Abstract
Human telomeres consist of thousands of base pairs of double-stranded TTAGGG repeats, organized by histone proteins into tightly spaced nucleosomes. The double-stranded telomeric repeats are also specifically bound by the telomeric proteins hTRF1 and hTRF2, which are essential for telomere length maintenance and for chromosome protection. An unresolved question is what role nucleosomes play in telomere structure and dynamics and how they interact and/or compete with hTRF proteins. Here we show that hTRF1 specifically induces mobility of telomeric nucleosomes. Moreover, Atomic Force Microscopy (AFM) imaging shows that hTRF1 induces compaction of telomeric DNA only in the presence of a nucleosome, suggesting that this compaction occurs through hTRF1–nucleosome interactions. Our findings reveal an unknown property of hTRF1 that has implications for understanding telomere structure and dynamics.
INTRODUCTION
Linear eukaryotic chromosomes end in specialized nucleoprotein complexes named telomeres (1,2). In humans, telomeres consist of 10–15 kb of double-stranded TTAGGG repeats, and 100–200 nt 3′ protrusions of the G-rich strand. The establishment of a capping structure at telomeres protects chromosomes from degradation and recombination, however the structure of telomeres remains elusive. The proteins hTRF1 and hTRF2 specifically bind human duplex telomeric DNA as homodimers (3–5), whereas hPOT1 recognizes the 3′ single-stranded overhangs (6). These three proteins, together with TIN2, hRAP1 and TPP1, form the complex named shelterin (7,8). It has been proposed that the shelterin complex stabilizes a lariat structure named t-loop (9), presumably deriving from invasion of the upstream telomeric double-stranded DNA by the single-stranded G-overhang (10). Telomeric DNA in higher eukaryotes is organized into a chromatin structure characterized by regularly and unusually closely spaced nucleosomes (11–13), separated by short linkers that in humans are only ∼10 bp long (11,12). Electron microscopy (14) and micrococcal nuclease (MNase) footprinting experiments (15) indicate that nucleosomes are also present in the terminal t-loop.
Due to the complex functions they perform, telomeres are likely to interconvert between different and still poorly defined conformations. Whereas the functions associated to telomeric proteins have been widely studied, the role played by nucleosomes in telomere structure and function is almost completely unexplored (16). Several in vitro studies have shown that nucleosomes assembled on telomeric DNA have particular features. As a consequence of the 6 bp sequence repeat of human telomeric DNA that is out of phase with the helical periodicity of DNA (13,17,18), telomeric nucleosomes are the least stable nucleosomes so far studied and occupy multiple isoenergetic positions without rotational phase (13,17,19,20). In our view, it seems very likely that the competition between histones and TRF proteins for telomeric DNA binding is involved in regulating telomere dynamics.
Recently, we showed that hTRF1 forms specific and stable ternary complexes with nucleosomes containing human telomeric repeats (21). hTRF1 binding causes alterations in the nucleosome structure, without dissociation of histone subunits (21). Similarly, hTRF2 interacts with telomeric chromatin (22), albeit with a lower affinity for nucleosomal binding sites than hTRF1 (A.G., M.S., S.C., unpublished results). Photobleaching experiments have shown that nucleosome dissociation is very slow (23), compared with that of H1 and several other binding proteins, including TRF1 and TRF2 (24,25). Biochemical probing experiments have shown that nucleosomes are dynamic complexes in equilibrium between wrapped and partially unwrapped states (26), and capable of spontaneous short-range movements along DNA (27,28). Significantly, as a consequence of the peculiar features of telomeric DNA sequences, telomeric nucleosomes are intrinsically more mobile than bulk nucleosomes (29). An attractive hypothesis arising from these observations is that hTRF1 binding may result in nucleosome repositioning rather than in nucleosome disruption or loss.
Here, we show that binding of hTRF1 causes the sliding of the telomeric nucleosome. Furthermore, TRF1 seems to interact with the mobilized nucleosome inducing the DNA to fold back. Our results reveal an unknown sequence-specific remodeling activity of hTRF1 that allows binding in the context of chromatin and provides an insight into the dynamic structure of telomeres.
MATERIALS AND METHODS
DNAs and proteins
The Biotin-TG580 DNA fragment was prepared by PCR with the primers 5′-Biotin-TGA ACC ATC ACC CTA ATC AAG-3′ and 5′-GTT GTG TGG AAT TGT GAG CG-3′, using as a template the plasmid pBIIKS-TG580Ade (29). The PCR product was then digested with AdeI and gel-purified. To obtain the DNA fragments hTel27 and TAND1, the plasmids pUC-hTEL27Ade and pUC-TAND1Ade (29) were cut with NheI. The linearized plasmids were dephosphorylated with calf intestinal phosphatase and the 5′ protruding ends were filled in with Klenow enzyme in the presence of 100 µM each of dATP, dCTP, dTTP and ddGTP. For the restriction enzyme (RE) assay, filling in of 5′ protruding ends was performed in the presence of 50 µCi of [α-32P]dCTP. Finally the plasmids were cut with AdeI and gel-purified.
Nucleosome and purified histone octamer were prepared from chicken erythrocytes as previously described (19). Recombinant, His6-tagged full-length hTRF1 was baculovirus expressed in Sf9 cells and purified as previously described (30).
