Abstract
P21, a cyclin-dependent kinase inhibitor, plays a pivotal role in the cell-cycle regulation in response to stress stimuli. P21 expression is highly regulated through transcriptional, post-transcriptional and post-translational mechanisms. Previously, we and others showed that p21 expression is regulated through p21 mRNA stability by RNPC1, a target of the p53 family and HuR, a member of the ELAV family RNA-binding proteins. HuR carries three highly conserved RNA recognition motifs (RRMs) whereas RNPC1 carries one. Here we found that the ability of RNPC1 to regulate p21 mRNA stability is dependent on HuR. We also found that RNPC1 and HuR physically interact, and the RRM domain in RNPC1 and RRM3 in HuR are necessary for their interaction. Interestingly, we found that RNPC1 and HuR, both of which can bind AU-rich elements (AREs) in p21 3′-UTR, preferentially bind the upstream and downstream AREs, respectively. Finally, we showed that the RNA-binding activity of HuR to p21 transcript was enhanced by RNPC1 in vitro and in vivo. Together, we hypothesize that RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability.
INTRODUCTION
P21, a cyclin-dependent kinases (CDKs) inhibitor, is a major mediator of p53 to regulate the cell-cycle transition from G1 to S and G2 to M (1–3). In addition, upon exposure to certain stress signals, p21 is highly induced, leading to irreversible growth arrest in the early stages of cellular senescence and differentiation (4–5). Thus, p21 expression is highly regulated in response to stress signals, through transcriptional, post-transcriptional and post-translational mechanisms (6). For example, p21 expression is upregulated by many transcription factors, including the p53 family (7,8). P21 expression is also regulated by protein turnover via multiple post-translational modifications, such as ubiquitination and phosphorylation (5,9). Furthermore, p21 expression is regulated by mRNA turnover (5). The rate of mRNA turnover is controlled by specific elements, such as AU-rich elements (AREs), which are usually located in the 3′-UTR and recognized by various RNA-binding proteins. Indeed, p21 mRNA stability is found to be regulated by a group of RNA-binding proteins that have an affinity to poly(C) and a family of RNA-binding proteins that contain RNA recognition motif (RRM) and have an affinity to AREs (5,10).
The Hu/ELAV family of RNA-binding proteins consists of HuB, HuC, HuD and HuR, all of which carry several RRMs and are known to bind the ARE-containing transcripts, including p21 transcript (11). HuB, HuC and HuD are expressed in terminally differentiated neurons and neuroendocrine tumors whereas HuR is ubiquitously expressed (12–19). Recent studies showed that upon UV irradiation, HuR is phosphorylated by p38 MAPK and PKC and translocated from nucleus to cytosol wherein p21 mRNA stability is regulated by HuR (9,20–26). RNPC1, also called RBM38, is a RNA-binding protein with one RRM domain and a target of the p53 family (27). RNPC1 is expressed as two isoforms, RNPC1a with 239 amino acids and RNPC1b with 121 amino acids (27). RNPC1 is also found to regulate p21 mRNA stability by binding to p21 3′-UTR (27). Thus, both HuR and RNPC1 bind to p21 3′-UTR and regulate p21 expression. Although a variety of RNA-binding proteins are known to regulate p21 mRNA stability via binding to p21 transcript, it is still unclear how these RNA-binding proteins cooperatively or antagonistically regulate p21 expression. Here we showed that HuR is necessary for RNPC1 to regulate p21 mRNA stability. In addition, we found that RNPC1 directly interacts with HuR and enhances its RNA-binding activity to p21 3′-UTR. These data suggest that RNPC1 cooperates with HuR to regulate p21 expression.
MATERIALS AND METHODS
Plasmids
To generate glutathione S-transferase (GST)- and histidine (His)-tagged RNPC1a, the coding region of RNPC1a was amplified by PCR and cloned into pGEX4T3, pRSet-A and pcDNA3.1/HisB vectors, respectively. To generate GST-, His- and Myc-tagged HuR, the coding region of HuR was amplified by PCR and cloned into pGEX4T3, pRSet-A, pcDNA3.1/HisB and pcDNA3/Myc, respectively. To generate constructs expressing HA-tagged RNPC1a lacking RNP1 or RNP2, two-step PCR reactions were performed. The first-step was performed to separately amplify two cDNA fragments. Fragment #1 was amplified with forward primer, 5′-GAA GCT T GC CGC CAT GGA GTA CCC ATA CGA CGT ACC AGA TTA CGC TAT GCT GCT GCA GCC CGC GCC G-3′ and reverse primer, 5′-GGC GTC GGT AGT GTG GTA CTT GGT GAA CGT GGT GTC C-3′ for RNPC1a(ΔRNP1), or 5′-AGC TGC CGC CCG GTC GGC GGA CTT GCC CGT CTG GCG GT-3′ for RNPC1a(ΔRNP2). Fragment #2 was amplified with forward primer, 5′-GGA CAC CAC GTT CAC CAA GTA CCA CAC TAC CGA CGC C-3′ for RNPC1a(ΔRNP1), or 5′-ACC GCC AGA CGG GCA AGT CCG CCG ACC GGG CGG CAG CT-3′ for RNPC1a(ΔRNP2) and reverse primer, 5′-GGA ATT CTC ACT GCA TCC TGT CAG GCT GC-3′. The second-step PCR reaction was performed using a mixture of fragments #1 and #2 as a template with the forward primer for fragment #1 and the reverse primer for fragment #2, and resulting fragments were separately cloned and confirmed by sequencing. A HindIII–EcoRI fragment containing the coding region for ΔRNP1 and ΔRNP2 was purified and cloned into pcDNA4.
Cell culture and siRNA transfection
RKO and MCF7 cell lines that inducibly express HA-tagged RNPC1a, RNPC1b, ΔRNP1 and ΔRNP2 were generated and cultured as previously reported (27). For siRNA knockdown, cells were transfected with scrambled siRNA or siRNA against HuR (5′-GGGATAAAGTAGCAGGACA-3′) using siLentFect™ Lipid Reagent (Bio-Rad) and harvested for analysis 48 h later.
Western blot analysis and immunoprecipitation
Cells were harvested by scrapping and extracted with 2× SDS/sample buffer. Proteins were separated in 8% or 12% SDS–PAGE, transferred to a nitrocellulose membrane and probed with indicated antibodies followed by ECL detection. The antibodies used in this study were: anti-HuR (H-280, Santa Cruz), anti-p53 (DO-1 and Pab1801; Santa Cruz), anti-p21 (C-19, H-164; Santa Cruz), anti-actin (Sigma), anti-Omni probe (for His-tag recognition: D-8, Santa Cruz), anti-HA (HA.11, Covance) and anti-Myc (9E10, Cell signaling). For immunoprecipitation, cell lysates were pre-treated with RNase A (0.5 mg/ml) and then incubated with 1 µg of antibody and protein A/G beads for 4 h. The immuncomplexes were washed and used for western blot analysis.
