimethylation of H3K4 on a tandem array of the mouse mammary tumour virus promoter and on the PSA promoter respectively (17,73). Therefore, in the case of steroid-hormone-induced gene expression it appears that H3K4 demethylation does not prevent, and may even promote transcription. One example of H3K4 demethylation being an activating histone mark has been provided by the autoimmune regulator, which binds to non-methylated H3K4 and promotes transcription (74). Further studies will have to be conducted to determine the complete role of H3R2 and H3K4 methylation in steroid-hormone-activated transcription.
As PRMT6 can regulate oestrogen-dependent transcription; we investigated whether it is also involved in the oestrogen-stimulated proliferation of ER + breast cancer cells. Reducing PRMT6 levels by siRNA interference significantly inhibited the oestrogen-dependent, but not oestrogen-independent, proliferation of MCF-7 cells (Figure 5). This demonstrates that PRMT6 is a fundamental component of the oestrogen-signalling pathway in breast cancer cells. This inhibition of proliferation was similar to that obtained by knocking down CARM1, which has previously been demonstrated to inhibit the oestrogen-stimulated proliferation of MCF-7 cells (57) (Figure 5). As PRMT6 and CARM1 can cooperate to stimulate ERα-dependent transcription (Figure 3), we investigated the effects of knocking down both PRMTs simultaneously on oestrogen-dependent proliferation. Reducing the levels of both PRMT6 and CARM1 together further reduced oestrogen-driven MCF-7 cell proliferation when compared to knocking down either PRMT alone. Therefore, both PRMT6 and CARM1 are required to stimulate oestrogen-dependent proliferation of breast cancer cells, and their roles are not redundant in this process.
As well as regulating transcriptional initiation, we have shown that PRMT6 can also affect aspects of RNA processing, specifically alternative splicing. The siRNA knockdown of PRMT6 altered the relative levels of alternatively spliced products of the endogenous VEGF and Syk genes (Figure 6). In both cases, PRMT6 knockdown leads to an increase in exon inclusion, and a decrease in exon skipping. Therefore, it would appear that one cellular function of PRMT6 is to increase skipping of alternative exons. The effects of PRMT6 on alternative splicing of both VEGF and Syk were steroid hormone independent, despite the transcription of VEGF being oestrogen-dependent (Supplementary Figure S6H). CARM1, the only other PRMT identified to have a direct role in alternative splicing, also regulates alternative splicing in a hormone-independent manner (31). Unlike several coactivators involved in coupling transcription and alternative splicing, such as PGC-1, CoAA and CAPERα/β, both PRMT6 and CARM1 have none of the domains characteristic of splicing factors such as RNA-recognition motifs and serine-arginine-rich domains. As PRMT6 requires its enzymatic activity to regulate the splicing of Syk (Figure 6G), the most likely method by which it regulates alternative splicing is by methylating other splicing proteins. In the study by Cheng et al. (31), CARM1 and PRMT6 were found to have different substrate specificities in relation to the methylation of various splicing-related proteins. Therefore, we investigated whether PRMT6 and CARM1 have non-redundant roles in the regulation of alternative splicing. Both PRMTs regulate the production of the VEGF165 isoform. However, only PRMT6 regulates the production of the VEGF189 alternatively spliced product (Figure 6B and Supplementary Figure S7E). In addition, CARM1 has no effect on the splicing of the Syk gene (Figures 6G and Supplementary Figure S8E). Therefore, CARM1 and PRMT6 play a similar yet non-redundant role in the regulation of alternative splicing. In conclusion, this study provides evidence that PRMT6 has a pleiotropic role in multiple aspects of gene expression, and is able to couple transcription and alternative splicing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Health and Medical Research Council of Australia project grant (to D.H.D.); ANZ Trustees PhD scholarship in Medical Research (to M.J.H.). Funding for open access charge: National Health and Medical Research Council of Australia.