Abstract
According to the recruitment model of transcriptional activation, an activator helps initiate transcription by bringing the RNA polymerase to a specific location on the DNA through interaction with components of the transcriptional machinery. However, it is difficult to isolate and define the activities of specific activator–target pairs experimentally through rearranging existing protein parts. Here we designed and constructed an RNA-based transcriptional activator to study specificity from both sides of the activator–target interface. Utilizing a well-characterized site-specific RNA aptamer for TFIIB, we were able to delineate some key features of this process. By rationally converting an inhibitory aptamer into the activation domain of the activator, we also introduced a new source of submolecular building blocks to synthetic biology.
INTRODUCTION
In eukaryotic organisms, genes encoding messenger RNA are transcribed by RNA polymerase II (Pol II) with the help of general transcription factors (GTFs) (1). To initiate transcription, the TATA-binding protein (TBP) first binds to DNA. Next, TFIIA and TFIIB bind to TBP and the core promoter, followed by TFIIF and Pol II. Finally, TFIIE and TFIIH join to complete the assembly of a Pre-Initiation Complex (PIC) (2,3). In addition, transcription of most genes requires activators, because the formation of chromatin makes the transcriptional ground state restrictive (4). There are two general mechanisms by which activators facilitate transcription: directly through interacting with members of the Pol II entourage or indirectly through altering chromatin structure (5,6). In either case, the location at which the activator binds to DNA determines which gene is activated. Therefore, a transcription activator requires a minimum of two domains, a DNA-binding domain and an activation domain. According to the recruitment model, the target of an activation domain is likely to be either a GTF or a subunit of the Pol II complex. Among the GTFs, TBP and TFIIB are most strongly implicated as the targets of activators (5).
Although the general scheme of transcriptional activation by recruitment has been delineated in broad outline, certain important details remain elusive due to experimental difficulties. For example, an activator often interacts with multiple GTFs, and its effect on a single factor is therefore difficult to isolate; artificial recruitment of a single factor through fusion to a DNA-binding domain does not yield any information about the site or sites on the factor contacted by activators (5). Many protein activators share a common amino-acid composition rather than exhibiting similarity in sequence or structure (7); many RNA sequences have been isolated based on their capability to activate transcription, but the mechanistic basis for this activity is unknown (8,9). Both observations raised questions regarding the specific features of surface topography that are essential for the function of an activation domain.
An understanding of the mechanism underlying a phenomenon should enable the design and construction of different systems that are able to reproduce that phenomenon. Therefore, deliberate creation of novel molecules with explicitly and strictly defined biological function is a reliable way to test our current knowledge. Following this principle, in the present study we implemented the mechanism of transcription activation by recruitment of a GTF using an RNA molecule assembled from refined and standardized parts, especially those derived from aptamers. To explore specificity inherent to both sides of the activator–target interface, we made use of a well-characterized site-specific aptamer as the activation domain of a synthetic activator.
RNA aptamers are generated in an in vitro process emulating Darwinian evolution (10,11). For many proteins, aptamers with a dissociation constant in the nanomolar range have been isolated. Because selection of an aptamer based on affinity for its target is performed outside the cellular and organismal milieu, the aptamer often interferes with the function of the protein when introduced into a living system (12). Consequently, aptamers are routinely used as inhibitors of protein activity. Here we attempted to rationally convert this passive role of aptamers into an active one by placing an aptamer in a designed molecular context, in which it functions as one of several intentionally chosen interacting sites.
In particular, we constructed a ‘transcription activator RNA (taRNA)’ in the yeast Saccharomyces cerevisiae, analogous to a protein-based activator. Using a set of modular parts in a combinatorial manner, we specifically implemented the mechanism of transcriptional activation by recruiting TFIIB to the promoter of reporter genes in the chromatin environment. For this purpose, an RNA aptamer for TFIIB (13), which is a potent inhibitor of transcription by default, was converted into the activation domain of the taRNA by design. With the help of several other constructs originally designed for the yeast three-hybrid system (14), we were able to show that this synthetic RNA molecule activated transcription at a level comparable to a protein activator. Comparing the results obtained by creating new RNA-based factors with those obtained by reorganizing existing protein-based factors allowed us to highlight some critical features of this mechanism.
MATERIALS AND METHODS
Yeast strain and media
The S. cerevisiae strain YBZ-1 (MATa, ura3-52, leu2-3, 112, his3-200, trp1-1, ade2, LYS2::(LexA op)-HIS3, ura3::(LexA op)-LacZ, LexA-MS2-MS2 coat (N55K)) was a gift from Professor Marvin Wickens (University of Wisconsin, Madison) (14). Media consisted of yeast nitrogen base (USBiological), 2% glucose, and synthetic drop-out supplements lacking histidine or histidine and uracil (USBiological). Transformation was performed according to standard protocol using lithium acetate. Yeast were cultured either on agar plates or in liquid medium at 30°C if not otherwise indicated. Growth rate in liquid media was measured by cell density through turbidity at O.D. 600.
