Abstract
Gene promoters are enriched in guanine clusters that potentially fold into quadruplex structures. Such quadruplexes were implicated in the regulation of gene expression, plausibly by interacting with transcription factors. We showed previously that homodimers of the myogenic transcription factor MyoD bound in vitro most tightly bimolecular quadruplexes of promoter sequences of muscle-specific genes. By contrast, MyoD-E47 heterodimers formed tighter complexes with d(CANNTG) E-box motifs that govern muscle gene expression. Here, we show that DNA quadruplexes enhance in vivo MyoD and E-box-driven expression of a firefly luciferase (FL) reporter gene. HEK293 cells were transfected with FL expressing p4RTK-FL vector alone or together with MyoD expressing pEMSV-MyoD plasmid, with quadruplexes of α7 integrin or sarcomeric mitochondrial creatine kinase (sMtCK) muscle gene promoters or with a combination thereof. Whereas MyoD elevated by ∼10-fold the levels of FL mRNA and protein, the DNA quadruplexes by themselves did not affect FL expression. However, together with MyoD, quadruplex DNA increased by ∼35-fold the amounts of FL mRNA and protein. Without affecting its expression, DNA quadruplexes bound MyoD in the cells. Based on these results, we propose models for the regulation of muscle gene transcription by direct interaction of MyoD with promoter quadruplex structures.
INTRODUCTION
Skeletal muscle develops during vertebrate embryogenesis from progenitor cells originating in the somites. These cells migrate into the limb bud where they proliferate, express myogenic determination factors and differentiate into skeletal muscle. In an initial determination stage, external signals released by surrounding cells induce differentiation of mesodermal cells into dividing myoblasts that become committed to the myogeneic lineage. The myoblasts are subsequently induced by other external signals to differentiate into non-dividing myocytes that ultimately fuse to generate syncitial myotubes (1–3). Four myogenic regulatory factors (MRFs): MyoD, Myf5, MRF4 and Myogenin (Myf4), control the development of muscle tissue (4,5). These basic helix–loop–helix (bHLH) master transcription factors induce timed expression of numerous muscle-specific genes. MyoD, Myf5 and MRF4 direct the transformation of mesenchimal cells into dividing myoblasts (6) whereas their subsequent progression into non-dividing myocytes and then to myotubes are mediated by Myf4 and MRF4 (7,8). The different MRFs appear to have distinct but partly intersecting functions as each transcription factor induces the expression of particular but partially overlapping sets of muscle-specific proteins (9,10).
Mammalian MRFs self-associate through their HLH sections to form homodimers or generate heterodimers with Class I bHLH E-proteins: HEB, E2-2, and two alternative splice products of E2A: E12 and E47 (11).
MyoD-E protein heterodimers initiate the expression of muscle proteins by binding through the basic segments of their bHLH domains to conserved E-box motifs, d(CANNTG), in regulatory regions of the muscle-specific genes (12). In contrast to the transcriptionally effective MyoD-E-protein heterodimers, homodimers of full-length MyoD displayed significantly lower affinity for E-box and acted as inefficient transcription factors (13,14). Although E-box elements are found in promoter and enhancer regions of both muscle-specific and non-muscle genes, MRF heterodimers selectively induce the expression of muscle genes but not of E-box regulated non-muscle genes (11). This specificity of gene activation was attributed to a number of factors such as the presence of three unique amino acids in the basic regions of MRFs: Ala114, Thr115 and Lys124 (12,15,16), the identity of the two variable residues d(CANNTG) within E-box (17), the effects of cis-acting suppressor elements (18) and the interaction of MRFs with additional transcription factors.
The activity and specificity of MRFs might also be determined by their differential interaction with non-canonical DNA structures in regulatory regions of muscle-specific genes. Of particular interest are quadruplex structures of guanine-rich promoter sequences. Promoters and 5′ untranslated tracts of genes are preferentially and highly enriched in clusters of contiguous guanine residues that can potentially form quadruplex structures (19–22). Indeed, in-depth studies of selected genes implicated tetrahelical DNA formations in the negative or positive regulation of the insulin gene (23–27), c-MYC (28–32), c-kit (33–35), bcl-2 (36,37), VEGF (38–40) and PDGF-A (41). Promoters and enhancers of several muscle-specific genes were shown to display a statistically disproportional high incidence of contiguous guanines clusters that readily formed G′4 unimolecular and G′2 bimolecular quadruplex structures (42). In addition, association into tetraplex of two non-adjacent guanine-rich hairpins within a single-strand raised the possibility that unimolecular G′2-like quadruplexes might also be formed (42) [see ref. (43) for a model] . Most interestingly, homodimeric MyoD and MRF4 were shown to bind in vitro the G′2 and G′2-like quadruplex structures in preference over double-stranded E-box whereas their respective heterodimers with E47 bound more tightly to E-box than to the tetraplex formations (44,45). Interestingly, however, whereas Myf4-E47 heterodimers also associated preferentially with E-box, homodimers of Myf4 in distinction from MyoD or MRF4 homodimers, bound tetraplex DNA weakly and non-preferentially (46). Yet when the Myf4 basic region was replaced by a MyoD basic region, the homodimeric chimerical Myf4 associated with quadruplex DNA tightly and in preference over E-box, suggesting that the basic region dictated the high affinity of MyoD homodimers for quadruplex DNA (46). Based on these in vitro finding we suggested that the distinct activities of MyoD or MRF4 versus Myf4 during myogenesis might be in part a result of the dissimilar affinities of their homodimers for quadruplex structures in regulatory regions of muscle-specific genes.
