Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that mediate gene expression at the post-transcriptional and translational levels by an imperfect binding to target mRNA 3′UTR regions. While the ab-initio computational prediction of miRNA–mRNA interactions still poses significant challenges, it is possible to overcome some of its limitations by carefully integrating into the analysis the paired expression profiles of miRNAs and mRNAs. In this work, we show how the choice of a proper probe annotation for microarray platforms is an essential requirement to achieve good sensitivity in the identification of miRNA–mRNA interactions. We compare the results obtained from the analysis of the same expression profiles using both gene and transcript based custom CDFs that we have developed for a number of different annotations (ENSEMBL, RefSeq, AceView). In all cases, transcript-based annotations clearly improve the effectiveness of data integration and thus provide a more reliable confirmation of computationally predicted miRNA–mRNA interactions.
INTRODUCTION
MicroRNAs (miRNAs) are a family of small non-coding RNAs, derived from hairpin precursors, abundant in animals, plants and viruses (1–8). miRNAs play central roles in cell differentiation, in the development of tissues and organs, in the pathogenesis of human diseases (9,10) and tumors (11–13). At the molecular level miRNAs influence the stability and translational efficiency of target RNA messengers (mRNAs), mainly by an imperfect binding to their 3′UTR regions (14). More than 800 miRNAs have been identified in human and mouse (15); computational predictions provide even higher figures (16). Recent works estimate that, on average, each miRNA can regulate ∼200 target genes (17–19), suggesting that a wide proportion of mammalian genes and biological processes respond to miRNA control mechanisms.
The computational prediction of miRNA targets is extremely challenging due to the lack of a sufficiently large group of experimentally validated targets to be used as a robust training set, and of high-throughput experimental methods for validating results (16). Tools like miRanda, TargetScan, PicTar, PITA and RNAhybrid (19–25), though based on different algorithms and philosophies, all suffer from the limited understanding of the molecular basis involving miRNA-target pairing, that probably, in turn, leads to a reduction of their predictions specificity (26,27). The integration of in-silico predictions with other genomic data may overcome the limits of computational predictors and facilitate the identification of functional interactions. In particular, the combination of target predictions with paired miRNA–mRNA expression profiles has been proposed as an efficient way to refine results obtained from methods based on sequences alone.
Although miRNAs may stabilize transcriptional regulation through complex feed-forward and feed-back loops (28), integrative approaches postulate that miRNAs down-regulate mRNAs and that the expression profiles of genuinely interacting miRNA–mRNA pairs are anti-correlated. The standard integrative approaches comprise three steps: (i) prediction of miRNA targets through sequence-based algorithms, (ii) quantification of target expression levels and (iii) assessment of the anti-correlation among miRNAs and their predicted targets. The anti-correlation can be quantified through a variational Bayesian model (29,30) or by computing a correlation coefficient among miRNA and mRNA expression signals (31–33).
Given that miRNA interactions depend on specific sequences in the 3′UTR regions of their targets and that alternative transcripts of a same gene may differ in such UTRs, integrative analyses of expression profiles must take into account the entire length of a transcript. This has been clearly shown by Legendre and colleagues (34) who studied 3′UTRs containing multiple EST-supported poly(A) sites, and looking for known miRNA targets and other phylogenetically conserved motifs, highlighted that motif-containing and motif-free isoforms were differentially represented in specific tissues. In addition, other studies demonstrated that the same miRNA target prediction algorithm produces significantly different results when applied to genes/transcripts defined by distinct annotations: for example, Rajewsky et al. (25) reported a 20% variability in the predicted regulatory relationships moving from RefSeq transcripts to UCSC ‘known genes’. Target identification, moreover, was affected by alternative adenylation and multiple polyA sites in terminal exons (25,35).
The choice of a transcript-based (TB) approach influences the analysis right from the quantification of target expression. It is well known that a considerable fraction of microarray probes can be (i) entirely mis-assigned (not associated to any gene/transcript in a recent genome annotation), (ii) non-gene-specific (i.e. matching multiple genes) or (iii) non-transcript-specific (matching multiple alternative transcripts of a gene). Several groups have explored the effects of using alternative microarray annotations to quantify gene expression (36,37) and proposed the adoption of custom Chip Definition Files (CDFs) (38–41). The importance of the annotation increases when we consider the integration of miRNA and mRNA expression data because of the role played by alternative 3′UTRs. Unfortunately, the computational procedures developed so far seem to overlook this aspect and adopt gene based CDFs to correlate miRNA-target profiles. The matter is further complicated by the ambiguous definition of a gene 3′UTR region, which may be taken as the union of all 3′UTRs of its transcripts or as the longest one (42).
In this work we investigate how different microarray probe annotations affect the integrative analysis of miRNA-mRNA expression. The analysis has been performed through a computational pipeline that (i) re-annotates microarray probes into GB (gene based) and TB custom CDFs; (ii) predicts miRNA targets starting from transcripts and miRNA sequences; (iii) integrates miRNA target predictions with paired miRNA–mRNA expression signals. In particular, we explore the degree of specificity of miRNA seed pairing to alternative 3′UTR splicing variants and then compare the miRNA–mRNA expression correlation obtained from GB and TB probe annotations. The entire procedure has been tested on paired expression data originally collected to investigate the role of perineural invasion pathway (PNI) in prostate cancer (43). Results clearly show that microarray probe annotations have a substantial impact on the integrative analysis and that TB annotations outperform their GB counterparts.