Construction of plasmids
The plasmids pIIIA/IRE-MS2 and pAD-IRP, were gifts from Professor Wickens. The plasmid pDB-sansA was derived from pIIIA/MS2-1 (14) by means of the following manipulations. First, the unique NotI site was destroyed by digesting with NotI, then the sticky ends were filled in using the Klenow fragment of DNA polymerase I, and the blunt ends were re-ligated. Second, the EcoRI fragment was removed and replaced with the following sequence containing a NotI site (bold and underlined): 5′-ACTTGAGGTCTGGGCTAAGCCCACT GATGAGTCGCTGAAATGCGACG AAACCTCGAGTCATACTCGCGGCCGCGAGGCGGCAGTATTCCGGTTCGCGCAGAAACA TGAGGATCACCCATGTCCTGTGCCAC AGCGGTGAAACATGAGGATCACCCATGTCCA CCAGCGTTCCGGAGTACTGCCGTGACTCGACGTCTAGCGA TGTGGTTTCGCTACTGATGAGTCCGTGAGGACGAAACGTCGAC-3′.
The plasmids encoding taRNA and its derivatives were constructed by inserting a NotI fragment into the pDB-sansA vector. Each plasmid and the RNA it encoded were named after the standardized aptamer or aptamer derivative being engrafted to the ‘DB-sansA’ scaffold (e.g. the taRNA is ‘DB-B4’ encoded by the plasmid ‘pDB-B4’). The positive control was derived from the RNA-based transcription activator m26-29 (9) with the insert sequence 5′-CGACTCTAGAGGATCGCTT CGGCGGCTAGAACTAGTGGATCCCCCGGGC GCGGAAGATTGTTCCCCCAAGTGGATGCCTAAACCTCATGCAT-3′. The sequences of other inserts are each listed below after the name of plasmid. pDB-B4: 5′-AGCTAATGTAGGATGCTGGGGTAGTCCA GCCCTAGAATAAGCGCTAGTACTACAAGCT-3′. pDB-B4mutS: 5′-AGCTAATGTAGGATGCTGGCTTCGGCCAGCCCTAGAATAAGCGCTAGTACTACAAGC T-3′. pDB-B4mutL: 5′-AGCTAATGTAGGATGC TGGGGTAGTCCAGCCCTA GCTTCGGCTAGTACTACAAGCT-3′. pDB-B4rev: 5′-AGCTTGTAGTACTAGCGCTT ATTCTAGGGCTGGACTACCCC AGCATCCTACATTAGCT-3′. pDB-B60: 5′-GGGAGAATTCAACTGCCATCTA GGCGGTGATCGCACAGACACGGGCACTG ATGCGGCTCCC-3′. pDB-TBP12: 5′-GCCGTGCCCGGTTTG GATAGGCACATAAGAC-3′. pDB- TBP101: 5′-AGAATTCAACTCTTCGGAG CCAAGGTAAACAATTCAGT TAGTGGAATGAAACTG-3′. pDB-FC: 5′-TCGCTCACGATACAGCACTGAT TGCGGTCGATGGTAGCG TTGATGGGCCACGCGCG A-3′. pDB-RA1: 5′-GAATTCAACTGCCTTCGGGCAT CGCGATACAAAATTAAGTTGAACGCGAGTTC-3′. Inserts were prepared by bi-directional extension of overlapping oligonucleotides. All synthetic oligonucleotides were purchased from Integrated DNA Technologies, Inc.
Secondary structure prediction and confirmation
Secondary structures of RNA constructs were predicted using the mfold program (v. 3.2) (15). To identify a stable stem as the point of integration, a series of derivatives were constructed based on the most thermodynamically stable predicted structure of an aptamer and tested for binding activity (16).
Assays for β-galactosidase activity
The filter assay was performed as described in (17). The colonies were lifted from the plate using nitrocellulose filters (Millipore). Cells were permeabilized by freezing the filter in liquid N2. The enzymatic reaction was initiated by overlying the filter on a piece of Whatman 3 MM paper saturated with X-gal containing Z buffer (60 mM Na2HPO4, 60 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, and 1mg/ml X-gal), and allowed to proceed for 30 min. For the quantitative liquid assay, a standard protocol (18) was followed using O-nitrophenyl-β-d-galactopyranoside (ONPG) as the substrate. Activity was calculated as Miller units and normalized to the Gal4-based positive control for the three-hybrid system (14). At least three independent cultures of each construct were measured.
Single cell analysis
Continuous optical measurements of individual localized cells were performed on a microfluidic gridded array, the LiveCell Array (Molecular Cytomics), with a standard microscope (Olympus) (19). A 500-μl aliquot of cell suspension was combined with a 500-µl aliquot of 250 μg/ml Concanavalin A-Tetramethylrhodamine (ConA-TAMRA) conjugate, a labeling reagent that allowed the cells’ position to be registered. The cells were washed with media and loaded onto the array. The microfluidic chamber enabled addition of the substrate 5-dodecanoylaminofluorescein di-β-D-galactopyranoside (C12FDG, Molecular Probes) in media to the localized cells. The measurements were taken with optical filter systems specific for fluorescein (C12FDG) and TAMRA.