Quantitative RT–PCR (qRT–PCR)
Total RNAs were purified with a Trizol reagent (Invitrogen). Reverse transcription was performed with iScript (Bio-Rad). PCR reaction was performed using Absolute Blue SyBr Green QPCR Master mix (Thermo) and realplex2 Mastercycler (Eppendorf). The primers used to amplify RNPC1 were forward primer 5′-ACGCCTCGCTCAGGAAGTA-3′ and reverse primer, 5′-GTCTTTGCAAGCCCTCTCAG-3′. The primers used to amplify HuR were forward primer 5′-CGCAGAGATTCAGGTTCTCC-3′ and reverse primer, 5′-GCCCCAGGTTGTAGATGAAA-3′. The primers used to amplify p21 and GAPDH were as described (27).
RNA-immunoprecipitation (RNA–IP)
RNA-IP was carried out as described (27). Briefly, cells were seeded at 5 × 106 per 100 mm plate and uninduced or induced to express RNPC1a for 24 h. Cell extracts were prepared with lysis buffer (10 mM HEPES, pH 7.0, 100 mM KCl, 5 mM MgCl2, 0.5% NP-40, 1 mM DTT) and then incubated with 2 µg of anti-HA, anti-HuR, or mouse IgG at 4°C for 4 h. The RNA-protein immunocomplexes were brought down by protein A/G beads, and RNA was then purified from the complexes for RT–PCR.
GST-pull down assay
Recombinant GST- and His-tagged RNPC1, HuR and their mutants were expressed in Escherichia coli BL21 (DE3) and purified by standard protocol (28). Ten picomoles of His-tagged RNPC1 (or HuR) and GST-fused HuR (or RNPC1) were mixed and incubated in a binding buffer (50 mM HEPES, pH 7.6, 50 mM NaCl, 5 mM EDTA, 0.1% NP-40 and glycerol) at 4°C for 2 h, followed by precipitation with glutathione-sepharose 4B beads. Beads were washed three times and resuspended in 1× SDS loading buffer. The interaction of HuR–RNPC1 was detected by western blot analysis.
RNA Electrophoretic Mobility Shift Assay (REMSA)
Various regions in p21 3′-UTR were PCR-amplified using primers containing T7 promoter sequence (5′-GGATCCTAATACGACTCACTATAGGGAG-3′). RNA probes were made from in vitro transcription by T7 RNA polymerase in the presence of α-32P-UTP. REMSA was performed with 200 nM of recombinant protein, 1 mg/ml of yeast tRNA and 50 000 CPM 32P-labeled RNA probe in a reaction buffer (10 mM Tris–Cl, pH 7.5, 25 mM KCl, 5 mM MgCl2, 1 mM DTT) for 20 min at 25°C. RNA/protein complexes were digested with 100 U RNaseT1 for 10 min at 37°C and then separated in 7% of native PAGE. RNA-protein complexes were visualized by autoradiography. For supershift assay, 3 µg of anti-HA antibody was pre-incubated with HA-tagged proteins for 30 min on ice prior to incubation with a RNA probe.
Cell fractionation
Cells were washed with ice-cold PBS for three times and harvested by scrapping. To isolate cytoplasmic fractions, cells were resuspended in lysis buffer for immunoprecipitaiton (20 mM Tris–Cl, pH 8.0, 125 mM NaCl, 2 mM EDTA, 0.5% NP-40, protease inhibitor cocktail) and pelleted by centrifugation (5000 rpm, 5 min, 4°C). Supernatants were saved and used as cytoplasmic fraction. To prepare nuclear fractions, pellets (nuclei) were washed two times with the lysis buffer without NP-40, re-suspended in the lysis buffer and sonicated for 20 s. Cell debris from nuclear extracts was removed by centrifugation.
RESULTS
HuR is necessary for RNPC1 to regulate p21 mRNA stability
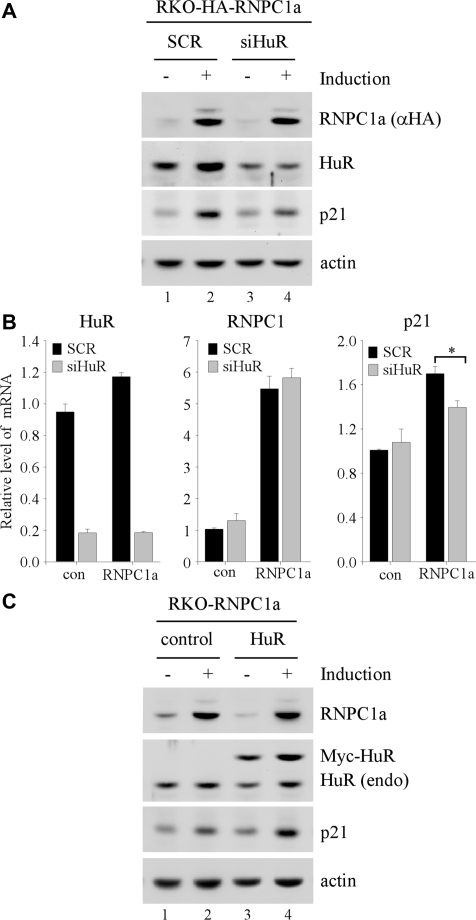
Recently, we and others showed that RNPC1 and HuR, both of which share a homology in their RNA-binding motifs, are capable of enhancing p21 mRNA stability via directly binding p21 3′-UTR (27,29). As a critical regulator of the cell cycle, p21 expression needs to be tightly controlled. Interestingly, RNPC1 can be induced by DNA damage in a p53-dependent manner (27), whereas HuR is ubiquitously expressed. Thus, we examined whether HuR is required for RNPC1 to regulate p21. To test this, the level of p21 was measured in RKO cells, which were transfected with scrambled siRNA or siRNA against HuR and then uninduced or induced to express HA-tagged RNPC1a for 2 days. We showed that upon induction of RNPC1a, the level of p21 protein was markedly increased (Figure 1A, compare lanes 1–2), consistent with previous report (27). We also showed that the level of HuR protein was decreased by siRNA against HuR but not scrambled siRNA (Figure 1A, compare lanes 1–2 with 3–4). Interestingly, upon knockdown of HuR, the extent of p21 induction by RNPC1a was significantly diminished (Figure 1A, compare lanes 1 and 3 with 2 and 4, respectively). Since the increased expression of p21 by RNPC1 is due to enhanced p21 mRNA stability (27), the level of p21 transcript along with HuR and RNPC1 was measured by qRT–PCR. We showed that the level of HuR transcript was decreased by ∼80% by siRNA against HuR compared to that by scrambled siRNA regardless of RNPC1 overexpression (Figure 1B, HuR column). In addition, we found that upon inducible expression of RNPC1, the level of RNPC1 transcript was increased by ∼5- to 6-fold (Figure 1B, RNPC1 column). We would like to mention that endogenous RNPC1 can be increased by more than 3-fold in response to DNA damage or p53 activation (27,30), suggesting that the level of RNPC1 expressed in the inducible RNPC1-producing RKO cells is within the physiological range. Consistent with the above result, we showed that the level of p21 transcript increased by RNPC1 was significantly diminished upon knockdown of HuR (Figure 1B, p21 column). However, knockdown of HuR alone had no obvious effect (Figure 1B, p21 column). Conversely, we examined whether overexpression of HuR is capable of enhancing RNPC1-mediated p21 induction. We showed that upon expression of Myc-tagged HuR in RKO cells, the extent of the increase in p21 protein by RNPC1 was enhanced (Figure 1C, compare lanes 1 and 3 with 2 and 4, respectively). In contrast, ectopic expression of HuR alone had no obvious effect on p21 expression (Figure 1C, compare lane 1 with 3), consistent with the idea that HuR needs to be activated in order to exert its effect on p21 mRNA stability (22).