MATERIALS AND METHODS
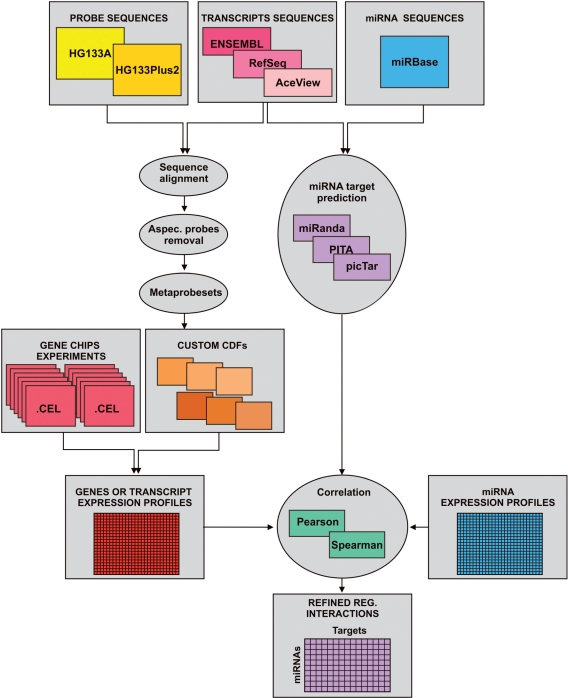
We have developed a computational pipeline (Figure 1) to compare the efficiency of GB and TB annotations in the integrative analysis of miRNA–mRNA data. Such pipeline is composed of three major steps: (i) re-annotation of microarray probes to design GB and TB custom CDFs; (ii) prediction of miRNA targets using the sequences of transcripts and miRNAs; (iii) integration of miRNA target predictions with paired miRNA–mRNA expression signals.
Figure 1.
Computational pipeline. Detailed scheme of the pipeline implemented for the construction of our custom CDFs and for the miRNA–mRNA expression profiles integration.
In the first step we used the sequences of Affymetrix microarray probes and those of transcripts and genes derived from several annotations (ENSEMBL, RefSeq, AceView) to build custom CDFs. We then obtained miRNA sequences from the mirBase database and used the miRanda, PITA and PicTar algorithms to predict their targets (both at the gene and at the transcript level). In the last step we evaluated gene and transcript expression profiles, and we integrated each with the corresponding miRNA expression signals to refine the predicted miRNA–mRNA interactions.
Transcript sequences and annotations
Transcript sequences and annotations were obtained from three databases, i.e. ENSEMBL (version 52), RefSeq (version 33) and AceView (UCSC hg18). Some RefSeq transcripts were associated to multiple UTRs of different extension; to remove redundancy, we defined a single 3′UTR as the region going from the first base after the end of the coding sequence to the first annotated polyA site.
Construction of custom CDFs
GB and TB custom CDFs have been built for a number of human Affymetrix arrays (i.e. HG95v2, HG133A 2.0, HG133plus2, and Human Exon 1.0 ST) using the ENSEMBL, RefSeq and AceView annotations. Specifically, the custom CDFs were generated (i) matching gene/transcript sequences with all the probes of the microarray, (ii) filtering out all non-specific probes, i.e. those matching more than one gene/transcript, (iii) grouping probes into meta-probe sets with at least four entries, and finally, (iv) discarding all those probes not belonging to any meta-probe set. Details on the number of specific probes and of recognized genes and transcripts are reported in Supplementary Table S1.
Prediction of miRNA targets
Human miRNA sequences were obtained from the miRBase::Sequences repository (version 12). We updated target predictions using three different algorithms, characterized by different target identification strategies (miRanda, PITA and PicTar). We ran miRanda and PITA over the ENSEMBL, RefSeq and AceView annotations; PicTar target predictions, based on RefSeq sequences, were downloaded as provided by PicTar developers since the software is not freely available.
The thresholds for miRanda and PITA were defined applying the two algorithms to artificial sequences generated through a permutational approach, i.e. shuffling 3′UTR sequences. miRanda scores were all smaller than 200 when applied to shuffled data, while PITA did not recognize any target at all. As a result, the threshold score of miRanda was set to 200 while targets predicted by PITA were further limited to those with the top 10% scores.
Integrative analysis of mRNA and miRNA expression data
We obtained from the GEO database matched mRNA and miRNA expression data of 57 prostate cancer samples [GSE7055 (43)] generated using Affymetrix HGU133A 2.0 microarrays and OSU-CCC MicroRNA custom arrays, respectively.
The Robust Multichip Average model with quantile normalization and HG133A 2.0 custom-CDFs were used to generate and normalize mRNA expression signals. miRNA expression levels were pre-processed using the approach adopted in the original publication (43). Briefly, spots having signal/background ratio below a specific threshold (calculated as the average of blank spots) were filtered out; each experiment was normalized dividing the expression values by their corresponding median level; replicate signals within the array were averaged. This procedure resulted in a matrix containing the expression levels of 426 miRNAs, 236 of which were human-specific.