RESULTS
Mechanism-driven choice and refinement of an aptamer
Based on the recruitment model of PIC assembly, we surveyed published aptamers to find a candidate that would function as an activation domain when tethered to a promoter. Five aptamers were identified: one for Pol II, two for TBP, and two for TFIIB. All of these aptamers showed inhibitory effects on transcription either in vitro or in vivo. Four of them were deemed unfit to act as an activator based on the following mechanistic information. First, the aptamer for the Pol II, FC (20), binds in the Pol II active center cleft and prevents the DNA template from entering (21). Therefore, FC would not be able to activate transcription even if it were used to bring Pol II to a promoter. Second, the two aptamers for TBP, AptTBP-12 and AptTBP-101, recognize two discrete sites respectively (16,22). Both inhibit transcription by preventing PIC formation, although in mechanistically distinctive ways attributable to their specific binding sites (16). Third, one of the aptamers for TFIIB, AptTFIIB-60, inhibits transcription by preventing the incorporation of TFIIB into the PIC (13).
Intriguingly, the other TFIIB aptamer, AptTFIIB-4, inhibitory as it is, does not affect TFIIB occupancy at the PIC, nor does it affect TBP and TFIIA levels on a template (13). Therefore, AptTFIIB-4 met our design criteria and was chosen as the putative activation domain. Because it binds only TFIIB and not any other factor, its target in the Pol II machinery can be precisely assigned. Because it does not prevent TFIIB from being incorporated into the PIC, when tethered to a promoter we presumed AptTFIIB-4 would activate transcription either through recruitment of TFIIB to the PIC or through stabilization of a pre-bound complex on DNA. Because AptTFIIB-4 alone is inhibitory, its tethered form must activate transcription through the recruitment of TFIIB rather than by stabilizing the pre-formed PIC. Viewed retrospectively, the assay for the effect of the aptamer on in vitro transcription (13) was analogous to in vivo ‘squelching’ experiments (23,24), thus excluding the possibility of activation through PIC stabilization. Taken together, this specific functional information allowed us to implement the predetermined mechanism from the bottom up.
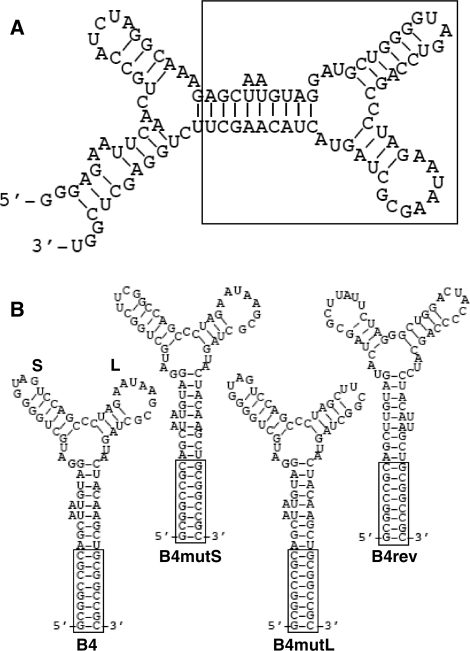
The first step of this implementation was to refine the ‘raw’ aptamer into a standardized portable submolecular module. As we did before for other aptamers (16), we made a series of mutations of AptTFIIB-4 to map the ‘true’ aptamer moiety and identify a stable stem as the point of integration (see Supplementary Data for details). This led to the proposed secondary structure depicted in Figure 1A, where the sequence and structure enclosed in the rectangular box is necessary and sufficient for TFIIB binding. This minimized version of the aptamer was composed of a three-way junction, exiting from which were two stem loops (‘S’ and ‘L’) and a stem with an open end (Figure 1B). To the open stem we added a ‘GC-clamp’ (25) by pairing the 5′ and 3′ termini, which served as a standardized interface with the rest of the composite molecule.
Figure 1.
AptTFIIB-4 and its derivatives. (A) Predicted secondary structure of AptTFIIB-4. The structure was generated by mfold and supported by mutational analysis. The box indicates the ‘B4’ aptamer moiety. (B) Standardized aptamer B4 and its derivatives. The ‘S’ and ‘L’ loops are indicated in B4. The boxes indicate the identical GC-clamp in each structure.
Molecular and genetic design
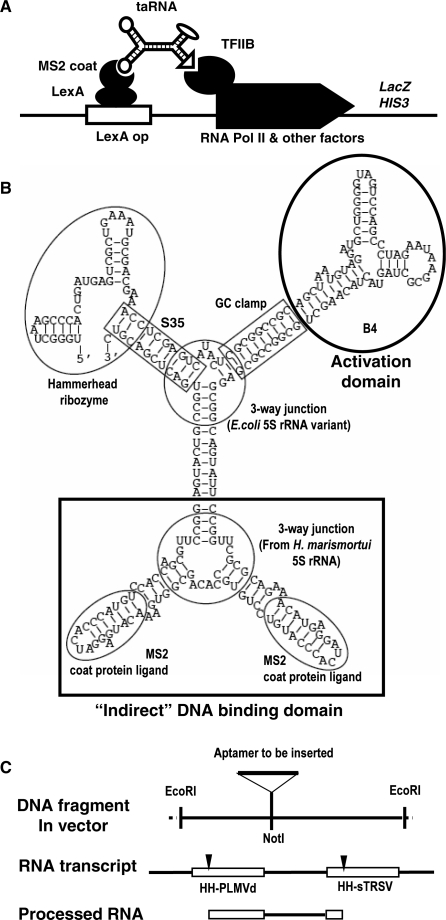
We converted the inhibitory AptTFIIB-4 into the activation domain of the taRNA by providing its minimized version (hereafter referred to as B4) with a designed molecular context (Figure 2A). To integrate it into the cellular regulatory network, the taRNA was produced by transcription from a synthetic gene. Both the taRNA and its expression system were designed using a modular approach, with each part previously tested individually in vivo. To design the composite taRNA molecule, we employed a method described elsewhere, which allows the use of a library of parts with a common junction at the submolecular level to simplify the prediction of function when the parts are combined (26). Specifically, the taRNA was composed of four functional elements—the B4 aptamer, two copies of the MS2 coat protein ligand, and a hybrid hammerhead ribozyme—connected in a single molecular entity through two three-way junctions (Figure 2B).