Figure 1.
HuR is necessary for RNPC1 to regulate p21 expression. (A) The level of p21, HuR, RNPC1a and actin was measured in RKO cells transfected with scrambled siRNA (SCR) or siRNA against HuR for 24 h, followed with or without induction of RNPC1a for 48 h. (B) The level of p21, RNPC1 and HuR transcripts was measured by qRT–PCR with total RNA purified from RKO cells treated as in (A) above. The level of GAPDH transcript was measured as an internal control for normalization. *P < 0.05 (Students’ t-test). (C) The level of p21, HuR, Myc-HuR, RNPC1a and actin was measured in RKO cells transfected with an empty vector or a vector expressing Myc-tagged HuR for 24 h, followed with or without induction of RNPC1a for 48 h.
RNPC1 physically interacts with HuR in vitro and in vivo
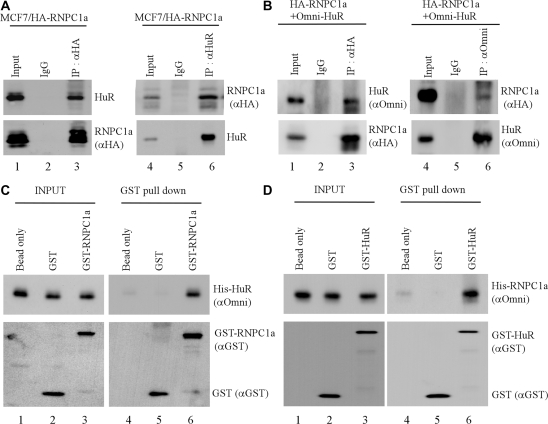
Considering that RRMs constitute a unique set of protein–protein interaction motifs (31), we examined whether HuR and RNPC1 physically interact and whether their interaction plays a role in the regulation of p21 mRNA stability. To test this, potential RNPC1–HuR complexes were immunoprecipitated by either anti-RNPC1 or anti-HuR, followed by western blot analysis. To eliminate the possibility that RNPC1 interacts with HuR indirectly via binding to the same RNA transcript, all assays were performed with cell extracts treated with RNase A to degrade RNAs prior to immunoprecipitation. First, potential interaction between RNPC1 and HuR was examined in MCF7 cell line that can inducibly express HA-tagged RNPC1. We showed that endogenous HuR was detected in RNPC1 but not control IgG immunocomplexes (Figure 2A, compare lanes 2–3). Conversely, HA-tagged RNPC1 was detected in HuR immunocomplexes (Figure 2A, compare lanes 5–6). Next, the interaction between RNPC1 and HuR was examined in RKO cells co-transfected with HA-tagged RNPC1a and His-tagged HuR. We showed that HuR was detected in RNPC1a immunocomplexes (Figure 2B, lane 3) whereas RNPC1 was detected in HuR immunocomplexes (Figure 2B, lane 6). To further examine the physical interaction between RNPC1 and HuR, in vitro GST-pull down assay was performed. We showed that His-tagged HuR bound to GST-RNPC1 but not GST beads (Figure 2C, compare lanes 5–6). Conversely, His-tagged RNPC1 bound to GST-HuR but not GST beads (Figure 2D, compare lanes 5–6). To verify the physical interaction between RNPC1 and HuR in vivo, we examined the interaction of endogenous RNPC1a with endogenous HuR in RKO cells. We showed that endogenous RNPC1 was detected in HuR immunocomplexes (Figure 2E, lane 3). Conversely, endogenous HuR was also detected in RNPC1a immunocomplexes (Figure 2E, lane 4). Together, our data demonstrate that RNPC1 directly interacts with HuR in vitro and in vivo.
Figure 2.
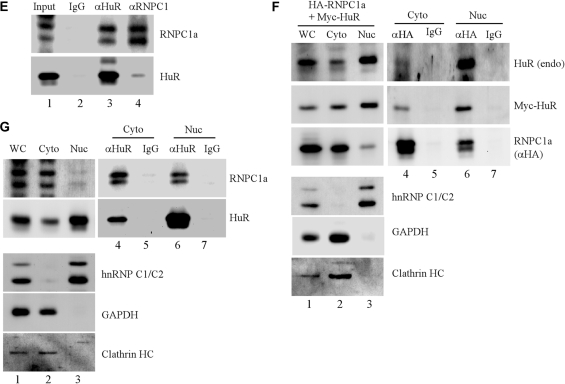
RNPC1 physically interacts with HuR in vitro and in vivo. (A) MCF7 cells were induced to express HA-tagged RNPC1a for 16 h, and then used for immunoprecipitation with anti-HA (lane 3), anti-HuR (lane 6), or mouse IgG (lanes 2 and 5). The immunoprecipitated complexes were analyzed by western blot analysis using anti-HA to detect HA-RNPC1a or anti-HuR to detect HuR. (B) RKO cells were transiently transfected with HA-tagged RNPC1a and His-tagged HuR for 48 h, and then used for immunoprecipitation with anti-HA to recognize HA-RNPC1 (lane 3), anti-Omni to recognize His-HuR (lane 6) or mouse IgG (lanes 2 and 5). The immunoprecipitated complexes were analyzed by western blot analysis using anti-HA to detect HA-RNPC1a or anti-Omni to detect His-HuR. (C) GST or GST-RNPC1a recombinant proteins were incubated with His-tagged HuR in a pull-down buffer containing glutathione sepharose beads for 2 h. Protein complexes on the beads were washed, and used for western blot analysis with anti-GST to detect GST and GST-RNPC1a or anti-Omni to detect His-HuR. Input controls were presented in lanes 1–3. (D) The experiment was performed as in (C) except that HuR was GST-tagged whereas RNPC1a was His-tagged. (E) Whole-cell extracts from RKO cells were used for immunoprecipitation with anti-HuR (lane 3), anti-RNPC1 (lane 4) or mouse IgG (lanes 2). The immunoprecipitated complexes were analyzed by western blot analysis with anti-HuR or anti-RNPC1. (F) Cytoplasmic and nuclear extracts were purified from RKO cells transiently transfected with HA-tagged RNPC1a and Myc-tagged HuR for 48 h and then used for immunoprecipitation with anti-HA to recognize HA-RNPC1 immunocomplexes (lane 4 and 6) or mouse IgG as a control (lanes 5 and 7). Whole cell lysates, cytoplasmic and nuclear extracts, and anti-HA and mouse IgG immunocomplexes were analyzed by western blot analysis using anti-HuR, anti-Myc to detect Myc-HuR, anti-HA to detect HA-RNPC1, anti-hnRNP C1/C2, anti-GAPDH and anti-clathrin heavy chain, respectively. (G) Cytoplasmic and nuclear extracts from RKO cells were used for immunoprecipitation with anti-HuR (lanes 4 and 6) or mouse IgG (lanes 5 and 7). Whole cell lysate, cytoplasmic and nuclear extracts and anti-HuR and mouse IgG immunocomplexes were analyzed by western blot analysis using anti-RNPC1, anti-HuR, anti-hnRNP C1/C2, anti-GAPDH and anti-clathrin heavy chain, respectively.
