Abstract
The continuing discoveries of potentially active small RNAs at an unprecedented rate using high-throughput sequencing have raised the need for methods that can reliably detect and quantitate the expression levels of small RNAs. Currently, northern blot is the most widely used method for validating small RNAs that are identified by methods such as high-throughput sequencing. We describe a new northern blot-based protocol (LED) for small RNA (∼15–40 bases) detection using digoxigenin (DIG)-labeled oligonucleotide probes containing locked nucleic acids (LNA) and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide for cross-linking the RNA to the membrane. LED generates clearly visible signals for RNA amounts as low as 0.05 fmol. This method requires as little as a few seconds of membrane exposure to outperform the signal intensity using overnight exposure of isotope-based methods, corresponding to ∼1000-fold improvement in exposure-time. In contrast to commonly used radioisotope-based methods, which require freshly prepared and hazardous probes, LED probes can be stored for at least 6 months, facilitate faster and more cost-effective experiments, and are more environmentally friendly. A detailed protocol of LED is provided in the Supplementary Data.
INTRODUCTION
Recent advances in high-throughput sequencing have led to the characterization of several important classes of small RNAs including microRNAs (miRNAs), endogenous small interfering RNAs (endo-siRNAs), piwi-interacting RNAs (piRNAs), transcription start-site associated RNAs (TSSa-RNAs) and unusually small RNAs (usRNAs) (1,2). The most convincing analytical method to validate small RNAs identified by high-throughput approaches is northern blot. Although northern blot is less sensitive than other analytical methods, it can readily reveal the presence of irrelevant products and can quantitate the expression level and size of both the small RNAs and their precursors (3). Several distinct northern blot protocols are currently used for small RNA detection. These methods primarily differ in the labeling and design of the probes used to detect RNA. The most popular probe-labeling protocol is based on incorporation of radio isotopes (32P). However, isotope labeling is often inconvenient, hazardous and is restricted by many institutions. As a safer alternative, non-isotopic-labeling methods using DIG-labeled probes are used to detect small RNAs (4,5). The DIG assay is comparable to isotope labeling-based methods in its sensitivity, and is safer than radioactive methods (5). Probe-design strategies also have significantly improved in the recent past. The traditional DNA oligonucleotide probes are increasingly replaced by LNA oligonucleotide probes that considerably improve the sensitivity in detecting small RNAs (3,6).
Cross-linking of the RNA to the membrane frequently improves the sensitivity of northern blots. However, conventional methods such as UV-cross-linking are generally not optimal for detection of small RNAs such as miRNAs and piRNAs that are shorter than 40 bases (7). A recently developed method using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) to cross-link RNAs to the membrane was demonstrated to have enhanced specificity over traditional cross-linking methods (7,8).
Thus, a hybrid approach based on the widely used 32P labeling, EDC-based cross-linking of RNA to membrane and LNA probes is likely to provide the most sensitive northern blot protocol for small RNAs (8). Since DIG-labeling is not known to outperform 32P-based protocols (5), substitution of 32P by DIG in such a hybrid approach is not expected to have any advantage over isotope-based protocols. However, we developed a highly sensitive protocol (L-E-D) that harnesses the advantages of LNA, EDC and DIG (Supplementary Figure S1) that is superior to 32P-based methods. LED was developed via a top-down optimization approach, in which its performance was assessed in detecting two different miRNAs (miR-21 and miR-16) over five distinct DIG-labeled LNA probe concentrations, eight different hybridization buffers, various temperatures and four types of membranes. We further compared the performance of LED and its equivalent 32P-based protocol using four different miRNAs (22–23 bases) across various RNA concentrations, and studied the specificity and sensitivity of LED using known concentrations of small RNAs.