Nucleosome reconstitution
For the RE assay, nucleosomes were assembled onto radiolabeled hTel27 and TAND1 DNA fragments by the octamer exchange method as described (20). For AFM imaging nucleosome were assembled by the salt dialysis technique (29,31). Briefly, 2 µg of hTel27 or TAND1 were mixed with 1.8 µg of purified histone octamer from chicken erythrocytes in 50 µl of 10 mM Tris–HCl (pH 7.6), 2.0 M NaCl, 0.5 mM EDTA, 0.5 mM PMSF, 1 mM benzamidine. Samples were stepwise dialyzed against buffers containing decreasing NaCl concentrations (1.2, 1.0, 0.8, 0.6, 0.2 M NaCl), and finally dialyzed overnight against 5 mM Tris–HCl (pH 7.6), 0.5 mM EDTA.
RE assay of nucleosome mobility
Radiolabeled nucleosomes (1 µg) were ligated to equimolar amounts of Biotin-TG580 by using 5 U of T4 DNA ligase at 4°C overnight. To prevent self-ligation, the DNA fragments have asymmetric protruding ends derived by cutting with AdeI. Moreover, the opposite ends have been dephosphorylated and filled in with a terminal dideoxynucleotide (see the section DNAs and proteins). Samples were then incubated for 5 h at 4°C on a rotating wheel with streptavidin-coated paramagnetic beads (M280 streptavidin-dynabeads), in order to remove unligated nucleosomes. After attachment, beads were washed three times with 100 µl WBN (20 mM Tris–HCl, pH 7.6, 1 mM EDTA, 0.15 M NaCl, 0.1% Nonidet-P40, 1 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA, 40 µg/ml oligonucleosomes). Bound DNAs and nucleoparticles (concentration 25–50 nM) were then resuspended in WBN and incubated with hTRF1 (final concentration 1 µM) for 1 h at 25°C on a rotating wheel. Finally, samples were washed three times with RE buffer, digested for 20 min with SmaI or BseGI, and run on a 4.5% native polyacrylamide gel. The percentage of mobilized nucleosome (% Mobil. Nuc.) has been calculated using the equation
where % Nuc. Ref. is the fraction of nucleosomal band in the reconstituted nucleosome or in the ligated complex before incubation at 25°C. As shown in Supplementary Figure S1, after ligation of the nucleosome assembled on hTel27 to Biotin-TG580 and before incubation at 25°C, the fraction of nucleosome released by SmaI (Supplementary Figure S1, lane 4) is equivalent to that present in the reconstituted sample (Supplementary Figure S1, lane 1).
AFM imaging
After ligation, samples were incubated with hTRF1 (final concentration 1 µM) at 25°C for 1 h, and crosslinked by dialysis against 0.1% glutaraldehyde, 10 mM Tris–HCl (pH 7.6), 1 mM EDTA for 3 h at 4°C. After fixation, samples were diluted ∼4-fold and immediately applied on spermidine (1 mM)-treated mica surface (SPD-mica). Imaging was performed using a Nanoscope IIIa equipped with E-scanner (Digital Instruments Inc., Santa Barbara, CA), operating in air under Tapping ModeTM at room temperature using canonical sharp silicon tips. Images have been recorded with a scanning rate of 3–4 Hz. Raw SFM images have been processed only for background removal (flattening). Volumes have been derived by area and height of the molecules, measured by using the microscope manufacturer's; image processing software. To evaluate contour lengths, AFM images were converted from Nanoscope format into TIF files and processed using SigmaScan Pro software (SPSS Inc., Chicago, IL). To measure the L1 parameter and the apparent contour length, we used the read-through method, assuming that DNA passes through the center of the protein (32). The histograms were fitted to Gaussian or multi-Gaussian functions by using the open source QtiPlot software.
RESULTS
hTRF1 binding induces the sliding of telomeric nucleosomes in a sequence-specific way
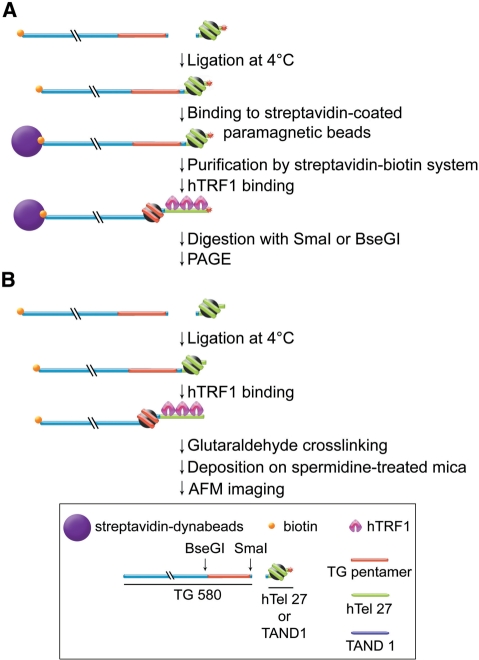
To investigate whether human TRF1 induces nucleosome mobilization, we used an experimental system that we previously designed to study nucleosome thermal repositioning [Figure 1A, ref. (29)]. The principle of this system is that a nucleosome reconstituted on a DNA fragment containing telomeric repeats is ligated to a naked DNA fragment and then incubated with the telomeric DNA binding factor hTRF1. The position of the nucleosome before and after hTRF1 binding is then investigated through restriction site mapping.
Figure 1.
Experimental system to study nucleosome mobility. (A) Schematic drawing of the experimental system used to study nucleosome mobility by restriction enzyme assay. (B) Schematic drawing of the experimental system used to study nucleosome mobility by AFM imaging. The box reports the key of symbols used in the figures.