In the present work we assessed the in vivo impact of the high binding affinity of homodimeric MyoD for quadruplex DNA structures of promoter sequences of muscle-specific genes. To this end we examined in living cultured cells the effect of the quadruplex structures on MyoD- and E-box-governed expression of firefly luciferase (FL) reporter gene. We show that transfection of G′2 tetraplex structures of promoter sequences of the muscle-specific genes α7 integrin or sarcomeric mitochondrial creatine kinase (sMtCK) into MyoD expressing human embryonic kidney 293 (HEK293) cells specifically enhanced the MyoD and E-box-driven transcription and expression of a reporter FL gene. We also report that without affecting the expression of MyoD, the quadruplex DNA structures, but not single-strands of the same sequence, existed in complexes with MyoD in the cells. Based on these results we propose a model for the role of quadruplex DNA structures in the regulation of the MyoD-dependent expression of muscle-specific genes.
MATERIALS AND METHODS
Plasmids
The following plasmids were used: p4RTK-FL vector containing four copies of an E-box motif that mediated MyoD-dependent expression of FL; pCMV-RL that encoded Renilla reniformis luciferase (RL) gene (Promega); pEMSV-MyoD vector that expressed a harbored MyoD encoding gene and pCMV2-Flag carrier plasmid. The plasmids were prepared and propagated as previously detailed (47).
Formation of G′2 bimolecular quadruplex and single-stranded DNA structures
Quadruplex structures were formed by oligomers that corresponded to guanine-rich promoter tracts of two MyoD-responsive muscle-specific genes (guanine clusters highlighted): α7 integrin; 5′-d(CATGGGGGCGGGAAGGGGCGGGGTCT)-3′ and sMtCK; 5′-d(CTGAGGAGGGGCTGGAGGGACCAC)-3′. These oligomers fold in the presence of K+ ions into G′2 bimolecular quadruplex structures (42). Briefly, 6 μg of either 8 M urea–10% polyacrylamide denaturing gel electrophoretically purified sMtCK DNA oligomer (48) or HPLC purified integrin DNA (Sigma/Genosys) were dissolved in 10 μl of 300 mM KCl in TE buffer (10 mM Tris–HCl buffer, pH 8.0, 1 mM EDTA), boiled for 5 min and incubated at 37°C for 16 h. Electrophoresis in 10% non-denaturing polyacrylamide gel of aliquots of each oligomer after the incubation showed that ∼40–50% of the DNA was converted into slowly migrating G′2 bimolecular quadruplex forms [see also ref. (36)]. Solutions of the quadruplex DNA structures in TE buffer, 300 mM KCl were stored at −20°C until used and remained stable for 1–2 months.
Single-strands of sMtCK or integrin DNA or of adenosine-substituted integrin sequence: A-integrin; 5′-d(CATGGGAGCGAGAAGGAAGAAGTCT)-3′ that could not form quadruplexes (unpublished results), were generated immediately prior to use by 10 min boiling of DNA oligomers solutions in water.
Transfection of cultured human cells
HEK293 cells were inoculated in 0.1% gelatin-coated 10 cm plates and grown to 60–70% confluence at 37°C and under 5% CO2 atmosphere in Dulbecco Modified Eagle’s Medium (DMEM) supplemented with 4.5 g/l d-glucose, 5.0 mM l-glutamine, 10% fetal calf serum, 83.3 U/ml each of penicillin and streptomycin and 0.2 mg/ml amphotericin B (Biological Industries, Israel). The cells were detached by Trypsin–EDTA, reseeded in gelatin-coated 6-well plates at 3–5 × 105 cells/well in a volume of 2.5 ml/well and immediately transfected by different combinations of three plasmids as detailed under ‘Results’ section: p4RTK-FL—an E-boxes containing MyoD-responsive FL reporter plasmid; pCMV-RL—a normalizing RL reporter plasmid; and pEMSV-MyoD—a MyoD expressing vector. The plasmids were co-transfected without or with G′2 quadruplex structures of integrin or sMtCK DNA sequences or with their single-stranded forms. Briefly, 6 µl of jetPEI DNA transfection reagent (Polyplus-Transfection) in 100 µl of 150 mM NaCl was added to an equal volume of 150 mM NaCl that contained 0.4 μg 4RTK-FL DNA, 0.05 μg pCMV-RL DNA and varying indicated amounts of pESMV-MyoD vector. pCMV2-Flag DNA was used to complete the total amount of DNA to 1.95 μg. To assess the effect of quadruplex DNA on MyoD-dependent FL expression, 1.2 μg of G′2 integrin or sMtCK DNA or their single-stranded forms were included in parallel transfection mixtures of the above plasmids. Following incubation at 37°C for 2 h, each well was supplemented with 2.0 ml of growth medium. The described procedure allowed for transfection efficiency of >90% as assessed by the parallel monitoring of GFP expression in cells that were similarly transfected with a GFP harboring vector. The cells were harvested 24 h post-transfection using Trypsin–EDTA and resuspended in 1.0 ml of cold growth medium. Aliquots of each cell sample were used to determine FL and RL activities and to conduct semi-quantitative RT-PCR measurements of the levels of their mRNA transcripts as detailed below.
Adjustment for variations in cell viability and transfection efficiency were made as described (47), except that the RL-corrected FL activity was normalized relative to FL activity of cells that were transfected with FL and RL reporter vectors and the pCMV2-Flag plasmid but without a MyoD expressing plasmid or quadruplex DNA.
Dual luciferase assay
A dual luciferase reporter assay system was applied as instructed by the manufacturer (Promega) to determine the activities of FL and RL in lysates of transfected HEK293 cells. Briefly, following cell lysis in passive lysis buffer (Promega), 40 μl of cell lysate were added to 50 μl luciferase reagent II. After a 2 s delay, FL activity was measured for 10 s in a Glomax 20/20 luminometer. To terminate the reaction, 50 μl of Stop and Glo reagent were added to quench the FL activity and after a 2 s delay the activity of RL was determined for 10 s.