MATERIALS AND METHODS
Total RNA from MCF-7 cells was loaded onto 15% SequaGel (National Diagnostics), electrophoresed and transferred to nylon membranes at 10–15 V (90 min) using Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad). Membranes were cross-linked to the RNA (60°C for 1–2 h) using freshly prepared cross-linking reagent (Doc-S). For 32P-based blots, LNA–DNA mixed oligonucleotide probes were end-labeled with [γ-32P] ATP by T4 polynucleotide kinase using KinaseMaxTM Kit (Ambion). For human miRNAs, pre-synthesized LNA-modified oligonucleotides were purchased from Exiqon (http://www.exiqon.com). For the KSHV miRNA, the probes were synthesized by Integrated DNA Technologies, IA. For LED blots, probes were labeled with the non-radioactive DIG, using End Tailing Kit (Roche Applied Science, Indianapolis, IN). Probe sequences used against the human miRNAs are TCAACATCAGTCTGATAAGCTA (miR-21), CGCCAATATTTACGTGCTGCTA (miR-16), TCCATCATTACCCGGCAGTATTA (miR-200c) and CAGACTCCGGTGGAATGAAGGA (miR-205). Pre-hybridization and hybridization were carried out using various hybridization buffers at different temperatures (Supplementary Table S1). For radioactive blots, hybridization buffers contained 106 cpm/ml of probe. For both methods, after hybridization the membranes were washed (37°C) twice using a low stringency buffer solution (2× SSC, 0.1% SDS), and a high stringency buffer solution (0.1× SSC, 0.1% SDS), for five and ten minutes, respectively. During optimization of northern blot analysis, photoemissions were detected using ChemiDoc-IT Imaging System (Figures 1, 2, Supplementary Figures S2 and S4). Since ChemiDoc-IT system is not compatible with 32P-based methods, to enable an unbiased comparison between the two methods (Figures 3 and 4), we used phosphor image screens to detect signals for both methods. To study the specificity of LED method, we used synthesized single-stranded kshv-miR-K12-1 and its mutants (M1, M2, M3) containing a 5′ phosphate to closely mimic miRNAs. The sequences are (mutations underlined): 5′-/Phos/AUUACAGAAACUGGGUGUAAGC-3′ (kshv-miR-K12-1), 5′-/Phos/AUUACAGAAACAGGGUGUAAGC-3′ (M1), 5′-/Phos/AUUACAGAAAGAGGGUGUAAGC-3′ (M2) and 5′-/Phos/AUUACAGAACGAGGGUGUAAGC-3′ (M3). The DNA-LNA mixed sequence (LNA underlined) 5′-GCTTACACCCAGTTTCCTGTAAT-3′ was used as the probe sequence against all four KSHV miRNA sequences. For specificity analysis, each lane in the gel (15% of polyacrylamide gel) was loaded with K12-1 synthetic RNA (0.2 fmol) mixed with MCF7 total RNA (5 µg). For sensitivity analysis, we used serially diluted amounts (0–0.4 fmol) of K12-1 miRNA that was spiked into MCF7 total RNA (5 µg).
Figure 1.
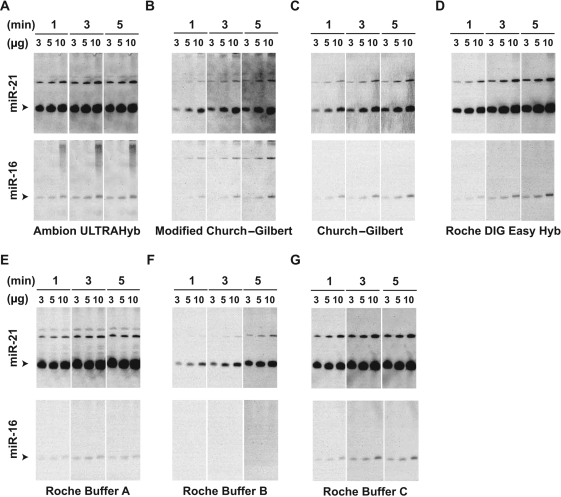
Effect of various hybridization buffers on the sensitivity of LED protocol in detecting miR-21 and miR-16. Seven different hybridization buffers (A–G) based on a probe concentration of 0.2 nM were used as indicated and detailed in supplementary document (Supplementary Table S1). Varying amounts of total RNA (3, 5 and 10 µg) were used to detect mature miR-21 and miR-16 (arrowheads) for each probe concentration, and the corresponding photo-luminescence was recorded over varying lengths (1, 3 and 5 min) of time. The upper bands may correspond to the precursor and primary transcripts of the miRNAs.