An in vitro reconstituted nucleosome on a terminally labeled DNA sequence (hTel27 or TAND1) was ligated to a 580-bp-long DNA sequence (Biotin-TG580) containing the TG-pentamer nucleosome positioning sequence (33) close to the ligation site. This DNA fragment binds the histone octamer with higher affinity than hTel27, a 207 bp DNA fragment containing 27 tandem TTAGGG repeats (29), and than TAND1, a 182-bp-long DNA fragment which we have used as a standard of average sequence DNA (17,34). The free energy difference of nucleosome formation of TG-pentamer is of −2.20 kcal/mol of nucleosome with respect to TAND1 and of −3.27 kcal/mol of nucleosome with respect to hTel27 [see ref. (17) and Supplementary Figure S2]. Therefore, the TG-pentamer should act as a nucleosome trap, preventing histone octamers from sliding back towards the original terminal position.
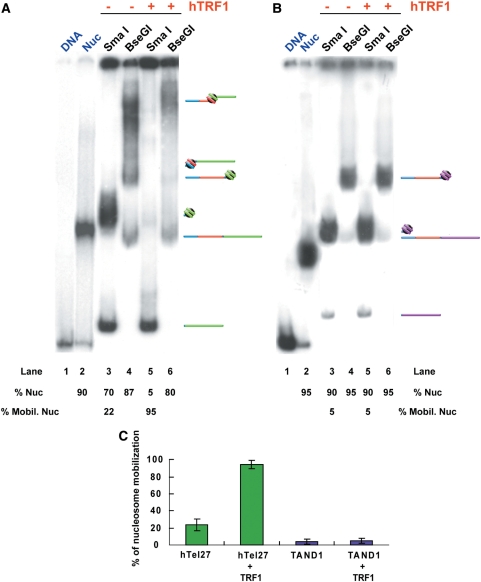
The ligation products (BioTG-Tel/Nuc and BioTG-TAND/Nuc) were then purified from unligated nucleosomes by binding to streptavidin-coated paramagnetic beads and then incubated with hTRF1. The nucleosome position before and after hTRF1 binding was determined by separate digestions with the REs SmaI and BseGI (RE digestion assay, detailed in Figure 1A). Figure 2A shows a RE digestion assay of the complex BioTG-Tel/Nuc after 1 h incubation at 25°C in presence and absence of hTRF1. Digestion with SmaI, which cuts immediately downstream of the TG-pentamer sequence, generates two bands corresponding respectively to a naked 233-bp-long DNA fragment and to the nucleosomal complex. Estimations of the relative fraction of the nucleosomal band before and after incubation at 25°C or with hTRF1 (see Figure 2 and Supplementary Figure S1) allows the percentage of nucleosome sliding to be calculated (29,35). After incubation at 25°C, the fraction of naked DNA released by SmaI increases with respect to that present in the reconstituted sample (Figure 2A, lanes 3 and 2), indicating that part of the nucleosome population shifted inward, leaving ∼70% of the nucleosomes still in the starting position. When hTRF1 was added to the reaction, the nucleosomal band almost completely disappears (Figure 2A, lane 5), suggesting that nucleosomes are repositioned by hTRF1 binding. It can be seen that after incubation with hTRF1, a fraction of the sample that remains in the well increases (see Figure 2A, lane 5 compared with lane 3), indicating that part of the nucleosome population slides onto the SmaI restriction site, thus inhibiting digestion. The invariance of the relative percentage of the 469 bp naked DNA band when the complex is cut with the enzyme BseGI, whose recognition site is located upstream of the TG-pentamer sequence, rules out the possibility that hTRF1 causes nucleosome dissociation (Figure 2A, lanes 4 and 6). The induction of nucleosome sliding by hTRF1 is further manifested by the increase of the low-mobility nucleosomal band that corresponds to a nucleosome centered on the fragment released by BseGI (Figure 2A, lanes 4 and 6). Moreover, the fraction of undigested complex that remains in the well is higher when the sample is incubated with hTRF1 (see Figure 2A, lanes 4 and 6); as for the SmaI digests, this indicates that digestion is inhibited by the sliding of the nucleosome onto the restriction site. To test that the hTRF1 remodeling activity is specific to telomeric nucleosomes, the RE assay was performed on the BioTG–TAND/Nuc complex (Figure 2B). In this case, incubation with hTRF1 has no effect on nucleosome position, since the nucleosome remains on the terminal TAND1 DNA (Figure 2B, lanes 3 and 5).
Figure 2.
Restriction enzyme assay of nucleosome mobility. (A) Gel mobility shift assay. From the left: hTel27 naked DNA (lane 1), hTel27 nucleosome (lane 2), BioTG-Tel/Nuc (lanes 3 and 4), BioTG-Tel/Nuc/TRF1 (lanes 5 and 6). (B) From the left: TAND1 naked DNA (lane 1), TAND1 nucleosome (lane 2), BioTG-TAND/Nuc (lanes 3 and 4), BioTG-TAND/Nuc/TRF1 (lanes 5 and 6). The percentage of nucleosomal band is indicated below the lanes (% Nuc). The proportion of mobilized nucleosomes is also indicated (% Mobil. Nuc). Bands in the gel correspond only to naked DNA and nucleosomal complexes (see drawings flanking the gels). Washes with RE buffer before enzymatic digestions (see Materials and Methods) result in hTRF1 dissociation. (C) Percentage of nucleosome mobilization from hTel27 (green bars) and TAND1 (blue bars) as a function of hTRF1 binding. For each DNA construct, at least three experiments have been considered. Values are reported as mean ± standard deviation (vertical bar).
