Abstract
All organic fluorophores undergo irreversible photobleaching during prolonged illumination. While fluorescent proteins typically bleach at a substantially slower rate than many small molecule dyes, lack of photostability remains an important limiting factor for experiments requiring large numbers of images of single cells. Screening methods focusing solely on brightness or wavelength are highly effective in optimizing both properties, but the absence of selective pressure for photostability in such screens leads to unpredictable photobleaching behavior in the resulting fluorescent proteins. Here, we describe an assay for screening libraries of fluorescent proteins for enhanced photostability. With this assay, we developed highly photostable variants of mOrange (a wavelength-shifted monomeric derivative of DsRed from Discosoma sp.) and TagRFP (a monomeric derivative of eqFP578 from Entacmaea quadricolor) that maintain most of the beneficial qualities of the original proteins and perform as reliably as Aequorea victoria GFP derivatives in fusion constructs.
Introduction
Substantial progress has recently been made in developing monomeric or dimeric fluorescent proteins covering the visual spectrum1-13, but while brightness and wavelength have been primary concerns, photostability has generally been an afterthought (with the notable exception of mTFP1 (ref. 12)). Consequently, many novel fluorescent protein variants have relatively poor photostability. The first-generation monomeric red fluorescent protein, mRFP1 (ref. 1), while reasonably bright, was less photostable than its ancestor, Discosoma sp. DsRed14. In subsequent generations of mRFP1 variants (the “mFruits”), we observed serendipitous enhancement in photostability in some variants2, leading us to believe that it would be possible to apply directed evolution strategies to this property as well.
To extend the utility of fluorescent proteins, having optimized them for many other properties, we have developed a new screening method that additionally assays photostability in a medium-throughput format. This selection scheme allows us to select simultaneously for the most photostable mutants that also maintain an acceptable level of fluorescence emission at the desired wavelength, minimizing the tradeoff of desirable properties that frequently results from single-parameter screens. We applied our photostability screening assay to the directed evolution of variants derived from the bright red monomeric red fluorescent protein TagRFP and the fast-bleaching monomeric orange fluorescent protein mOrange. The resulting variants, TagRFP-T and mOrange2, are 9-fold and 25-fold more photostable than their respective ancestors, and both were found to make excellent fusion partners when expressed in mammalian cells.
Results
Photostability assay and rationale
To photobleach large numbers of bacterial colonies, we utilized a solar simulator, which produces a collimated beam approximately 10cm in diameter with light intensities of 95 or 141 mW/cm2 with 525-555 or 548-588 nm bandpass filters respectively (see Methods for details). This intensity, while approximately 100-fold lower than that produced by unattenuated arc lamp illumination and 105-fold lower than instantaneous intensities during confocal laser illumination, is sufficient to photobleach the photolabile fluorescent protein mOrange to 50% initial intensity after approximately 10 minutes. This reasonably short time allowed us to quickly screen bacterial libraries of up to 100,000 clones on plates. Heating of plates was minimized by placing them on a custom-built water-cooled aluminum block. At wavelengths necessary to photobleach orange and red fluorescent proteins, we found no substantial decrease in bacterial viability after 2 hours of illumination.
Evolution of a brighter photostable red monomer
To create a better red monomer, we initially undertook a rational design approach, drawing on analysis of mCherry's enhanced photostability and mOrange's higher quantum yield relative to mRFP1. Six generations of directed evolution with constant photostability selection led to the novel variant “mApple,” which, though substantially brighter than mCherry, displayed complex photoswitching behavior (see Fig. 1, Tables 1 and 2, and Supplementary Fig. 1 and Supplementary Note 1 online). This behavior was more pronounced with continuous wide-field than with laser-scanning illumination and could be largely eliminated by excitation at alternate wavelengths or by intermittent illumination. However, given our later results using the brighter TagRFP as starting material, we chose not to pursue mApple any further.
