Abstract
Insight into protein structure and function is best obtained through a synthesis of experimental, structural and bioinformatic data. Here, we outline a framework that we call MUSE (mutual information, unigenic evolution and structure-guided elucidation), which facilitated the identification of previously unknown residues that are relevant for function of the GIY-YIG homing endonuclease I-BmoI. Our approach synthesizes three types of data: mutual information analyses that identify co-evolving residues within the GIY-YIG catalytic domain; a unigenic evolution strategy that identifies hyper- and hypo-mutable residues of I-BmoI; and interpretation of the unigenic and co-evolution data using a homology model. In particular, we identify novel positions within the GIY-YIG domain as functionally important. Proof-of-principle experiments implicate the non-conserved I71 as functionally relevant, with an I71N mutant accumulating a nicked cleavage intermediate. Moreover, many additional positions within the catalytic, linker and C-terminal domains of I-BmoI were implicated as important for function. Our results represent a platform on which to pursue future studies of I-BmoI and other GIY-YIG-containing proteins, and demonstrate that MUSE can successfully identify novel functionally critical residues that would be ignored in a traditional structure-function analysis within an extensively studied small domain of ∼90 amino acids.
INTRODUCTION
The explosion of sequence and structural data has rapidly accelerated the pace of protein structure and function studies. Bioinformatic approaches that predict function based on amino-acid conservation (1,2), homology modelling studies (3) and identification of co-evolving residues (4,5) are among methods commonly used to address structure and function questions. There are, however, many protein families for which mechanistic insight is lacking. The GIY-YIG homing endonuclease family is one such example. Homing endonucleases are site-specific yet sequence-tolerant DNA endonucleases that are distinguishable from other DNA endonucleases in their ability to bind long target sequences and tolerate multiple substitutions within their binding site (6). They function primarily as mobile genetic elements, initiating the movement of their coding sequence and surrounding DNA by binding and cleaving a target site (the homing site) in genomes that lack the endonuclease (7). Homing endonucleases are phylogenetically widespread, and have traditionally been categorized into one of four large families based on conserved amino-acid motifs, the LAGLIDADG, HNH, His-Cys box and GIY-YIG families (6). The PE-(D/E)-XK and Vsr-like enzymes are only recently described and have fewer family members (8,9). Much effort has been devoted towards re-engineering LAGLIDADG endonucleases to cleave novel target sequences with clinical relevance in the human genome (10–13). Similar studies could in principle be performed on any endonuclease family, necessitating a detailed understanding of mechanism.
Within the four largest endonuclease families, the GIY-YIG endonucleases are the least understood in terms of mechanism. The prototypical GIY-YIG family endonuclease is I-TevI, encoded with the genome of Escherichia coli phage T4 (14). Studies on I-TevI revealed that the enzyme has a two-domain structure, composed of a N-terminal catalytic domain containing the class-defining GIY-YIG motif that is connected to a C-terminal DNA-binding domain by a flexible linker (15). Substantial experimental evidence suggests that the DNA-binding domain tethers the catalytic domain on its substrate to perform two sequential nicking reactions that generate a staggered 2-nt 3′ overhang (16). The catalytic domain has no measurable DNA-binding activity when expressed independently, and a critical function of the linker region in I-TevI, and its isoschizomer I-BmoI, is to correctly position the domain on substrate (17–19). Early bioinformatic studies revealed that the GIY-YIG domain is not exclusive to homing endonucleases (20), illustrated by the presence of the domain in the UvrC nucleotide excision repair protein (21), restriction enzymes (22,23), and retrotransposable elements (24). Structural, biochemical and bioinformatic studies have shown that the GIY-YIG domain is ∼90 amino acids with an α/β-fold composed of a central three-stranded antiparallel β-sheet flanked by three helices (21,25) (Figure 1). Four highly conserved residues in the GIY-YIG domain, Y17, R27, E74 and N87 (numbered according to the I-BmoI sequence, Figure 1B) comprise a putative active site cleft (Figure 1C), with a single divalent metal ion coordinated by the glutamic-acid residue in both the I-TevI and UvrC structures. Mutation of any of these residues abolishes DNA cleavage activity in a number of GIY-YIG enzymes (20–22,26,27).
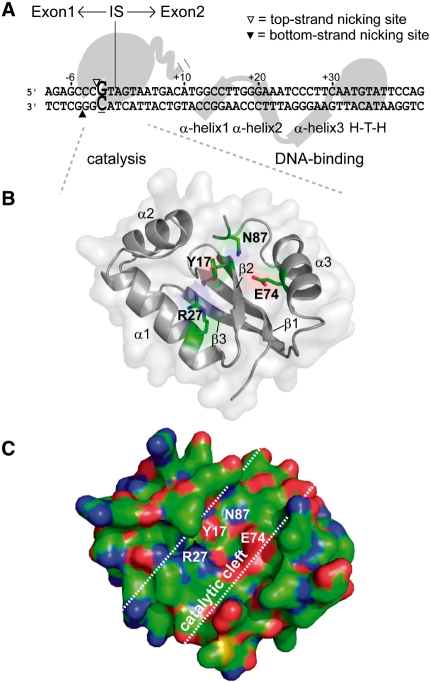
Figure 1.
I-BmoI is a modular GIY-YIG homing endonuclease. (A) Schematic representation of I-BmoI interactions with intronless thyA substrate based on biochemical data (27,32). Top- and bottom-strand nicking sites are shown as open and filled triangles, respectively, and the critical −2 GC base pair is shown in enlarged, bold-type font. The intron insertion site is indicated by a vertical line, with exon 1 sequence upstream (−) and exon 2 sequence downstream (+). (B) Homology model of the I-BmoI catalytic domain (residues 1–88). Highlighted are four highly conserved residues in GIY-YIG alignments that are critical for function, and secondary structure elements of the domain. Subsequent illustrations of the catalytic domain will be shown from this view (front) or a 180-degree rotation (back). (C) Surface representation of a front view of the I-BmoI homology model highlighting the putative catalytic cleft. The side chains of Y17, R27, E74 and N87 are surface exposed, lie along the base of the cleft, and are situated in close proximity to one another. Patches of charge are shown in colour, blue being positive, red being negative and green being hydrophobic.
In spite of a wealth of bioinformatic, biochemical and structural data, the mechanism by which GIY-YIG homing endonucleases introduce a double-strand break (DSB) in substrate is unknown (28). The mechanism must involve repositioning of a (presumably) single active site within the catalytic domain on substrate to perform two sequential nicking reactions, with the bottom (non-coding) strand nicked before the top (coding) strand (27,29). This mechanism is likely to be distinct from other enzymes that contain the GIY-YIG domain, including the restriction enzyme Cfr42I that functions as a tetramer (30), Eco29kI that functions as a dimer (31), or the UvrC proteins that nick only a single-strand adjacent to a damaged base (21). In an effort to gain insight into the mechanism by which GIY-YIG homing endonucleases introduce a DSB, we have been studying I-BmoI (Figure 1), (32). Like I-TevI, I-BmoI is a two-domain endonuclease with an extended recognition sequence. Both enzymes cleave at the same positions within their respective intronless substrates, but I-BmoI requires only a critical G-C base pair at position −2 of intronless substrate for cleavage (33). As a model GIY-YIG homing endonuclease, I-BmoI has a number of advantages over I-TevI, including the fact that the wild-type (WT) enzyme can be overexpressed and purified in quantities that are difficult to obtain with I-TevI. Moreover, I-BmoI is ∼750-fold less active than I-TevI, suggesting that early steps in the reaction pathway are more amenable to in vitro analysis (27,33).
Here, we present a unified experimental framework that will provide a platform on which to base future structure and function studies of GIY-YIG homing endonucleases, and other GIY-YIG-containing enzymes. Our framework, which we term MUSE, synthesizes data from three distinct experimental approaches; mutual information analyses that identify co-evolving residues in the GIY-YIG domain, a unigenic evolution strategy that uses a functional genetic selection to identify hypo- and hyper-mutable residues, and interpretation of the data using paralog-specific sequence alignments and structural models of the GIY-YIG domain. While none of the approaches used in our study are individually novel, the synthesis of data from all three methods facilitated the identification of residues that are unlikely to have been identified as important for function using any one of the approaches in isolation. Mutational analyses of the positions revealed phenotypic differences relative to WT I-BmoI in functional assays, validating that MUSE can successfully identify previously unrecognized residues with the GIY-YIG domain as relevant for function.
MATERIALS AND METHODS
Strain and plasmid construction
Strains and plasmids used in this study are listed in Supplementary Table S1, and oligonucleotides are listed in Supplementary Table S2. To construct strain BW25141(λDE3) for use in unigenic evolution experiments, E. coli BW25141 was lysogenized using the λDE3 lysogenization kit (Novagen). The toxic plasmid backbone, p11-lacY-wtx1 (34), was used to construct pToxBmoHS and pToxBmoIn+ by inserting the corresponding intronless homing site (HS) and intron-containing target site (In+), respectively. To construct pToxBmoHS, oligonucleotides DE-395 and DE-396 were annealed and ligated into the XbaI and SphI sites of p11-lacY-wtx1. The 51-bp intronless homing site corresponds to positions −10 to +41 of the exon1–exon2 junction relative to the intron insertion site (32). To construct pToxBmoIn+, oligonucleotides DE-429 and DE-430 were annealed and ligated into the XbaI and SphI sites of p11-lacY-wtx1. The 51-bp intron-containing site corresponds to the final 10 bp of the 3′-end of the intron plus the first 41 bp of the 5′-end of exon 2 (32). pIBmoIE, which is a pUC57 derivative containing a codon optimized I-BmoI gene (IDT DNA), was used as a template for cloning the I-BmoI gene (optimized I-BmoI sequence is presented in Supplementary Figure S1). Primers DE-331 and DE-384 were used to amplify and clone the codon optimized I-BmoI into the NdeI and XhoI sites of pACYCDuet-1 to generate pACYCIBmoI. This plasmid was subsequently used as a template to generate pACYCR27A using the Quikchange XL site-directed mutagenesis kit (Stratagene) with primers DE-419 and its reverse complement, DE-420. All plasmid constructs were verified by sequencing.