Results for at least three experiments for each construct are summarized in Figure 2C, which reports the percentages of mobilized nucleosomes calculated by measuring the relative intensities of the nucleosomal and the naked DNA bands after SmaI digestion. The results show that at 25°C, ∼25% of telomeric nucleosomes slide inwards (Figure 2C). Significantly, nucleosome sliding in the presence of hTRF1 increases to ∼95%. It is also very likely that this value is slightly underestimated, since in calculating the percentage of mobilized nucleosomes we do not take into account the increase of material in the well of the gel, which is likely to derive from mobilized nucleosomes that hinder RE digestion. Instead, nucleosomes assembled on the TAND1 DNA remain mostly in the starting position, independently from hTRF1 presence. In conclusion, hTRF1 binding induces nucleosome repositioning away from the telomeric DNA repeats.
AFM imaging shows that hTRF1-induced nucleosome mobilization is coupled to DNA compaction
As a complementary method to study nucleosome mobility, we used the single-molecule imaging technique AFM (36). By measuring a series of parameters—contour length, volume, distance from the DNA ends—AFM allows the estimation of nucleosome positions relative to DNA (29) and to obtain information on shape and position of the ternary complexes (37). The experimental strategy is shown in Figure 1B. After incubation with hTRF1, the complexes were fixed by glutaraldehyde crosslinking and samples deposited on spermidine-treated mica and imaged by AFM (see Figure 3 for representative AFM fields).
Figure 3.
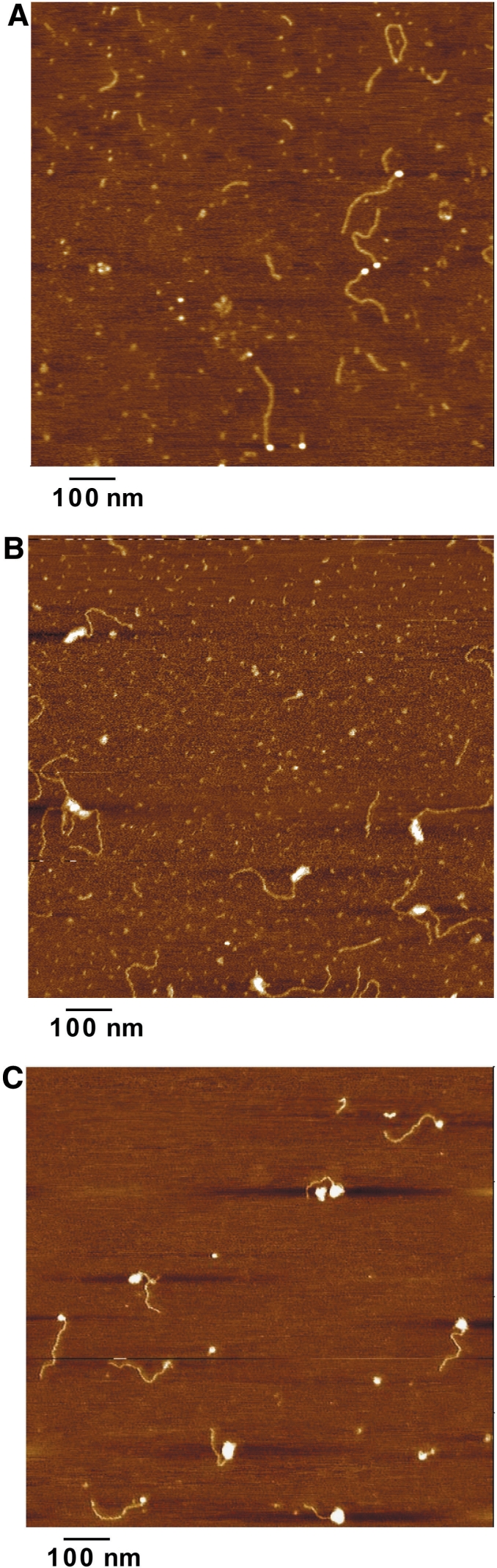
Representative AFM images of the analyzed complexes. (A) TG-Tel/Nuc (TG580/hTel27 construct with nucleosomal complexes). (B) TG-Tel/TRF1(hTRF1 bound to the naked TG580/hTel27 DNA). (C) The mixed population obtained incubating the nucleosomal construct with hTRF1, comprising the ternary complex,TG-Tel/Nuc/TRF1.