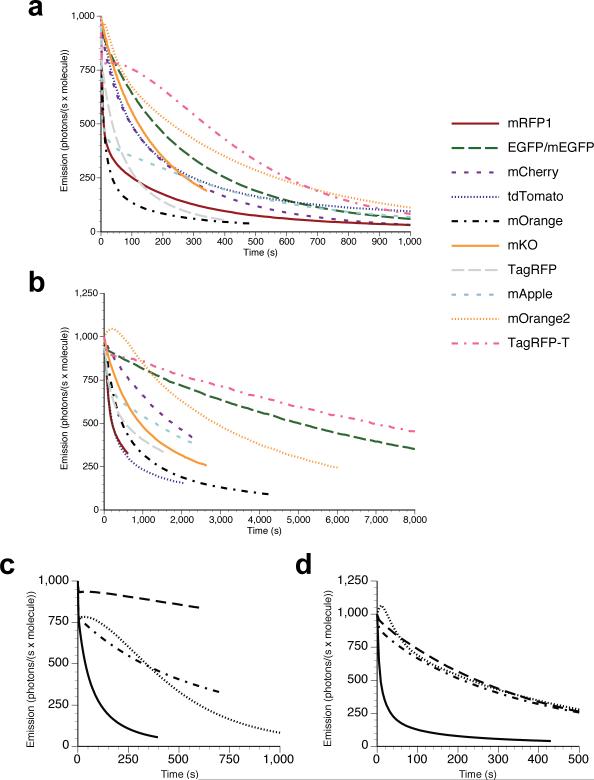
Figure 1. Comparison of photobleaching curves.
(a) Arc lamp photobleaching curves for mRFP1, EGFP, mCherry, tdTomato, mOrange, mKO, TagRFP, mApple, mOrange2, and TagRFP-T, as measured in purified protein (see Methods) and plotted as intensity versus normalized total exposure time with an initial emission rate of 1000 photons/s per molecule; (b) normalized laser scanning confocal microscopy bleaching curves for the same proteins (except for EGFP which in this case is the monomeric A206K variant) fused to histone 2B and imaged in live cells. The time axis represents normalized total imaging time for an initial scan-averaged emission rate of 1000 photons/s per molecule; (c) arc lamp photobleaching curves for normoxic TagRFP (solid line) and TagRFP-T (dotted line) and O2-free TagRFP (dot-dashed line) and TagRFP-T (dashed line). All photobleaching curves were measured under continuous illumination without neutral density filters and are plotted as intensity versus normalized total exposure time with an initial emission rate of 1000 photons/s per molecule; (d) arc lamp photobleaching curves for normoxic mOrange (solid lines) and mOrange2 (dotted lines) and (O2-free) mOrange (dot-dashed line) and mOrange2 (dashed line).
Table 1.
Physical and optical properties of novel photostable fluorescent protein variants.
| Fluorescent protein | Excitation maximum (nm) | Emission maximum (nm) | Extinction coefficient (M−1 × cm−1) | Fluorescence quantum yield | Brightnessa | pKa | t1/2 for maturation at 37°C | t1/2 bleach (arc lamp)b (s) | t1/2 bleach (O2-free)c (s) | t1/2 bleach (confocal)d (s) |
|---|---|---|---|---|---|---|---|---|---|---|
| mRFP1 | 584 | 607 | 50,000 | 0.25 | 13 | 4.5 | < 1 h | 8.7 | NDe | 210 |
| mCherry | 587 | 610 | 72,000 | 0.22 | 16 | < 4.5 | 15 min | 96 | ND | 1800 |
| mOrange | 548 | 562 | 71,000 | 0.69 | 49 | 6.5 | 2.5 h | 9.0 | 250 | 460 |
| DsRed | 558 | 583 | 75,000 | 0.79 | 59 | 4.7 | 10 h | 326 | ND | ND |
| tdTomato | 554 | 581 | 138,000 | 0.69 | 95 | 4.7 | 60 min | 98 | ND | 210 |
| mKO | 548 | 559 | 51,600 | 0.60 | 31 | 5.0 | 4.5 h | 122 | ND | 930 |
| TagRFPf | 555 | 584 | 98,000 | 0.41 | 40 | 3.1 | 100 min | 37 | 323 | 550 |
| EGFP/mEGFP | 488 | 507 | 56,000 | 0.60 | 34 | 6.0 | ND | 174 | ND | 5000 |
| mOrange2 | 549 | 565 | 58,000 | 0.60 | 35 | 6.5 | 4.5 h | 228 | 228 | 2900 |
| mApple | 568 | 592 | 75,000 | 0.49 | 37 | 6.5 | 30 min | 4.8 | ND | 1300 |
| TagRFP-T | 555 | 584 | 81,000 | 0.41 | 33 | 4.6 | 100 min | 337 | >>600 | 6900 |
Brightness of fully mature protein, (EC × QY) / 1000
Time (s) to bleach to 50% emission intensity under arc-lamp illumination, at an illumination level that causes each molecule to emit 1000 photons/s initially, as measured in our lab. See ref. 16 for more details.