Figure 2.
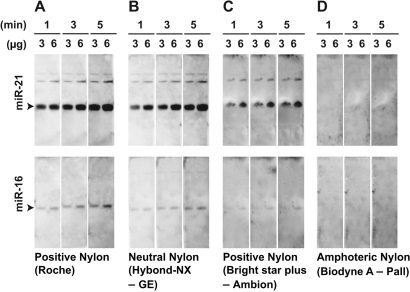
Evaluation of four different nylon membranes for LED protocol. Duration of photo-exposure (1, 3 and 5 min) and amount of total RNA (3 and 6 µg) are indicated. Among the tested membranes (A–D), positively charged and neutral nylon membranes purchased from Roche (A) and GE Healthcare (B), yielded the strongest signals.
Figure 3.
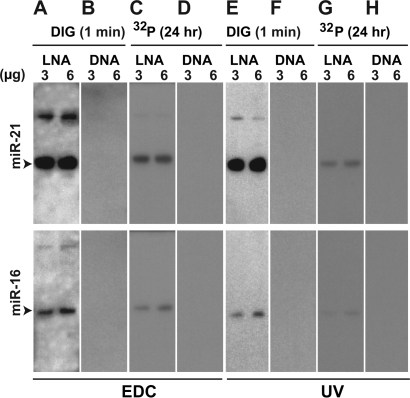
Systematic evaluation of the contribution of probe type, RNA–membrane cross-linking and probe labeling to LED performance. LNA probes, EDC-based cross-linking and DIG labeling in LED protocol were systematically substituted by DNA probes, UV-based cross-linking and 32P labeling to generate the following eight combinations for detecting miR-21 and miR-16: LNA-EDC-DIG (A) corresponds to LED protocol (A), DNA-EDC-DIG (B), LNA-EDC-32P (C), DNA-EDC-32P (D), LNA-UV-DIG (E), DNA-UV-DIG (F), LNA-UV-32P (G) and DNA-UV-32P (H). Phosphor image screens were used for all protocols to eliminate any bias due to the imaging system (‘Materials and Methods’ section). Duration of photo-exposure (1 min and 24 h) and amount of total RNA (3 and 6 µg) are indicated.
Figure 4.
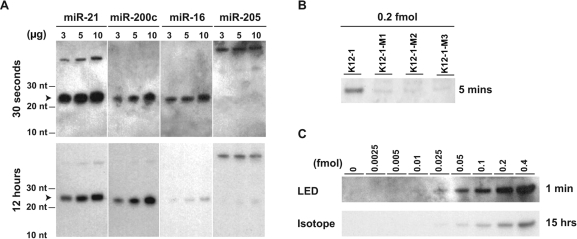
Performance assessment of LED using human miRNAs, miR-21 (22 bases), miR-200c (23 bases), miR-16 (22 bases) and miR-205 (22 bases) and a viral miRNA miR-K12-1 (23 bases). (A) Comparison of LED (top panel) to analogous method that uses 32P-labeling (bottom panel). LED northern blots yield notably strong signals for three miRNAs expressed at varying levels in MCF7 breast cancer cell line. An exposure time as short as 30 s is sufficient for LED to generally detect miRNAs. The mature miRNA, miR-205, that is not expressed in MCF7 is used as a negative control. For both methods, phosphor image screens were used to detect the signals to eliminate any biases from imaging. (B) Evaluation of specificity of LED using K12-1 viral miRNA that is absent in MCF-7, and using three different mutants of K12-1. Data suggests single-nucleotide specificity for LED. (C) Quantification of the absolute sensitivity of LED using serially diluted K12-1 miRNA (top panel) and its comparison to the method using 32P-labeling (bottom panel). The following amounts of K12-1 RNA are spiked into 5 µg of total RNA from MCF-7 in each lane (left to right): 0, 0.00019, 0.00038, 0.00075, 0.00188, 0.00375, 0.0075, 0.015 and 0.03 µg.