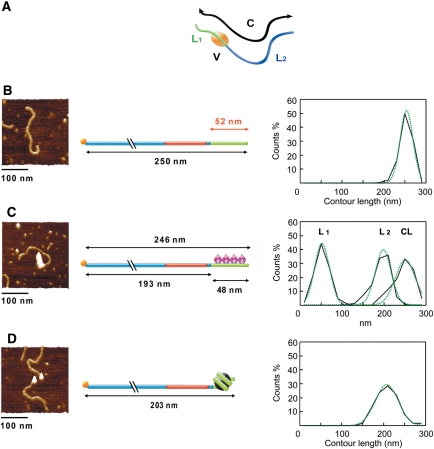
Although we have previously successfully studied thermal repositioning by AFM imaging (29), studying hTRF1-dependent nucleosome mobilization by AFM raises additional difficulties because of the presence of at least three nucleoprotein complexes: the nucleosomal TG580/hTel27 construct (TG-Tel/Nuc), hTRF1 bound to the naked TG580/hTel27 DNA (TG-Tel/TRF1) and the ternary complex with hTRF1 bound to the nucleosomal construct (TG-Tel/Nuc/TRF1). Therefore, to characterize the different complexes, populations of TG-Tel (naked TG580/hTel27 DNA), TG-Tel/TRF1, TG-Tel/Nuc, were analyzed separately as well as the mixed population obtained by incubating the nucleosomal construct with hTRF1 to form the ternary complex TG-Tel/Nuc/TRF1. We measured several parameters for every molecule: the contour length, C, the distances between the ends of the construct and one border of the nucleoprotein complex, L1 and L2, and the volume of the nucleoprotein complex, V (Figure 4A). In order to estimate a reliable value for DNA lengths, we fitted all the obtained distributions with either Gaussian or multi-Gaussian functions (32). In this way, it is possible to make a comparison in terms of mean values of distributions, corresponding to the most probable value of contour length. Figure 4 reports the values from the measurements of the contour length of the protein-free construct TG-Tel, of the complex TG-Tel/TRF1 and of the complex TG-Tel/Nuc; in Figure 4C are also reported the L1 and L2 values of the TG-Tel/TRF1 complex. Data are presented as interpolations of the frequency histograms and Gaussian fittings. The corresponding frequency histograms are shown in Supplementary Figure S3. The measured mean value for the contour length of the protein-free construct TG-Tel is 250 ± 12 nm (Figure 4B), corresponding to a DNA helical rise of 0.32 nm/bp (38). The mean value of the contour length of the complex TG-Tel/TRF1 is 246 ± 23 nm (Figure 4C), a value only slightly smaller than that of the naked DNA, indicating that the binding of hTRF1 does not significantly shorten the construct, in agreement with previous observations (39). Furthermore, L1 and L2 lengths confirm the specific binding of hTRF1 to TTAGGG repeats. The L1 mean value of TG-Tel/TRF1 is 48 ± 17 nm, very close to the theoretical value of 52 nm corresponding to the length of the 27 telomeric repeats. Consistent with these results is the obtained mean value of the L2 parameter, 193 ± 17 nm, in good agreement with the theoretical length of the TG580 sequence (Figure 4C).
Figure 4.
AFM measurement of molecules parameters. (A) Schematic representation of dimensions measured on the AFM images: L1 is the distance between the end of the construct (telomeric portion) and the end of the nucleoprotein complex; L2 is the distance between the end of the construct (TG580 portion) and the edge of the nucleoprotein complex; C is the contour length of the construct; V is the volume of the protein complex. (B–D) Summary of the measured parameters for TG-Tel DNA (B), TG-Tel/TRF1 (C) and TG-Tel/Nuc (D), respectively. Left panel: representative AFM images (surface plots, rotation 0°, pitch 70°). Middle panel, model of the molecules with the mean values of the measured lengths. Right panel, distributions of the contour lengths expressed as percentage of total measurements and plotted as interpolation of the frequency histograms (black solid line) and Gaussian fitting (green dashed line). In particular, in the right panel of (C) the contour length, L1 and L2 distributions of TG-Tel/TRF1 are superimposed in the same plot area.
Differently from hTRF1 binding to telomeric DNA, the formation of a nucleosome reduces the contour length of the DNA molecule, consistent with the wrapping of 147 bp of DNA around the histone octamer. In the case of the TG-Tel/Nuc complex, the mean value of the contour length is 203 ± 25 nm (Figure 4D), 47 nm shorter than that of the naked DNA.
We then measured the area and the height of the ellipsoid representing the nucleoprotein complexes, and calculated the volumes. The mean value of nucleosome volumes corresponding to TG-Tel/Nuc samples is of 579 ± 164 nm3. This finding is in good agreement with the x-ray crystallography volume of nucleosome (532 nm3) (40) and with previous AFM studies on mononucleosome (41). In the case of the TG-Tel/TRF1 complex, the presence of 27 telomeric repetitions gives rise to an extremely broad distribution of volumes deriving from multiple hTRF1 binding. For this reason the mean value corresponding to this distribution is statistically irrelevant. Considering that the molecular weight of the proteins and their volume obtained by AFM are linearly correlated (42), we inferred the volume of the hTRF1 homodimer bound to DNA (MW: 110 kDa) on the basis of AFM data for the nucleosome (MW: 200 kDa). Consequently, the estimated volume of hTRF1 homodimer is 318 nm3.
In order to analyze the mixed population of complexes obtained after incubation of hTRF1 with TG-Tel/Nuc, we only considered the molecules fulfilling the following parameters to be ternary complexes: (i) contour lengths <203 nm, indicating the presence of the nucleosome onto the construct, (ii) volumes of the nucleoprotein complex >897 nm3, indicating the presence of the nucleosome and of at least one hTRF1 homodimer.