To evaluate the impact of GB and TB annotations on the integration of miRNA–mRNA expression data, we used only those probe sets of the HG133A 2.0 GB-CDF that measured genes having at least one transcript represented by a probe set in the HG133A 2.0 TB-CDF. In addition, we filtered out genes having a single transcript (the choice of the type of annotation does not affect the quantification of their expression signal). The filtering procedure resulted in 1715 genes and 1818 transcripts using ENSEMBL, 621 genes and 746 transcripts with RefSeq and 12 184 genes and 4599 transcripts considering the AceView annotation. These genes and transcripts were then used as targets to predict miRNA-target interactions with miRanda, PITA and PicTar (the latter is limited to RefSeq sequences only; see Supplementary Table S2).
We calculated the correlation among all miRNA and mRNA expression profiles using both parametric (Pearson) and non-parametric (Spearman) coefficients. To quantify the impact of different annotations we defined the delta correlations (Δc) as the differences between the correlation levels of a miRNA-gene pair and all of the corresponding miRNA-transcript pairs. The significance of the Δc was assessed comparing the observed Δc with Δc*, the distribution of Δc calculated by randomly permuting 100 times the mRNA expression levels. Specifically, since the maximum absolute value of Δc* resulted equal to 0.2, a |Δc| > 0.2 was considered as an indication of a significant impact of the type of probe annotation (GB or TB) on the correlation of miRNA–mRNA expression data.
Functional enrichment
We calculated the functional enrichment of target genes obtained through TB and GB approaches using the hypergeometric distribution (Fisher exact test) and the GSEA (44,45). The hypergeometric test was performed in DAVID (46) with KEGG (47) and Biocarta pathways (EASE score less than 0.1), while we used the Java application of the Broad Institute (http://www.broadinstitute.org/gsea/) for the GSEA.
RESULTS AND DISCUSSIONS
Analysis of 3′UTR alternative transcripts
The transcript annotation databases, ENSEMBL (v.52), AceView (UCSC hg18) and RefSeq (v.33), provided a total of 54 617, 260 113 and 46 417 transcripts (respectively), resulting in 39 680, 210 003 and 33 518 sequences with annotated 3′UTR regions. The distribution of the number of transcripts per gene according to ENSEMBL, RefSeq and AceView showed that a significant fraction of genes (30% for ENSEMBL, 20% for RefSeq and 29% for AceView) have at least two alternative transcripts (Figure 2A and Table 1). Moreover, as different transcripts may share the same 3′UTR, transcripts with the same 3′UTR have been considered as putative targets of the same set of miRNAs. We define a 3′UTR equivalence class as a set of transcripts of a gene sharing exactly the same 3′UTR sequence. A significant fraction of genes (71% for ENSEMBL, 36% for RefSeq and 94% for AceView) has two or more equivalence classes (Figure 2B and Table 2); such variability in the proportions may be ascribed to differences in terms of annotations of the human genome (48). For instance, predicted alternative transcripts with different 3′UTRs are more numerous and longer in ENSEMBL than in RefSeq. On the contrary AceView, that has been developed to provide a strictly cDNA-supported view of the human transcriptome and to summarize all quality-filtered cDNA data from GenBank, dbEST and RefSeq, is characterized by a larger number of alternative transcripts within the same gene, most of which have different 3′UTR sequences (Figure 2B).
Figure 2.
The 3′UTRs transcript variability. (A) Distribution of the number of transcripts per gene. (B) Distribution of the number of different 3′UTRs per gene (hereafter called 3′UTR equivalence classes) with at least two alternative transcripts. (C) Distribution of the average number of 3′UTR equivalence classes per gene targeted by miRNAs using ENSEMBL, RefSeq and AceView databases with miRanda target prediction algorithm. (D) Distribution of the average number of 3′UTR equivalence classes per gene targeted by miRNAs using ENSEMBL, RefSeq and AceView databases with the PITA target prediction algorithm.
Table 1.
Distribution and cumulative distribution of the number of genes with transcript variants according to ENSEMBL (v 52), RefSeq (v.33) and AceView (UCSC hg18)
| Number of transcript variants within a gene | ENSEMBL |
RefSeq |
AceView |
|||
|---|---|---|---|---|---|---|
| Frequency (%) | Cumulative frequency (%) | Frequency (%) | Cumulative frequency (%) | Frequency (%) | Cumulative frequency (%) | |
| 1 | 69.9 | 69.9 | 80.4 | 80.4 | 70.8 | 70.8 |
| 2 | 13.7 | 83.6 | 13.3 | 93.7 | 5.0 | 75.8 |
| 3 | 7.3 | 90.9 | 3.5 | 97.2 | 3.1 | 78.9 |
| 4 | 3.9 | 94.8 | 1.4 | 98.6 | 2.7 | 81.6 |
| 5 | 2.0 | 96.8 | 0.5 | 99.1 | 2.3 | 83.9 |
| >5 | 3.2 | 100 | 0.9 | 100 | 16.1 | 100 |
Table 2.