RESULTS
Optimization of DIG-labeled LNA-based probe concentration
Intuitively, an increase in probe concentrations would enhance the sensitivity of LED, but higher probe levels could also adversely affect northern blots. For instance, higher probe concentrations will increase background signal contributions from non-specifically bound DIG-labeled probes on the membrane. To determine the optimal probe concentration, we tested the dependence of starting probe concentration on sensitivity of LED in detecting miR-21 and miR-16 (Supplementary Figure S2), based on a commonly used buffer (ULTRAhyb by Ambion). For both miRNAs, the sensitivity is prominently improved as the probe concentration is increased from low (0.05 nM) to high (0.2 or 0.5 nM) concentrations. However, further increase (>0.5 nM) in probe concentration did not seem to greatly improve the sensitivity in detecting miR-21. In contrast, LED signals for the more difficult-to-detect RNA, miR-16, considerably improved upon increasing the probe concentration from 0.5 nM (up to 2 nM). These experiments indicate that the probe level of ∼0.2 nM is sufficient to detect both miRNAs, and hence this concentration level was used for additional optimization experiments. Further improvements in detection sensitivity can also be achieved by fine-tuning probe concentrations for RNAs of interest, as illustrated by the different levels of improvements that were realized for miR-21 and miR-16.
Effects of hybridization buffers on detection of small RNAs
Among the first seven hybridization buffers that we tested (Supplementary Table S1), ULTRAhyb (Ambion), DIG easy buffer (Roche) and buffers A and C (Roche) were among the most sensitive miRNA-detection approaches (Figure 1). The performances of the buffers indicate that some buffers could be more useful than others in certain situations. For example, using as little as 3 µg of total RNA, the difficult-to-detect miRNA, miR-16, is readily visible in the blots using Ambion ULTRAhyb, DIG easy buffer and buffer C. In comparison, buffer A is more sensitive than others in detecting miR-21 while ULTRAhyb, DIG Easy Hyb, buffer C, Church–Gilbert (9) and modified Church–Gilbert buffer (10) yield readily detectable, linear, concentration-dependent signals for both miR-21 and miR-16. The Church–Gilbert buffers are also useful in detecting miRNAs due to their linear detection range, and provide a cost- and time-advantage over other tested buffers since they can be readily prepared from a few reagents.
Since ULTRAhyb, and buffers B/C contain formamide that reduces the melting temperature for probe–RNA hybridization and hence may be a factor in their performance, we also tested modified Church–Gilbert protocol with 50% formamide at a reduced temperature (37°C). The formamide containing protocol did not yield a better signal than ULTRAhyb (Supplementary Figure S3) and did not appear to have any notable improvement over the protocol without formamide. The evaluations of eight different buffers allowed us to select a set of three buffers [ULTRAhyb, Buffer A and modified (10) Church–Gilbert] that could be further optimized and analyzed. ULTRAhyb and Buffer A were selected due to their high sensitivity, while the modified Church–Gilbert protocol was chosen due to its ease of availability and reduced cost. We note that buffer C and DIG easy Hyb, is likely as useful as buffer A, yet we selected buffer A for further tests because buffer A appeared to be more sensitive in detecting the low abundance, putative primary miR-16 transcript (Figure 1, upper most band).