Considering that the majority of HuR protein is localized in nucleus and HuR can be translocated from nucleus to cytoplasm upon exposure to a stress, we examined in which cell compartment (i.e. nucleus and cytoplasm) RNPC1 and HuR physically interact. First, the level of HuR and RNPC1 was examined in cytoplasmic and nuclear extracts purified from RKO cells co-transfected with HA-tagged RNPC1a and Myc-tagged HuR. We showed that the majority of endogenous HuR protein was localized in nucleus whereas a small amount in cytoplasm (Figure 2F, HuR panel, lanes 1–3), consistent with previous report (29). In addition, Myc-tagged HuR was similarly distributed in cells as endogenous HuR (Figure 2F, Myc-HuR panel, lanes 1–3). However, a majority of RNPC1a was localized in cytoplasm (Figure 2F, RNPC1 panel, lanes 1–3), consistent with previous report (27). As a control, GAPDH and clathrin heavy chain were primarily localized in cytoplasm whereas hnRNP C1 and C2 were primarily expressed in nucleus. Next, cytoplasmic and nuclear extracts containing potential HuR-RNPC1 complexes were used for immunoprecipitation by either anti-RNPC1 or anti-HuR, followed by western blot analysis. We showed that both endogenous and Myc-tagged HuR were detected in RNPC1a-immunocomplexes from either cytoplasmic or nuclear extracts (Figure 2F, lanes 4 and 6). Conversely, we showed that endogenous RNPC1a was detected in endogenous HuR immunocomplexes from either cytoplasmic or nuclear extracts (Figure 2G, lanes 4 and 6). These results indicate that RNPC1 interacts with HuR regardless of cellular compartment.
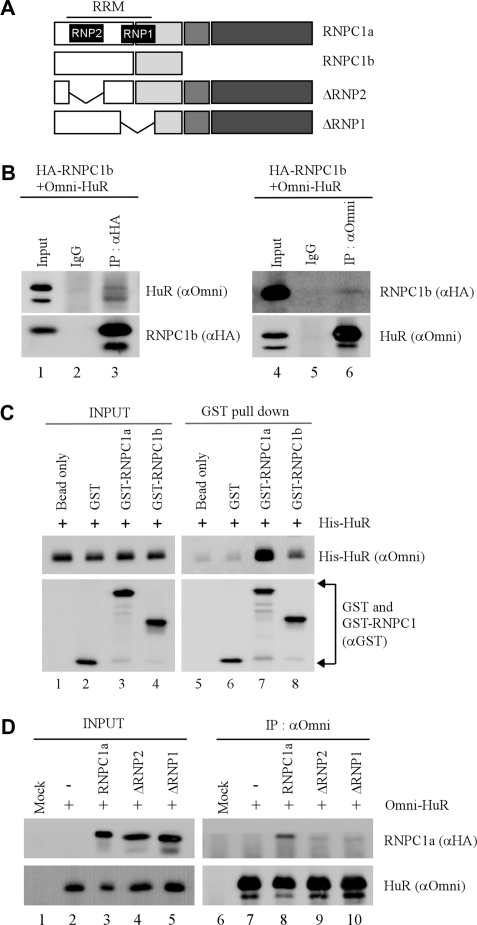
Next, we wanted to examine whether RNPC1b, which lacks most of the C-terminal region in RNPC1a, interacts with HuR. To test this, the interaction between RNPC1b and HuR was examined in RKO cells transfected with HA–RNPC1b and His–HuR. We showed that like RNPC1a (Figure 2), RNPC1b was detected in HuR immunocomplexes (Figure 3B, lane 6). Conversely, we showed that His-HuR protein was detected in RNPC1b-immunocomplexes (Figure 3B, lane 3). To examine the affinity of HuR to RNPC1a and RNPC1b, in vitro GST-pull down assay was performed. We showed that His-tagged HuR had a much stronger affinity to GST-tagged RNPC1a than to GST-tagged RNPC1b (Figure 3C, compare lanes 7–8). This suggests that the N-terminal region in RNPC1 is sufficient for, but the C-terminal region in RNPC1a facilitates, the interaction between RNPC1 and HuR. Since the RRM domain in RNPC1 consists of two sub-domains, RNP1 and RNP2 (27), we determined which sub-domain in RNPC1 is required for its interaction with HuR. We found that while RNPC1a was detected in HuR immunocomplexes, ΔRNP1 and ΔRNP2 were not immunoprecipitated with HuR (Figure 3D, compare lanes 8–10). This suggests that both RNP1 and RNP2 sub-domains are required for the interaction between RNPC1 and HuR.
Figure 3.
RRM in RNPC1 is necessary for interacting with HuR. (A) Schematic presentation of RNPC1a, RNPC1b, ΔRNP1 and ΔRNP2. The location of RRM domain and RNP sub-domains are indicated. (B) RKO cells were transfected with His-tagged HuR for 24 h and then induced to express HA-tagged RNPC1b for another 24 h. Potential HuR–RNPC1b complexes were immunoprecipitated with anti-HA (lane 3), anti-Omni (lane 6) or mouse IgG (lanes 2 and 5), which were then used for western blot analysis using anti-HA to detect HA-RNPC1b or anti-Omni to detect His-HuR. (C) GST, GST-RNPC1a or GST-RNPC1b recombinant proteins was incubated with His-tagged HuR in a pull-down buffer containing glutathione sepharose beads for 2 h. Protein complexes on the beads were washed, and used for western blot analysis with anti-GST to detect GST, GST-RNPC1a and GST-RNPC1b or anti-Omni to detect His-HuR. Input controls were presented in lanes 1–4. (D) RKO cells were transfected with His-tagged HuR along with HA-tagged RNPC1a, ΔRNP1, or ΔRNP2 for 48 h, and then used for immunoprecipitation with anti-Omni. The immunoprecipitated His–HuR complexes were analyzed by western blot analysis with anti-HA to detect HA-RNPC1 or anti-Omni to detect His–HuR. Input control was presented in lanes 1–5.