Quantitative RT–PCR measurement of relative mRNA levels
Total cell RNA was isolated from the HEK293 cells 24 h after transfection using a total RNA purification kit (Norgen Biotech). Remaining residues of cell and plasmid DNA were digested by DNase (Turbo DNA-free kit, Ambion). Spectrophotometrically measured (Nanovue, GE) RNA samples of 0.5 or 0.75 μg each were reverse-transcribed (Verso cDNA kit, Thermo Fisher Scientific) using anchored oligo dT primers. To verify that all the amplification products were copies of the RNA template and not of contaminating DNA, every set of reactions included a no RT negative control reaction mixture. Quantitative RT-PCR measurements of relative mRNA levels were conducted in a Rotor Gene 6000 analyzer (Corbett Life Science). Reaction mixtures contained in a final volume of 20 μl; 5 μl of 1 : 10 diluted cDNA, 1 × ABsolute Blue SYBR Green mix (Thermo Fisher Scientific) and 170 μM of each primer. The FL forward and reverse primers were, respectively, 5′-d(CTCACTGAGACTACATCAGC)-3′ and 5′-d(TCCAGATCCACAACCTTCGC)-3′ and the respective RL forward and reverse primers were 5′-d(GGAATTATAATGCTTATCTACGTGC)-3′ and 5′-d(CTTGCGAAAAATGAAGACCTTTTAC)-3′. Mixtures were incubated at 95°C for 15 min, followed by 40 cycles of: 10 s at 95°C, 15 s at 60°C and 20 s at 72°C. Three independent qRT-PCR measurements of RL-normalized amounts of FL cDNA were made for each of the two starting amounts of 0.5 and 0.75 μg RNA (a total of six measurements of each mRNA isolate). Relative amounts of FL mRNA were calculated by the comparative CT method and expressed as levels relative to FL mRNA measured in cells that were co-transfected with the pCMV2-flag p4RTK-FL, pCMV-RL and plasmids but without a pEMSV-MyoD expression vector.
Western analysis
HEK293 cells were co-transfected with p4RTK-FL, pCMV-RL and pEMSV-MyoD plasmids without or with G′2 quadruplex integrin DNA as detailed above. The cells were harvested and lysed 24 h post transfection and equal amounts of cell lysate protein were resolved by 10% SDS–PAGE and transferred to nitrocellulose membrane. Amounts of expressed MyoD were assessed by blotting with rabbit polyclonal anti MyoD primary antibody followed by goat anti rabbit IgG secondary antibody (both Santa Cruz products). Horseradish peroxidase activity was monitored using a Super We Pico chemiluminescence substrate (Pierce).
Immunoprecipitation
To assess the extent of interaction of MyoD with G′2 quadruplex DNA within the transfected cells, HEK293 cells were co-transfected as described above with a pEMSV-MyoD plasmid and 0.6 μg 5′-32P labeled (49) G′2 quadruplex sMtCK or integrin DNA or with the single-stranded forms of these oligomers or of A-integrin DNA. The cells were harvested 24 h post-transfection and lysed in 50 μl passive lysis buffer and 25 μl aliquots were incubated at 4°C overnight with either 10 μl of rabbit polyclonal anti MyoD or non-immune rabbit IgG (both Santa Cruz products). Twenty microliters of 1:1 slurry of protein A-coupled sepharose beads (Santa Cruz) that was thrice washed with phosphate buffered saline (PBS) were added to the mixtures and the reaction volume was completed to 1 ml with PBS. Following incubation under rotation at 4°C for 1 h, the mixtures were centrifuged for 3 min at 900g and 4°C, the supernatant was decanted and the beads were similarly washed three more times with 1 ml PBS each time. Relative amounts of immunoprecipitated MyoD-5′-32P DNA complexes were determined by measuring Cerenkov counts in the precipitates.
RESULTS
Quadruplex DNA enhances in vivo MyoD-directed expression of a reporter FL gene
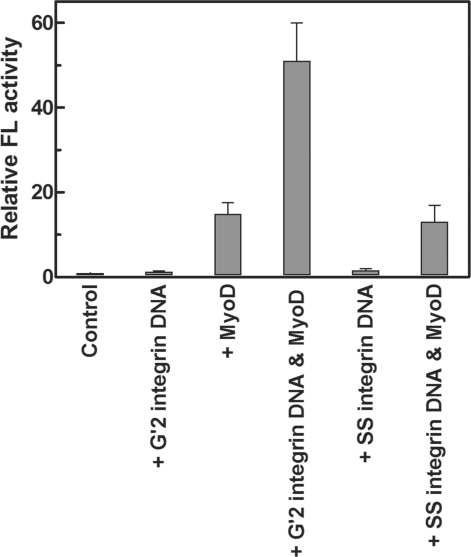
Previously published results indicated that MyoD homodimers formed in vitro tighter complexes with bimolecular quadruplex structures of guanine-rich DNA tracts of muscle gene regulatory regions than with double-stranded E-box motif. By contrast, MyoD-E47 heterodimers displayed higher affinity for E-box than for the quadruplex structures of muscle-gene sequences (44,45). Based on these observations, we first inquired whether such quadruplex DNA structures could affect the capacity of MyoD to induce gene expression in vivo. To monitor E-box and MyoD-dependent expression of a reporter FL gene, HEK293 cells in culture were transfected with a MyoD-responsive p4RTK-FL vector that contained four copies of promoter E-box motif upstream to an FL gene. Cells that were transfected with the p4RTK-FL reporter vector together with pCMV-RL plasmid and pCMV2-Flag DNA exhibited a very low basal level of FL expression (Figure 1). As shown, this low level of expression did not change significantly when G′2 quadruplex α7 integrin DNA was co-transfected with the p4RTK-FL reporter vector FL. However, as expected for the MyoD and E-box responsive FL gene, the measured FL activity was increased ∼15-fold when a MyoD expressing plasmid, pESMV-MyoD was co-transfected with the p4RTK-FL reporter vector. More interestingly, in cells that were co-transfected with the p4RTK-FL and pESMV-MyoD plasmids together with G′2 quadruplex integrin DNA, the measured FL activity increased ∼50- or ∼3.5-fold, respectively, relative to its basal or MyoD-induced levels (Figure 1). Parallel results indicated that single-stranded integrin DNA that was co-transfected into cells by itself or together with MyoD did not enhance the basal or MyoD induced expression of FL (Figure 1), suggesting that the elevation of FL activity was exerted by the quadruplex structure of the DNA rather than by its specific sequence.