With arc lamp illumination, equilibrated under O2-free conditions. See Methods.
Time (s) to bleach to 50% emission intensity measured during laser scanning confocal microscopy, at an average illumination level over the scanned area that causes each molecule to emit an average 1000 photons/s initially, as measured in our lab. A 543nm laser line was used for all proteins except mEGFP, which was bleached with a 488nm laser. See Supplementary Methods for detailed description of normalization.
ND, not determined.
All measurements were performed in our lab.
Table 2.
Mutations of new photostable fluorescent protein variants.
| protein | Mutations |
|---|---|
| mApple | mOrange-R17H/G40A/T66M/A71V/V73I/K92R/V104I/V105I/T106H/T108N/E117V/S147E/G159S/M163K/T174A/S175A/G196D/T202V |
| mOrange2 | mOrange-Q64H/F99Y/E160K/G196D |
| TagRFP-T | TagRFP-S158T |
While the recently developed orange-red monomer TagRFP13 exhibits remarkable brightness, we have found that its photostability is still far from optimal. In both our standard arc lamp photobleaching and laser scanning confocal assays, we determined that TagRFP bleaches approximately 3-fold faster than mCherry (see Fig. 1a, and Table 1). Thus, we chose this protein as another starting point for improvement of photostability. We first attempted rational design of a mutant library guided by the crystal structure of the closely-related protein eqFP611 (ref. 13). With the rationale that chromophore-interacting residues could influence photostability, we performed saturation mutagenesis of Ser158 and Leu199, two residues proximal to the TagRFP chromophore. We then screened this library in bacteria with our solar simulator-based assay, using the 540/30 nm bandpass filter and exposure times of 120 minutes per plate, imaging the plates before and after bleaching to select those colonies that displayed high absolute brightness and a high ratio of post-bleach to pre-bleach fluorescence emission.
From this directed library, we identified one clone, TagRFP S158T (designated “TagRFP-T”), which had a photobleaching half-time of 337 seconds by our standard assay, making it approximately 9-fold more photostable than TagRFP (see Fig. 1a-c and Table 1). TagRFP-T, which was further modified by appending GFP-like N and C termini, possesses identical excitation and emission wavelength, quantum yield, and maturation time to TagRFP, with only a slightly lower extinction coefficient (81,000 versus 98,000 M−1 × cm−1) and a higher fluorescence pKa (4.6 versus 3.1). We expect that the benefit of increased photostability should offset the small decrease in brightness and increase in acid sensitivity in most applications. Additionally, TagRFP-T matures to apparent completion and has virtually no emission in the green region of the spectrum (Supplementary Fig. 1 online), making it suitable for co-imaging with green fluorescent proteins. We verified that TagRFP-T remains monomeric by gel filtration (data not shown). Because the S158T mutation is internal, we anticipated that TagRFP-T would perform nearly identically as TagRFP when used as a fusion tag. Indeed, live cell imaging confirmed that TagRFP-T does not interfere with localization of any fusions tested (see Fig. 2).
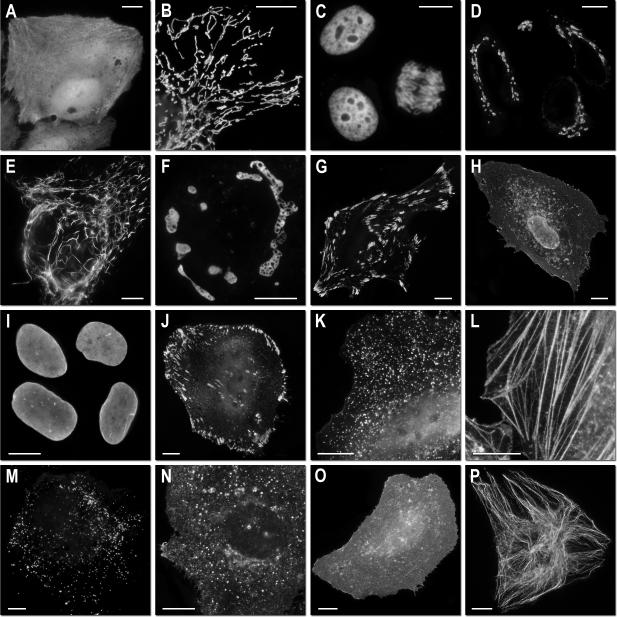
Figure 2. Fluorescence imaging of TagRFP-T subcellular targeting fusions.