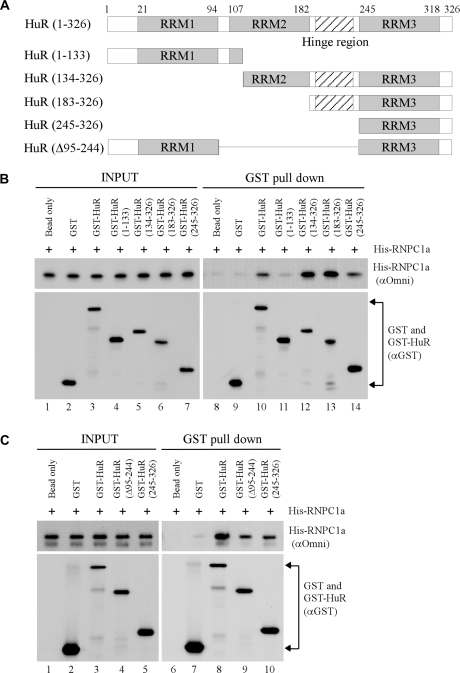
Similarly, the domain in HuR necessary for interacting with RNPC1 was determined. To test this, various HuR deletion mutants were made as GST-fusion proteins for GST pull down assay (Figure 4A). We found that His-tagged RNPC1 bound to GST-HuR (1–326), GST-HuR (134–326), GST-HuR (183–326) and weakly to GST-HuR (245–326), but little if any to GST-HuR (1–133) and GST (Figure 4B, lanes 9–14). Furthermore, we showed that the binding affinity of HuR (Δ95-244) and HuR (245–326) to RNPC1 was comparable (Figure 4C, lanes 9–10). This suggests that RRM3 in HuR is sufficient for, but the hinge region facilitates, the interaction between HuR and RNPC1.
Figure 4.
RRM 3 in HuR is sufficient for interacting with RNPC1. (A) Schematic presentation of HuR and various deletion mutants. (B and C) GST or various GST-HuR proteins were incubated with His-tagged RNPC1a in a pull-down buffer containing glutathione sepharose beads for 2 h. Protein complexes on the beads were washed and then used for western blot analysis with anti-GST to detect GST and various GST-tagged HuR proteins or anti-Omni to detect His-tagged RNPC1. Input controls were presented in lanes 1–7 (B) and lanes 1–5 (C).
RNPC1 binds to AREs in p21 3′-UTR and enhances the RNA-binding activity of HuR in vitro and in vivo
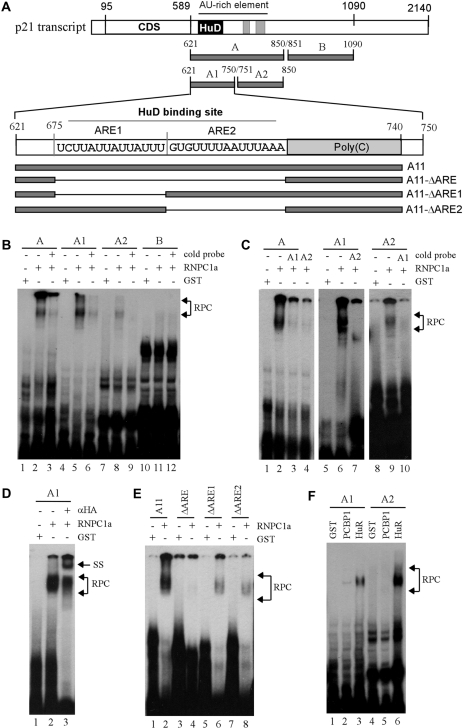
Previously, we showed that RNPC1 directly binds to p21 3′-UTR and regulates the stability of luciferase transcript fused to the region between nt 571–727 (27). However, the binding site in p21 3′-UTR for RNPC1 was not mapped. Thus, REMSA was performed with purified recombinant RNPC1 and 32P-labeled RNA probes corresponding to various regions in p21 3′-UTR (Figure 5A). We showed that RNPC1 bound strongly to A and A1 fragments, weakly to A2 fragment, but not to B fragment (Figure 5B, lanes 2, 5, 8 and 11). The specificity of the RNPC1 binding to these fragments was verified by competition assay using an excess amount of their own unlabeled probes (Figure 5B, compare lanes 2, 5, 8 and 11 with lanes 3, 6, 9 and 12, respectively). We also showed that the binding of RNPC1 to probe A was inhibited by cold probe A1 and A2 (Figure 5C, compare lane 2 with 3–4) whereas the binding of RNPC1 to probe A1 was inhibited by cold probe A2 (Figure 5C, compare lanes 6–7). Conversely, the binding of RNPC1 to probe A2 was inhibited by cold probe A1 (Figure 5C, compare lanes 9–10). In addition, we showed that HA-tagged RNPC1 in the RNPC1–RNA complexes were recognized and supershifted by anti-HA (Figure 5D, compare lanes 2–3). To further map the region in fragment A1, four well-defined RNA probes were generated (Figure 5A): probe A11, which is derived from nt 621–740 and contains two ARE elements and a poly(C)-rich element; probe A11-ΔARE, which is a derivative of probe A11 but lacks both ARE1 and ARE2 elements; and probes A11-ΔARE1 and A11-ΔARE2, which lack ARE1 and ARE2, respectively. We showed that RNPC1 bound to A11, weakly to A11-ΔARE1 and A11-ΔARE2, but not to A11-ΔARE (Figure 5E). This suggests that a transcript containing one ARE can be recognized by RNPC1, but the affinity of RNPC1 to a transcript with two AREs is markedly increased.
Figure 5.