Genetic selection
To generate strains harbouring the toxic plasmid for unigenic evolution, BW25141(λDE3) was transformed with one of the three toxic (reporter) plasmids (p11-lacY-wtx1, pToxBmoHS, or pToxBmoIn+) and plated on LB plates containing 100 µg/ml ampicilin and 0.2% glucose. For each strain, a single colony was picked to inoculate 500 ml LB plus 100 µg/ml ampicilin and 0.2% glucose to generate electrocompetent cells. Typically, 50 µl of electrocompetent cells were transformed with 100 ng of the expression plasmid (pACYCIBmoI or pACYCR27A). The transformations were allowed to recover in 500 µl of SOC media at 37°C for 5 min, then diluted into 2 ml 37°C SOC and shaken at 37°C for 75 min. We found that addition of IPTG was not necessary to induce I-BmoI expression, as an IPTG concentration of 0.1 mM led to toxic effects. After incubation, transformations were diluted 1000-fold in SOC, and 100-µl aliquots were spread on plates containing LB plus 25 µg/ml chloramphenicol to estimate number of transformants, or plates containing LB plus 25 µg/ml chloramphenicol and 10 mM arabinose to observe the number of colonies surviving the selection. Survival rate was calculated by dividing the number of colonies observed on chloramphenicol plus arabinose plates by colonies observed on chloramphenicol only plates.
Construction of mutagenized I-BmoI libraries
I-BmoI mutant libraries were generated by error-prone PCR from pACYCIBmoI using primers DE-490 and DE-491. The forward primer (DE-490) was designed such that only the ATG start codon was included in the primer, and the reverse primer (DE-491) was designed such that no part of the I-BmoI gene was present in the primer. Three mutagenic libraries were generated using identical PCR conditions in parallel 50-µl reactions containing 80 ng of pACYCIBmoI as template, 20 pmol of each primer (DE-490, DE-491), 0.2 mM dATP, 0.2 mM dGTP, 1 mM dCTP, 1 mM dTTP, 0.5 mM MgCl2, 0.5 mM MnCl2 and 2.5 U of Taq polymerase (NEB) in the presence of 1 × PCR buffer (10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris–HCl pH 8.8, 2 mM MgSO4, 0.1% Triton X-100). A total of 30 PCR cycles were run as follows: 94°C for 60 s, 46.5°C for 60 s and 72°C for 60 s. Mutagenic PCR products were digested with NdeI and XhoI and ligated into pACYCR27A (used as a ligation target due to the fact that re-ligated singly cut R27A I-BmoI would be non-functional in the selection). The ligated pools were independently transformed into DH5α, grown in 3 ml LB plus 25 µg/ml chloramphenicol for 16 h at 37°C, and miniprepped (QIAGEN) to generate the mutant I-BmoI libraries.
I-BmoI unigenic evolution and selection of variants
The three mutagenic libraries were subjected to unigenic evolution to determine survival percentage and to obtain clones required for sequence analysis. We sequenced a total of 167 selected clones picked from LB plus chloramphenicol and arabinose plates, of which 87 independent clones (36, 34 and 17 clones from pools 1, 2 and 3, respectively) were identified (the rest discarded due to redundancy of DNA or amino-acid sequence). These clones contained a total of 460-nt substitutions corresponding to 271 amino-acid substitutions. We also sequenced 62 unselected clones from LB plus chloramphenicol only plates. The unselected clones harboured a total of 760-nt substitutions, 577 amino-acid substitutions and were used to establish baseline mutation frequencies. The EoS value was calculated as described (A. Fernandes et al., submitted for publication).
Construction and purification of site-directed mutants
We created a library of site-directed mutants using the Quikchange® XL Site-Directed Mutagenesis Kit (Stratagene) to generate point mutants in the pACYCIBmoI backbone. For purification purposes, a subset of these mutants were sub-cloned into the pTYB1 vector and were expressed and purified as previously described with one change to the protocol (27). Once the clarified lysate has been loaded, the five column volume wash with Buffer A contained a final concentration of 1 mM ATP (Bioshop Canada Inc.) to help remove bound chaperones. The concentrations of purified WT I-BmoI and I-BmoI mutants were determined by a standard Bradford assay in duplicate using an Ultrospec 2100 pro (Biochrom Ltd).
Characterization of I-BmoI variants
A set of I-BmoI variants identified from the unigenic evolution study and the library of site-directed mutants were run through the genetic selection (as described above) to determine their survival versus WT. Cleavage assays were subsequently performed with I-BmoI mutants that were amenable to purification. The cleavage activities of WT and I-BmoI mutants were determined using titrations with 10 nM pBmoHS and 2-fold serial dilutions of I-BmoI from 700 nM to 1.37 nM in 10-µl volumes for 5 min at 37°C in 50 mM Tris pH 7.9, 50 mM NaCl, 2 mM MgCl2 and 1 mM DTT. Reactions were stopped by addition of 4 µl stop dye (100 mM EDTA pH 8.0, 25% glycerol and 0.2% bromophenol blue) and heated at 90°C for 5 min. Stopped reaction were run on 1% agarose gels, stained with ethidium bromide (OmniPur) and analysed on an AlphaImager™3400 (Alpha Innotech). Percent linear DNA is defined as the percentage of total DNA (circular, nicked and linear) converted to linear product. Nicking assays were conducted as above, with a standard protein concentration of 175 nM and 10-µl aliquots of a reaction pool were stopped at 15 s, 30 s, 45 s, 1 min, 1 min 30 s, 2 min, 3 min and 5 min. The rate constants for the first strand (circular to nicked) and second strand (nicked to linear) steps were calculated as described in Supplementary Figure S4.
Structure-based alignment
Sequences similar to the endonuclease domain of I-TevI (GI: 29345254) were identified by BLAST in the non-redundant protein database and in the metagenomic protein database at NCBI. All proteins with an E-value less than 0.1 were aligned to the I-TevI catalytic domain structure (PDB ID: 1LN0) using the block align feature of Cn3D (http://www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml). The defaults of this feature were changed to not perform global alignments and to allow changes to the block structure of existing alignments. Long gaps were not allowed. Totally 174 full-length sequences were identified. Partial sequences, sequences that required large gaps or that were missing the presumed catalytic residues (R27, E75 and N90, I-TevI numbering) were excluded from the alignment. The alignment included the I-BmoI sequence (GI: 12958590). The alignment contained four bacteriophage sequences, 97 sequences from the marine metagenome, 23 bacterial sequences and 50 eukaryotic sequences (Supplementary Figure S2). The sequences were culled to remove the shorter member of a redundant pair with a threshold of 90% using JalView (35), leaving 146 sequences. To test the quality of the alignment and for contamination by paralogous GIY-YIG sequences, the UvrC sequence from Thermotoga maritima (GI: 8134799) was included in the initial alignment. Using the Cn3D block align method, the sequences were sorted by matches to the position sensitive scoring matrix. The T. maritima UvrC sequence was the third worst scoring protein by this measure, suggesting that UvrC proteins were effectively excluded from the alignment. Second, we examined the alignment for an excess of local covariation that can detect as little as 10% contamination of the alignment (R.J. Dickson et al., submitted). This often results when paralogous gene families are incorporated into one alignment. No excess local covariation was identified, suggesting that contamination by UvrC or other parologous GIY-YIG type endonuclease domains was rare. Based on the alignment, we built a homology model of the I-BmoI GIY-YIG domain (residues 1–88) using MODELLER (36) and SWISS-MODEL (37) with the I-TevI structures (1KM0 and 1LN0) as templates. There was no significant difference between the two structural models.
Co-variation analyses
Three semi-independent methods, MIp (38), ΔZp and ΔZpx (R.J. Dickson et al., submitted), were applied to the multiple sequence alignment to identify pairs or small groups of positions that showed non-independent evolution in the GIY-YIG domain, using scoring cutoffs of 4.5 (MIp), 3.5 (ΔZpx) and 1.5 (ΔZp), respectively. Mutual information values were assigned to residues using the I-BmoI reference sequence. Distances were calculated using the closest non-hydrogen atom of co-evolving residues in the I-BmoI homology model.
RESULTS
Improved alignment of the GIY-YIG domain
Previous multiple sequence alignments of the GIY-YIG domain have included sequences of proteins with diverse functions, diluting the phylogenetic signal from GIY-YIG domains specific to homing endonucleases (20,23). To gain better insight into residues that are conserved amongst potential GIY-YIG homing endonucleases, we collected sequences by BLAST and aligned them to the I-TevI catalytic domain structure with Cn3D. BLAST searches were dominated by matches to the nucleotide excision repair protein UvrC, all of which were subsequently discarded. In addition, sequences that contained obvious insertions and deletions, or sequences that lacked residues equivalent to the functionally critical R27, E74 and N87 (I-BmoI numbering), were removed resulting in a final alignment of 146 sequences.