Distribution and cumulative distribution of the number of 3′UTR equivalence classes per gene with at least two alternative 3′UTRs according to ENSEMBL (v 52), RefSeq (v.33) and AceView (UCSC hg18)
| Number of 3′UTRs within a gene | ENSEMBL |
RefSeq |
AceView |
|||
|---|---|---|---|---|---|---|
| Frequency (%) | Cumulative frequency (%) | Frequency (%) | Cumulative frequency (%) | Frequency (%) | Cumulative frequency (%) | |
| 1 | 29.0 | 29.0 | 64.0 | 64.0 | 6.0 | 6.0 |
| 2 | 45.7 | 74.7 | 30.8 | 94.8 | 17.7 | 23.7 |
| 3 | 15.7 | 90.4 | 4.2 | 99.0 | 10.8 | 34.5 |
| 4 | 5.7 | 96.1 | 0.9 | 99.9 | 9.5 | 44.0 |
| 5 | 2.5 | 98.6 | 0.02 | 99.92 | 8.2 | 52.2 |
| >5 | 1.4 | 100 | 0.08 | 100 | 47.8 | 100 |
We used the miRanda, PITA and PicTar algorithms to evaluate the specificity of miRNA target predictions with respect to 3′UTR equivalence classes. We computed for each putative miRNA-gene pair the percentage of equivalence classes recognized by the miRNA. Figure 2 C and D and Supplementary Figure S2A shows the distribution of the average percentage of 3′UTR equivalence classes per miRNA over all its putative target genes using miRanda, PITA and PicTar, respectively. These findings indicate that the heterogeneity of alternative 3′UTRs results in miRNAs highly specific in their targeting 3′UTR equivalence classes. Indeed, while using ENSEMBL and RefSeq approximately half of all 3′UTR equivalence classes of a protein-coding gene are recognized by a specific miRNA; with AceView this quantity drops to <20%, indicating a greater miRNA specificity.
Considering that 26% of genes have more than one transcript (taking the average over the three annotation databases), GB data integration could be deceptive for a significant proportion of protein-coding genes. Indeed, 71% of ENSEMBL genes with more than one transcript exhibit more than one 3′UTR equivalence class; a GB data integration would have been ambiguous for at least 18% of them (9 and 23% for RefSeq and AceView, whose proportions of genes having more than one 3′UTR equivalence class are 36 and 94%, respectively). As an example, the BAIAP2 gene (brain-specific angiogenesis inhibitor-1, ENSG00000175866, a secretin receptor family member whose expression is induced by p53) is associated to three different transcripts which differ in their 3′ region (3′CDS and 3′UTR) and encode different isoforms of an insulin receptor tyrosine kinase substrate of the secretin receptor family (49). Figure 3 shows the alternative transcripts of BAIAP2 that are characterized by the same 5′ region and their regulating miRNAs. Among the 95 miRNAs regulating BAIAP2, only 7 (7%) are shared by all transcripts, while 45% of them (9% for ENST00000321300, 36% for ENST00000321280 and none for ENST00000321238) are transcript-specific. This evidence supports the hypothesis that using GB instead of TB annotation for miRNA–mRNA data integration could lead to misleading results.
Figure 3.
Differences between gene and transcript-based annotation approaches. miRNAs regulating the three alternative transcripts of the gene BAIAP2. Triangles represent transcripts and circles miRNAs. The Venn diagram highlights the fact that only 7% of the 95 miRNAs regulating BAIAP2 are shared by all transcripts, while 45% of them (9% for ENST00000321300, 36% for ENST00000321280 and none for ENST00000321238) are transcript-specific.
Construction of custom CDFs
Several groups have explored the effect of using alternative microarray annotations to improve the estimation of expression values. For instance, Dai and colleagues periodically update several custom CDFs for various Affymetrix platforms (38). In their annotation pipeline, however, probes of a given meta-probe set may match different transcripts. As such transcripts may have different expression profiles, the use of non-specific probes in the process of signal quantification could bias the expression value estimates by increasing expression variability. To overcome this limitation, we have developed an alternative annotation scheme and, as suggested by Moll et al. (41), eliminated all non-specific probes. In particular, we reconstructed TB custom CDFs for the most commonly used Affymetrix arrays using ENSEMBL, RefSeq and AceView sequences. Table 3 and Supplementary Table S1 shows the details, in terms of number of genes, probes and transcripts contained in the custom CDF for the HGU133A 2.0 platform based on the three different annotation databases.
Table 3.