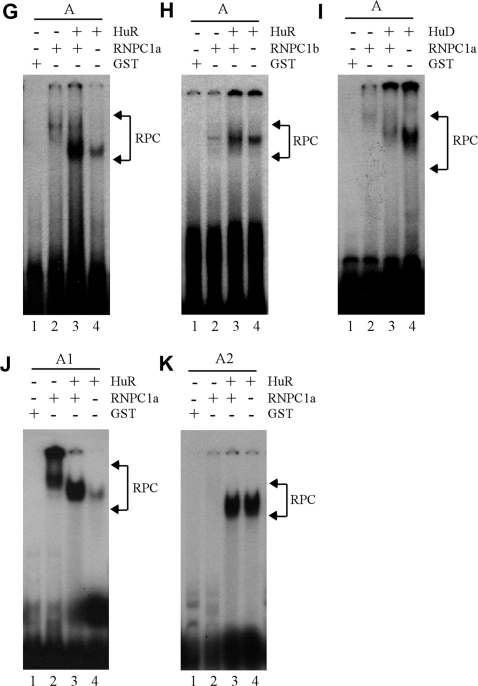
Identification of AREs as a RNPC1-binding site in p21 3′-UTR. (A) Schematic presentation of p21 3′-UTR along with the location of AREs, poly(C)-rich region, HuD-binding site and RNA probes used for REMSA. (B) REMSA was performed by mixing 32P-labeled probe A, A1, A2, or B with recombinant GST or GST-HA-RNPC1a. An excess amount of unlabeled cold probe was used for competition in lanes 3, 6, 9 and 12. (C) The experiment was performed as in (B) except probes used. For competition assay, an excess amount of unlabeled cold probe (A1 or A2) was added to a reaction mixture containing RNPC1 and probe A, A1 or A2. (D) For supershift assay, 3 µg of anti-HA antibody was added to a reaction mixture containing probe A1 and GST-HA-RNPC1. (E) The experiment was performed as in (B) except probes used. (F) HuR has a higher affinity to probe A2 than A1 whereas PCBP1 only weakly binds to probe A1. The experiment was performed with GST-tagged HuR or PCBP1 mixed with 32P-labeled probe A1 or A2. (G and H) RNPC1 cooperates with HuR to bind p21 transcript. GST-tagged RNPC1a or RNPC1b was mixed with GST-tagged HuR and 32P-labeled probe A for REMSA analysis. (I) RNPC1 inhibits the binding of HuD to p21 transcript. The experiment was performed as in (G–H) except that GST-tagged HuD was used. (J and K) The experiments were performed as in (G) except the probes used.
Fragment A1 contains a region of poly(C), which can be recognized by poly(C)-binding proteins, including PCBP1 (32). In addition, a region overlapping with the AREs in fragment A2 was found to be recognized by HuR (11). Thus, the binding of HuR and PCBP1 to fragments A1 and A2 was measured. We showed that HuR had a stronger affinity to the AREs in fragment A2 than that in A1 (Figure 5F, compare lane 3 with 6). We also showed that PCBP1 had a weak affinity to fragment A1 but not to fragment A2 (Figure 5F, lanes 2 and 5). This is not surprising since fragment A2 does not contain a poly(C)-rich element. These data indicate that RNPC1 and HuR have their own preferential AREs in p21 3′-UTR: RNPC1 to the upstream AREs within nt 621–750 versus HuR to downstream AREs within nt 751-850. Next, we examined whether RNPC1 and HuR modulate each other’s RNA-binding activity to p21 3′-UTR. To test this, the binding of HuR and RNPC1 to fragment A was examined. We showed that while RNPC1a and HuR bound to fragment A, a combination of HuR and RNPC1a markedly enhanced the RNA-binding affinity of HuR to fragment A (Figure 5G, lanes 2-4). In addition, we showed that RNPC1b, which had a weak interaction with HuR (Figure 3) and had a weak RNA-binding activity to the AREs in fragment A, were also capable of enhancing the RNA-binding activity of HuR, albeit to a lesser extent (Figure 5H). Since AREs in fragment A1 can be recognized by HuD (29), we examined the effect of RNPC1 on HuD binding to the AREs in fragment A. We showed that while HuD had a strong affinity to the AREs in fragment A, its RNA-binding activity was markedly decreased when RNPC1 was present (Figure 5I, lanes 2–4), suggesting that RNPC1 and HuD compete to recognize the same AREs. Considering that RNPC1 has a stronger affinity to AREs in fragment A1 than in A2 whereas HuR has a stronger affinity to A2 than to A1, we examined whether the binding of RNPC1 to RNA is necessary for enhancing the RNA-binding activity of HuR. To test this, the binding of HuR and RNPC1 to fragment A1 or A2 was examined. We showed that a combination of HuR and RNPC1a markedly enhanced the RNA-binding affinity of HuR to fragment A1 but not to fragment A2 (Figure 5J–K, lanes 3–4). This suggests that the enhanced RNA-binding activity of HuR to p21 transcript is dependent on the binding of RNPC1 to the same transcript.
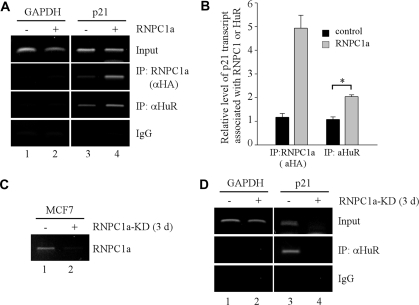
To examine whether RNPC1 is capable of enhancing the RNA-binding activity of HuR in vivo, RNA immunoprecipitation assay was performed with cell extracts purified from RKO cells uninduced or induced to express RNPC1 for 24 h. We showed that p21 transcript was detected in RNPC1a but not control IgG immunoprecipitates (Figure 6A, compare lanes 3–4). We also showed that p21 transcript was detected in HuR immunoprecipitates (Figure 6A, lane 3), consistent with the fact that HuR is ubiquitously expressed (22). Interestingly, the level of p21 transcript associated with HuR was markedly increased in the presence of RNPC1 (Figure 6A, HuR panel, compare lanes 3–4). To quantify the level of p21 transcript associated with RNPC1 and HuR, qRT–PCR was performed and showed that upon expression of RNPC1a, the level of p21 transcript associated with RNPC1 was increased by 5-fold (Figure 6B). Importantly, we found that the amount of p21 transcript associated with HuR was further increased by ∼2-fold when RNPC1 was present (Figure 6B). To further test this, RNA immunoprecipitation assay was performed with cell extracts purified from MCF7 cells uninduced or induced to knockdown RNPC1a for 3 days. We showed that upon induction, the level of RNPC1 transcript was markedly reduced (Figure 6C, compare lanes 1–2), leading to marked decrease of p21 transcript (Figure 6D, input panel), consistent with previous report (27). In addition, we showed that upon knockdown of RNPC1a, the level of p21 transcript associated with HuR was markedly reduced (Figure 6D, HuR panel, compare lanes 3–4). These results suggest that upon knockdown of RNPC1, the loss of HuR binding to p21 mRNA is at least in part due to decreased RNA-binding activity of HuR as well as decreased amounts of p21 mRNA available for HuR to interact. Together, our data indicate that the RNA-binding activity of HuR can be enhanced by RNPC1 in vitro and in vivo.
Figure 6.
RNPC1 enhances the RNA-binding activity of HuR in vivo. (A) RKO cells were uninduced or induced to express HA-tagged RNPC1a for 16 h, and then used for immunoprecipitation with anti-HA to capture RNPC1–RNA complexes, anti-HuR to capture HuR–RNA complexes, or mouse IgG as a control. Total RNAs were purified from immunocomplexes and subjected to RT–PCR to measure the level of p21 transcript associated with RNPC1a or HuR. The level of GAPDH was also measured as a control. (B) The level of p21 transcript associated with RNPC1- or HuR-immunocomplexes was quantified by qRT–PCR. *P < 0.01 (Students’ t-test). (C) The level of RNPC1a was measured by RT–PCR in MCF7 cells uninduced (−) or induced (+) to knock down RNPC1a for 3 days. (D) MCF7 cells were uninduced or induced to knockdown RNPC1a for 3 days, and then used for immunoprecipitation with anti-HuR to capture HuR–RNA complexes or mouse IgG as a control. Total RNAs were purified from immunocomplexes and subjected to RT–PCR to measure the level of p21 transcript associated with HuR. The level of GAPDH was also measured as a control.