Figure 1.
G′2 quadruplex α7 integrin DNA cooperates with MyoD to enhance the in vivo expression of an E-box driven FL reporter gene. HEK293 cells were transfected with FL expressing p4RTK-FL vector and normalizing RL expressing pCMV-RL plasmid alone or together with either 0.3 μg pEMSV-MyoD expression plasmid, with muscle-gene promoter quadruplexes or single-strands of α7 integrin DNA or with a combination thereof (see ‘Materials and Methods’ section). The cells were harvested and lysed 24 h post-transfection and RL-normalized FL activities were determined. The shown data represent average results of four independent experiments.
Quadruplex DNA enhances in vivo MyoD-dependent FL gene transcription
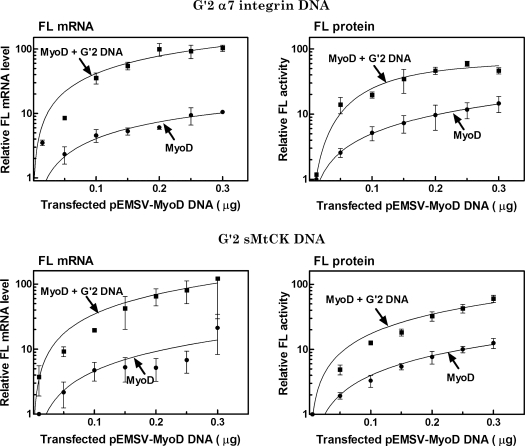
We next examined the molecular basis for the in vivo enhancement by quadruplex DNA of MyoD-directed FL expression. Levels of FL mRNA and activity were determined in parallel in HEK293 cells that were co-transfected with the p4RTK-FL reporter plasmid and with increasing amounts of the MyoD expressing plasmid pESMV-MyoD without or with constant amounts of G′2 quadruplex structures of promoter sequences of the muscle-specific α7 integrin or sMtCK genes. Multiple independent qRT-PCR determinations of RL-normalized FL mRNA were conducted in parallel to measurements of the amounts of RL-normalized FL protein activity (see ‘Materials and Methods’ section). Results presented in Figure 2 indicated that the levels of FL mRNA and protein activity were progressively elevated in cells that were transfected with increasing amounts of the MyoD expressing vector. Cells that were transfected with 0.2–0.3 μg of pESMV-MyoD approached plateau levels of FL mRNA and protein activity that were ∼10-fold higher than their respective basal amounts. Larger progressive increase in the level of FL mRNA were recorded in cells that were also co-transfected with G′2 quadruplex integrin or sMtCK DNA. As seen in Figure 2 the maximum amounts of FL mRNA that accumulated in the presence of the quadruplex DNA structures were, respectively, ∼100- or ∼10-fold higher than their basal or MyoD induced levels. In parallel, the FL protein activity was also increased in the presence of MyoD and quadruplex integrin or sMtCK DNA to levels that were, respectively, ∼50- or ∼5-fold higher than the basal or MyoD-induced FL activities (Figure 2). These results indicated, therefore, that quadruplex DNA elevated the in vivo expression of FL protein by enhancing the MyoD-dependent transcription of the FL reporter gene.
Figure 2.
G′2 quadruplex structures of α7 integrin or sMtCK DNA enhance in vivo MyoD-dependent expression of FL reporter gene. HEK293 cells were transfected with FL expressing p4RTK-FL vector and a normalizing RL expressing pCMV-RL plasmid alone or together with the indicated increasing amounts of the MyoD expressing pEMSV-MyoD plasmid without or with 1.2 μg of co-transfected G′2 quadruplex integrin or sMtCK DNA. Following growth for 24 h, the cells were lysed and RL-normalized FL activity and levels of RL-normalized FL mRNA were determined as described under ‘Materials and Methods’ section. The presented results are averages of three or four independent experiments.
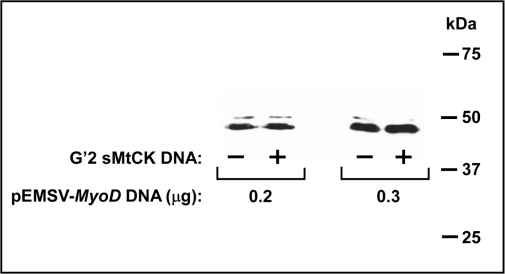
Quadruplex DNA might have enhanced FL gene transcription indirectly by increasing the amounts of expressed MyoD in the cells. To examine this possibility, HEK293 cells were co-transfected with the p4RTK-FL and pCMV-RL reporter plasmids and with 0.2 or 0.3 μg pESMV-MyoD vector DNA without or together with G′2 quadruplex sMtCK DNA. The cells were harvested and lysed 24 h post-transfection and the amounts of expressed MyoD were assessed by western blotting with anti MyoD antibody. Results presented in Figure 3 indicated that cells that were transfected with 0.3 μg pESMV-MyoD plasmid DNA expressed higher amounts of MyoD than cells that were exposed to 0.2 μg of the vector. However, each level of MyoD remained unchanged whether or not the cells were also co-transfected with quadruplex DNA. These results indicated that quadruplex DNA did not act through elevation of the amount of MyoD but rather, it directly enhanced the MyoD-dependent transcription of the FL reporter gene.