Optimization of hybridization-temperature conditions
Based on the three selected buffers, we next studied the effect of hybridization temperature on sensitivity (Supplementary Figure S4). Consistent with the manufacturer-recommended conditions for DNA/RNA probes (∼40–60°C), our results show that there is not a large difference between sensitivities at 37 and 60°C for ULTRAhyb using LNA-based probes. However, background effects clearly improve upon increasing temperature (37–60°C). We also found that the modified Church–Gilbert protocol performs best at ∼60°C and its sensitivity is comparable to that of ULTRAhyb and Buffer A for detecting miR-21. Although the sensitivity of buffer A was comparable between the optimal temperature conditions of 37°C and 50°C, the performance of buffer A manifested greater variability than both ULTRAhyb and the modified Church–Gilbert protocol (Figure 1 and Supplementary Figure S4). Specifically, in contrast to previous experiments using buffer A, the band intensities during the temperature-optimization experiments for buffer A are weaker and are linear with RNA concentration. Such inconsistencies (additional data not shown) of buffer A and the increased sensitivity of ULTRAhyb over the modified Church–Gilbert protocol in detecting miR-16 led us to choose ULTRAhyb (37°C) as the preferred buffer solution for further evaluation of LED. Although we selected the ULTRAhyb buffer for further tests due to its overall performance, in our experience the modified Church–Gilbert protocol generally generates less background than ULTRAhyb, and is clearly an effective buffer for detecting abundant miRNAs.
Effect of membranes on LED performance
Since we relied on our experience in using positively charged Nylon membrane (Roche) for previous experiments, we sought to systematically test whether different membranes could significantly affect LED performance. Among the four different membranes tested (Figure 2), we found that the selected positively charged membrane (Roche) and the neutral Nylon membrane (Hybond-NX, GE) performed best (Figure 2A and B). Surprisingly, another widely used positively charged membrane (Bright star plus, Ambion) yielded relatively weak signals with moderate background levels (Figure 2C) in LED protocol. The amphoteric (Figure 2C) membrane performed poorly, and did not yield any signals for either miR-21 or miR-16. The results confirmed that the selected membrane (Roche) for LED is a reasonable choice, and also highlighted the influence of different membranes and membrane sources on northern blots used for small RNA detection.
Comparison of LED to other methods
To better understand the contribution of LNA probes, EDC cross-linking and DIG labeling to the LED protocol, we systematically tested their effects on the performance of northern blots in detecting miR-21 and miR-16. Specifically, one by one, we substituted LNA probes, EDC cross linking and DIG labeling by DNA probes, UV cross-linking and 32P labeling, respectively (Figure 3). As expected, the LNA-based protocols are considerably more sensitive than DNA-based northern blots (Figure 3: A versus B, C versus D, E versus F and G versus H). In comparison to 32P labeling, the use of DIG also significantly improves performance of the blot (Figure 3: A versus C and E versus G). Although the use of EDC cross-linking consistently improves the sensitivity of the method (Figure 3: A versus E and C versus G), it increases the background in DIG labeling-based method. Nevertheless, the complete LED protocol using EDC appears to perform the best among the various methods that we tested.
We further compared the sensitivity of LED to that of an equivalent method (7,8), which uses both EDC cross-linking of RNA to membrane and LNA probes, but uses 32P-labeled probes instead of DIG-labeled probes. In addition to using LNA probes and EDC cross-linking, the selected 32P-based protocol also uses identical hybridization buffer, hybridization temperature and LNA probe concentration to that of LED. Thus, the selected 32P-based protocol arguably represents the most sensitive northern blot method that is publicly available. We selected four miRNAs that are expressed at varying levels (miR-21—high, miR-16—medium, miR-200c—medium and miR-205—undetectably low) in the MCF-7 cell line. Serial dilution of RNA was performed to assess the performance of the methods at various RNA concentrations.
LED yielded stronger signals than the corresponding radioactive method for the three well- expressed miRNAs at all three different concentrations (Figure 4A). These results appear significant when considering the 1400-fold difference in exposure times of the two methods (30 s versus 12 h). LED required only a small amount (3 µg) of total RNA for detecting the three miRNAs and manifested an approximately linear detection range as was also observed in previous experiments.