Figure 2.
Molecular and genetic design of taRNA. (A) Schematic diagram showing the function of the taRNA. (B) The most stable structure of the taRNA as predicted by mfold. Each structural and functional unit is indicated by a circle or a rectangle, with the name of the element next to it. The three-way junction connecting the two MS2 coat protein ligands was derived from the 5S rRNA of H. marismortui (47). The other three-way junction was a stable 5S rRNA variant named ‘System F’ (48). (C) Schematic diagram depicting the EcoRI segment in the pDB series of plasmids before and after subcloning, and the RNA transcribed from the synthetic gene before and after further processing by hammerhead ribozyme cleavage. The DNA and RNA segments are aligned to show their relationship. The two ribozymes were taken from peach latent mosaic viroid (PLMVd) and tobacco ringspot virus satellite RNA (sTRSV), respectively (31). The cleavage sites of the ribozymes are indicated by arrowheads.
Protein activators achieve their sequence-specific association with DNA either directly through a DNA-binding domain [e.g. yeast Gal4 (27)] or indirectly through an adaptor protein [e.g. VP16 of herpes simplex virus (28)]. For the taRNA we took the latter strategy: instead of a direct DNA–RNA interface, we employed a LexA-MS2 coat protein fusion construct that had been previously used to localize RNA to DNA through more specific DNA–protein and protein–RNA recognition (29). As an indirect DNA association domain, we used a dimeric configuration of the coat protein ligand to improve its avidity to the MS2 coat protein.
To express the taRNA we designed a transcriptional unit driven by RNA polymerase III (Pol III), as TFIIB is not involved in Pol III transcription and its aptamer would not affect the production of the taRNA. In addition to the coding region, the synthetic gene comprised the promoter and terminator of the RNase P RNA gene RPR1 (30). The same promoter–terminator system was used previously to express the aptamer FC, but failed to generate a detectable growth phenotype (20), suggesting that the taRNA thus produced would be kept at an appropriate level without causing systemic effects (e.g. squelching). However, the RPR1 promoter is intragenic, and in the FC construct the leader sequence was not cleaved off (20). Also carried on the transcript is part of the terminator sequence including a stretch of uridines. To eliminate these flanking sequences, we used two cis-acting hammerhead ribozymes known to be functional in vivo (31) (also see Supplementary Data).
After self-cleavage, the 3′ half of the 5′ ribozyme and the 5′ half of the 3′ ribozyme would form a new ‘hybrid hammerhead ribozyme’, which is part of the taRNA shown in Figure 2B. Because it is not poly-adenylated, the taRNA would be retained inside the nuclei as we demonstrated previously (12). Inside cells most RNA molecules are degraded by exonucleases that digest single-stranded RNA. Enhanced stability has been demonstrated by confining the termini of an RNA in a double-stranded stem (termed S35) (32). The Stem III of the hybrid ribozyme was such an S35 (Figure 2B). The hammerhead ribozyme cleaves the RNA backbone to yield a 5′ hydroxyl and a 2′, 3′ cyclic phosphodiester; under certain conditions it might ligate these products using the bond energy retained in the non-hydrolyzed cyclic product to re-form a phosphodiester (33).
Sequences encoding all parts of the taRNA except the activation domain were embedded in the yeast/Escherichia coli shuttle vector pDB-sansA, which produced an RNA construct with DNA-binding (DB) activity conferred by the protein adaptor, but without an activation domain (hence ‘sansA’). Figure 2C depicts a section of the vector between the two EcoRI sites, in alignment with the corresponding RNA transcript before and after ribozyme cleavage. The taRNA gene was generated by inserting the B4 sequence through subcloning into a NotI site to form a ‘DB-B4’ construct. The two resulting NotI sequences would form a GC-clamp to insulate the incoming aptamer from the rest of the molecule to ensure its correct folding. Alternative aptamers and positive or negative control units were also added to the ‘DB-sansA’ scaffold through this standardized procedure.
Activity of the transcription activator RNA
The taRNA activity was measured through the transcription of reporter genes under the control of LexA operators, with the help of a LexA-MS2 coat protein fusion adaptor. The yeast strain YBZ-1, designed for the three-hybrid system, contains both the reporters and the adaptor. Importantly, the reporter genes HIS3 and lacZ are in the chromatin environment (14). To verify the predetermined function of the activation domain in the taRNA, we employed a positive control along with a battery of negative controls. For the positive control, we used the RNA-based transcription activator m26-29 (9). This RNA was selected for its capability of activating transcription, but its target and mechanism are unknown. Our first negative control was the antisense sequence of B4 (B4rev)—a ‘DB-B4rev’ construct was a cloning byproduct of the taRNA. As shown in Figure 1B, B4rev was able to form a secondary structure similar to B4. We also generated two other negative controls, B4mutS and B4mutL, by replacing one of the two loops of B4 with a UUCG-tetraloop. The ‘empty’ vector also served as a negative control as it would produce the ‘DB-sansA’ RNA analogous to an activation domain deletion.