Figure 3.
G′2 quadruplex sMtCK DNA does not affect the level of MyoD expression in cells. HEK293 cells were transfected with FL expressing p4RTK-FL vector, normalizing RL expressing pCMV-RL plasmid and the indicated amounts of the MyoD expressing pEMSV-MyoD plasmid without or with 1.2 μg G′2 quadruplex sMtCK DNA. The cells were harvested and lysed 24 h post-transfection and the amounts of expressed MyoD were assessed by western analysis as described under ‘Materials and Methods’ section. Shown is an autoradiogram of the immune blotted denaturing gel with molecular sizes of marker proteins indicated at the right hand.
MyoD preferentially forms complexes with quadruplex DNA in the cells
The observed direct stimulation of MyoD-dependent gene transcription by quadruplex DNA suggested that MyoD interacted directly in vivo with the quadruplex DNA. Data indicated that MyoD homodimers formed in vitro tight complexes with G′2 quadruplex DNA structures (44–46). To inquire whether MyoD that was expressed in cells also interacted preferentially with the transfected quadruplex DNA, p4RTK-FL and pEMSV-MyoD plasmids were co-transfected into HEK293 cells together with 5′-32P-labeled G′2 quadruplex forms of α7 integrin or sMtCK DNA oligomers. Control cells were similarly treated except that the cells were transfected with 32P labeled single-stranded heat denatured integrin or sMtCK DNA or with single-stranded adenosine-substituted A-integrin DNA. Following growth for 24 h, the cells were lysed and the cell lysates were incubated with either rabbit polyclonal anti MyoD antibody or with non-immune rabbit IgG. Formed immune complexes were adsorbed onto protein A-coupled sepharose beads, washed and monitored for Cerenkov counts of co-precipitated labeled DNA (see ‘Materials and Methods’ section). Table 1 summarizes the results of multiple independent replicates of this experiment. Listed are average relative net Cerenkov counts in MyoD-5′-32P DNA complexes that were precipitated by anti MyoD antibody after subtraction of non-specifically adsorbed radioactivity in non-immune IgG precipitates of the corresponding DNA. As seen, the anti MyoD antibody precipitated 100- and 20-fold higher amounts of G′2 quadruplex integrin DNA than single-stranded integrin or A-integrin DNA. In a similar vein, the amounts of G′2 sMtCK DNA that were immune-precipitated by anti MyoD were 7-fold higher than the amounts of similarly treated single-stranded sMtCK (Table 1). Measured molar ratios of single-stranded to quadruplex integrin or sMtCK DNA that were recovered from the cells were, respectively, 0.85 or 0.70. The roughly similar initial amounts of the two forms of both DNA sequences indicated that the preferential immune precipitation of the G′2 quadruplex structures was not due to their preferred accumulation or preservation in the transfected cells. In contrast to anti MyoD antibodies, anti E-47 protein antibodies failed to co-precipitate either single-stranded or quadruplex integrin DNA (data not shown). This result suggested that the immune precipitated MyoD was homodimeric (see ‘Discussion’ section). In sum, our results implied that at least a portion of the cellular MyoD existed in vivo in specific complexes with the integrin or sMtCK DNA tetraplexes. The selectivity of the in vivo binding of quadruplex DNA by MyoD was underlined by the significantly lower amounts of complexes that MyoD formed with single-stranded integrin or sMtCK DNA sequences.
Table 1.
Transfected G′2 quadruplexes of integrin and sMtCK DNA sequences, but not their single-stranded forms are co-immunoprecipitated with MyoD in cell lysates
| DNA | Relative amounts of 32P-DNA co-immunoprecipitated by anti MyoD |
|---|---|
| G′2 integrin | 1.0 |
| ss integrin | 0.01 |
| ss A-integrin | 0.05 |
| G′2 sMtCK | 1.0 |
| ss sMtCK | 0.14 |
HEK293 cells were co-transfected with a pEMSV-MyoD vector and 5′-32P labeled G′2 quadruplex sMtCK or integrin DNA or with the single-stranded forms of these sequences or of adenosine-substituted A-integrin. Following growth for 24 h, the cells were lysed, and MyoD and associated labeled DNA were immunoprecipitated with either anti MyoD antibodies or with control non-immune IgG. Listed are net values of relative radioactivity of labeled DNA that was precipitated by anti MyoD after subtraction of the background radioactivity in precipitates of non-immune IgG. Each presented value is an average of two or three independent determinations. Net Cerenkov counts in co-immunoprecipitated G′2 DNA ranged in different experiments between 2200 and 8000.