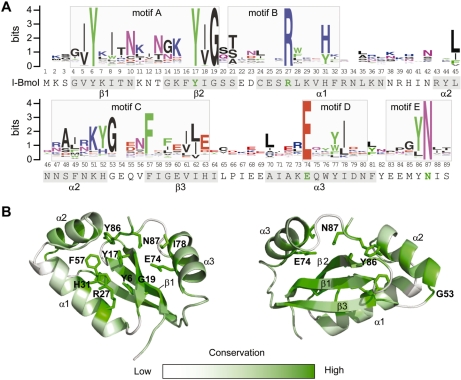
The GIY-YIG domain has previously been separated into five conserved regions that are characterized by highly conserved residues, termed motifs A through E (20). In our new alignment (Figure 2A), the information content associated with the highly conserved residues has not changed significantly, with the exceptions of increases associated with the tyrosine residues of motif A (Y6 and Y17) and the phenylalanine residue of motif C (F57). There are, however, differences outside of the highly conserved residues that are apparent in our alignment. In particular, there are obvious increases in the information content of positions that intervene the GIY and YIG elements of motif A (I8, N10 and K15), and the stretch of residues (S48, K51, H52, G53) that precede the phenylalanine (F57) of motif C. In contrast, there is a decrease in the information content of the proline residue (position 84) that precedes the conserved tyrosine (Y86) and asparagine (N87) residues of motif E (Figure 2A), attributable to the removal of UvrC sequences from the alignment that predominantly possess a proline at this position.
Figure 2.
Alignment of the GIY-YIG domain. (A) Multiple sequence alignment of 146 sequences represented as a sequence logo (50). Positions are numbered according to the I-BmoI sequence that is shown below the alignment (conserved functionally critical residues are shown in green). Predicted secondary structure elements of the I-BmoI GIY-YIG domain are indicated on the sequence by shading, and motif assignments are identified on the alignment with shaded boxes. (B) Front (left) and rear (right) views of the homology model with the degree of conservation mapped onto the structure. Conserved positions are shown in dark green, with the side chains of highly conserved residues indicated. Variable positions are shown in white.
To display sequence conservation within the GIY-YIG domain, the entropy for each position in the alignment was calculated and subsequently displayed on the I-BmoI homology model, with conserved positions (low entropy, high information content) coloured green and non-conserved positions (high entropy, little information content) coloured white (Figure 2B). As expected, the highly conserved residues that characterize the GIY-YIG domain (Y6, Y17, R27, E74 and N87) all surround the proposed catalytic surface. Of particular interest is H31 that displays high information content, and lies in close proximity to the essential R27 in α-helix1, with both residues stacked against each other and orientated towards the presumed catalytic surface (Figure 2B). In general, the majority of conserved residues are similarly positioned, with the exception of a block of moderately conserved residues spanning positions 47–53 (located in α-helix 2, orientated away from the catalytic surface). Strikingly, many of the non-conserved residues occupy positions distinct from the proposed catalytic surface, including residues in α-helix 1 and the loop connecting α-helix 1 and α-helix 2. Collectively, these data highlight the conservation of residues that surround the proposed catalytic surface of the GIY-YIG domain, whereas non-conserved residues tend to be on the periphery of the domain.
Identification of co-evolving positions in the I-BmoI catalytic domain
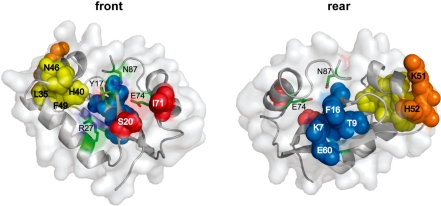
Additional insight into functionally important residues of the I-BmoI catalytic domain was obtained by applying mutual information analyses to detect amino-acid positions that co-evolve, or co-vary, with each other. Such analyses can detect non-independent evolution of residues that co-vary with other residues because of functional constraints. Indeed, most residues that co-vary tend to be within contact distance of each other (4,38). We applied three different methods to analyse co-variation in the GIY-YIG domain alignment, MIp, ΔZpx and ΔZp (R.J. Dickson et al., submitted), and found that the highest scoring amino-acid pair by all methods was S20-I71 (Table 1). While S20 and I71 are not within predicted contact distance (6.3 Å) in the homology model of the I-BmoI GIY-YIG domain, they occupy a surface of the domain that can be envisioned as a gateway to the catalytic cleft formed by the C-terminal end of β-sheet 2 and the N-terminal end of α-helix 3 (Figure 3). In addition, we identified a set of four residues, L35-H40-N46-F49, that co-evolve with each other (Table 1). This set of residues was of interest because the H40Y mutation in I-TevI reduces catalytic activity relative to the WT protein (39), and the interaction between these residues may be required to position the H40 residue appropriately within the active site (Figure 3). Another intriguing residue identified by mutual information analyses was K7 that coevolves with three other residues, T9, F16 and E60 (Table 1). All four residues lie in adjacently positioned β-sheets (Figure 3), suggesting that interaction between these residues is critical for folding of the GIY-YIG domain. Similarly, other sets of coevolving residues such as K51-H52 and H63-K73-W76 (Table 1), are all within contact distance of each other and likely have roles in folding or stability of the domain.
Table 1.
Co-evolving residues in the I-BmoI GIY-YIG domain
| Pair | MIp | ΔZpx | ΔZp | Distance (Å) |
|---|---|---|---|---|
| S20-I71 | 8 | 5.7 | 5.5 | 6.3 |
| H63-K73 | 4.8 | 3.8 | 3.5 | 3.1 |
| K51-H52 | 4.7 | 4.3 | 2 | 1.3 |
| K7-E60 | 4.1 | 3.6 | 2 | 3 |
| L35-N46 | 5.2 | 3.4 | 1.8 | 3.7 |
| K7-F16 | 3.9 | 3.7 | 1.8 | 4.2 |
| K73-W76 | 6 | 4.6 | 1.8 | 3.3 |
| K7-T9 | 3.2 | 3 | 1.8 | 3.8 |
| Q75-D79 | 3.8 | 4.1 | 1.8 | 3.1 |
| L35-F49 | 3.5 | 3.2 | 1.7 | 3.6 |
| N10-S48 | 4.6 | 3.9 | 1.6 | 3 |
| L35-H40 | 4.7 | 3.4 | 1.5 | 3.5 |
Figure 3.
Co-evolving residues of the I-BmoI catalytic domain. Position of four sets of co-evolving residues mapped onto the I-BmoI homology model, colour-coded by co-evolving residues (yellow, L35, H40, N46, F49; red S20, I71; blue K7, T9, F16, E60; orange K51, H52). Front (left) and rear (right) views are shown, with functionally critical residues highlighted in light green.
Unigenic evolution identifies mutable positions in I-BmoI
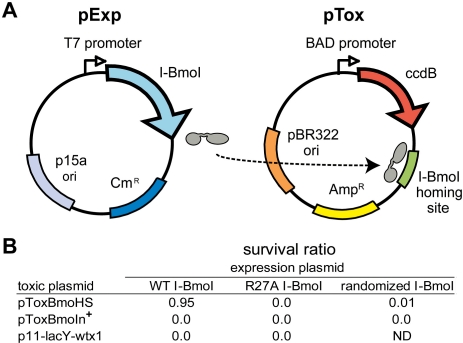
The above results provided a phylogenetic and structural framework for the identification and analysis of potential functionally critical residues in the catalytic domain of I-BmoI. To gain experimental insight into residues that are functionally relevant across the full length of I-BmoI, we adapted the unigenic evolution method (40). In this method, a genetic selection is used to isolate functional variants of I-BmoI after random mutagenesis, facilitating identification of amino-acid positions that were either tolerant (hypermutable) or intolerant (hypomutable) of substitutions. The selection utilizes a two-plasmid system that relies on I-BmoI expression from one plasmid (pExp) to cleave a second, toxic plasmid (pTox) that contains the cognate I-BmoI homing site (34) (Figure 4). Cells survive plating on selective media only if the toxic plasmid has been cleaved by a functional I-BmoI. As shown in Figure 4, we observed a survival ratio of 0.95 when WT I-BmoI was expressed from pExp, and pTox contained the I-BmoI homing site (pToxBmoHS). In contrast, survival ratios of 0 were observed when WT I-BmoI was used with the pTox backbone (p11-lacY-wtx1) containing no homing site, and when a catalytically inactive I-BmoI variant, R27A, was used in combination with pToxBmoHS. Furthermore, the survival ratio was <0.0001 when pTox contained the intron-containing I-BmoI substrate (pToxBmoIn+), which I-BmoI cleaves poorly (33). These results validate the genetic selection system, showing that cells survive only when I-BmoI can cleave the toxic plasmid that contains the I-BmoI homing site.
Figure 4.
The two-plasmid genetic selection. (A) Schematic of the expression plasmid (pExp) and toxic (reporter) plasmid (pTox). (B) Verification of the genetic selection using variants of pExp and pTox. Survival rates are expressed as the ratio of colonies on chloramphenicol + arabinose plates to colonies on chloramphenicol only plates. WT I-BmoI, pExp expressing WT I-BmoI; R27A I-BmoI, pExp expressing an inactive R27A I-BmoI; randomized I-BmoI, library of I-BmoI variants; p11-lacY-wtx1, parental pTox without I-BmoI target site; ND, not determined.
We used error-prone PCR to generate three independent libraries of I-BmoI variants in pExp, mutagenzied over the entire I-BmoI coding region. The libraries were transformed into the selection strain carrying pToxBmoHS, and survivors identified by plating on selective media with an average survival ratio of ∼0.01 for the three pools. We identified and sequenced 87 independent I-BmoI variants that survived the selection, containing an average of 5.28-nt substitutions (or 3.11 amino-acid substitutions) per clone (Supplementary Figure S3). To determine the baseline mutation frequencies inherent to the error-prone PCR method, we sequenced 62 independent clones plated on non-selective media, which contained an average of 12.26-nt substitutions per clone (Supplementary Figure S3). As expected, the average number of substitutions in the pool of unselected clones was much greater than the number in the selected pool, and the distribution of the number of changes between the selected and unselected clones differed.