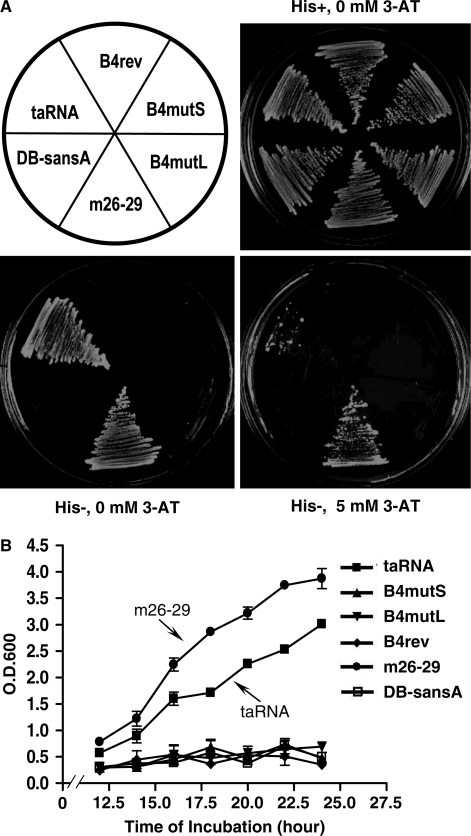
After transformation, the expression of the RNA constructs in each yeast strain was confirmed by RT–PCR (Supplementary Data), and the transformants expressing the taRNA and the controls were plated at 30°C on media with or without histidine. As shown in Figure 3A, all strains grew equally well on the plate with histidine, indicating that, as predicted, taRNA at this level of expression had no significant systemic effect. In contrast, on the plate without histidine, only strains expressing the taRNA or the positive control DB-m26-29 were able to survive. To confirm this result, we performed two more assays. First, we added increasing amounts of 3-aminotriazole (3-AT), a competitive inhibitor of His3p activity, to the his- medium to test the strength of the taRNA. As shown in Figure 3A, in media containing 5 mM 3-AT, some yeast colonies expressing the taRNA were still able to grow. Second, we measured the growth rate of these strains in liquid media. As shown in Figure 3B, the taRNA was able to sustain a growth rate comparable to DB-m26-29, although none of the negative controls grew without histidine.
Figure 3.
Activity of the taRNA as measured by the HIS3 reporter gene transcription. (A) Growth on agar plates. Each strain is identified by the molecular construct it harbors and its position on the plates is indicated in the drawing (upper left). The upper right plate is a control plate (ura−, his+) demonstrating that all strains are viable and grow at a normal rate in the absence of selection for HIS3. The two lower plates lack histidine. The lower right plate also contains 5 mM 3-AT. (B) Growth curves measured in liquid media lacking histidine. Values represent the average of three independent cultures of each strain. Error bars show standard deviations.
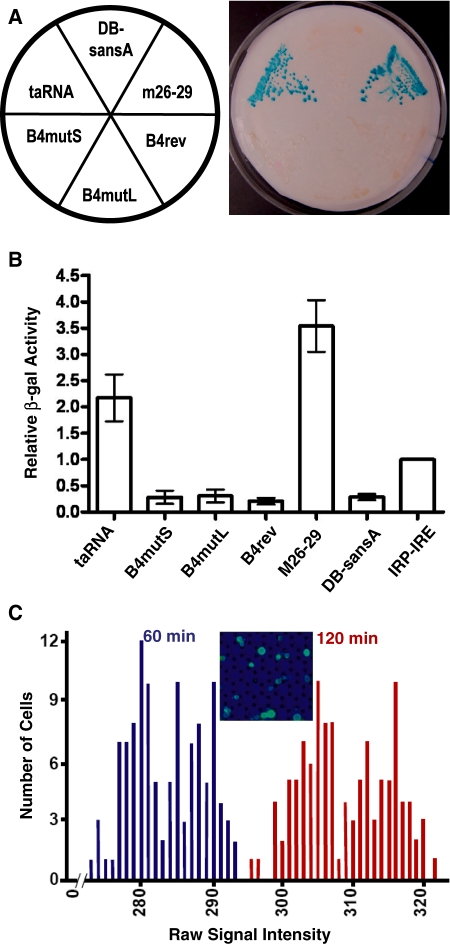
As an alternative and independent reporter gene we used lacZ, the activity of whose product is easier to quantify. First, we measured the β-galactosidase activity using a qualitative filter assay (17). As shown in Figure 4A, permeabilized cells in yeast colonies expressing the taRNA or the positive control were able to convert X-gal to produce a blue color, but all the negative controls lacked this capability. To quantify the specific activity of β-galactosidase, we next used a liquid assay with cell lysates and ONPG (18). As a benchmark, we compared the activity of taRNA to the positive control used in the three-hybrid screening, in which a Gal4 activation domain is tethered to DNA using the binding of the iron regulatory protein 1 (IRP1) to the iron response element (IRE) (29). As shown in Figure 4B, the taRNA has activity more than 2-fold greater than this control.
Figure 4.