N-terminal fusion constructs (linker amino acid length indicated after fusion protein name): (a) TagRFP-T-N1 (N-fusion cloning vector; expression in nucleus and cytoplasm with no specific localization); (b) TagRFP-T-mitochondria-7 (human cytochrome C oxidase subunit VIII); (c) TagRFP-T-H2B-6 (N-terminus; human, showing two interphase nuclei and one nucleus in early anaphase); (d) TagRFP-T-Golgi-7 (N-terminal 81 amino acids of human β-1,4-galactosyltransferase); (e) TagRFP-T-vimentin-7 (human); (f) TagRFP-T-Cx43-7 (rat α-1 connexin-43); (g) TagRFP-T-zyxin-7 (human); C-terminal fusion constructs: (h) TagRFP-T-annexin (A4)-12 (human; illustrated with ionomycin-induced translocation to the plasma and nuclear membranes); (i) TagRFP-T-lamin B1-10 (human); (j) TagRFP-T-vinculin-23 (human); (k) TagRFP-T-clathrin light chain-15 (human); (l) TagRFP-T-β-actin-7 (human); (m) TagRFP-T-peroxisomes-2 (peroximal targeting signal 1; PTS1); (n) TagRFP-T-endosomes-15 (human RhoB GTPase with an N-terminal c-Myc epitope tag); (o) TagRFP-T-farnesyl-5 (20-amino acid farnesylation signal from c-Ha-Ras); (p) TagRFP-T-β-tubulin-6 (human). All TagRFP-T fusion vectors were expressed in HeLa (ATCC; CCL-2) cells. Scale bars are 10 μm.
Photobleaching of TagRFP and TagRFP-T under oxygen-free conditions revealed that TagRFP-T's photobleaching remains oxygen-sensitive (see Fig. 1c and Table 1). However, the oxygen-free bleaching half-time for TagRFP is similar to the ambient oxygen bleaching half-time for TagRFP-T. We next compared TagRFP and TagRFP-T as fusions to H2B expressed in living cells under confocal illumination (see Fig. 1b and Table 1). TagRFP-T had a photobleaching half-time approximately 9-fold greater than that of TagRFP, consistent with the results on purified proteins under continuous widefield illumination.
Evolution of a photostable orange monomer
We next attempted to engineer a photostable variant of mOrange, which is the brightest of the previously engineered mRFP1 variants, but exhibits relatively fast bleaching. Because substitutions at position 163 improved photostability during the evolution of mCherry and mApple, we initially tested the M163Q mutant of mOrange, but found that improved photostability was accompanied by undesirable decreases in quantum yield and maturation efficiency. The M163K mutant of mOrange exhibited enhanced photostability and matured very efficiently, but suffered from increased acid sensitivity (pKa ~7.5). Because another orange fluorescent protein, mKO (derived from Fungia concinna)6, is both highly photostable16 and possesses a methionine at the position equivalent to 163, we reasoned that other pathways must exist for increasing photostability.
To explore alternative photostability-enhancement evolution pathways, we used iterative random and directed mutagenesis with selection using the solar simulator. Initially, a randomly mutagenized library of mOrange was screened by photobleaching with 525-555 nm for 15 to 20 minutes per plate (a time sufficient to bleach mOrange to ~25% of its initial brightness) and selecting the brightest post-bleach clones by eye. This screen identified a single clone, mOrange F99Y, which had approximately two-fold improved photostability (data not shown). Saturation mutagenesis of residue 99 and residues 97 and 163, which we imagined could have synergistic interactions with residue 99, did not yield further improvements.
We then constructed a randomly mutagenized library of mOrange F99Y, and screened with a longer illumination time of 40 minutes per plate. This round of screening identified the additional mutation Q64H, which conferred a remarkable ~10-fold increase in photostability over the F99Y single mutant. Again, saturation mutagenesis of residues 64 and 99 along with surrounding residues 97 and 163 failed to produce clones that were improved over the original clone identified in the random screen. Additionally, we found that the Q64H mutation alone did not confer substantially enhanced photostability, but rather required the presence of the F99Y mutation (data not shown). Two further rounds of directed evolution with continued selection for photostability (540/30 nm filter, 40 minutes per plate) improved the folding efficiency with mutations E160K and G196D, giving the final clone, “mOrange2” (see Table 2).