Number of unique probes covering genes and transcripts for the construction of our custom CDFs for the Affymetrix platform HG133A 2.0
| Platform ID | ENSEMBL | RefSeq | AceView |
|---|---|---|---|
| Number of genes | 12 136 | 12 011 | 12 184 |
| Number of unique probes covering genes | 186 624 | 185 315 | 193 901 |
| Number of transcripts | 6583 | 8842 | 4599 |
| Number of unique probes covering transcripts | 86 756 | 124 420 | 51 227 |
Integrative analysis of paired miRNA–mRNA expression profiles
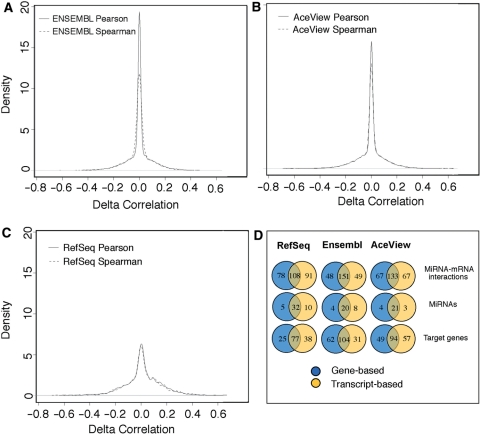
We have used the computational pipeline of Figure 1 for the analysis of paired miRNA–mRNA expression data from 57 prostate cancer samples (43) in order to evaluate the impact of TB annotations on the identification of miRNA targets. The comparative evaluation of GB and TB approaches focused only on those genes having at least two alternative transcripts, i.e. on those cases where the TB annotation should improve data integration. In particular, we evaluated the distributions of differences between correlation estimates (Δc), i.e. the impact of the annotation adopted for expression signal quantification (GB or TB), as a function of the algorithm used for the prediction of miRNA targets (miRanda, PITA or PicTar) and of the type of correlation (parametric or non-parametric coefficients). Figure 4A–C shows the distribution of Δc using the miRanda algorithm for the prediction of miRNA targets and similar results are reported in Supplementary Figures S1A and S2B when using PITA and PicTar, respectively. Δc distributions are centered on zero for all annotation databases and for all type of correlations, but are interestingly characterized by strong kurtosis levels (fat distribution tails), suggesting the presence of feed-forward and -back transcriptional regulation (28). The significance threshold for Δc has been assessed through a permutational approach and set equal to 0.2 (see ‘Materials and methods’ section for details and Supplementary Figure S3 for the distributions of delta correlations of real and randomly permuted data). This threshold allowed us to select those genes/transcripts whose correlation coefficient with at least one miRNA is affected by the choice of the annotation (GB or TB). Specifically, 7% of ENSEMBL and AceView gene/transcripts and 14% of RefSeq ones resulted in delta correlations exceeding the threshold of 0.20 (|Δc| > 0.2 at a FDR < 0.1, Supplementary Figure S4). Among this remaining fraction of interactions, we further considered only those miRNA–mRNA pairs having the top 1% anti-correlation coefficients. As expected, the adoption of GB or TB annotations severely affects the number of miRNA–mRNA interactions, as well as the number of relevant miRNAs and target genes involved in putative interactions (Figure 4D), irrespective of the considered database. Although the aim of Legendre et al. study (34) was different from our goal (they do not perform integrative analysis with microarray data and do not evaluate the impact that chip annotation has on correlation calculation), their findings on few specific miRNAs were concordant with our results. For instance, among the 248 most significant anti-correlated mRNA–miRNA pairs identified using miRanda and ENSEMBL, only 151 (60%) are shared between GB and TB lists. Forty-eight miRNA–mRNA pairs, on the other hand, are specific to the GB and 49 to the TB annotations, respectively. Differences in the top 200 anti-correlated interactions identified through the PITA and miRanda are reported in Supplementary Figure S1 (panel B).
Figure 4.
miRNA–mRNA correlations. Differences in parametric and non-parametric correlation estimates obtained using transcript and gene-based annotations, respectively, using ENSEMBL (A), AceView (B) RefSeq (C) sequences and miRanda algorithm. (D) Number of miRNA–mRNA interactions, miRNAs and target genes involved in the putative 1% most relevant interactions detected using gene- and transcript-based annotations.
Figure 5 shows some examples of miRNA–mRNA interactions together with their GB and TB delta correlations; this highlights the bias introduced by the GB annotation. As an example, all three transcripts of the BAIAP2 are putative targets of miR-328, but the anti-correlation of expression signals is not significant (e.g. −0.06) using the GB approach. Using TB annotations, on the other hand, ENST00000321300 and miR-328 show a significant negative correlation (e.g. −0.3), whereas expression data indicate no correlation between ENST00000321280 and ENST00000321280 and miR-328 (−0.02 and 0.03, respectively).
Figure 5.
Differences between gene and transcript-based annotation. Selected cases of correlations obtained between mRNA and miRNA with a gene-based (black bars) and a transcript-based (red bars) annotation (only for genes with more than one 3′UTR alternative transcript). The symbol of the gene is reported at the top of the panel, the name of the miRNA targeting at least one transcript is reported in the x-axis and the numbers 1, 2 and 3 on the red bars represent the alternative transcripts of the same gene.