Activity of the taRNA as measured by β-galactosidase activity. (A) Qualitative filter lift assay of permeabilized cells from a ura-, his- plate using X-gal. (B) Quantitative colorimetric assay of cell lysate using ONPG. The average activity of each strain is shown normalized relative to the Gal4 three-hybrid positive control (IRE-MS2 + AD-IRP) (29). Error bars show standard deviations. (C) Optical array analysis of living single cells using C12FDG. A total of 114 cells were measured one and two hours after adding the substrate. The inset shows a section of the array.
Furthermore, to gauge the population diversity, we determined the β-galactosidase activity in living cells using optical arrays of single cells (19) with the chromogenic substrate C12FDG. The substrate contains a lipophilic tail enabling its entry into the cell. Once processed by the enzyme, this tail is lost, and a fluorescent product is trapped inside the cell. This assay allowed examination of cells individually under identical conditions. As shown in Figure 4C, the taRNA cells exhibited minimal heterogeneity. The standard deviations associated with the optical signal responses across the population were less than 2% of the measured signal intensity.
Specificity of the transcription activator RNA
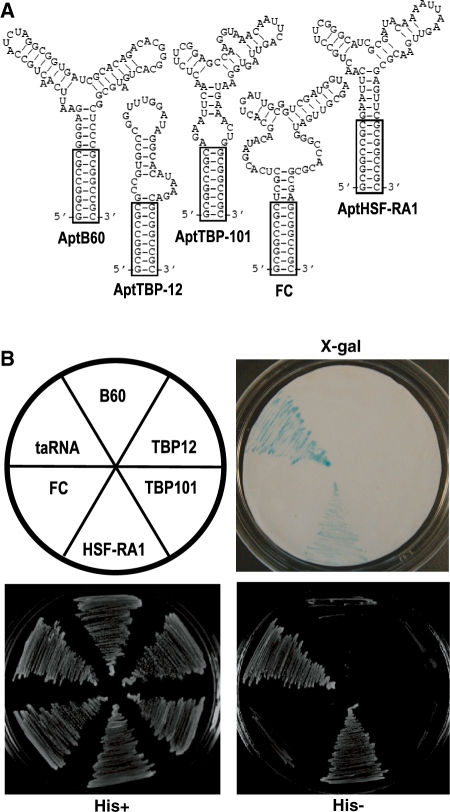
Whereas B4 was the main focus of the present study, we used other aptamers for components of the Pol II machinery to help clarify and corroborate our mechanistic claims by comparing and contrasting their effects with those of B4’s. As shown in Figure 5A, we refined four other aptamers for TBP, TFIIB, or Pol II by trimming their sequences and converting them to the standardized form. Our modular design allowed rapid and easy addition of these standardized aptamers to the DB-sansA scaffold. As predicted, none of these aptamers were able to function as an activation domain (Figure. 5B). While the failure of FC, which binds the active cleft of Pol II, provides a straightforward illustration of the importance of the binding site on the target (21), the other three constructs afforded more subtle insights. Both TBP and TFIIB are primary targets of transcription activators, yet simply recruiting them to the promoter was not enough to activate transcription. In the case of TBP, fusion with a DNA-binding domain was able to activate (34–36), but recruitment through a tethered aptamer was not. Presumably the fusion protein was incorporated into the holoenzyme, but aptamer binding prevented TBP from doing so, even when the binding site on TBP is the DNA-binding surface (16). A more interesting case is TFIIB. Both B4 and AptTFIIB-60 bind to TFIIB with similar affinity, and their contact sites overlap on the c-terminal core domain (cIIB) at or close to the linker region (13). Transcription activators such as Pho4 have also been shown to bind cIIB (37). However, the two aptamers for TFIIB behaved differently both in vitro and in vivo. Taken together, this collection of constructs demonstrated that the specific site of contact and the mode of contact with the target were important for activation to occur.
Figure 5.
Specificity of the taRNA. (A) Secondary structures of five refined and standardized aptamers. Boxes indicate the identical GC-clamp in each structure. (B) Capability or lack thereof of each aptamer to function as an activation domain, as assayed by HIS3 reporter gene transcription and β-galactosidase activity.
Finally, we attempted to use these newly acquired mechanistic insights to build another aptamer-based transcription activator with a specified target. Previously, we isolated an RNA aptamer for the heat-shock factor (HSF) (38). HSF recognizes the heat-shock elements on DNA and upon heat shock activates transcription by recruiting other factors or complexes to the promoter. The aptamer, AptHSF-RA1, binds to the HSF DNA-binding domain and the linker region but not the domains involved in trimerization and activation. Therefore, we reasoned that AptHSF-RA1 might be used to recruit HSF to a non-heat-shock promoter. Not being occluded by the aptamer, the activation domain of the HSF would remain functional. Indeed, when we expressed a DB-RA1 construct (AptHSF-RA1 in the pDB-sansA vector) in yeast, we observed transcription activation of both reporter genes at a moderate level, lower than that produced by the taRNA but significantly above the background level (Figure 5B). However, this activity was indifferent to temperature change. Apparently, recruiting HSF this way was not able to fully recapitulate the natural molecular interactions and modification at the promoter (39). Nonetheless, the activity we observed lends further support to the mechanism of recruitment illustrated by the B4-derived taRNA.