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Tsai MJ, O'M;alley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu. Rev. Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 2.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 3.McKenna NJ, O'M;alley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 4.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 5.Stallcup MR, Kim JH, Teyssier C, Lee Y.-H, Ma H, Chen D. The roles of protein–protein interactions and protein methylation in transcriptional activation by nuclear receptors and their coactivators. J. Steroid Biochem. Mol. Biol. 2003;85:139–145. doi: 10.1016/s0960-0760(03)00222-x. [DOI] [PubMed] [Google Scholar]
- 6.Chen YH, Kim JH, Stallcup MR. GAC63, a GRIP1-dependent nuclear receptor coactivator. Mol. Cell. Biol. 2005;25:5965–5972. doi: 10.1128/MCB.25.14.5965-5972.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X, Li H, Chen JD. The human homologue of the yeast DNA repair and TFIIH regulator MMS19 Is an AF-1-specific coactivator of estrogen receptor. J. Biol. Chem. 2001;276:23962–23968. doi: 10.1074/jbc.M101041200. [DOI] [PubMed] [Google Scholar]
- 8.Kim JH, Li H, Stallcup MR. CoCoA, a nuclear receptor coactivator which acts through an N-terminal activation domain of p160 coactivators. Mol. Cell. 2003;12:1537–1549. doi: 10.1016/s1097-2765(03)00450-7. [DOI] [PubMed] [Google Scholar]
- 9.Lee YH, Campbell HD, Stallcup MR. Developmentally essential protein flightless I is a nuclear receptor coactivator with actin binding activity. Mol. Cell. Biol. 2004;24:2103–2117. doi: 10.1128/MCB.24.5.2103-2117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 11.Yao TP, Ku G, Zhou N, Scully R, Livingston DM. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc. Natl Acad. Sci. USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen D, Ma H, Hong H, Koh SS, Huang S.-M, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 13.Koh SS, Chen D, Lee Y.-H, Stallcup MR. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J. Biol. Chem. 2001;276:1089–1098. doi: 10.1074/jbc.M004228200. [DOI] [PubMed] [Google Scholar]
- 14.Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- 15.Wolf S. The protein arginine methyltransferase family: an update about function, new perspectives and the physiological role in humans. Cell. Mol. Life Sci. 2009;66:2109–2121. doi: 10.1007/s00018-009-0010-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 2002;3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma H, Baumann CT, Li H, Strahl BD, Rice R, Jelinek MA, Aswad DW, Allis CD, Hager GL, Stallcup MR. Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr. Biol. 2001;11:1981–1985. doi: 10.1016/s0960-9822(01)00600-5. [DOI] [PubMed] [Google Scholar]
- 18.Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson BL, Henschen-Edman A, Mackay DR, Stallcup MR, Aswad DW. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry. 2001;40:5747–5756. doi: 10.1021/bi002631b. [DOI] [PubMed] [Google Scholar]
- 19.Lee YH, Stallcup MR. Minireview: protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol. Endocrinol. 2009;23:425–433. doi: 10.1210/me.2008-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chevillard-Briet M, Trouche D, Vandel L. Control of CBP co-activating activity by arginine methylation. EMBO J. 2002;21:5457–5466. doi: 10.1093/emboj/cdf548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc. Natl Acad. Sci. USA. 2005;102:3611–3616. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naeem H, Cheng D, Zhao Q, Underhill C, Tini M, Bedford MT, Torchia J. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol. Cell. Biol. 2007;27:120–134. doi: 10.1128/MCB.00815-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng Q, Yi P, Wong J, O'M;alley BW. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol. Cell. Biol. 2006;26:7846–7857. doi: 10.1128/MCB.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee YH, Koh SS, Zhang X, Cheng X, Stallcup MR. Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities. Mol. Cell. Biol. 2002;22:3621–3632. doi: 10.1128/MCB.22.11.3621-3632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monroy MA, Schott NM, Cox L, Chen JD, Ruh M, Chrivia JC. SNF2-related CBP activator protein (SRCAP) functions as a coactivator of steroid receptor-mediated transcription through synergistic interactions with CARM-1 and GRIP-1. Mol. Endocrinol. 2003;17:2519–2528. doi: 10.1210/me.2003-0208. [DOI] [PubMed] [Google Scholar]