Enrichment analyses
An optimal approach should identify, among the supported miRNA–mRNA interactions, a significant proportion of targets and miRNAs with a known role in the pathological processes under examination. As such, we verified the functional enrichment of the most significant anti-correlated mRNA-miRNA pairs among those predicted by miRanda on the ENSEMBL database and confirmed using GB and TB annotations (Figure 4). Specifically, we performed gene set enrichment analysis on GB and TB lists of targets and a literature search on the identified miRNAs. The list of targets obtained through the TB annotation led to highly enriched metabolic pathways using both the hypergeometric and the GSEA approaches (Table 4 and Supplementary Table S3). Both enrichment statistics identified several oncogenes involved in pancreatic and prostate cancer pathways, like CDC42 (associated to human miR-214), EGFR (associated to human miR-134), RELA (associated to miR-205), SMAD4 (associated to miR-200c), CREB3L2 (associated to miR-187), ERBB4 (associated to miR-31) and several other genes involved in the ErbB and GnRH signaling pathways. ERBB2, ERBB4 and EGFR have been implicated in the development of many types of human cancer (50), while the gonadotropin-releasing hormone (GnRH) receptor activation has been demonstrated to inhibit cell proliferation in vitro and in vivo in prostate cancer (51–53) and GnRH agonists have been used as therapeutical treatment in prostate cancer clinical trials since the early 1980s (54). Interestingly, only some of these interesting pathways were found enriched using the list of targets obtained by the GB annotation.
Table 4.
Enriched metabolic pathways of the GB and TB significantly anti-correlated miRNA–mRNA interactions (using ENSEMBL and miRanda)
| TB |
GB |
||
|---|---|---|---|
| Pathway | P-value | Pathway | P-value |
| Pancreatic cancer | 0.016 | Wnt signaling pathway | 0.05 |
| Adherens junction | 0.016 | GnRH signaling pathway | 0.1 |
| Dorso-ventral axis formation | 0.018 | beta-Alanine metabolism | 0.15 |
| ErbB signaling pathway | 0.024 | MAPK signaling pathway | 0.18 |
| Neuroregulin receptor degredation protein-1 Controls ErbB3 receptor recycling | 0.06 | ||
| Regulation of actin cytoskeleton | 0.07 | ||
| Wnt signaling pathway | 0.09 | ||
| Adipocytokine signaling pathway | 0.1 | ||
| Prostate cancer | 0.1 | ||
| GnRH signaling pathway | 0.1 | ||
| Focal adhesion | 0.1 | ||
Among the miRNAs shared by both approaches, 20 out of 32 are highly involved in prostate carcinogenesis [such as miR-221, miR-222 (55,56) and miR-145 (57,58)], in bladder cancer, [miR-23a, miR-23b and miR-205 (59)] and in testis cancer [miR-373 (60)]. Among the miRNAs identified only through the TB annotation, 29% (8 out of 28) are still cancer related, e.g. miR-106a and miR-106b are known to be involved in prostate cancer (58,61), miR-223 is involved in bladder cancer (59), miR-200 in hepatocellular carcinoma (62), miR-15b in chronic lymphocytic leukemia (63,64) and miR-17 in lung cancer and lymphomas (65). Finally, among the miRNAs identified using the GB annotation, only miRNA (miR-184) has been reported to be involved in prostate cancer development (58,66). These results become even more intriguing if considering that correlation and enrichment analyses have been performed on a subset of all possible transcripts, i.e. those belonging to genes with more than one alternative 3′UTRs. This suggests that the use of a GB annotation results in a significant loss of information about post-transcriptional regulation, thus impairing the effectiveness of integrative analyses in the identification of real miRNA targets.
CONCLUSIONS
The ENCODE consortium recently completed the characterization of 1% of the human genome showing a striking picture of its complex molecular activity. While the human genome sequencing revealed a number of protein-coding genes lower than previously estimated (<21 000, according to ENSEMBL), ENCODE identified an extensive transcriptional activity of the genome and highlighted the complexity of the RNA transcriptome (67). At the same time, the miRNA revolution in cell-biology and functional genetics has deeply changed the scenario of gene expression regulation, assigning an increasing importance to post-transcriptional mechanisms in development, physiology and disease. Thus, in the light of these new insights, the definition of gene should be somehow revised (67). Recently Gerstein et al. (67) proposed an alternative definition of gene as the ‘union of genomic sequences encoding a coherent set of potentially overlapping functional products’. This definition pays particular attention to 5′ and 3′ UTRs whose key roles in translation, regulation, stability and localization of mRNAs is widely accepted. Neglecting UTRs from the definition of a gene, one can avoid the problem of multiple 5′ and 3′ ends. Most of the longer protein-coding transcripts identified by ENCODE, differ only in their UTRs (67), thus reinforcing the Gerstein’s new suggested definition of gene. This is particularly important when studying post-transcriptional regulation, where the 3′UTRs is the key region for a miRNA–mRNA seed pairing.