The highly desirable increase in photostability achieved in mOrange2 is balanced by a modest decrease in quantum yield (0.60 versus 0.69) and extinction coefficient (58,000 versus 72,000 M−1 × cm−1), together corresponding to a 30% decrease in brightness compared to mOrange. It also exhibits slightly shifted excitation and emission peaks (549nm and 565nm) and an increased maturation half-time (4.5 hours versus 2.5 hours). However, its photostability under arc lamp illumination is over 25-fold greater than that of mOrange (Fig. 1d), making it nearly twice as photostable as mKO6, the previously most photostable known orange monomer16, approximately 6-fold more photostable than TagRFP13, and about 1.3-fold more photostable than EGFP16 (see Supplementary Fig. 1, Table 1, and Table 2). During laser scanning confocal imaging, mOrange2 is approximately 6-fold more photostable than mOrange and 3-fold more photostable than mKO (see Fig. 1b). Curiously, the brightness and maturation time of mOrange2 are quite similar to those for mKO. mOrange2 remains acid-sensitive with a pKa of 6.5, making it undesirable for targeting to acidic compartments, but attractive as a possible marker for exocytosis or other pH-variable processes17. As with TagRFP-T, we verified that mOrange2 remained monomeric using gel filtration (data not shown). We subsequently investigated the role of the key photostability-enhancing mutations present in mOrange2, tested it in a wide range of fusion constructs, and compared its performance with mKO and tdTomato (see Fig. 3 and Supplementary Note 2 online).
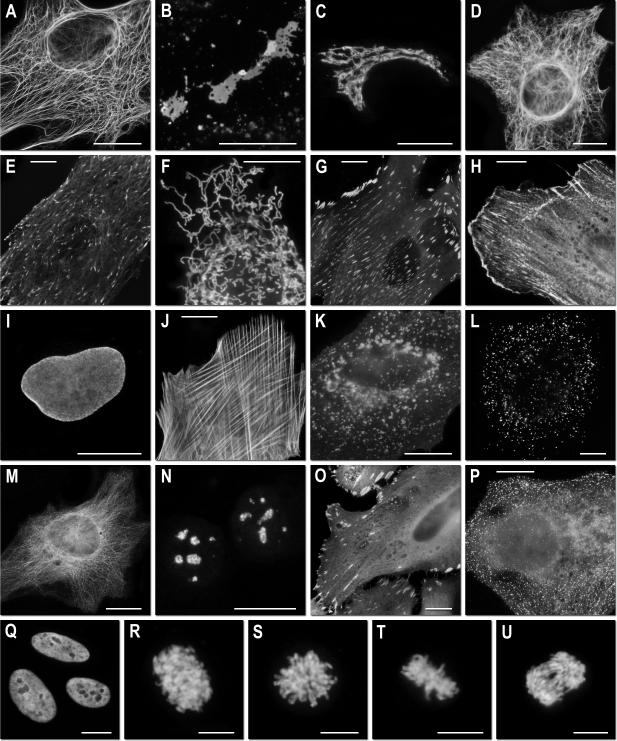
Figure 3. Fluorescence imaging of mOrange2 subcellular targeting fusions.
Widefield fluorescence images of mOrange2 chimeras in N- and C-terminal fusions. N-terminal fusion constructs (linker amino acid length indicated after fusion protein name): (a) mOrange2-Keratin-17 (human cytokeratin 18); (b) mOrange2-Cx26-7 (rat β-2 connexin-26); (c) mOrange2-Golgi-7 (N-terminal 81 amino acids of human β-1,4-galactosyltransferase); (d) mOrange2-vimentin-7 (human); (e) mOrange2-EB3-7 (human microtubule-associated protein; RP/EB family); (f) mOrange2-mitochondria-7 (human cytochrome C oxidase subunit VIII); (g) mOrange2-paxillin-22 (chicken); (h) mOrange2-α-actinin-19 (human non-muscle); C-terminal fusion constructs: (i) mOrange2-Lamin B1-10 (human); (j) mOrange2-β-Actin-7 (human); (k) mOrange2-lysosomes-20 (rat lysosomal membrane glycoprotein 1); (l) mOrange2-peroxisomes-2 (peroximal targeting signal 1); (m) mOrange2-β-tubulin-6 (human); (n) mOrange2-Fibrillarin-7 (human); (o) mOrange2-vinculin-23 (human); (p) mOrange2-Clathrin Light Chain-15 (human). (q–u) Laser scanning confocal images of HeLa cells expressing mOrange2-H2B-6 (N-terminal fusion; human) progressing through (q) interphase; (r) prophase; (s) prometaphase; (t) metaphase; (u) early anaphase. The cell line used for expressing mOrange2 fusion vectors was Gray fox lung fibroblast cells (FoLu) in panels (e) and (j), and human cervical adenocarcinoma cells (HeLa) in the remaining panels. Scale bars are 10 μm.