DISCUSSION
Global bioinformatics genomic analyses (19–22) as well as direct evidence gathered for selected genes (21–39) strongly implied that guanine-rich tracts in promoter regions of numerous genes might fold into quadruplex structures and that these tetraplexes function in the regulation of gene expression. Yet, little is presently known on the interaction of transcription factors with such quadruplex DNA formations. A recently described case is the binding of nucleolin to a quadruplex formation in the c-myc promoter (50) and another is the interaction of members of the MRF family of myogenic transcription factors with quadruplex structures of muscle-cell gene promoter sequences. We have shown in the past that homodimers of the master myogenic transcription factor MyoD preferentially form in vitro tight complexes with G′2 bimolecular quadruplex structures of guanine-rich tracts of promoter and enhancer regions of muscle-specific genes. By contrast, heterodimers of MyoD with E47 protein associated with double-stranded E-box motif more tightly than with the quadruplex DNA formations (44–46). Based on these observations we suggested that the attraction of MyoD to bimolecular-like quadruplexes in the 5′-ends of muscle-specific genes might serve to modulate their transcription (43). In the present work, we undertook to examine this proposition in living cells by assessing the effect of quadruplex DNA on MyoD-dependent gene expression. Essentially, MyoD-directed expression of an FL reporter gene was measured at the RNA and protein levels in HEK293 cells that were transfected with G′2 quadruplex structures of promoter DNA sequences of the muscle-specific genes α7 integrin and sMtCK. Initial results showed that whereas MyoD was essential for the production E-box-driven FL protein, quadruplex integrin DNA by itself failed to induce FL synthesis. However, upon transfection of MyoD expressing cells with tetraplex integrin DNA FL synthesis was, respectively, enhanced ∼50- and ∼3.5-fold relative to its basal and MyoD-induced levels (Figure 1). A failure of a single-stranded form of the integrin DNA sequence to stimulate in vivo FL synthesis either by itself or together with MyoD indicated that a tetrahelical conformation of the DNA was essential for the enhancement of FL gene expression (Figure 1). The introduction of G′2 quadruplex structures of integrin or sMtCK DNA into MyoD expressing HEK293 cells was also shown to correspondingly elevate the amounts of FL mRNA (Figure 2). The primary level of action of these tetraplex DNA structures appeared, therefore, to be the stimulation of FL transcription. One mechanism that might have been responsible for the enhancement of transcription by tetrahelical DNA could be an increase in MyoD expression by the quadruplex DNA structures. This possibility was excluded, however, by a direct demonstration that similar amounts of MyoD were expressed in cells with or without co-transfected G′2 quadruplex sMtCK DNA (Figure 3). The preference of homodimeric MyoD to form in vitro tight complexes with quadruplex DNA (44–46) raised the alternative possibility that the observed in vivo enhancement of MyoD-dependent gene transcription by tetrahelical DNA was an outcome of its direct binding to MyoD. Results summarized in Table 1 indeed indicated that anti MyoD antibody precipitated from cell lysates complexes of MyoD with transfected 5′-32P-labeled G′2 quadruplex integrin or sMtCK DNA complexes. By contrast, similarly transfected labeled single-stranded integrin or sMtCK DNA failed to combine with anti MyoD and to form similar amounts of immune co-precipitates (Table 1). Although anti E-47 antibodies failed to similarly co-precipitate bound quadruplex integrin DNA (see ‘Results’ section), the possibility that the anti MyoD antibodies co-precipitated complexes of quadruplex DNA with heterodimers of MyoD with other E-proteins could not be excluded. However, based on the high binding affinity of MyoD homodimers for G′2 quadruplex DNA versus the much lower affinity of MyoD-E47 heterodimers for such DNA structures (41), it was likely that all or most of the bound protein was homodimeric.
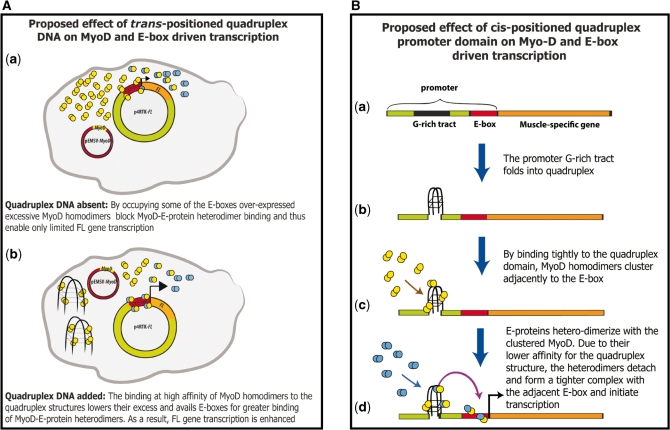
In this study the G′2 quadruplex DNA structures were positioned in trans to the four promoter E-box elements that drove transcription of the reporter FL gene. A proposed mechanism of enhancement of MyoD-directed FL gene expression by a trans-positioned quadruplex DNA structures is schematically illustrated in Figure 4A. Introduction of the pEMSV-MyoD vector into the cells entails over-expression of MyoD molecules that mostly form homodimers. By virtue of their excessive amount, and despite their lower relative affinity for the E-boxes in the reporter p4RTK-FL plasmids (13,14,44,45), the transcriptionally ineffective MyoD homodimers might occupy a portion of these DNA elements which are thus deprived of some of the transcriptionally active MyoD-E47 heterodimers (Figure 4A(a)). As a result, MyoD by itself enhances to only a limited degree the expression of the reporter FL gene (Figures 1 and 2). However, when integrin or sMtCK G′2 quadruplexes are introduced into the cell (Figure 4A(b)), they trap at a very high affinity the MyoD homodimers (44,45) and decrease the probability of their association with the E-box. As a result, more MyoD-E-protein heterodimers are allowed to bind to the E-boxes and the expression of the reporter FL gene is increased (Figures 1 and 2).
Figure 4.
Proposed models for the enhancement of transcription by quadruplex DNA. (A) Model for a possible mode of action of quadruplex structures positioned in trans to MyoD and E-box-driven reporter gene. (B) Model for a possible mechanism of action of promoter quadruplex structures positioned in cis to E-box-driven muscle-specific gene. Yellow circles, MyoD; Blue circles, E-protein. See the text for detailed discussion.