Detection limits of LED
To evaluate the specificity of LED, we spiked (Methods) human total RNA from MCF-7 with the synthetic miR-K12-1 miRNA of the rarely occurring Kaposi sarcoma-associated herpesvirus (KSHV) and three miR-K12-1 synthetic mutants containing 1–3 point mutations. The viral miRNA was chosen since it is present only in KSHV-infected cells and has little similarity to any known human miRNA, thus reducing the possibility of cross-hybridization of the probe to other human miRNAs. The specificity analysis indicates that LED can differentiate between closely related miRNA family members (Figure 4B) and minor differences (1–2 bases) between small RNA family members are sufficient for quantifying them using LED. LED specificity is similar to what is expected for LNA probes (11), suggesting that the optimizations did not negatively impact the overall detection specificity. Finally, to quantify the absolute sensitivity of LED, we used LED to detect the synthetic miR-K12-1 at various concentrations. LED yields visible signals for RNA amounts as low as 0.05 fmol, and compares favorably to the sensitivity of the radioactive method using up to 15 h of exposure (Figure 4C).
DISCUSSION
The continuing discoveries of novel classes of small RNAs in numerous model organisms have raised the need for improved methods that are sensitive, safe, fast and reliable in quantifying small RNA expression. Traditional northern blot techniques using conventional DNA or RNA probes generally do not perform adequately in detecting small RNAs (<∼40 bases) due to their small sizes and sequence compositions (e.g. few G/C bases). Northern blot methods for small RNA detection have constantly improved in sensitivity, particularly with the introduction of LNA-DNA mixed hybrid probes and EDC-based cross-linking of small RNAs to membrane. We combined the strengths of LNA, DIG and EDC cross-linking, and optimized several key parameters to develop a useful small RNA detection protocol that is detailed in a step-by-step manner in the supplementary material (Doc-S). Although we focused our report on LED detection of miRNAs (∼22 bases), the most widely studied class of small RNAs, we also note that LED can detect some usRNAs (2) that are even smaller (∼16 bases) than miRNAs. In addition to miRNAs, LED will be likely useful for the detection of piRNAs and siRNAs, because of the presence of terminal 5′ phosphates that are amenable to approaches based on EDC cross-linking (8).
With relatively short exposure times (<60 s), LED outperforms (Figure 4A and C) isotope-labeling protocols that use much longer exposure periods (12–15 h). A small amount (∼3 µg) of total RNA is sufficient for LED to detect miRNAs as demonstrated for miR-21, miR-16 and miR-200c (Figure 4A). Analysis using known quantities of serially diluted synthetic RNAs indicates that a range of 0.01–0.025 fmol of RNA marks a lower detection limit of LED. However, for experiments based on total RNA from cells, the overall sensitivity may vary in a probe sequence-dependent manner due to the competitive inhibition by non-specific RNAs with partial complementarity to the probe. Effects due to such non-specific interactions are unavoidable for hybridization-based techniques such as northern blots. Similarly, although LED manifests single-nucleotide specificity as expected for LNA-based methods (11), specificity could vary depending on probe sequence, though such variations are arguably minor (11).
Several features important to LED emerged during optimization of the protocol. Experiments indicate that while a probe concentration of 0.2 nM is sufficient to detect miRNAs, a concentration of 0.5 nM or higher can significantly improve the sensitivity. However, the added sensitivity due to increasing probe concentration seems to plateau as the concentration is increased to 1 nM or higher. Thus, while sensitivity marginally improves with concentrations over 0.5 nM, the experimental costs increase significantly because LNA probes are relatively expensive, ∼50-fold more than DNA probes. We also note that selecting appropriate amounts of LNA probes has additional advantages since re-synthesis of customized LNA probes for novel RNAs is generally time consuming (∼1 month). Another important observation was that some buffers could in some instances be more useful than others, particularly the modified Church–Gilbert protocol (60°C), which is a much cheaper alternative to the commercially available buffers.
In addition to being a non-radioactive method and a faster method, LED has the advantage of detecting low-abundance small RNAs such as miR-16, or other RNAs that are difficult-to-detect due to factors such as sequence composition and length. It is important to note that the various parameters that we report in the detailed protocol (Doc-S) can be fine-tuned to obtain better sensitivity in detection of difficult-to-detect small RNAs. For example, while the probe concentration of 0.2 nM is sufficient to detect all four miRNAs, it is clear from the optimizations (Supplementary Figure S2) that higher probe concentrations (1–2 nM) are more appropriate for the detection of miR-16. Similarly, the choice of buffer can improve the detection of such low abundance RNAs and the associated background, as indicated by the detection of miR-16 (Figure 1) using the DIG Easy Hyb buffer (Roche).