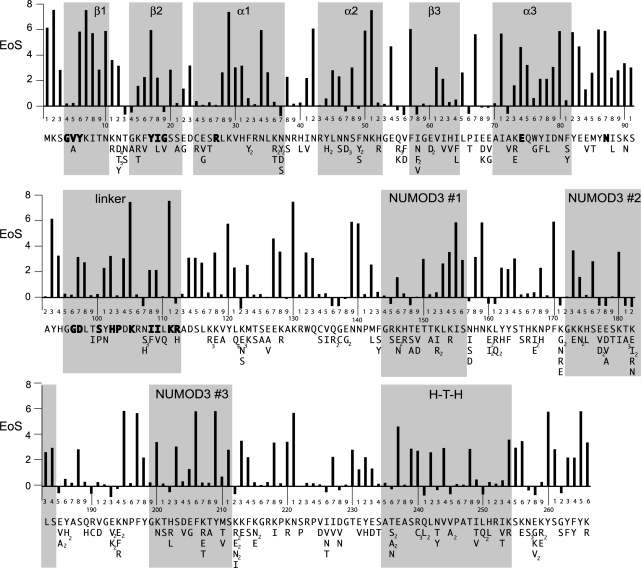
By mapping the mutable positions from the selected clones onto the I-BmoI sequence, we found that amino-acid positions tolerant to substitutions were distributed throughout the length of the coding region (Figure 5). To gain further insight into tolerated mutations within the GIY-YIG catalytic domain, the mutable residues from positions 1–88 were mapped onto a homology-based 3D model of the I-BmoI catalytic domain (Figure 6). Interestingly, the majority of the mutations mapped to the periphery or the backside of the catalytic domain (opposite the proposed catalytic surface). Also, few mutations were observed near the proposed catalytic surface and no substitutions were observed in the four functionally critical residues (Y17, R27, E74 and N87). Some positions tolerated the same substitution more than once, as evidenced by the appearance of the same mutation in independent clones that contained an alternate set of accompanying mutations (Supplementary Table S3). For example, residue N12 tolerated 10 amino-acid substitutions (five to aspartate, four to threonine and one to tyrosine) in different clones, indicating that the position is highly mutable. The significance of mutations in positions outside of the I-BmoI catalytic domain were difficult to interpret due to the absence of similarity between the DNA-binding domain of I-BmoI and that of I-TevI, for which there is an available structure. However, the I-BmoI and I-TevI linker regions, encompassing residues 95–112 of I-BmoI, show some degree of similarity (18). In particular, alanine-scanning mutants in this region of I-TevI functionally divided the linker into deletion intolerant and deletion tolerant regions (17,20). The equivalent deletion intolerant region of the I-BmoI linker appeared to contain few mutations among the selected clones (Figure 5), with some amino-acid positions exhibiting high statistical confidence that the lack of mutation was not due to random chance (see below).
Figure 5.
Unigenic evolution analysis of I-BmoI. Shown is a summary of the mutations found in the 87 selected clones and the EoS value for each position. Non-synonymous substitutions found at each position are indicated beneath the corresponding I-BmoI sequence over the entire length of the protein (multiple independent occurrences of the same mutation are shown in subscript). Shown above each line of sequence is a graph of the evidence of selection (EoS) at each amino-acid position of I-BmoI. Regions of modelled or predicted secondary structure are indicated by grey rectangles, and bold residues indicate the GIY and YIG motifs, functionally critical residues, or residues that are identical between I-BmoI and I-TevI in the linker domain.
Figure 6.
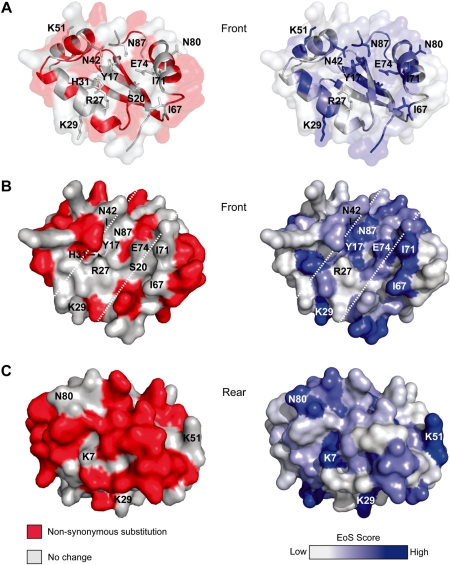
Mapping positions that tolerate non-synonymous substitutions and EoS data onto the I-BmoI homology model. For the left of each panel, amino-acid positions that tolerate non-synonymous substitutions are shown in red and positions where no change was found are in white. On the right of each panel, the EoS score is shown as gradient. Positions with high EoS values are blue, and low EoS values are white. (A) Ribbon representations of a front view of the catalytic domain, with functionally critical residues identified by the present and previous studies indicated. (B) Surface representation of a front view of the I-BmoI catalytic domain, with the putative catalytic cleft bounded by white dashed lines. (C) Surface representation of a rear view of the catalytic domain.
Statistical analysis of unigenic evolution data reveals residues of potential functional importance
More rigorous interpretation of the unigenic evolution data to identify hypo- and hyper-mutable residues requires statistical analyses to determine the probability that any codon will undergo a synonymous or non-synonymous change, taking into account the expected substitution frequencies based on the bias of the mutagenic PCR and the base composition of individual codons. Previous methods utilized a statistic, H, varying between −1 and 1, to determine the mutability of an individual amino-acid position (40,41). We found that H both over- and under-estimates the mutability of positions because it lacks the power to determine if a significant difference exists between the selected and unselected clones at each position (A. Fernandes et al., submitted for publication). For instance, the number of hypomutable residues in a given protein was vastly overestimated by assigning a value of −1 for any residue that had no non-synonymous changes, whereas there will be many positions that do not contain sequence changes simply because the mutagenic method is not saturating.
To take this, and other issues into consideration, we developed a new method for analysing unigenic evolution data called EoS (Evidence of Selection) (A. Fernandes et al., submitted for publication). The EoS value assesses whether the observed frequency of substitutions for any given codon in the selected clones is statistically different from the expected frequency of mutations based on data for the same codon from the unselected pool of clones. Importantly, the EoS value explicitly represents both selection and the power to detect selection for each residue, and is plotted as the log2 ratio of the probability of non-random changes per codon versus the probability of random changes per codon. Thus, an EoS value of 8 represents a 1 in 256 probability that the observed spectrum of mutations occurred at random. Moreover, the fact that some residues possess low EoS scores does not necessarily indicate that these residues are not of potential functional significance, only that the sample size is insufficient to determine their significance.
For example, A/T rich codons are more likely to be mutated due to the bias of Taq polymerase, whereas G/C rich codons are less likely to be mutated. Under the EoS method, a lysine codon (AAA) for which there were no non-synonymous changes in the selected clones would be flagged as significant only if substitutions were observed at the same codon in the unselected clones. As an example, consider position K130. Six non-synonymous substitutions were present in the unselected clones at this position, whereas no substitutions were observed in the selected clones at this position (Supplementary Table S3). Because the spectrum of observed substitutions in the selected clones was significantly different from the observed and expected spectrum of mutations in the unselected clones, K130 is assigned a high EoS value of 8.03, thus providing strong evidence that the position is intolerant to substitution. In contrast, K171 has a low EoS value of −0.65 because a similar spectrum of mutations was observed in the both the selected and unselected clones at this position (Supplementary Table S3).
Using the EoS value as a guide, many positions throughout the length of I-BmoI appear to have potential functional importance (Figure 5). Of particular interest were residues in the catalytic domain that have a high EoS value. For example, K29, N42, N50, K51, F57, I67, I71, N80 and Y82 all have EoS values greater than 6. Apart from F57, none of these positions have been implicated by previous studies to be of potential functional importance. We mapped the EoS values of residues onto the I-BmoI homology model, and compared this representation to that of the mutable amino-acid residues mapped onto the model (Figure 6). Residues that line the catalytic surface were refractory to mutation in the selected clones (Figure 6A and B, and coloured white), whereas the EoS values indicate that the majority of these residues are predicted to be functionally critical (Figure 6A and B, and coloured blue). Many residues with high EoS scores were located away from the active site surface (Figure 6B and C), consistent with structural roles (for instance, K51, which strongly co-evolves H52; Table 1). Again, it is important to note that residues with a low EoS value (coloured white in Figure 6) do not imply that this position is not of potential significance, only that we lack the statistical power to draw such a conclusion given the observed spectrum of mutations (for example, R27).
Inspection of EoS values for the remainder of the I-BmoI sequence revealed that many residues in the linker and C-terminal region appear to be of potential functional significance (Figure 5). For instance, two residues in the linker region that are identical between I-TevI and I-BmoI, K105 and K111, possesses EoS values >8. These residues in I-BmoI are excellent candidates for future mutagenic studies because they correspond to the deletion intolerant region of the I-TevI linker, where mutations drastically reduced or abolished cleavage activity (17,20).