The observed ability of quadruplex DNA structures to enhance gene transcription in trans raises the possibility that such DNA tetraplexes are involved in the positive regulation of gene transcription from genomic DNA. It is tempting, therefore, to speculate on the way that muscle gene expression might be affected by a quadruplex domain which forms adjacently to a promoter E-box. A provisional model illustrated in the right-hand panel of Figure 4B assumes that G′2-like quadruplex structures are transiently formed by guanine-rich tracts that are positioned cis to adjacent E-boxes in muscle-gene promoters. By virtue of their high affinity for these structures (44,45), MyoD homodimers are proposed to be attracted to the quadruplex domains in gene promoters. The resulting clustering of the MyoD homodimers concentrates them and might thus increases the probability of association with E-proteins, enhancing the efficiency of MyoD-E-proteins heterodimerization. Moreover, having the E-box elements close by, binding of the transcriptionally competent heterodimers to their DNA target is also likely to be more efficient and gene transcription might then be enhanced. Promoter regions of several muscle-specific genes were found to include guanine-rich clusters that are positioned in close proximity to E-box motifs. These G-rich tracts were shown to readily fold in vitro into G′4 unimolecular, G′2 bimolecular and G′2-like quadruplex structures (42). Binding of MyoD-E-proteins heterodimers to promoter E-box elements is required for the initiation of muscle gene transcription and for the ensuing induction of a myogenic cellular phenotype (51). We suggest that DNA at promoter regions of muscle-specific genes becomes unwound prior to transcription thus allowing the guanine-rich tracts (Figure 4B(a)) to fold into tetraplex structures (Figure 4B(b)). With the tetrahelical DNA domains being a preferred target for MyoD homodimers, their binding to these DNA structures gathers them in close proximity to the E-box elements (C). This clustering of MyoD might advance its association with the constitutive E-proteins and increase heterodimer formation. Having a low binding affinity for quadruplex DNA and a much higher affinity for E-box (44), the heterodimers might then detach from the tetrahelical DNA domain and associate with the neighboring E-box motifs thus initiating transcription (Figure 4B(d)). Future investigations of the effect of cis positioned quadruplex DNA formations on MyoD-dependent E-box-driven gene transcription should put this model to a test.
FUNDING
United States–Israel Binational Science Foundation and the Israel Science Foundation (to M.F.). Funding for open access charge: United States–Israel Binational Science Foundation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr Eyal Bengal for valuable discussions.
REFERENCES
- 1.Buckingham M. Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 2001;11:440–448. doi: 10.1016/s0959-437x(00)00215-x. [DOI] [PubMed] [Google Scholar]
- 2.Francis-West PH, Antoni L, Anakwe K. Regulation of myogenic differentiation in the developing limb bud. J. Anat. 2003;202:69–81. doi: 10.1046/j.1469-7580.2003.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tajbakhsh S. Stem cells to tissue: molecular, cellular and anatomical heterogeneity in skeletal muscle. Curr. Opin. Genet. Dev. 2003;13:413–422. doi: 10.1016/s0959-437x(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 4.Rudnicki MA, Jaenisch R. The MyoD family of transcription factors and skeletal myogenesis. Bioessays. 1995;17:203–209. doi: 10.1002/bies.950170306. [DOI] [PubMed] [Google Scholar]
- 5.Pownall ME, Gustafsson MK, Emerson C.P., Jr Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- 6.Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- 7.Rawls A, Morris JH, Rudnicki M, Braun T, Arnold HH, Klein WH, Olson EN. Myogenin's; functions do not overlap with those of MyoD or Myf-5 during mouse embryogenesis. Dev. Biol. 1995;172:37–50. doi: 10.1006/dbio.1995.0004. [DOI] [PubMed] [Google Scholar]
- 8.Zhang W, Behringer RR, Olson EN. Inactivation of the myogenic bHLH gene MRF4 results in up-regulation of myogenin and rib anomalies. Genes Dev. 1995;9:1388–1399. doi: 10.1101/gad.9.11.1388. [DOI] [PubMed] [Google Scholar]
- 9.Bergstrom DA, Penn BH, Strand A, Perry RL, Rudnicki MA, Tapscott SJ. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell. 2002;9:587–600. doi: 10.1016/s1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- 10.Chanoine C, Della Gaspera B, Charbonnier F. Myogenic regulatory factors: redundant or specific functions? Lessons from Xenopus. Dev. Dyn. 2004;231:662–670. doi: 10.1002/dvdy.20174. [DOI] [PubMed] [Google Scholar]
- 11.Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis RL, Cheng PF, Lassar AB, Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990;60:733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- 13.Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 14.Lassar AB, Davis RL, Wright WE, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 15.Brennan TJ, Chakraborty T, Olson EN. Mutagenesis of the myogenin basic region identifies an ancient protein motif critical for activation of myogenesis. Proc. Natl Acad. Sci. USA. 1991;88:5675–5679. doi: 10.1073/pnas.88.13.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Black BL, Molkentin JD, Olson EN. Multiple roles for the MyoD basic region in transmission of transcriptional activation signals and interaction with MEF2. Mol. Cell Biol. 1998;18:69–77. doi: 10.1128/mcb.18.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blackwell TK, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 18.Weintraub H, Genetta T, Kadesch T. Tissue-specific gene activation by MyoD: determination of specificity by cis-acting repression elements. Genes Dev. 1994;8:2203–2211. doi: 10.1101/gad.8.18.2203. [DOI] [PubMed] [Google Scholar]