DISCUSSION
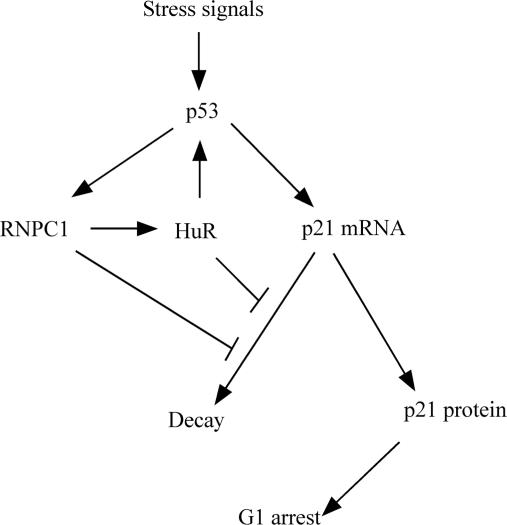
Here, we showed that knockdown of HuR inhibits the extent of p21 mRNA stability induced by RNPC1. We also provided evidence that RNPC1 directly interacts with, and then enhances the RNA-binding activity of, HuR. Furthermore, we showed that the ability of RNPC1 to induce p21 expression is enhanced by ectopic expression of HuR. This suggests that activation of HuR by RNPC1 contributes to p21 expression. Considering that HuR is capable of enhancing p53 translation in response to UV irradiation (33), p53-RNPC1–HuR forms a feedback–feedforward loop to coordinately regulate p21 expression and thus the cell-cycle control (Figure 7). Together, we hypothesize that in response to stress signals, p53 is activated and induces RNPC1, which modulates the RNA-binding activity of HuR, which in turn increases p53 expression via translation and cooperates with RNPC1 to regulate p21 mRNA stability.
Figure 7.
Regulation of p21 by an integrated network of the p53 pathway.
Previously, we showed that RNPC1 binds to p21 3′-UTR (27), but it is not clear whether AREs are the sequence to which RNPC1 binds. Within nt 621–750, there are two AREs, designated upstream AREs, which can be recognized by HuD (11). In addition, within nt 751–850, there are two additional AREs, designated downstream AREs, which can be recognized by HuR (29). Interestingly, we showed that RNPC1 has a much higher affinity to the upstream AREs than to the downstream AREs. Consistent with this, we showed that RNPC1 competes with HuD to bind the upstream AREs. In addition, we showed that RNPC1 promotes, rather than competes with, HuR to bind the downstream AREs and the binding of RNPC1 to RNA is required for enhancing the RNA-binding activity of HuR. These data suggest that although RRM has a propensity to bind an ARE sequence (11,27,29), the RRM in RNPC1 has a unique property that is similar to HuD, but different from HuR. Furthermore, it appears that while RNPC1 is able to recognize a transcript with one ARE, the affinity of RNPC1 to the one with two AREs is markedly increased (Figure 5E). Considering that RNPC1 has only one RRM (27) and both sub-domains (RNP1 and RNP2) in RNPC1 RRM are essential for RNA binding (data not shown), we postulate that RNPC1 may exist as a dimer. Thus, when one RRM from RNPC1 dimer binds to ARE, the binding of other RRM from the dimer to ARE is stabilized. Similarly, when RNPC1 and HuR form a heterodimer, the binding of RNPC1 to one ARE element in A1 probe enhances the binding of HuR to the other ARE element (Figure 5J). However, due to weak or no binding of RNPC1 to the ARE in A2 probe, the RNA-binding activity of HuR cannot be enhanced even if HuR and RNPC1 form a heterodimer (Figure 5K). Nevertheless, it is still not clear why the interaction of RNPC1 with HuR does not enhance the RNA-binding activity of RNPC1, which merits further investigation.
HuR is a ubiquitously expressed member of the Hu/ELAV family and primarily localized in nucleus at a normal condition (22). However, in response to a stress signal, such as UV irradiation, HuR is translocated from nucleus to cytosol wherein HuR binds to its target RNAs and then regulates their stability or translation (34). Not surprisingly, the translocation of HuR from nucleus to cytosol is tightly controlled. This includes: enhanced cytoplasmic translocation of HuR via methylation at residue R217 by CARM1 and phosphorylation at residue S221 by PKC; and nuclear retention of HuR via phosphorylation at residue S202 by CDK1 and at residue S242 by an unknown kinase (22). Here, we showed that HuR activity is modulated by RNPC1 via protein–protein interaction. Given the fact that in response to stress signals, RNPC1 expression is induced and HuR is activated through translocation, the enhanced RNA-binding activity of HuR by RNPC1 represents a coordinated effort to regulate key DNA damage responsive genes, such as p21, under a stress condition.
RRM domain is known to serve as a domain for protein–protein interaction as well as a domain for RNA binding (31). Here, we provided evidence that RNPC1 directly interacts with HuR via the RRM domain in RNPC1 and RRM3 in HuR. In addition, we found that the C-terminal unique region in RNPC1a contributes to its interaction with HuR since RNPC1b, which lacks most of the C-terminal region, is weak in interacting with HuR (Figure 3C). Consistent with this, the extent of enhanced HuR RNA-binding activity is stronger by RNPC1a than that by RNPC1b (Figure 5G–H). These data further support our hypothesis that upon interaction with HuR, the binding of RNPC1 to the upstream AREs stabilizes the binding of HuR to the downstream AREs in p21 3′-UTR.
Our hypothesis for how RNPC1 and HuR coordinately regulate p21 mRNA stability may explain similar phenomenon for the regulation of HIF-1α by HuR and RNA-binding protein PTB. Both HuR and PTB are known to promote translation of HIF-1α (35), but the mechanism is still uncertain. It is possible that HuR and PTB may enhance each other’s RNA-binding affinity to HIF-1α mRNA and thus cooperatively upregulate HIF-1α translation. Similarly, if RNPC1 were found to bind HIF-1α transcript, it is possible that RNPC1 may enhance the RNA-binding activity of HuR to HIF-1α and thus cooperatively regulate HIF-1α translation. Finally, we would also like to note that the extent of the increased level of p21 mRNA is less than that of p21 protein (Figure 1), suggesting that other mechanism may be involved in p21 expression, especially protein translation. Since RNA-binding protein is known to regulate protein translation and p21 is subject to translational control (36), we examined the possibility that RNPC1 may regulate p21 translation. Indeed, we found that in the presence of RNPC1, the extent of p21 mRNA associated with polysomes is increased (data not shown). Furthermore, we found that the level of newly synthesized p21 protein is increased in the presence of RNPC1 (data not shown). Considering that p21 translational regulation by RNPC1 possibly together with HuR has a significant implication in the cell-cycle control, further study is warranted to address this issue.