Genetic analysis of site-directed mutants
The data generated by mutual information and unigenic evolution facilitated the identification of amino acids of potential functional importance that could be tested through mutational analyses. We focussed on amino-acid positions within the N-terminal catalytic domain, because these positions could be interpreted within the context of known and modelled GIY-YIG domain structures and previous mutagenic studies, and because mutations in the catalytic domain would not affect the DNA-binding activity of I-BmoI (15,32). Thus, we could be confident that any phenotype we observed for site-directed mutants was due to a defect related to the function of the catalytic domain. We selected amino-acid positions for mutagenesis based on one of the following criteria: (i) the position had a high EoS value, (ii) the residue(s) was identified as co-evolving with another residue, or (iii) the residue possessed high information content in the alignment and had not been previously analysed by mutational studies. For instance, H31 was chosen for mutagenesis because it possessed high information content, had a moderate EoS value of 3.52, and has not previously been targeted by mutagenesis studies of GIY-YIG homing endonucleases. In contrast, position 71, an isoleucine in I-BmoI, displayed very low information content, yet had a high EoS value of 6.37. Furthermore, I71 co-evolves with S20, thus both residues were chosen for further analyses. The rationale for choosing other amino-acid positions for mutagenesis is provided in Supplementary Table S4. In addition to generating site-specific mutants, we chose two clones identified in the unigenic evolution study that contained single mutations within the catalytic domain (N12D and H52R) for further analyses. These clones are representative of amino-acid positions within the catalytic domain that are tolerant to change.
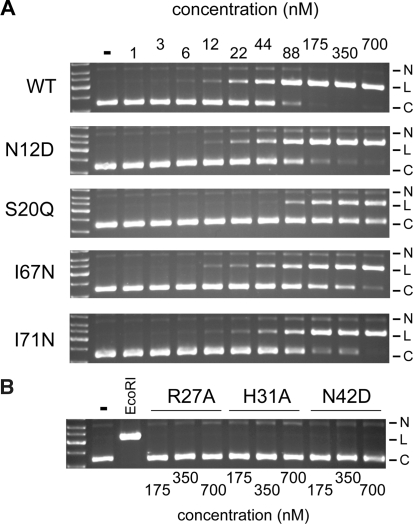
To determine if the identified residues were indeed critical for I-BmoI function, we individually analysed the mutants using the genetic selection, allowing us to calculate a survival ratio that could be directly compared to that for WT I-BmoI. As shown in Table 2, the mutants could be divided into four classes. Class I mutants, Y17F, Y17H, S20A, H31F, H31Y, N42D and N42A, showed the most dramatic effect as none survived the selection. Class II mutants were severely compromised in their survival, with ratios <0.07, and included H31A and I67N. Class III mutants, S20Q, H52R, I71N and I71A, exhibited moderate survival ratios of between 0.14 and 0.43. The class IV mutants, N12D, S48A, K51L, H52Y and Y86F all showed survival ratios essentially equivalent to WT I-BmoI, and thus were considered to have little effect on I-BmoI activity. We considered the possibility that a lack of survival in the selection could be due to the generation of a hyperactive mutant or an altered specificity mutant, resulting in a toxic endonuclease that would cause cell death. For all mutants, however, we observed no reduction in viable cells when cultures were plated on non-selective media during the selection protocol, suggesting that none of the mutants were toxic. Furthermore, we purified the H31A and N42D mutants and found by in vitro cleavage assays that both mutants were severely compromised for cleavage activity (see below and Figure 7), suggesting that the lack of survival in the genetic selection was due to loss of endonuclease activity.
Table 2.
Survival ratios of I-BmoI variants with mutations in the catalytic domain
| Mutant | Survival ratio relative to WTa | |
|---|---|---|
| Class I | Y17F | 0 |
| Y17H | 0 | |
| S20A | 0 | |
| H31F | 0 | |
| H31Y | 0 | |
| N42A | 0 | |
| N42D | 0 | |
| Class II | H31A | 0.003 ± 0.002 |
| I67N | 0.06 ± 0.04 | |
| Class III | S20Q | 0.12 ± 0.02 |
| H52Rb | 0.43 ± 0.04 | |
| I71A | 0.22 ± 0.03 | |
| I71N | 0.20 ± 0.03 | |
| Class IV | N12Db | 1.06 ± 0.03 |
| S48A | 1.02 ± 0.03 | |
| K51L | 0.86 ± 0.10 | |
| H52Y | 0.96 ± 0.05 | |
| Y86F | 0.99 ± 0.04 | |
aValues shown represent averages and standard deviation of at least three independent trials, normalized against the survival ratio for a WT I-BmoI selection performed in parallel.
bThe H52R and N12D mutants were isolated from the pool of selected clones.
Figure 7.
Cleavage activity of WT I-BmoI and site-directed mutants. (A) Shown are representative cleavage assays using 2-fold serial dilutions of the WT and mutant I-BmoI proteins, from 700 nM on the right to 1 nM on the left. The second lane from the left (-) of each gel image is unreacted substrate. (B) For the R27A, H31A and N42D mutants, only the three highest protein concentrations were tested. Substrate linearized by EcoRI is shown in the third lane from the left. Circular (C), linear (L) and nicked (N) plasmid forms are indicated to the right of each gel image.
Cleavage assays with key site-directed mutants
To better understand the effect of individual mutations on endonuclease activity, we next purified key site-directed mutants (N12D, S20Q, H31A, N42D, I67N and I71N) for use in in vitro cleavage assays (Figures 7 and 8). The H31Y and I71A mutants were insoluble and not studied further, and apart from the N12D mutant, mutants with similar survival ratios to WT I-BmoI in the genetic assay were not purified for cleavage assays (Table 2). We first analysed the activity of the mutant proteins relative to WT I-BmoI over a wide range of protein concentrations in cleavage assays using a circular plasmid substrate that contained a single I-BmoI homing site. As shown in Figure 7A, the N12D, S20Q, I67N and I71N mutants exhibited a range of activities relative to WT I-BmoI. The N12D mutant was as active as WT I-BmoI, whereas I61N and I71N displayed intermediate cleavage activities. The S20Q mutant was the least active, and we estimate that this mutant retained ∼30% cleavage activity of WT I-BmoI. In contrast, the H31A and N42D mutants were severely compromised for cleavage activity, with no linear product visible at the highest protein concentrations tested (Figure 7B).
Figure 8.
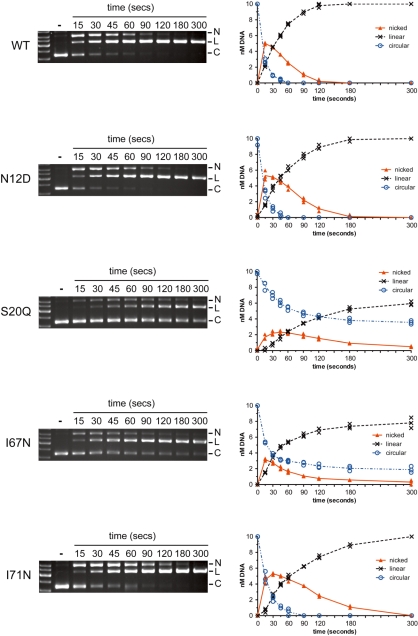
Nicking assays with WT and mutant proteins. Shown are representative agarose gels of time-course assays, with time points in seconds indicated above the gels. The second lane of each gel from the left contains unreacted substrate (−). Circular (C), linear (L) and nicked (N) plasmid forms are indicated to the right of each gel. Beside each gel image is a graphical representation of the disappearance of substrate and appearance of products formed over time. Data points from three independent experiments are plotted, with a continuous line drawn through the average.
To gain additional insight into the observed phenotypes, we performed time-course cleavage assays with WT and mutant proteins to detect the appearance of a nicked intermediate as well as the linear product (Figure 8). These assays were performed with limiting metal ion, as we have previously shown that this reaction condition can effectively distinguish the two sequential nicking reactions that generate a DSB (27). With circular plasmid substrates used in this assay, the first nicking reaction generates a nicked intermediate with slow mobility, and the subsequent nicking reaction generates a linear product. The profile of the nicking assays suggested that the I-BmoI cleavage reaction followed the reaction scheme shown in Equation (1):
| (1) |
where C, N and L are intact plasmid substrate, nicked intermediate and linear product resulting from a DSB. The rate constant for the first nicking reaction is k1, and k2 is the rate constant for the second nicking reaction.
As shown in Figure 8 and Table 3, the reaction profiles and rate constants of the WT protein and N12D mutant were very similar, with conversion of the nicked intermediate to linear product complete by ∼120 s. The reaction profiles of the S20Q and I67N mutants were very different from WT and N12D, with k1 constants indicative of a slower first nicking reaction. Under conditions used in these assays, neither reaction was complete at the end of the time course, making it difficult to determine k2 for the S20Q and I67N mutants using Equation (1) (Supplementary Figure S4). In contrast, Figure 8 illustrates that the I71N mutant displayed a similar first nicking step to the WT and N12D proteins, but that the conversion to linear product was slower as indicated by kinetic analysis (Table 3), consistent with a nicked intermediate accumulating over an extended period of time relative to WT protein. Consequently, conversion to linear product was delayed, but still complete by the end of the time course. Collectively, these results clearly implicate residues identified by MUSE as functionally important, as mutation of these residues generates phenotypes that are distinct from the WT protein.
Table 3.
DNA cleavage by WT I-BmoI and variants identified by MUSE
| protein | k1 (s−1)a | k2 (s−1)b |
|---|---|---|
| WT | 0.080 ± 0.0033 | 0.034 ± 0.00069 |
| N12D | 0.073 ± 0.0045 | 0.024 ± 0.00081 |
| I71N | 0.045 ± 0.0028 | 0.015 ± 0.00051 |
| S20Q | 0.012 ± 0.003 | N.D. |
| I67N | 0.036 ± 0.0005 | N.D. |
ak1 is the rate constant for the first nicking step that generates nicked intermediate from circular substrate. For S20Q and I67N, k1 is valid only at the initial time point (Supplementary Figure S4).
bk2 is the rate constant for the second nicking step that generates a linear product.