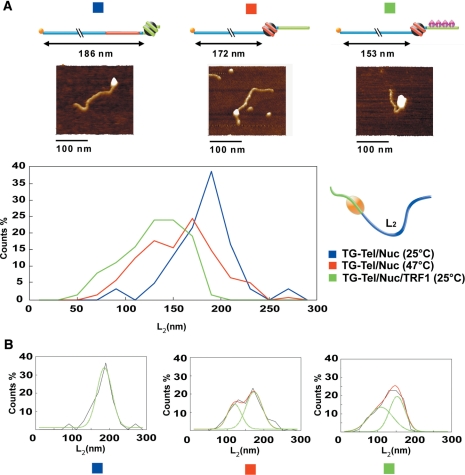
The specific binding of hTRF1 to telomeric DNA at one end of the construct (Figure 4C), allows us to identify the nucleosome position by measuring the distance L2 from the opposite DNA end to the border of the nucleoprotein complex (Figure 4A). Figure 5 shows the distributions of nucleosome positions after incubation at 25°C (in presence and in absence of hTRF1) and at 47°C. Nucleosome positions are expressed as L2 values and are reported as interpolated data, multi-Gaussian fitting and single-Gaussian fitting (see also Supplementary Figure S4 that reports L2 values in histogram form, and Supplementary Figure S5 in which are summarized the frequencies of the various forms found in the AFM experiments). From the comparison of the L2 distribution of TG-Tel/Nuc/TRF1 (Figure 5A, green line and Figure 5B, right panel; L2 most probable value: 153 ± 21 nm) and the L2 distribution of TG-Tel/Nuc (Figure 5A, blue line and Figure 5B, left panel; L2 most probable value: 186 ± 21 nm) it emerges that hTRF1 induces nucleosome repositioning onto adjacent sequences, in agreement with the data from the RE assay reported in Figure 2. Interestingly, nucleosome repositioning induced by hTRF1 binding at 25°C is higher than thermal repositioning obtained by incubating at 47°C (Figure 5A, red line and Figure 5B, central panel; L2 most probable value: 172 ± 23 nm). From the distributions of nucleosome positions reported in Figure 5A, it emerges that ∼98% of nucleosomes move inward as a consequence of hTRF1 binding, in very good agreement with the conclusions drawn from biochemical analysis.
Figure 5.
Nucleosome mobility analysis by AFM imaging. (A) L2 distribution, expressed as percentage of total measurements. Blue line: TG-Tel/Nuc at 25°C, 163 molecules measured. Red line: TG-Tel/Nuc at 47°C, 152 molecules measured. Green line: TG-Tel/Nuc/TRF1, 165 molecules measured. In the upper panel, representative AFM images of the corresponding constructs are identifiable by the same color code used for the graph (surface plots, rotation 0°, pitch 70°). The figure also shows a schematic drawing of the parameter L2 used to evaluate the nucleosome mobilization. (B) Nucleosome positions expressed as L2 length distributions, represented as the interpolation of frequency histogram (black solid line), multi-Gaussian fitting (red solid line) and single Gaussian fitting (green dashed line). From the left, TG-Tel/Nuc at 25°C (mean value 186 ± 21 nm), TG-Tel/Nuc at 47°C (first peak 172 ± 23 nm; second peak 119 ± 19 nm), TG-Tel/Nuc/TRF1 (first peak 153 ± 21 nm; second peak 108 ± 32 nm).
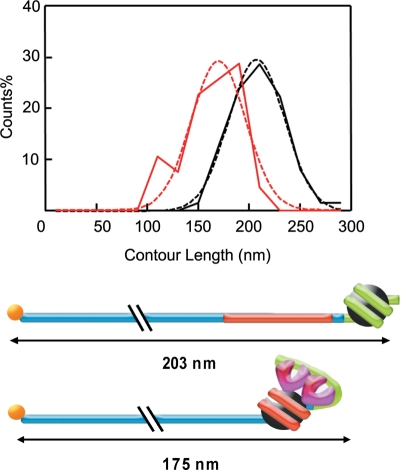
Finally, a noteworthy result emerges from the analysis of the contour lengths of the complexes TG-Tel/Nuc/TRF1. hTRF1 binding to the nucleosomal complex causes a sharp decrease of the mean value of the contour length, that is 37 nm shorter in the case of the ternary complex (171 ± 26 nm) than that of TG-Tel/Nuc (203 ± 25 nm) (Figure 6 and Supplementary Figure S6). This effect is not an artifact due to the experimental methodology, since hTRF1 binding on naked DNA results in only a small decrease of the contour length (Figure 4C). We attribute these results to the formation of additional interactions of hTRF1 with the mobilized nucleosome that lead to DNA compaction.
Figure 6.
DNA condensation induced by hTRF1 binding. Top, distribution of contour lengths of TG-Tel/Nuc (black lines) and TG-Tel/Nuc/Trf1 (red lines) expressed as interpolation of frequency histogram (solid lines) and Gaussian fitting (dashed lines). Bottom, models for the DNA compaction induced by hTRF1 binding with the mean values of the measured contour lengths.
DISCUSSION
Nucleosome mobility and remodeling are key events in the regulation of biological processes such as transcriptional activation and repression, replication, differentiation, cell cycle progression (43). Several regulative proteins are able to recognize their binding sites on the nucleosome (44,45), yet generally this event is not coupled to nucleosome repositioning (46,47). Nucleosome mobility usually requires ATP-dependent chromatin remodeling complexes, which alter nucleosome structure and positioning and favor access to DNA of regulative proteins or enzymatic complexes. Despite the dynamic nature of telomeres, ATP-dependent remodeling has not been associated to telomere function in higher eukaryotes. Only recently have chromatin remodeling complexes been implicated in telomere positioning effects and in telomere elongation in budding and fission yeast (48,49), suggesting that ATP-dependent remodeling enzymes are involved in telomere dynamics, although their role in telomere function of higher eukaryotes is yet to be demonstrated. Nevertheless, differently from other chromosome regions, telomeric chromatin possesses inherent dynamical properties that can be ascribed to intrinsic features imparted by the peculiar sequence of telomeric DNA (20,29).