DISCUSSION
In this study, we took a forward engineering approach to synthesize a transcription activator to implement the mechanism of transcription activation by recruitment of a GTF. By comparing the expected and observed behavior of the designed molecules, we were able not only to validate this mechanism, but also to investigate the specificity between an activator and its target. In the future, the taRNA will be used as a model to study the events that occur in the process of transcription activation after the B4 associates with TFIIB.
Our work was greatly facilitated by existing constructs originally designed for the yeast three-hybrid screening (14). However, there is a fundamental difference between our work and the three-hybrid system. The general utility of the three-hybrid system lies in the fact that transcriptional activation in this system relies on the physical rather than the biological properties of the RNA (29). Numerous types of RNA–protein interactions, including the binding specificity of aptamers, have been analyzed using this system, regardless of the normal function of the RNA molecule (40,41). In contrast, here the taRNA was designed to provide the biological function and integrate itself into the functional context of PIC formation. This difference can be appreciated by comparing our work with a recently published study (42). In this elegant piece of work, as well as similar works from the same group (25,43), an RNA aptamer was also involved and the target of the aptamer was also a transcription factor. However, the B4 aptamer in our construct is functionally not analogous to the anti-NF-κB aptamer, but instead analogous to the GAL4 activation domain in their system.
In order to regulate biological processes, proteins and other molecules associate with each other through a complex network of interactions. Modification of the network connectivity forms the basis of experimental perturbation and therapeutic intervention. Traditional methods modify such connectivity by blocking or abolishing molecular interactions. Following this strategy, RNA aptamers have been used as protein antagonists for more than a decade (44). An alternative and possibly more effective strategy is to introduce new links between non-interacting molecules. This concept is rarely used because bridging two molecules specifically and selectively is substantially more difficult than blocking one molecule. A recent study converted a monomeric aptamer into a non-covalent homodimer functioning as an agonist by inducing the targets to multimerize (45). Differing from this approach, our implementation of the aptamer-based taRNA can be viewed as a molecular surgery that ‘rewired’ the connectivity of an existing regulatory network to bypass a native activator.
By transfiguring an inhibitory RNA aptamer into an activation domain and constructing covalent composites using a modular and combinatorial procedure, we have expanded the utility of aptamers and introduced a new type of building blocks, or ‘synthons,’ for use in synthetic biology. Like proteins, RNA can form complex structures with sophisticated functions, although these are used only occasionally by contemporary organisms. Most cellular functions are actualized by proteins because proteins possess two distinctive characteristics: first, a single protein molecule is capable of bearing more than three sites recognizable specifically by other molecules, collectively forming a scale-free network (46); second, proteins can be genetically encoded, and their biosynthesis and degradation can be regulated by environmental and developmental cues. To realize the potential of RNA, we designed novel molecules to be ‘protein-like’ in these two fundamental aspects. This type of RNA construct is easier to design than novel protein molecules and has much less immunogenicity when used in vivo. Based on these features, we envision that more engineered biological systems will make use of composite RNA aptamers to empower design-based predictive modification of organisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Concept Award from the US Department of Defense (#BC075466 to H.S.) Funding for open access charge: College of Arts and Sciences, University at Albany, State University of New York.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the other members of the Shi Lab for insightful discussions and Drs R. Zitomer and K. Nishikawa for critical comments on the manuscript.
REFERENCES
- 1.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woychik NA, Hampsey M. The RNA polymerase II machinery: structure illuminates function. Cell. 2002;108:453–463. doi: 10.1016/s0092-8674(02)00646-3. [DOI] [PubMed] [Google Scholar]
- 3.Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, Cramer P. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 4.Struhl K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- 5.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 6.Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 7.Ma J, Ptashne M. A new class of yeast transcriptional activators. Cell. 1987;51:113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- 8.Saha S, Ansari AZ, Jarell KA, Ptashne M. RNA sequences that work as transcriptional activating regions. Nucleic Acids Res. 2003;31:1565–1570. doi: 10.1093/nar/gkg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buskirk AR, Kehayova PD, Landrigan A, Liu DR. In vivo evolution of an RNA-based transcriptional activator. Chem. Biol. 2003;10:533–540. doi: 10.1016/s1074-5521(03)00109-1. [DOI] [PubMed] [Google Scholar]
- 10.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 11.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 12.Shi H, Hoffman BE, Lis JT. RNA aptamers as effective protein antagonists in a multicellular organism. Proc. Natl Acad. Sci. USA. 1999;96:10033–10038. doi: 10.1073/pnas.96.18.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sevilimedu A, Shi H, Lis JT. TFIIB aptamers inhibit transcription by perturbing PIC formation at distinct stages. Nucleic Acids Res. 2008;36:3118–3127. doi: 10.1093/nar/gkn163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein DS, Buter N, Stumpf C, Wickens M. Analyzing mRNA-protein complexes using a yeast three-hybrid system. Methods. 2002;26:123–141. doi: 10.1016/S1046-2023(02)00015-4. [DOI] [PubMed] [Google Scholar]