- 19.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huppert L. Hunting G-quadruplexes. Biochimie. 2008;90:1140–1148. doi: 10.1016/j.biochi.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 23.Hammond-Kosack MC, Docherty K. A consensus repeat sequence from the human insulin gene linked polymorphic region adopts multiple quadriplex DNA structures in vitro. FEBS Lett. 1992;301:79–82. doi: 10.1016/0014-5793(92)80214-2. [DOI] [PubMed] [Google Scholar]
- 24.Hammond-Kosack MC, Kilpatrick MW, Docherty K. Analysis of DNA structure in the human insulin gene-linked polymorphic region in vivo. J. Mol. Endocrinol. 1992;9:221–225. doi: 10.1677/jme.0.0090221. [DOI] [PubMed] [Google Scholar]
- 25.Hammond-Kosack MC, Kilpatrick MW, Docherty K. The human insulin gene-linked polymorphic region adopts a G-quartet structure in chromatin assembled in vitro. J. Mol. Endocrinol. 1993;10:121–126. doi: 10.1677/jme.0.0100121. [DOI] [PubMed] [Google Scholar]
- 26.Lew A, Rutter WJ, Kennedy GC. Unusual DNA structure of the diabetes susceptibility locus IDDM2 and its effect on transcription by the insulin promoter factor Pur-1/MAZ. Proc. Natl Acad. Sci. USA. 2000;97:12508–12512. doi: 10.1073/pnas.97.23.12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Connor AC, Frederick KA, Morgan EJ, McGown LB. Insulin capture by an insulin-linked polymorphic region G-quadruplex DNA oligonucleotide. J. Am. Chem. Soc. 2006;128:4986–4991. doi: 10.1021/ja056097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seenisamy J, Rezler EM, Powell TJ, Tye D, Gokhale V, Joshi CS, Siddiqui-Jain A, Hurley LH. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J. Am. Chem. Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- 30.Lemarteleur T, Gomez D, Paterski R, Mandine E, Mailliet P, Riou JF. Stabilization of the c-myc gene promoter quadruplex by specific ligands' inhibitors of telomerase. Biochem. Biophys. Res. Commun. 2004;323:802–808. doi: 10.1016/j.bbrc.2004.08.150. [DOI] [PubMed] [Google Scholar]
- 31.Duquette ML, Pham P, Goodman MF, Maizels N. AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation. Oncogene. 2005;24:5791–5798. doi: 10.1038/sj.onc.1208746. [DOI] [PubMed] [Google Scholar]
- 32.Kumar N, Patowary A, Sivasubbu S, Petersen M, Maiti S. Silencing c-MYC expression by targeting quadruplex in P1 promoter using locked nucleic acid trap. Biochemistry. 2008;47:13179–13188. doi: 10.1021/bi801064j. [DOI] [PubMed] [Google Scholar]
- 33.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Todd AK, Haider SM, Parkinson GN, Neidle S. Sequence occurrence and structural uniqueness of a G-quadruplex in the human c-kit promoter. Nucleic Acids Res. 2007;35:5799–5808. doi: 10.1093/nar/gkm609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dexheimer TS, Sun D, Hurley LH. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 2006;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan N, Avino A, Tauler R, Gonzalez C, Eritja R, Gargallo R. Solution equilibria of the i-motif-forming region upstream of the B-cell lymphoma-2 P1 promoter. Biochimie. 2007;89:1562–1572. doi: 10.1016/j.biochi.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 38.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun D, Liu WJ, Guo K, Rusche JJ, Ebbinghaus S, Gokhale V, Hurley LH. The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex-interactive agents. Mol. Cancer Ther. 2008;7:880–889. doi: 10.1158/1535-7163.MCT-07-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo K, Gokhale V, Hurley LH, Sun D. Intramolecularly folded G-quadruplex and i-motif structures in the proximal promoter of the vascular endothelial growth factor gene. Nucleic Acids Res. 2008;36:4598–4608. doi: 10.1093/nar/gkn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin Y, Rezler EM, Gokhale V, Sun D, Hurley LH. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007;35:7698–7713. doi: 10.1093/nar/gkm538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yafe A, Etzioni S, Weisman-Shomer P, Fry M. Formation and properties of hairpin and tetraplex structures of guanine-rich regulatory sequences of muscle-specific genes. Nucleic Acids Res. 2005;33:2887–2900. doi: 10.1093/nar/gki606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dexheimer TS, Fry M, Hurley LH. DNA quadruplexes and gene regulation. In: Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. Cambridge, UK: RSC Publishing; 2006. pp. 180–207. [Google Scholar]
- 44.Etzioni S, Yafe A, Khateb S, Weisman-Shomer P, Bengal E, Fry M. Homodimeric MyoD preferentially binds tetraplex structures of regulatory sequences of muscle-specific genes. J. Biol. Chem. 2005;280:26805–26812. doi: 10.1074/jbc.M500820200. [DOI] [PubMed] [Google Scholar]
- 45.Shklover J, Etzioni S, Weisman-Shomer P, Yafe A, Bengal E, Fry M. MyoD uses overlapping but distinct elements to bind E-box and tetraplex structures of regulatory sequences of muscle-specific genes. Nucleic Acids Res. 2007;35:7087–7095. doi: 10.1093/nar/gkm746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yafe A, Shklover J, Weisman-Shomer P, Bengal E, Fry M. Differential binding of quadruplex structures of muscle-specific genes regulatory sequences by MyoD, MRF4 and myogenin. Nucleic Acids Res. 2008;36:3916–3925. doi: 10.1093/nar/gkn340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khateb S, Weisman-Shomer P, Hershco-Shani I, Ludwig AL, Fry M. The tetraplex (CGG)n destabilizing proteins hnRNP A2 and CBF-A enhance the in vivo translation of fragile X premutation mRNA. Nucleic Acids Res. 2007;35:5775–5788. doi: 10.1093/nar/gkm636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fry M, Loeb LA. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl Acad. Sci. USA. 1994;91:4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short Protocol Mol. Biol. 2002;1:3–25. [Google Scholar]
- 50.Gonzalez V, Guo K, Hurley L, Sun D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 2009;284:23622–23635. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]