The absence of bands and high levels of background on the membrane are among the major problems observed with LED. Such difficulties are generally resolvable (Supplementary Table S2). Nevertheless, specific small RNAs can pose significant challenge to obtaining a good signal due to various factors such as low abundance and low purification yields of small RNAs, and poor probe–RNA hybridization. In such difficult cases, we recommend using synthetic oligos of the RNAs of interest to first evaluate whether the designed probe is sufficient to accurately detect the signal. If the probe is deemed insufficient, various probes containing different LNA-spiking patterns should be tested to improve probe sensitivity. Redesigning the probe may also help minimize cross-hybridization of the probe to non-specific RNAs, thus reducing irrelevant bands on the membrane. For a few small RNAs, we have successfully used a stretch of eight LNA bases at 5′-positions 8–15 of small RNA probes, with comparable or better performance than currently marketed probes. We also note that membranes can be re-probed in LED protocol. We have successfully re-probed membranes up to three times, based on washing the membrane in near-boiling 0.1% aqueous SDS solution.
In conclusion, LED is a highly sensitive northern blot method that considerably reduces both time and labor involved in northern blot assays, and provides an environmentally safe detection method for small RNAs. In comparison to traditional 32P-labeled probes that are freshly prepared before each experiment, DIG-labeled LNA probes can be synthesized in advance and used for many experiments because they can be stored (−80°C) for at least 6 months. Moreover, for comparable results, the exposure time needed for LED is ∼1000-fold less than its equivalent radio-isotopic method (Figure 4). Such advantages not only enhance research, but also help reduce overall costs and promote more environmentally friendly laboratory practices.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM079756 to B.J., CA12193 to Y.C./P.S.M., MH60774 to A.P.M.); American Cancer Society (RSG-09-054-01 to B.J.); ACS Research Professorships (RP-09-094-01 to Y.C., RP-09-096-01 to P.S.M.). Funding for open access charge: National Institutes of Health and American Cancer Society.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank members of the laboratories of YC/PSM, M.N. and B.J. for helpful discussions.
REFERENCES
- 1.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Z, Kim SW, Lin Y, Moore PS, Chang Y, John B. Characterization of viral and human RNAs smaller than canonical MicroRNAs. J. Virol. 2009;83:12751–12758. doi: 10.1128/JVI.01325-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varallyay E, Burgyan J, Havelda Z. MicroRNA detection by northern blotting using locked nucleic acid probes. Nat. Protoc. 2008;3:190–196. doi: 10.1038/nprot.2007.528. [DOI] [PubMed] [Google Scholar]
- 4.Holtke HJ, Kessler C. Non-radioactive labeling of RNA transcripts in vitro with the hapten digoxigenin (DIG); hybridization and ELISA-based detection. Nucleic Acids Res. 1990;18:5843–5851. doi: 10.1093/nar/18.19.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramkissoon SH, Mainwaring LA, Sloand EM, Young NS, Kajigaya S. Nonisotopic detection of microRNA using digoxigenin labeled RNA probes. Mol. Cell Probes. 2006;20:1–4. doi: 10.1016/j.mcp.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Varallyay E, Burgyan J, Havelda Z. Detection of microRNAs by northern blot analyses using LNA probes. Methods. 2007;43:140–145. doi: 10.1016/j.ymeth.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 8.Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Church GM, Gilbert W. Genomic sequencing. Proc. Natl Acad. Sci. USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engler-Blum G, Meier M, Frank J, Muller GA. Reduction of background problems in nonradioactive northern and Southern blot analyses enables higher sensitivity than 32P-based hybridizations. Anal. Biochem. 1993;210:235–244. doi: 10.1006/abio.1993.1189. [DOI] [PubMed] [Google Scholar]
- 11.Valoczi A, Hornyik C, Varga N, Burgyan J, Kauppinen S, Havelda Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32:e175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.