N.D., not determined.
DISCUSSION
Detailed insight into protein structure and function can be obtained by synthesizing data from multiple experimental, computational and structural approaches. Here, we elaborate an experimental framework to identify previously unknown functionally relevant residues of the GIY-YIG endonuclease I-BmoI that takes into account evidence other than strict conservation of residues in a multiple sequence alignment. Our goal was to use MUSE to identify key amino acids in I-BmoI for proof-of-principle experiments, highlighting the utility of the MUSE framework. We anticipate that our data will form a platform on which to pursue future structure and function studies of GIY-YIG homing endonucleases and other proteins containing the GIY-YIG domain, and that the MUSE approach will be generally applicable to a broad range of proteins.
Application of MUSE to GIY-YIG homing endonucleases
Past studies on GIY-YIG enzymes have utilized alignments and structural data to identify a set of absolutely conserved residues, subsequent mutation of which abolished cleavage activity. The residues were chosen for mutational analyses on the assumption that conserved residues are important for function, and likely are components of the enzyme’s active site. Such approaches, however, provided limited mechanistic insight and would miss residues that are not universally conserved amongst GIY-YIG enzymes, but nonetheless may be functionally critical. Furthermore, alignments of GIY-YIG containing proteins are dominated by the nucleotide excision repair protein, UvrC, which has a different set of functional constraints than GIY-YIG homing endonucleases. Thus, one aspect of the MUSE approach was to assemble an alignment of GIY-YIG proteins that closely resembled known homing endonucleases, with the goal of enhancing the information specific to GIY-YIG homing endonucleases. For instance, it is known that ∼140 well-aligned sequences will provide sufficient information for covariation analyses to detect co-evolving residues (4). As discussed below, the strongest co-evolving amino-acid pair detected by covariation analyses was S20-I71. One of these residues, I71, has not previously been identified for mutational analyses because this position is highly variable in multiple sequence alignments, yet our data indicate that I71 is a functionally significant residue.
That I-BmoI is a site-specific endonuclease was another critical factor in this study, facilitating the use of a genetic selection where survival in the assay was dependent on endonuclease function. Survival could also be influenced by the solubility or stability of I-BmoI variants. We found, however, that very low levels of I-BmoI expression were required for survival in the selection, suggesting that solubility was not likely a factor. Furthermore, use of a functional genetic selection allowed us to screen through a large population of I-BmoI variants mutagenized over the entire coding region, thus avoiding biases introduced by localized mutagenesis of specific residues or regions of the protein. Using the EoS method for analysis of unigenic evolution data, we obtained sufficient power to identify residues of potential functional significance for a protein the length of I-BmoI (266 residues) by sequencing a relatively small number of selected (87) and unselected (62) clones. In addition to identifying residues of potential importance, MUSE is also expected to identify positions that are tolerant to mutation (hypermutable). For instance, N12 was mutated 10 times in the selected clones as opposed to 7 times in the unselected clones, clearly implying a tolerance to mutation that is not expected to drastically affect I-BmoI function. Indeed, our in vitro analyses of the N12D mutant (the most common mutation at that position) indicate essentially WT levels of activity, validating that the MUSE framework has the power to distinguish between residues in I-BmoI the catalytic domain that are functionally important and those that are not.
Residues within the GIY-YIG domain of I-BmoI that are important for function
The proposed catalytic mechanism for the GIY-YIG domain is based on the predicted function of a set of conserved residues (Y17, R27, E74 N87), each with distinct roles (21,25). Y17 is thought to act as a general base to activate a nucleophilic water; R27 may stabilize the 5′ phosphate of the cleavage intermediate, or contact substrate; E74 coordinates a divalent metal ion that likely functions as the Lewis acid; and N87 is thought mainly to have a structural role in maintaining the active site architecture. Mutation of any of these residues to alanine abolishes cleavage activity, an uninformative phenotype as limited mechanistic insight is gained from catalytically inactive mutants. Hence, many unanswered questions remain regarding the catalytic mechanism of the GIY-YIG domain. For instance, the path of substrate DNA has only been inferred from the position of the conserved residues in the catalytic domain. In this regard, it is worth noting that an I-BmoI R27A mutant displayed a loss of hypersensitivity in footprinting experiments compared to WT protein (27), suggesting that the R27A mutant possesses a DNA contact defect that would not be expected if R27 functioned as a catalytic residue. Thus, additional types of evidence other than structural data are needed to definitively assign functional roles to the presumed set of catalytic residues. Moreover, the significance of residues that lie very close to the proposed active site of I-TevI and UvrC are unknown, and have largely been ignored because they are not conserved in multiple sequence alignments. In the following sections, we discuss the potential structural and functional significance of residues identified by MUSE for which subsequent mutagenesis revealed effects on I-BmoI cleavage activity.
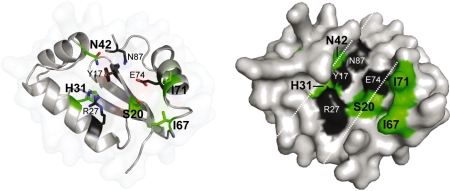
S20 and I71
The S20-I71 pair was chosen for further study because this pair was the highest scoring set of co-evolving residues (Table 1 and Supplementary Table S4). In the GIY-YIG domain alignment, S20 and I71 are both poorly conserved positions (but are conserved between I-BmoI and I-TevI), and are most commonly varied to glutamine and asparagine residues, respectively. In the I-BmoI homology model and the I-TevI structure, the residues are located in the proposed catalytic surface (Figure 9). In the I-TevI structure, S20 lies within hydrogen-bonding distance of the metal-coordinating residue E75, and could potentially position E75 or stabilize the interaction of E75 with divalent metal ion. Intriguingly, I71 can be structurally aligned with L116 of the His-Cys box homing endonuclease I-PpoI, where the residue is inserted into the minor groove of its homing site substrate (42,43). Further evidence for significant roles of S20 and I71 stemmed from the unigenic evolution data, as S20 possessed an EoS score of 3.25 and I71 possessed a score of 6.37. Neither position was mutated in the selected clones, whereas both positions were mutated in the unselected clones (Supplementary Table S3). We made S20A and S20Q mutations and found that the S20A mutant did not survive the genetic selection, whereas S20Q had a survival ratio of 0.063. Cleavage assays with S20Q revealed that at high protein concentrations the enzyme retained ∼30% activity of WT protein, and a similar reduction in cleavage activity was observed when the equivalent residue in UvrC (K32) was mutated (21).
Figure 9.
Summary of functionally relevant residues identified by MUSE. Shown are ribbon (left) and surface (right) representations of the I-BmoI catalytic domain with the residues identified by MUSE (S20, H31, N42, I67 and I71) highlighted in green, and previously identified functionally critical residues (Y17, R27, E74 and N87) shown in black. The catalytic cleft is highlighted as in Figure 1C.
Both I71 mutants (I71A and I71N) survived the genetic selection, but with reduced ratios relative to WT I-BmoI. We purified the I71N mutant for further analysis, and found that it had slightly reduced activity relative to WT protein. Importantly, time-course cleavage assays revealed that the nicked plasmid intermediate persisted longer than for WT protein (Figure 8). Because I-BmoI generates a DSB by two independent and sequential nicking reactions (27), the phenotype of the I71N mutant implies a defect in the second nicking reaction. While the mechanistic basis of this defect will require future study, it is possible that I71 functions in substrate recognition, and that the I71N mutation greatly reduces substrate interactions that effect the second nicking reaction.
One intriguing question regarding the S20 and I71 residues is why do they co-evolve? It is worth noting that the predicted distance separating the residues (∼6 Å) could reflect a limitation of the homology model of the I-BmoI catalytic domain, as these residues are ∼3.5 Å apart in the I-TevI structure. In the 146 sequences used for mutual information analyses, position 20 is most commonly a serine, and position 71 is most commonly an isoleucine. However, there are almost equal occurrences of Q20-N71, Q20-D71 and S20-L71 pairs among the sequences. We attempted to rescue the S20Q mutant by making a second site mutation in position I71, generating a S20Q/I71N double mutant, reasoning that this double mutant represented a tolerated amino-acid pairing at positions 20 and 71. However, the S20Q/I71N double mutant did not survive the genetic selection, suggesting that this combination of residues is not tolerated in the I-BmoI background, and that changes at other positions would be required to restore activity. Both S20 and I71 are in close proximity to the proposed metal-binding residue, E74 (E75 in I-TevI), implying that certain combinations of residues may be favoured for correct positioning of the metal-binding residue within the active site (Figure 9).
H31
H31 was chosen for further analysis due to its high information content in the alignment, the observation that this position has a relatively high EoS score of 3.52, and a lack of mutations in the selected clones (but multiple mutations were present in the unselected clones; Supplementary Table S3). H31 is well conserved amongst GIY-YIG endonucleases, but is replaced by a tyrosine in the equivalent position in UvrC (Y43). In the homology model and I-TevI structure (Figure 9), H31 packs against the side chain of R27 and is within hydrogen bonding distance of both H40 and Y6, but no potential functional role was assigned to H31 based on the I-TevI structure. In the UvrC structure, Y43 was proposed to act as a general base to deprotonate a nucleophilic water molecule (21). To test the importance of H31 in I-BmoI, we made three mutations, H31A, H31F and H31Y, reasoning that the H31F would be structurally similar but chemically inert, while H31Y might retain limited function. Intriguingly, the H31F and H31Y mutants did not survive the genetic selection, while the H31A mutation had a very low survival ratio (Table 2). Unfortunately, the H31Y mutant proved to be insoluble and could not be studied in vitro, whereas no cleavage activity could be detected in vitro with the H31A mutant using plasmid-based cleavage assays. The H31A mutant may retain an extremely low level of activity that is difficult to detect using non-radioactive substrates, but that may be sufficient to permit very a low level of survival in the genetic selection. These data provide the first evidence that H31 may play an important role in the active site of I-BmoI and other GIY-YIG endonucleases, either functioning as a base, by stabilizing the active site architecture by forming a hydrogen bond network with Y6 and H40, or by contacting substrate.