FUNDING
Funding for open access charge: National Institutes of Health (Grant CA076069).
Conflict of interest statement. None declared.
REFERENCE
- 1.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 2.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 3.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 4.Weinberg WC, Denning MF. P21Waf1 control of epithelial cell cycle and cell fate. Crit. Rev. Oral Biol. Med. 2002;13:453–464. doi: 10.1177/154411130201300603. [DOI] [PubMed] [Google Scholar]
- 5.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 7.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 8.Zhu J, Jiang J, Zhou W, Chen X. The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res. 1998;58:5061–5065. [PubMed] [Google Scholar]
- 9.Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl Acad. Sci. USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joseph B, Orlian M, Furneaux H. p21(waf1) mRNA contains a conserved element in its 3′-untranslated region that is bound by the Elav-like mRNA-stabilizing proteins. J. Biol. Chem. 1998;273:20511–20516. doi: 10.1074/jbc.273.32.20511. [DOI] [PubMed] [Google Scholar]
- 12.Barami K, Iversen K, Furneaux H, Goldman SA. Hu protein as an early marker of neuronal phenotypic differentiation by subependymal zone cells of the adult songbird forebrain. J. Neurobiol. 1995;28:82–101. doi: 10.1002/neu.480280108. [DOI] [PubMed] [Google Scholar]
- 13.Chung S, Jiang L, Cheng S, Furneaux H. Purification and properties of HuD, a neuronal RNA-binding protein. J. Biol. Chem. 1996;271:11518–11524. doi: 10.1074/jbc.271.19.11518. [DOI] [PubMed] [Google Scholar]
- 14.Dalmau J, Furneaux HM, Cordon-Cardo C, Posner JB. The expression of the Hu (paraneoplastic encephalomyelitis/sensory neuronopathy) antigen in human normal and tumor tissues. Am. J. Pathol. 1992;141:881–886. [PMC free article] [PubMed] [Google Scholar]
- 15.Good PJ. A conserved family of elav-like genes in vertebrates. Proc. Natl Acad. Sci. USA. 1995;92:4557–4561. doi: 10.1073/pnas.92.10.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 17.Marusich MF, Furneaux HM, Henion PD, Weston JA. Hu neuronal proteins are expressed in proliferating neurogenic cells. J. Neurobiol. 1994;25:143–155. doi: 10.1002/neu.480250206. [DOI] [PubMed] [Google Scholar]
- 18.Okano HJ, Darnell RB. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J. Neurosci. 1997;17:3024–3037. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szabo A, Dalmau J, Manley G, Rosenfeld M, Wong E, Henson J, Posner JB, Furneaux HM. HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and Sex-lethal. Cell. 1991;67:325–333. doi: 10.1016/0092-8674(91)90184-z. [DOI] [PubMed] [Google Scholar]
- 20.Lafarga V, Cuadrado A, Lopez de Silanes I, Bengoechea R, Fernandez-Capetillo O, Nebreda AR. p38 Mitogen-activated protein kinase- and HuR-dependent stabilization of p21(Cip1) mRNA mediates the G(1)/S checkpoint. Mol. Cell. Biol. 2009;29:4341–4351. doi: 10.1128/MCB.00210-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim HH, Yang X, Kuwano Y, Gorospe M. Modification at HuR(S242) alters HuR localization and proliferative influence. Cell Cycle. 2008;7:3371–3377. doi: 10.4161/cc.7.21.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HH, Gorospe M. Phosphorylated HuR shuttles in cycles. Cell Cycle. 2008;7:3124–3126. doi: 10.4161/cc.7.20.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HH, Abdelmohsen K, Lal A, Pullmann R., Jr, Yang X, Galban S, Srikantan S, Martindale JL, Blethrow J, Shokat KM, et al. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 2008;22:1804–1815. doi: 10.1101/gad.1645808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doller A, Pfeilschifter J, Eberhardt W. Signalling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR. Cell Signal. 2008;20:2165–2173. doi: 10.1016/j.cellsig.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 25.Doller A, Akool el S, Huwiler A, Muller R, Radeke HH, Pfeilschifter J, Eberhardt W. Posttranslational modification of the AU-rich element binding protein HuR by protein kinase Cdelta elicits angiotensin II-induced stabilization and nuclear export of cyclooxygenase 2 mRNA. Mol. Cell. Biol. 2008;28:2608–2625. doi: 10.1128/MCB.01530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doller A, Huwiler A, Muller R, Radeke HH, Pfeilschifter J, Eberhardt W. Protein kinase C alpha-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2. Mol. Biol. Cell. 2007;18:2137–2148. doi: 10.1091/mbc.E06-09-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shu L, Yan W, Chen X. RNPC1, an RNA-binding protein and a target of the p53 family, is required for maintaining the stability of the basal and stress-induced p21 transcript. Genes Dev. 2006;20:2961–2972. doi: 10.1101/gad.1463306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith DB, Corcoran LM. Hoboken, NJ: John Wiley & Sons, Inc.; 1994. Current Protocols in Molecular Biology. [Google Scholar]
- 29.Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook N, Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 2000;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X, Liu G, Zhu J, Jiang J, Nozell S, Willis A. Isolation and characterization of fourteen novel putative and nine known target genes of the p53 family. Cancer Biol. Ther. 2003;2:55–62. doi: 10.4161/cbt.180. [DOI] [PubMed] [Google Scholar]
- 31.Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 32.Giles KM, Daly JM, Beveridge DJ, Thomson AM, Voon DC, Furneaux HM, Jazayeri JA, Leedman PJ. The 3′-untranslated region of p21WAF1 mRNA is a composite cis-acting sequence bound by RNA-binding proteins from breast cancer cells, including HuR and poly(C)-binding protein. J. Biol. Chem. 2003;278:2937–2946. doi: 10.1074/jbc.M208439200. [DOI] [PubMed] [Google Scholar]
- 33.Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martindale JL, Atasoy U, Keene JD, Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl Acad. Sci. USA. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorospe M. HuR in the mammalian genotoxic response: post-transcriptional multitasking. Cell Cycle. 2003;2:412–414. [PubMed] [Google Scholar]
- 35.Galban S, Kuwano Y, Pullmann R., Jr, Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, Liu JO, et al. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol. Cell. Biol. 2008;28:93–107. doi: 10.1128/MCB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Chen X. Posttranscriptional regulation of p53 and its targets by RNA-binding proteins. Curr. Mol. Med. 2008;8:845–849. doi: 10.2174/156652408786733748. [DOI] [PMC free article] [PubMed] [Google Scholar]