The data presented in Figures 2 and 5 demonstrate that hTRF1 binding induces sliding of telomeric nucleosomes. This ATP-independent chromatin remodeling is telomere-specific, since hTRF1 is unable to shift nucleosomes formed on a reference DNA sequence. Moreover, nucleosome mobilization by hTRF1 is much greater than thermal repositioning (Figure 5A). The most probable explanation for these results is that hTRF1 binding enhances the intrinsic mobility of telomeric nucleosomes. According to this finding, although the histone octamer could slide randomly along telomeric DNA in a both forward and backward direction, the binding of hTRF1 onto telomeric DNA prevents the histone octamer to slide backwards, establishing a directional nucleosomal movement towards internal sequences of the construct. It is also possible that hTRF1 plays an active role in telomeric chromatin remodeling. In addition to promoting nucleosome sliding, hTRF1 binding induces alterations of nucleosome structure (21) that could represent the first step in nucleosome remodeling and hence contribute to nucleosome sliding. Finally, the AFM imaging presented here shows that hTRF1 not only shifts telomeric nucleosomes, but also causes a marked decrease in the DNA contour length in the complexes. An explanation for the observed compaction is that hTRF1 binding induces telomeric DNA to fold back and interact with the mobilized nucleosome, maybe through non-specific interactions between the acidic N-terminal domain of hTRF1 and the basic residues of histone tails (Figure 5B).
What do these results suggest about the interplay between hTRF1 and nucleosomes in vivo? In vitro hTRF1 interacts with telomeric nucleosomes in a number of different ways: it can bind to nucleosomal binding sites, induce nucleosome sliding, and form a bridge between telomeric DNA and nucleosomes. These features suggest that hTRF1 has the ability to regulate telomeric chromatin structure, whilst preserving the packaging of telomeric DNA into nucleosomes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fondazione Istituto Pasteur—Fondazione Cenci-Bolognetti (grant to M.S. and Postdoctoral Research Fellowship to S.P.); Sapienza University of Rome (grant to S.C.); Ministero dell’Istruzione, dell’Università e della Ricerca (grant PRIN97 to M.S. and S.C.). Funding for open access charge: Fondazione Istituto Pasteur-Fondazione Cenci-Bolognetti.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Thanks are due to P. De Santis for helpful discussion and to L. Chapman for preparation of hTRF1.
REFERENCES
- 1.Zakian VA. Telomeres: beginning to understand the end. Science. 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]
- 2.de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 3.Court R, Chapman L, Fairall L, Rhodes D. How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep. 2005;6:39–45. doi: 10.1038/sj.embor.7400314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilaud T, Koering CE, Binet-Brasselet E, Ancelin K, Pollice A, Gasser SM, Gilson E. The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res. 1996;24:1294–1303. doi: 10.1093/nar/24.7.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 6.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 7.Liu D, O'C;onnor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 8.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 9.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 10.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 11.Tommerup H, Dousmanis A, de Lange T. Unusual chromatin in human telomeres. Mol. Cell Biol. 1994;14:5777–5785. doi: 10.1128/mcb.14.9.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lejnine S, Makarov VL, Langmore JP. Conserved nucleoprotein structure at the ends of vertebrate and invertebrate chromosomes. Proc. Natl Acad. Sci. USA. 1995;92:2393–2397. doi: 10.1073/pnas.92.6.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fajkus J, Kovarik A, Kralovics R, Bezdek M. Organization of telomeric and subtelomeric chromatin in the higher plant Nicotiana tabacum. Mol. Gen. Genet. 1995;247:633–638. doi: 10.1007/BF00290355. [DOI] [PubMed] [Google Scholar]
- 14.Nikitina T, Woodcock CL. Closed chromatin loops at the ends of chromosomes. J. Cell Biol. 2004;166:161–165. doi: 10.1083/jcb.200403118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu P, de Lange T. No overt nucleosome eviction at deprotected telomeres. Mol. Cell Biol. 2008;28:5724–5735. doi: 10.1128/MCB.01764-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pisano S, Galati A, Cacchione S. Telomeric nucleosomes: forgotten players at chromosome ends. Cell Mol. Life Sci. 2008;65:3553–3563. doi: 10.1007/s00018-008-8307-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filesi I, Cacchione S, De Santis P, Rossetti L, Savino M. The main role of the sequence-dependent DNA elasticity in determining the free energy of nucleosome formation on telomeric DNAs. Biophys. Chem. 2000;83:223–237. doi: 10.1016/s0301-4622(99)00143-x. [DOI] [PubMed] [Google Scholar]
- 18.Anselmi C, Bocchinfuso G, De Santis P, Savino M, Scipioni A. Dual role of DNA intrinsic curvature and flexibility in determining nucleosome stability. J. Mol. Biol. 1999;286:1293–1301. doi: 10.1006/jmbi.1998.2575. [DOI] [PubMed] [Google Scholar]
- 19.Cacchione S, Cerone MA, Savino M. In vitro low propensity to form nucleosomes of four telomeric sequences. FEBS Lett. 1997;400:37–41. doi: 10.1016/s0014-5793(96)01318-x. [DOI] [PubMed] [Google Scholar]
- 20.Rossetti L, Cacchione S, Fua M, Savino M. Nucleosome assembly on telomeric sequences. Biochemistry. 1998;37:6727–6737. doi: 10.1021/bi9726180. [DOI] [PubMed] [Google Scholar]