N42
N42 was chosen for mutational studies because it possessed a high EoS score of 6.49 (Figure 5 and Supplementary Table S4). In the GIY-YIG domain alignment, position 42 has little information content, although there is a tendency for this position to be a polar residue. N42 is located in a loop connecting α-helix1 and α-helix2, and is orientated so that the side chain points towards the predicted active site surface (Figure 9). Interestingly, the amino group of N42 is within hydrogen bonding distance of the hydroxyl group of the functionally critical Y17. We made two mutations that would disrupt this interaction, N42A and N42D, and found that both mutants did not survive the genetic selection, and that the N42D mutant displayed no cleavage activity in vitro. N42 may be critical because it functions to correctly position Y17 within the active site of the enzyme.
I67
Like I71, I67 is not conserved amongst GIY-YIG endonucleases, with this position displaying no information content (Figure 2, Supplementary Table S4). In the homology model of the I-BmoI catalytic domain, I67 is positioned in a loop connecting α-helix 3 and β-sheet 3, with its side chain pointed towards the active site surface (Figure 9). Mutation to asparagine reduced in vitro cleavage activity to approximately half that of WT I-BmoI, with defects in both the first and second nicking reactions (Figure 8). We envision that I67 may be involved in substrate interactions, similar to a role for I71.
Residues outside of the catalytic domain predicted to be critical for function
GIY-YIG homing endonucleases are modular proteins, with the N- and C-terminal domains connected by a flexible linker. Past studies on the I-TevI linker have revealed that the linker is required to correctly position the N-terminal GIY-YIG domain on substrate for efficient cleavage (17,18,44). Remarkably, the linker can extend or retract to correctly position the catalytic domain on substrates that contain insertions or deletions that move the preferred cleavage sites from their WT position. This property of the linker has led to the proposal that I-TevI, and perhaps other GIY-YIG endonucleases, generates a DSB by a conformational change mechanism, whereby the linker is a critical component in repositioning the catalytic domain between the bottom- and top-strand nicking reactions. Interestingly, I-BmoI can also reposition the catalytic domain to cleave substrates with +5 and +10 insertions (18), suggesting that the I-BmoI linker functions analogously to the I-TevI linker in spite of limited amino-acid similarity between the two proteins in the linker region. Intriguingly, our unigenic evolution data revealed that many residues within the I-BmoI linker region are predicted to be functionally significant. Notably, K105 and K111, conserved between I-TevI and I-BmoI, have EoS scores greater than 8. These residues correspond to the deletion intolerant region of the I-TevI linker, where 2- or 3-amino-acid deletions abolish cleavage activity, although the functional basis for this phenotype is unknown (17,19). Furthermore, two additional sets of residues in the I-BmoI linker, centered on Y120 and K130, also have high EoS scores. Our data clearly implicate the I-BmoI linker as important for function, and identify a set of residues for future mutagenesis and functional studies.
Apart from a repeated nuclease-associated modular DNA-binding domain motif (NUMOD3) (45), I-BmoI shares little amino-acid similarity with I-TevI in the C-terminal DNA-binding domain, even though the enzymes are isoschizomers and bind the same stretch of thymidylate synthase sequence in Bacillus mojavensis and phage T4, respectively (32). The NUMOD3 motif corresponds structurally to a minor-groove binding α-helix that was first identified in the co-crystal of the I-TevI DNA-binding domain with its homing site substrate (46), and later in the structure of an unrelated HNH endonuclease, I-HmuI (47). In the I-TevI structure, the α-helix is positioned along the minor groove of its DNA substrate, and only one residue in the α-helix (S191) makes a single hydrogen bond contact to the phosphate backbone. Immediately preceding the α-helix, however, is H182 that makes two base-specific hydrogen bonds. Based on sequence predictions, I-BmoI possesses three NUMOD3 motifs, with H147, H175 and H202 corresponding to the critical H182 of I-TevI. The EoS scores for the three histidine residues in I-BmoI are extremely low (Figure 5), and each position had multiple substitutions in the selected clones (Supplementary Table S3), suggesting that these residues perform different functions than the equivalent H182 of I-TevI. Similarly, residues that comprise the predicted I-BmoI helix-turn-helix (HTH) motif at the C-terminal end of the DNA-binding all have low EoS scores (Figure 5). In I-TevI, the analogous HTH motif makes extensive hydrophobic contacts with thymine residues in the substrate, providing specificity to the I-TevI substrate interaction. The tolerance of the I-BmoI HTH to mutation implies that it may function differently than the analogous HTH motif of I-TevI, and a detailed study of the sequence requirements for DNA binding by both HTH motifs would provide an intriguing comparative study.
CONCLUSION
Using a unified experimental approach that synthesizes three distinct types of data, we have identified previously unknown functionally relevant residues in the GIY-YIG homing endonuclease I-BmoI. Our results will form a platform for future studies of I-BmoI and other GIY-YIG-domain containing proteins because residues identified by MUSE, when mutated, generate distinct phenotypes that will provide mechanistic insight. Many of the positions identified by MUSE are non-conserved, and have escaped detection by traditional analyses such as strict conservation in multiple sequence alignments, providing a cautionary tale against using only a single methodology for structure and function studies. We anticipate that the MUSE framework will be generally applicable to a wide range of protein families, requiring ∼140 well-aligned paralogous sequences, an enzymatic activity that forms the basis for a genetic selection and, although not essential, a structural model on which to interpret the MUSE data. For instance, the MUSE framework can be applied to any homing endonuclease or DNA endonuclease without difficulty, and would greatly aid in the re-design of endonucleases against novel target sequences. Unigenic evolution screens have been applied to eukaryotic proteins, including human Pin1 (a prolyl isomerase) and yeast TFIIB (48,49), and each of these proteins could be also analysed within the MUSE framework.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Sciences and Engineering Research Council of Canada Discovery Grant to D.R.E.; B.P.K was supported by an Ontario Graduate Scholarship from the Government of Ontario. Funding for open access charge: Natural Sciences and Engineering Research Council of Canada.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Megan Davey for discussion regarding cleavage assays.
REFERENCES
- 1.Capra JA, Singh M. Predicting functionally important residues from sequence conservation. Bioinformatics. 2007;23:1875–1882. doi: 10.1093/bioinformatics/btm270. [DOI] [PubMed] [Google Scholar]
- 2.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson JD, Laskowski RA, Thornton JM. Predicting protein function from sequence and structural data. Curr. Opin. Struct. Biol. 2005;15:275–284. doi: 10.1016/j.sbi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Martin LC, Gloor GB, Dunn SD, Wahl LM. Using information theory to search for co-evolving residues in proteins. Bioinformatics. 2005;21:4116–4124. doi: 10.1093/bioinformatics/bti671. [DOI] [PubMed] [Google Scholar]
- 5.Tillier ER, Lui TW. Using multiple interdependency to separate functional from phylogenetic correlations in protein alignments. Bioinformatics. 2003;19:750–755. doi: 10.1093/bioinformatics/btg072. [DOI] [PubMed] [Google Scholar]
- 6.Stoddard BL. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 7.Belfort M, Derbyshire V, Cousineau B, Lambowitz A. In: Mobile DNA II. Craig N, Craigie R, Gellert M, Lambowitz A, editors. New York: ASM Press; 2002. pp. 761–783. [Google Scholar]
- 8.Zhao L, Bonocora RP, Shub DA, Stoddard BL. The restriction fold turns to the dark side: a bacterial homing endonuclease with a PD-(D/E)-XK motif. EMBO J. 2007;26:2432–2442. doi: 10.1038/sj.emboj.7601672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dassa B, London N, Stoddard BL, Schueler-Furman O, Pietrokovski S. Fractured genes: a novel genomic arrangement involving new split inteins and a new homing endonuclease family. Nucleic Acids Res. 2009;37:2560–2573. doi: 10.1093/nar/gkp095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grizot S, Smith J, Daboussi F, Prieto J, Redondo P, Merino N, Villate M, Thomas S, Lemaire L, Montoya G, et al. Efficient targeting of a SCID gene by an engineered single-chain homing endonuclease. Nucleic Acids Res. 2009;37:5405–5419. doi: 10.1093/nar/gkp548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosen LE, Morrison HA, Masri S, Brown MJ, Springstubb B, Sussman D, Stoddard BL, Seligman LM. Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nucleic Acids Res. 2006;34:4791–4800. doi: 10.1093/nar/gkl645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat R.J., Jr, Stoddard BL, Baker D. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnould S, Chames P, Perez C, Lacroix E, Duclert A, Epinat JC, Stricher F, Petit AS, Patin A, Guillier S, et al. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J. Mol. Biol. 2006;355:443–458. doi: 10.1016/j.jmb.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 14.Quirk SM, Bell-Pedersen D, Belfort M. Intron mobility in the T-even phages: high frequency inheritance of group I introns promoted by intron open reading frames. Cell. 1989;56:455–465. doi: 10.1016/0092-8674(89)90248-1. [DOI] [PubMed] [Google Scholar]