- 26.Chen D, Huang S.-M, Stallcup MR. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J. Biol. Chem. 2000;275:40810–40816. doi: 10.1074/jbc.M005459200. [DOI] [PubMed] [Google Scholar]
- 27.Teyssier C, Ou C.-Y, Khetchoumian K, Losson R, Stallcup MR. Transcriptional intermediary factor 1α mediates physical interaction and functional synergy between the coactivator-associated arginine methyltransferase 1 and glucocorticoid receptor-interacting protein 1 nuclear receptor coactivators. Mol. Endocrinol. 2006;20:1276–1286. doi: 10.1210/me.2005-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim JH, Yang CK, Heo K, Roeder RG, An W, Stallcup MR. CCAR1, a key regulator of mediator complex recruitment to nuclear receptor transcription complexes. Mol. Cell. 2008;31:510–519. doi: 10.1016/j.molcel.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auboeuf D, Honig A, Berget SM, O'M;alley BW. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science. 2002;298:416–419. doi: 10.1126/science.1073734. [DOI] [PubMed] [Google Scholar]
- 30.Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, Berget SM, O'M;alley BW. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERα and CAPERβ. Mol. Cell. 2005;17:429–439. doi: 10.1016/j.molcel.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 31.Cheng D, Cote J, Shaaban S, Bedford MT. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell. 2007;25:71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 32.Rajan P, Gaughan L, Dalgliesh C, El-Sherif A, Robson CN, Leung HY, Elliott DJ. The RNA-binding and adaptor protein Sam68 modulates signal-dependent splicing and transcriptional activity of the androgen receptor. J. Pathol. 2008;215:67–77. doi: 10.1002/path.2324. [DOI] [PubMed] [Google Scholar]
- 33.Auboeuf D, Dowhan DH, Kang YK, Larkin K, Lee JW, Berget SM, O'M;alley BW. Differential recruitment of nuclear receptor coactivators may determine alternative RNA splice site choice in target genes. Proc. Natl Acad. Sci. USA. 2004;101:2270–2274. doi: 10.1073/pnas.0308133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark EL, Coulson A, Dalgliesh C, Rajan P, Nicol SM, Fleming S, Heer R, Gaughan L, Leung HY, Elliott DJ, et al. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res. 2008;68:7938–7946. doi: 10.1158/0008-5472.CAN-08-0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Auboeuf D, Dowhan DH, Dutertre M, Martin N, Berget SM, O'M;alley BW. A subset of nuclear receptor coregulators act as coupling proteins during synthesis and maturation of RNA ranscripts. Mol. Cell. Biol. 2005;25:5307–5316. doi: 10.1128/MCB.25.13.5307-5316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herrmann F, Bossert M, Schwander A, Akgun E, Fackelmayer FO. Arginine methylation of scaffold attachment factor A by heterogeneous nuclear ribonucleoprotein particle-associated PRMT1. J. Biol. Chem. 2004;279:48774–48779. doi: 10.1074/jbc.M407332200. [DOI] [PubMed] [Google Scholar]
- 37.Smith WA, Schurter BT, Wong-Staal F, David M. Arginine methylation of RNA helicase a determines its subcellular localization. J. Biol. Chem. 2004;279:22795–22798. doi: 10.1074/jbc.C300512200. [DOI] [PubMed] [Google Scholar]
- 38.Cote J, Boisvert FM, Boulanger MC, Bedford MT, Richard S. Sam68 RNA binding protein is an in vivo substrate for protein arginine N-methyltransferase 1. Mol. Biol. Cell. 2003;14:274–287. doi: 10.1091/mbc.E02-08-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nichols RC, Wang XW, Tang J, Hamilton BJ, High FA, Herschman HR, Rigby WF. The RGG domain in hnRNP A2 affects subcellular localization. Exp. Cell Res. 2000;256:522–532. doi: 10.1006/excr.2000.4827. [DOI] [PubMed] [Google Scholar]