Integrative approaches that aim at improving miRNA target identification through the integration of miRNA and mRNA expression profiles seem to underestimate this problem. GB annotation (which ignores the issue of alternative transcripts) is usually adopted to quantify mRNA expression and to calculate miRNA–mRNA expression correlation. Here, we evaluated the impact of using a GB annotation approach rather than a more appropriate TB one. Using prostate cancer as a case study, we demonstrated how TB array annotation shows more consistent results with the pathological state investigated, even when limiting the analysis to genes with multiple alternative transcripts. We identified a considerable number of miRNA–mRNA interactions whose GB anti-correlations show strong biases due to the presence of alternative 3′UTR transcripts with highly different expression profiles. Furthermore, the TB approach was able to predict new putative miRNA–mRNA interactions involving known oncogenes such as EGFR, RELA and ERBB4 whose regulators, i.e. miR-134, miR-205 and miR-31, respectively, could represent valid candidates for further experimental validations. Unfortunately, the use of a TB annotation lead to loss of information in terms of filtered non-specific probes, thus reducing the possibility of an exhaustive exploration of the transcriptome regulation. In this perspective, alternative technologies such as new generation deep-sequencers, Affymetrix Exon Arrays or custom arrays, such as Combimatrix, Agilent, Nimblegen (68), would provide a wider coverage.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charges: Fondazione Cassa di Risparmio di Padova e Rovigo (Progetti Eccellenza 2006, ‘A computational approach to the study of skeletal muscle genomic expression in health and disease’), University of Padova (CPDR070805 and CPDA07591), MIUR (PRIN 2007Y84HTJ), University of Modena (Finanziamento Linee Strategiche di Sviluppo dell’Ateneo, Medicina Molecolare e Rigenerativa, 2008) and Fondazione Cassa di Risparmio di Modena (Bando ricerca, 2007).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 2.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 5.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 6.Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 7.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 8.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 9.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 10.Zhang C. MicroRNomics: a newly emerging approach for disease biology. Physiol. Genomics. 2008;33:139–147. doi: 10.1152/physiolgenomics.00034.2008. [DOI] [PubMed] [Google Scholar]
- 11.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hobert O. miRNAs play a tune. Cell. 2007;131:22–24. doi: 10.1016/j.cell.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 13.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bentwich I. Prediction and validation of microRNAs and their targets. FEBS Lett. 2005;579:5904–5910. doi: 10.1016/j.febslet.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 17.Gennarino VA, Sardiello M, Avellino R, Meola N, Maselli V, Anand S, Cutillo L, Ballabio A, Banfi S. MicroRNA target prediction by expression analysis of host genes. Genome Res. 2009;19:481–90. doi: 10.1101/gr.084129.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 20.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 22.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhn DE, Martin MM, Feldman DS, Terry A.V., Jr, Nuovo GJ, Elton TS. Experimental validation of miRNA targets. Methods. 2008;44:47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 25.Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38(Suppl.):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 26.Didiano D, Hobert O. Molecular architecture of a miRNA-regulated 3′ UTR. Rna. 2008;14:1297–1317. doi: 10.1261/rna.1082708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 29.Huang JC, Babak T, Corson TW, Chua G, Khan S, Gallie BL, Hughes TR, Blencowe BJ, Frey BJ, Morris QD. Using expression profiling data to identify human microRNA targets. Nat. Methods. 2007;4:1045–1049. doi: 10.1038/nmeth1130. [DOI] [PubMed] [Google Scholar]
- 30.Huang JC, Morris QD, Frey BJ. Bayesian inference of MicroRNA targets from sequence and expression data. J. Comput. Biol. 2007;14:550–563. doi: 10.1089/cmb.2007.R002. [DOI] [PubMed] [Google Scholar]
- 31.Xin F, Li M, Balch C, Thomson M, Fan M, Liu Y, Hammond SM, Kim S, Nephew KP. Computational analysis of microRNA profiles and their target genes suggests significant involvement in breast cancer antiestrogen resistance. Bioinformatics. 2009;25:430–434. doi: 10.1093/bioinformatics/btn646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang YP, Li KB. Correlation of expression profiles between microRNAs and mRNA targets using NCI-60 data. BMC Genomics. 2009;10:218. doi: 10.1186/1471-2164-10-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruike Y, Ichimura A, Tsuchiya S, Shimizu K, Kunimoto R, Okuno Y, Tsujimoto G. Global correlation analysis for micro-RNA and mRNA expression profiles in human cell lines. J. Hum. Genet. 2008;53:515–523. doi: 10.1007/s10038-008-0279-x. [DOI] [PubMed] [Google Scholar]