- 15.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi H, Fan X, Sevilimedu A, Lis JT. RNA aptamers directed to discrete functional sites on a single protein structural domain. Proc. Natl Acad. Sci. USA. 2007;104:3742–3746. doi: 10.1073/pnas.0607805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breeden L, Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harbor Symp. Quant. Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- 18.Ralser M, Goehler H, Wanker EE, Lehrach H, Krobitsch S. Generation of a yeast two-hybrid strain suitable for competitive protein binding analysis. BioTechniques. 2005;39:165–166. doi: 10.2144/05392BM01. 168. [DOI] [PubMed] [Google Scholar]
- 19.Biran I, Walt DR. Optical imaging fiber-based single live cell arrays: a high-density cell assay platform. Anal. Chem. 2002;74:3046–3054. doi: 10.1021/ac020009e. [DOI] [PubMed] [Google Scholar]
- 20.Thomas M, Chedin S, Carles C, Riva M, Famulok M, Sentenac A. Selective targeting and inhibition of yeast RNA polymerase II by RNA aptamers. J. Biol. Chem. 1997;272:27980–27986. doi: 10.1074/jbc.272.44.27980. [DOI] [PubMed] [Google Scholar]
- 21.Kettenberger H, Eisenfuhr A, Brueckner F, Theis M, Famulok M, Cramer P. Structure of an RNA polymerase II-RNA inhibitor complex elucidates transcription regulation by noncoding RNAs. Nat. Struct. Mol. Biol. 2006;13:44–48. doi: 10.1038/nsmb1032. [DOI] [PubMed] [Google Scholar]
- 22.Fan X, Shi H, Adelman K, Lis JT. Probing TBP interactions in transcription initiation and reinitiation with RNA aptamers that act in distinct modes. Proc. Natl Acad. Sci. USA. 2004;101:6934–6939. doi: 10.1073/pnas.0401523101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 24.Farrell S, Simkovich N, Wu Y, Barberis A, Ptashne M. Gene activation by recruitment of the RNA polymerase II holoenzyme. Genes Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- 25.Cassiday LA, Maher L.J., 3rd Yeast genetic selections to optimize RNA decoys for transcription factor NF-kappa B. Proc. Natl Acad. Sci. USA. 2003;100:3930–3935. doi: 10.1073/pnas.0736013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu D, Shi H. Composite RNA aptamers as functional mimics of proteins. Nucleic Acids Res. 2009;37:e71. doi: 10.1093/nar/gkp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma J, Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- 28.Cress WD, Triezenberg SJ. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 29.SenGupta DJ, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl Acad. Sci. USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JY, Rohlman CE, Molony LA, Engelke DR. Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol. Cell. Biol. 1991;11:721–730. doi: 10.1128/mcb.11.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 32.Thompson JD, Ayers DF, Malmstrom TA, McKenzie TL, Ganousis L, Chowrira BM, Couture L, Stinchcomb DT. Improved accumulation and activity of ribozymes expressed from a tRNA- based RNA polymerase III promoter. Nucleic Acids Res. 1995;23:2259–2268. doi: 10.1093/nar/23.12.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buzayan JM, Feldstein PA, Segrelles C, Bruening G. Autolytic processing of a phosphorothioate diester bond. Nucleic Acids Res. 1988;16:4009–4023. doi: 10.1093/nar/16.9.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chatterjee S, Struhl K. Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature. 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 35.Klages N, Strubin M. Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature. 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 36.Xiao H, Friesen JD, Lis JT. Recruiting TATA-binding protein to a promoter: transcriptional activation without an upstream activator. Mol. Cell. Biol. 1995;15:5757–5761. doi: 10.1128/mcb.15.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu WH, Hampsey M. An activation-specific role for transcription factor TFIIB in vivo. Proc. Natl Acad. Sci. USA. 1999;96:2764–2769. doi: 10.1073/pnas.96.6.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao X, Shi H, Sevilimedu A, Liachko N, Nelson HC, Lis JT. An RNA aptamer that interferes with the DNA binding of the HSF transcription activator. Nucleic Acids Res. 2006;34:3755–3761. doi: 10.1093/nar/gkl470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorger PK, Pelham HR. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988;54:855–864. doi: 10.1016/s0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- 40.Zhang B, Kraemer B, SenGupta D, Fields S, Wickens M. Yeast three-hybrid system to detect and analyze RNA-protein interactions. Methods Enzymol. 2000;318:399–419. doi: 10.1016/s0076-6879(00)18066-8. [DOI] [PubMed] [Google Scholar]
- 41.Konig J, Julius C, Baumann S, Homann M, Goringer HU, Feldbrugge M. Combining SELEX and the yeast three-hybrid system for in vivo selection and classification of RNA aptamers. RNA. 2007;13:614–622. doi: 10.1261/rna.334307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wurster SE, Bida JP, Her YF, Maher L.J., 3rd Characterization of anti-NF-kappaB RNA aptamer-binding specificity in vitro and in the yeast three-hybrid system. Nucleic Acids Res. 2009;37:6214–6224. doi: 10.1093/nar/gkp670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cassiday LA, Maher L.J., 3rd In vivo recognition of an RNA aptamer by its transcription factor target. Biochemistry. 2001;40:2433–2438. doi: 10.1021/bi002376v. [DOI] [PubMed] [Google Scholar]
- 44.Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: an emerging class of therapeutics. Annu. Rev. Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 45.Dollins CM, Nair S, Boczkowski D, Lee J, Layzer JM, Gilboa E, Sullenger BA. Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem. Biol. 2008;15:675–682. doi: 10.1016/j.chembiol.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 47.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 48.Diamond JM, Turner DH, Mathews DH. Thermodynamics of three-way multibranch loops in RNA. Biochemistry. 2001;40:6971–6981. doi: 10.1021/bi0029548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.