- 40.Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer U.-M. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007;21:3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iberg AN, Espejo A, Cheng D, Kim D, Michaud-Levesque J, Richard S, Bedford MT. Arginine methylation of the histone H3 tail impedes effector binding. J. Biol. Chem. 2008;283:3006–3010. doi: 10.1074/jbc.C700192200. [DOI] [PubMed] [Google Scholar]
- 42.Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Luscher B, Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 43.Boulanger MC, Liang C, Russell RS, Lin R, Bedford MT, Wainberg MA, Richard S. Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J. Virol. 2005;79:124–131. doi: 10.1128/JVI.79.1.124-131.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Invernizzi CF, Xie B, Richard S, Wainberg MA. PRMT6 diminishes HIV-1 Rev binding to and export of viral RNA. Retrovirology. 2006;3:93. doi: 10.1186/1742-4690-3-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Invernizzi CF, Xie B, Frankel FA, Feldhammer M, Roy BB, Richard S, Wainberg MA. Arginine methylation of the HIV-1 nucleocapsid protein results in its diminished function. AIDS. 2007;21:795–805. doi: 10.1097/QAD.0b013e32803277ae. [DOI] [PubMed] [Google Scholar]
- 46.Chen SL, Dowhan DH, Hosking BM, Muscat G.EO. The steroid receptor coactivator, GRIP-1, is necessary for MEF-2C-dependent gene expression and skeletal muscle differentiation. Genes Dev. 2000;14:1209–1228. [PMC free article] [PubMed] [Google Scholar]
- 47.Lennon G, Auffray C, Polymeropoulos M, Soares MB. The I.M.A.G.E. Consortium: an Integrated Molecular Analysis of Genomes and Their Expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 48.Singh G, Cooper TA. Minigene reporter for identification and analysis of cis elements and trans factors affecting pre-mRNA splicing. Biotechniques. 2006;41:177–181. doi: 10.2144/000112208. [DOI] [PubMed] [Google Scholar]
- 49.Wellmann S, Taube T, Paal K, Graf v. Einsiedel H, Geilen W, Seifert G, Eckert C, Henze G, Seeger K. Specific reverse transcription-PCR quantification of vascular endothelial growth factor (VEGF) splice variants by LightCycler technology. Clin. Chem. 2001;47:654–660. [PubMed] [Google Scholar]
- 50.Wang L, Duke L, Zhang PS, Arlinghaus RB, Symmans WF, Sahin A, Mendez R, Dai JL. Alternative splicing disrupts a nuclear localization signal in spleen tyrosine kinase that is required for invasion suppression in breast cancer. Cancer Res. 2003;63:4724–4730. [PubMed] [Google Scholar]
- 51.Li X, Wong J, Tsai SY, Tsai MJ, O'M;alley BW. Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification. Mol. Cell. Biol. 2003;23:3763–3773. doi: 10.1128/MCB.23.11.3763-3773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun J, Nawaz Z, Slingerland JM. Long-range activation of GREB1 by estrogen receptor via three distal consensus estrogen-responsive elements in breast cancer cells. Mol. Endocrinol. 2007;21:2651–2662. doi: 10.1210/me.2007-0082. [DOI] [PubMed] [Google Scholar]
- 53.Subramanian K, Jia D, Kapoor-Vazirani P, Powell DR, Collins RE, Sharma D, Peng J, Cheng X, Vertino PM. Regulation of estrogen receptor α by the SET7 lysine methyltransferase. Mol. Cell. 2008;30:336–347. doi: 10.1016/j.molcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu X, Murdoch FE, Curran EM, Welshons WV, Fritsch MK. Transcription factor accessibility and histone acetylation of the progesterone receptor gene differs between parental MCF-7 cells and a subline that has lost progesterone receptor expression. Gene. 2004;328:143–151. doi: 10.1016/j.gene.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 55.Frankel A, Yadav N, Lee J, Branscombe TL, Clarke S, Bedford MT. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J. Biol. Chem. 2002;277:3537–3543. doi: 10.1074/jbc.M108786200. [DOI] [PubMed] [Google Scholar]