- 34.Legendre M, Ritchie W, Lopez F, Gautheret D. Differential repression of alternative transcripts: a screen for miRNA targets. PLoS Comput. Biol. 2006;2:e43. doi: 10.1371/journal.pcbi.0020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl. 1):S12 11–14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Draghici S, Khatri P, Eklund AC, Szallasi Z. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 2006;22:101–109. doi: 10.1016/j.tig.2005.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mecham BH, Klus GT, Strovel J, Augustus M, Byrne D, Bozso P, Wetmore DZ, Mariani TJ, Kohane IS, Szallasi Z. Sequence-matched probes produce increased cross-platform consistency and more reproducible biological results in microarray-based gene expression measurements. Nucleic Acids Res. 2004;32:e74. doi: 10.1093/nar/gnh071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferrari F, Bortoluzzi S, Coppe A, Sirota A, Safran M, Shmoish M, Ferrari S, Lancet D, Danieli GA, Bicciato S. Novel definition files for human GeneChips based on GeneAnnot. BMC Bioinformatics. 2007;8:446. doi: 10.1186/1471-2105-8-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu J, Lee JC, Salit ML, Cam MC. Transcript-based redefinition of grouped oligonucleotide probe sets using AceView: high-resolution annotation for microarrays. BMC Bioinformatics. 2007;8:108. doi: 10.1186/1471-2105-8-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moll AG, Lindenmeyer MT, Kretzler M, Nelson PJ, Zimmer R, Cohen CD. Transcript-specific expression profiles derived from sequence-based analysis of standard microarrays. PLoS ONE. 2009;4:e4702. doi: 10.1371/journal.pone.0004702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 43.Prueitt RL, Yi M, Hudson RS, Wallace TA, Howe TM, Yfantis HG, Lee DH, Stephens RM, Liu CG, Calin GA, et al. Expression of microRNAs and protein-coding genes associated with perineural invasion in prostate cancer. Prostate. 2008;68:1152–1164. doi: 10.1002/pros.20786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 46.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 47.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Larsson TP, Murray CG, Hill T, Fredriksson R, Schioth HB. Comparison of the current RefSeq, Ensembl and EST databases for counting genes and gene discovery. FEBS Lett. 2005;579:690–698. doi: 10.1016/j.febslet.2004.12.046. [DOI] [PubMed] [Google Scholar]
- 49.Okamura-Oho Y, Miyashita T, Yamada M. Distinctive tissue distribution and phosphorylation of IRSp53 isoforms. Biochem. Biophys. Res. Commun. 2001;289:957–960. doi: 10.1006/bbrc.2001.6102. [DOI] [PubMed] [Google Scholar]
- 50.Holbro T, Hynes NE. ErbB receptors: directing key signaling networks throughout life. Annu. Rev. Pharmacol. Toxicol. 2004;44:195–217. doi: 10.1146/annurev.pharmtox.44.101802.121440. [DOI] [PubMed] [Google Scholar]
- 51.Bahk JY, Hyun JS, Lee H, Kim MO, Cho GJ, Lee BH, Choi WS. Expression of gonadotropin-releasing hormone (GnRH) and GnRH receptor mRNA in prostate cancer cells and effect of GnRH on the proliferation of prostate cancer cells. Urol. Res. 1998;26:259–264. doi: 10.1007/s002400050054. [DOI] [PubMed] [Google Scholar]
- 52.Dondi D, Limonta P, Moretti RM, Marelli MM, Garattini E, Motta M. Antiproliferative effects of luteinizing hormone-releasing hormone (LHRH) agonists on human androgen-independent prostate cancer cell line DU 145: evidence for an autocrine-inhibitory LHRH loop. Cancer Res. 1994;54:4091–4095. [PubMed] [Google Scholar]
- 53.Halmos G, Arencibia JM, Schally AV, Davis R, Bostwick DG. High incidence of receptors for luteinizing hormone-releasing hormone (LHRH) and LHRH receptor gene expression in human prostate cancers. J. Urol. 2000;163:623–629. [PubMed] [Google Scholar]
- 54.Tolis G, Ackman D, Stellos A, Mehta A, Labrie F, Fazekas AT, Comaru-Schally AM, Schally AV. Tumor growth inhibition in patients with prostatic carcinoma treated with luteinizing hormone-releasing hormone agonists. Proc. Natl Acad. Sci. USA. 1982;79:1658–1662. doi: 10.1073/pnas.79.5.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi XB, Tepper CG, White RW. MicroRNAs and prostate cancer. J. Cell Mol. Med. 2008;12:1456–1465. doi: 10.1111/j.1582-4934.2008.00420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafre SA, Farace MG. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J. Biol. Chem. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 57.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 58.Schaefer A, Jung M, Kristiansen G, Lein M, Schrader M, Miller K, Stephan C, Jung K. MicroRNAs and cancer: Current state and future perspectives in urologic oncology. Urol. Oncol. 2008 doi: 10.1016/j.urolonc.2008.10.021. Dec 29. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 59.Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, et al. Micro-RNA profiling in kidney and bladder cancers. Urol. Oncol. 2007;25:387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 60.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Adv. Exp. Med. Biol. 2007;604:17–46. doi: 10.1007/978-0-387-69116-9_2. [DOI] [PubMed] [Google Scholar]
- 61.Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 63.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 64.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 66.Lin SL, Chiang A, Chang D, Ying SY. Loss of mir-146a function in hormone-refractory prostate cancer. Rna. 2008;14:417–424. doi: 10.1261/rna.874808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, Korbel JO, Emanuelsson O, Zhang ZD, Weissman S, Snyder M. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007;17:669–681. doi: 10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
- 68.Ghindilis AL, Smith MW, Schwarzkopf KR, Roth KM, Peyvan K, Munro SB, Lodes MJ, Stover AG, Bernards K, Dill K, et al. CombiMatrix oligonucleotide arrays: genotyping and gene expression assays employing electrochemical detection. Biosens. Bioelectron. 2007;22:1853–1860. doi: 10.1016/j.bios.2006.06.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.