- 21.Galati A, Rossetti L, Pisano S, Chapman L, Rhodes D, Savino M, Cacchione S. The human telomeric protein TRF1 specifically recognizes nucleosomal binding sites and alters nucleosome structure. J. Mol. Biol. 2006;360:377–385. doi: 10.1016/j.jmb.2006.04.071. [DOI] [PubMed] [Google Scholar]
- 22.Baker AM, Fu Q, Hayward W, Lindsay SM, Fletcher TM. The Myb/SANT domain of the telomere-binding protein TRF2 alters chromatin structure. Nucleic Acids Res. 2009;37:5019–5031. doi: 10.1093/nar/gkp515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattern KA, Swiggers SJ, Nigg AL, Lowenberg B, Houtsmuller AB, Zijlmans JM. Dynamics of protein binding to telomeres in living cells: implications for telomere structure and function. Mol. Cell Biol. 2004;24:5587–5594. doi: 10.1128/MCB.24.12.5587-5594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, Misteli T. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 27.Pennings S, Meersseman G, Bradbury EM. Mobility of positioned nucleosomes on 5S rDNA. J. Mol. Biol. 1991;220:101–110. doi: 10.1016/0022-2836(91)90384-i. [DOI] [PubMed] [Google Scholar]
- 28.Flaus A, Owen-Hughes T. Dynamic properties of nucleosomes during thermal and ATP-driven mobilization. Mol. Cell Biol. 2003;23:7767–7779. doi: 10.1128/MCB.23.21.7767-7779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pisano S, Marchioni E, Galati A, Mechelli R, Savino M, Cacchione S. Telomeric nucleosomes are intrinsically mobile. J. Mol. Biol. 2007;369:1153–1162. doi: 10.1016/j.jmb.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 30.Bianchi A, Stansel RM, Fairall L, Griffith JD, Rhodes D, de Lange T. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 1999;18:5735–5744. doi: 10.1093/emboj/18.20.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mechelli R, Anselmi C, Cacchione S, De Santis P, Savino M. Organization of telomeric nucleosomes: atomic force microscopy imaging and theoretical modeling. FEBS Lett. 2004;566:131–135. doi: 10.1016/j.febslet.2004.04.032. [DOI] [PubMed] [Google Scholar]
- 32.Rivetti C, Guthold M, Bustamante C. Wrapping of DNA around the E.coli RNA polymerase open promoter complex. EMBO J. 1999;18:4464–4475. doi: 10.1093/emboj/18.16.4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shrader TE, Crothers DM. Artificial nucleosome positioning sequences. Proc. Natl Acad. Sci. USA. 1989;86:7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 35.Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- 36.Bustamante C, Zuccheri G, Leuba SH, Yang G, Samori B. Visualization and analysis of chromatin by scanning force microscopy. Methods. 1997;12:73–83. doi: 10.1006/meth.1997.0449. [DOI] [PubMed] [Google Scholar]
- 37.Shin M, Song M, Rhee JH, Hong Y, Kim YJ, Seok YJ, Ha KS, Jung SH, Choy HE. DNA looping-mediated repression by histone-like protein H-NS: specific requirement of Esigma70 as a cofactor for looping. Genes Dev. 2005;19:2388–2398. doi: 10.1101/gad.1316305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rivetti C, Guthold M, Bustamante C. Scanning force microscopy of DNA deposited onto mica: equilibration versus kinetic trapping studied by statistical polymer chain analysis. J. Mol. Biol. 1996;264:919–932. doi: 10.1006/jmbi.1996.0687. [DOI] [PubMed] [Google Scholar]
- 39.Amiard S, Doudeau M, Pinte S, Poulet A, Lenain C, Faivre-Moskalenko C, Angelov D, Hug N, Vindigni A, Bouvet P, et al. A topological mechanism for TRF2-enhanced strand invasion. Nat. Struct. Mol. Biol. 2007;14:147–154. doi: 10.1038/nsmb1192. [DOI] [PubMed] [Google Scholar]
- 40.Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984;311:532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- 41.Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl Acad. Sci. USA. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider SW, Larmer J, Henderson RM, Oberleithner H. Molecular weights of individual proteins correlate with molecular volumes measured by atomic force microscopy. Pflugers Arch. 1998;435:362–367. doi: 10.1007/s004240050524. [DOI] [PubMed] [Google Scholar]
- 43.Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 44.Rhodes D. Structural analysis of a triple complex between the histone octamer, a Xenopus gene for 5S RNA and transcription factor IIIA. EMBO J. 1985;4:3473–3482. doi: 10.1002/j.1460-2075.1985.tb04106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Angelov D, Charra M, Seve M, Cote J, Khochbin S, Dimitrov S. Differential remodeling of the HIV-1 nucleosome upon transcription activators and SWI/SNF complex binding. J. Mol. Biol. 2000;302:315–326. doi: 10.1006/jmbi.2000.4069. [DOI] [PubMed] [Google Scholar]
- 46.Anderson JD, Thastrom A, Widom J. Spontaneous access of proteins to buried nucleosomal DNA target sites occurs via a mechanism that is distinct from nucleosome translocation. Mol. Cell Biol. 2002;22:7147–7157. doi: 10.1128/MCB.22.20.7147-7157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Z, Zheng C, Thiriet C, Hayes JJ. The core histone N-terminal tail domains negatively regulate binding of transcription factor IIIA to a nucleosome containing a 5S RNA gene via a novel mechanism. Mol. Cell Biol. 2005;25:241–249. doi: 10.1128/MCB.25.1.241-249.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu EY, Steinberg-Neifach O, Dandjinou AT, Kang F, Morrison AJ, Shen X, Lue NF. Regulation of telomere structure and functions by subunits of the INO80 chromatin remodeling complex. Mol. Cell Biol. 2007;27:5639–5649. doi: 10.1128/MCB.00418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.