- 56.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 57.Frietze S, Lupien M, Silver PA, Brown M. CARM1 Regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res. 2008;68:301–306. doi: 10.1158/0008-5472.CAN-07-1983. [DOI] [PubMed] [Google Scholar]
- 58.Tischer E, Mitchell R, Hartman T, Silva M, Gospodarowicz D, Fiddes JC, Abraham JA. The human gene for vascular endothelial growth factor. Multiple protein forms are encoded through alternative exon splicing. J. Biol. Chem. 1991;266:11947–11954. [PubMed] [Google Scholar]
- 59.Zhang X, Shrikhande U, Alicie BM, Zhou Q, Geahlen RL. Role of the protein tyrosine kinase Syk in regulating cell–cell adhesion and motility in breast cancer cells. Mol. Cancer Res. 2009;7:634–644. doi: 10.1158/1541-7786.MCR-08-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yagi S, Suzuki K, Hasegawa A, Okumura K, Ra CS. Cloning of the cDNA for the deleted Syk kinase homologous to ZAP-70 from human basophilic leukemia cell line (KU812) Biochem. Biophys. Res. Com. 1994;200:28–34. doi: 10.1006/bbrc.1994.1409. [DOI] [PubMed] [Google Scholar]
- 61.Latour S, Chow L.ML, Veillette A. Differential intrinsic enzymatic activity of Syk and Zap-70 protein-tyrosine kinases. J. Biol. Chem. 1996;271:22782–22790. doi: 10.1074/jbc.271.37.22782. [DOI] [PubMed] [Google Scholar]
- 62.Rowley RB, Bolen JB, Fargnoli J. Molecular cloning of rodent p72Syk. Evidence of alternative mRNA splicing. J. Biol. Chem. 1995;270:12659–12664. doi: 10.1074/jbc.270.21.12659. [DOI] [PubMed] [Google Scholar]
- 63.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 64.Monsalve M, Wu Z, Adelmant G, Puigserver P, Fan M, Spiegelman BM. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell. 2000;6:307–316. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 65.Auboeuf D, Dowhan DH, Li X, Larkin K, Ko L, Berget SM, O'M;alley BW. CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol. Cell. Biol. 2004;24:442–453. doi: 10.1128/MCB.24.1.442-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strahl BD, Briggs SD, Brame CJ, Caldwell JA, Koh SS, Ma H, Cook RG, Shabanowitz J, Hunt DF, Stallcup MR, et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- 67.Wagner S, Weber S, Kleinschmidt MA, Nagata K, Bauer U.-M. SET-mediated promoter hypoacetylation is a prerequisite for coactivation of the estrogen-responsive pS2 gene by PRMT1. J. Biol. Chem. 2006;281:27242–27250. doi: 10.1074/jbc.M605172200. [DOI] [PubMed] [Google Scholar]
- 68.Miranda TB, Webb KJ, Edberg DD, Reeves R, Clarke S. Protein arginine methyltransferase 6 specifically methylates the nonhistone chromatin protein HMGA1a. Biochem. Biophys. Res. Com. 2005;336:831–835. doi: 10.1016/j.bbrc.2005.08.179. [DOI] [PubMed] [Google Scholar]
- 69.Sgarra R, Lee J, Tessari MA, Altamura S, Spolaore B, Giancotti V, Bedford MT, Manfioletti G. The AT-hook of the chromatin architectural transcription factor high mobility group A1a is arginine-methylated by protein arginine methyltransferase 6. J. Biol. Chem. 2006;281:3764–3772. doi: 10.1074/jbc.M510231200. [DOI] [PubMed] [Google Scholar]
- 70.Zou Y, Webb K, Perna AD, Zhang Q, Clarke S, Wang Y. A mass spectrometric study on the in vitro methylation of HMGA1a and HMGA1b proteins by PRMTs: methylation specificity, the effect of binding to AT-rich duplex DNA, and the effect of C-terminal phosphorylation. Biochemistry. 2007;46:7896–7906. doi: 10.1021/bi6024897. [DOI] [PubMed] [Google Scholar]
- 71.Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J, Escoubet-Lozach L, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim J, Jia L, Tilley WD, Coetzee GA. Dynamic methylation of histone H3 at lysine 4 in transcriptional regulation by the androgen receptor. Nucleic Acids Res. 2003;31:6741–6747. doi: 10.1093/nar/gkg909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Org T, Chignola F, Hetenyi C, Gaetani M, Rebane A, Liiv I, Maran U, Mollica L, Bottomley MJ, Musco G, et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 2008;9:370–376. doi: 10.1038/embor.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.