- 15.Derbyshire V, Kowalski JC, Dansereau JT, Hauer CR, Belfort M. Two-domain structure of the td intron-encoded endonuclease I-TevI correlates with the two-domain configuration of the homing site. J. Mol. Biol. 1997;265:494–506. doi: 10.1006/jmbi.1996.0754. [DOI] [PubMed] [Google Scholar]
- 16.Bell-Pedersen D, Quirk SM, Bryk M, Belfort M. I-TevI, the endonuclease encoded by the mobile td intron, recognizes binding and cleavage domains on its DNA target. Proc. Natl Acad. Sci. USA. 1991;88:7719–7723. doi: 10.1073/pnas.88.17.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q, Dansereau JT, Puttamadappa SS, Shekhtman A, Derbyshire V, Belfort M. Role of the interdomain linker in distance determination for remote cleavage by homing endonuclease I-TevI. J. Mol. Biol. 2008;379:1094–1106. doi: 10.1016/j.jmb.2008.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Q, Derbyshire V, Belfort M, Edgell DR. Distance determination by GIY-YIG intron endonucleases: discrimination between repression and cleavage functions. Nucleic Acids Res. 2006;34:1755–1764. doi: 10.1093/nar/gkl079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dean AB, Stanger MJ, Dansereau JT, Van Roey P, Derbyshire V, Belfort M. Zinc finger as distance determinant in the flexible linker of intron endonuclease I-TevI. Proc. Natl Acad. Sci. USA. 2002;99:8554–8561. doi: 10.1073/pnas.082253699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kowalski JC, Belfort M, Stapleton MA, Holpert M, Dansereau JT, Pietrokovski S, Baxter SM, Derbyshire V. Configuration of the catalytic GIY-YIG domain of intron endonuclease I-TevI: coincidence of computational and molecular findings. Nucleic Acids Res. 1999;27:2115–2125. doi: 10.1093/nar/27.10.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Truglio JJ, Rhau B, Croteau DL, Wang L, Skorvaga M, Karakas E, Dellavecchia MJ, Wang H, Van Houten B, Kisker C. Structural insights into the first incision reaction during nucleotide excision repair. EMBO J. 2005;24:885–894. doi: 10.1038/sj.emboj.7600568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lagerback P, Carlson K. Amino acid residues in the GIY-YIG endonuclease II of phage T4 affecting sequence recognition and binding as well as catalysis. J. Bacteriol. 2008;190:5533–5544. doi: 10.1128/JB.00094-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunin-Horkawicz S, Feder M, Bujnicki JM. Phylogenomic analysis of the GIY-YIG nuclease superfamily. BMC Genomics. 2006;7:98. doi: 10.1186/1471-2164-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pyatkov KI, Arkhipova IR, Malkova NV, Finnegan DJ, Evgen'e;v MB. Reverse transcriptase and endonuclease activities encoded by Penelope-like retroelements. Proc. Natl Acad. Sci. USA. 2004;101:14719–14724. doi: 10.1073/pnas.0406281101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Roey P, Meehan L, Kowalski JC, Belfort M, Derbyshire V. Catalytic domain structure and hypothesis for function of GIY-YIG intron endonuclease I-TevI. Nat. Struct. Biol. 2002;9:806–811. doi: 10.1038/nsb853. [DOI] [PubMed] [Google Scholar]
- 26.Ibryashkina EM, Zakharova MV, Baskunov VB, Bogdanova ES, Nagornykh MO, Den'm;ukhamedov MM, Melnik BS, Kolinski A, Gront D, Feder M, et al. Type II restriction endonuclease R.Eco29kI is a member of the GIY-YIG nuclease superfamily. BMC Struct. Biol. 2007;7:48. doi: 10.1186/1472-6807-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carter JM, Friedrich NC, Kleinstiver B, Edgell DR. Strand-specific contacts and divalent metal ion regulate double-strand break formation by the GIY-YIG homing endonuclease I-BmoI. J. Mol. Biol. 2007;374:306–321. doi: 10.1016/j.jmb.2007.09.027. [DOI] [PubMed] [Google Scholar]
- 28.Van Roey P, Derbyshire V. In: Homing Endonucleases and Inteins. Belfort M.SB, Wood DW, Derbyshire V, editors. Berlin: Springer-Verlag; 2005. pp. 67–83. [Google Scholar]
- 29.Mueller JE, Smith D, Bryk M, Belfort M. Intron-encoded endonuclease I-TevI binds as a monomer to effect sequential cleavage via conformational changes in the td homing site. EMBO J. 1995;14:5724–5735. doi: 10.1002/j.1460-2075.1995.tb00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gasiunas G, Sasnauskas G, Tamulaitis G, Urbanke C, Razaniene D, Siksnys V. Tetrameric restriction enzymes: expansion to the GIY-YIG nuclease family. Nucleic Acids Res. 2008;36:938–949. doi: 10.1093/nar/gkm1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ibryashkina EM, Sasnauskas G, Solonin AS, Zakharova MV, Siksnys V. Oligomeric structure diversity within the GIY-YIG nuclease family. J. Mol. Biol. 2009;387:10–16. doi: 10.1016/j.jmb.2009.01.048. [DOI] [PubMed] [Google Scholar]
- 32.Edgell DR, Shub DA. Related homing endonucleases I-BmoI and I-TevI use different strategies to cleave homologous recognition sites. Proc. Natl Acad. Sci. USA. 2001;98:7898–7903. doi: 10.1073/pnas.141222498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgell DR, Stanger MJ, Belfort M. Importance of a single base pair for discrimination between intron-containing and intronless alleles by endonuclease I-BmoI. Curr. Biol. 2003;13:973–978. doi: 10.1016/s0960-9822(03)00340-3. [DOI] [PubMed] [Google Scholar]
- 34.Chen Z, Zhao H. A highly sensitive selection method for directed evolution of homing endonucleases. Nucleic Acids Res. 2005;33:e154. doi: 10.1093/nar/gni148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eswar N, Marti-Renom A, Webb B, Madhusudhan MS, Eramian D, Shen M, Pieper U, Sali A. Comparative protein structure modeling with MODELLER. Curr. Prot. Bioinform. 2006;15:5.6.1–5.6.30. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 38.Dunn SD, Wahl LM, Gloor GB. Mutual information without the influence of phylogeny or entropy dramatically improves residue contact prediction. Bioinformatics. 2008;24:333–340. doi: 10.1093/bioinformatics/btm604. [DOI] [PubMed] [Google Scholar]
- 39.Wu W, Wood DW, Belfort G, Derbyshire V, Belfort M. Intein-mediated purification of cytotoxic endonuclease I-TevI by insertional inactivation and pH-controllable splicing. Nucleic Acids Res. 2002;30:4864–4871. doi: 10.1093/nar/gkf621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deminoff SJ, Tornow J, Santangelo GM. Unigenic evolution: a novel genetic method localizes a putative leucine zipper that mediates dimerization of the Saccharomyces cerevisiae regulator Gcr1p. Genetics. 1995;141:1263–1274. doi: 10.1093/genetics/141.4.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Behrsin CD, Brandl CJ, Litchfield DW, Shilton BH, Wahl LM. Development of an unbiased statistical method for the analysis of unigenic evolution. BMC Bioinformatics. 2006;7:150. doi: 10.1186/1471-2105-7-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flick KE, Jurica MS, Monnat R.J., Jr, Stoddard BL. DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI. Nature. 1998;394:96–101. doi: 10.1038/27952. [DOI] [PubMed] [Google Scholar]
- 43.Galburt EA, Chadsey MS, Jurica MS, Chevalier BS, Erho D, Tang W, Monnat R.J., Jr, Stoddard BL. Conformational changes and cleavage by the homing endonuclease I-PpoI: a critical role for a leucine residue in the active site. J. Mol. Biol. 2000;300:877–887. doi: 10.1006/jmbi.2000.3874. [DOI] [PubMed] [Google Scholar]
- 44.Bryk M, Belisle M, Mueller JE, Belfort M. Selection of a remote cleavage site by I-TevI, the td intron-encoded endonuclease. J. Mol. Biol. 1995;247:197–210. doi: 10.1006/jmbi.1994.0133. [DOI] [PubMed] [Google Scholar]
- 45.Sitbon E, Pietrokovski S. New types of conserved sequence domains in DNA-binding regions of homing endonucleases. Trends Biochem. Sci. 2003;28:473–477. doi: 10.1016/S0968-0004(03)00170-1. [DOI] [PubMed] [Google Scholar]
- 46.Van Roey P, Waddling CA, Fox KM, Belfort M, Derbyshire V. Intertwined structure of the DNA-binding domain of intron endonuclease I-TevI with its substrate. EMBO J. 2001;20:3631–3637. doi: 10.1093/emboj/20.14.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen BW, Landthaler M, Shub DA, Stoddard BL. DNA binding and cleavage by the HNH homing endonuclease I-HmuI. J. Mol. Biol. 2004;342:43–56. doi: 10.1016/j.jmb.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 48.Behrsin CD, Bailey ML, Bateman KS, Hamilton KS, Wahl LM, Brandl CJ, Shilton BH, Litchfield DW. Functionally important residues in the peptidyl-prolyl isomerase Pin1 revealed by unigenic evolution. J. Mol. Biol. 2007;365:1143–1162. doi: 10.1016/j.jmb.2006.10.078. [DOI] [PubMed] [Google Scholar]
- 49.Zeng X, Zhang D, Dorsey M, Ma J. Hypomutable regions of yeast TFIIB in a unigenic evolution test represent structural domains. Gene. 2003;309:49–56. doi: 10.1016/s0378-1119(03)00492-x. [DOI] [PubMed] [Google Scholar]
- 50.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.