Evaluation of reversible photoswitching
Because of concerns that our screening method might select for photoswitching behavior, we tested our novel variants as well as other commonly used fluorescent proteins using both widefield and confocal imaging. Nearly all had some degree of reversible photoswitching, which was observed as an recovery of up to 100% of pre-bleach fluorescence intensity when the fluorescent protein was bleached to ~50% of its initial intensity and then observed again after 1 to 2 minutes without illumination. In fact, several commonly used Aequorea GFP variants including EGFP, Cerulean, and Venus, displayed reversible photoswitching18 more severe than observed for our novel variants. A summary table of the results of these experiments along with representative traces for TagRFP, TagRFP-T, EGFP, and Cerulean are shown in Supplementary Note 3 online. These results suggest that our screen is not selecting specifically for photoswitching, which is no worse in the new proteins (except for mApple) than in well-established fluorescent proteins.
While our observation of reversible photoswitching in such a broad range of fluorescent proteins certainly raises concerns about the potential for previously undetected experimental artifacts, it is beyond the scope of this study to determine how common or severe this phenomenon may be. Of particular concern is the implication that fluorescence recovery after photobleaching (FRAP) experiments may be prone to artifacts that would confound data interpretation. We performed a limited evaluation of this possibility using H2B fusions to EGFP and EYFP expressed in mammalian cells and imaged on a laser-scanning confocal microscope. When these proteins were bleached to near completion with full laser power and then observed for recovery, we observed a negligible amount of reversible photoswitching (data not shown). However, further in-depth investigation is warranted to rule out such an effect in other fluorescent proteins and under more varied experimental conditions.
Discussion
We have demonstrated a novel scheme for applying photostability as a selection criterion during directed evolution of fluorescent proteins. Using a high-intensity light source, we are able to photobleach entire 10 cm plates of bacteria expressing the fluorescent proteins of interest and select those that maintain the most brightness. This approach allowed us to screen libraries containing up to 100,000 clones reliably with no observed false-positive hits. While the precise kinetics of photobleaching for a given fluorescent protein are strongly dependent on illumination intensity and temporal regimen, we have found that improvements in photostability at ~0.1 W/cm2 usually qualitatively predict improved performance under typical conditions for wide-field and laser scanning microscopy. The exceptions are mApple's reversible photoswitching (Supplementary Note 1 online) and tdTomato's poor performance under laser scanning confocal illumination (Fig. 1b). Also, while our screen utilized bacteria to express fluorescent protein libraries, all proteins produced from these studies behaved similarly when later tested in purified form or expressed in mammalian cells, consistent with our previous experience.
Photobleaching using an array of LEDs was performed during the evolution of mTFP1 to select against unacceptable photolability or photoswitching, resulting in a protein with a bleaching half-time 110 seconds12. Our work applies photostability as a primary criterion to improve multiple fluorescent proteins, and successfully demonstrates that high photostability is a selectable phenotype. Moreover, a solar simulator benefits from the strong mercury lines at 546, 577, and 579 nm and allows greater flexibility in the choice of excitation wavelength than would be possible with LEDs.
While it is difficult to draw strong conclusions from the photostability mutations in mOrange2, specific regions proximal to the chromophore appear to influence the modes of photobleaching it is able to undergo. DsRed, when illuminated by a 532nm pulsed laser, undergoes decarboxylation of Glu215, as well as cis-to-trans isomerization of the chromophore19. Such chromophore isomerization has been implicated in the photoswitching behavior of Kindling fluorescent protein (KFP)20, 21 and Dronpa5, 22, as well as predecessors to mTFP1 (refs. 12 and 23). Decarboxylation of the corresponding glutamate (position 222) in Aequorea GFP also leads to changes in optical properties24-26. However, our observation that oxidation plays a large role in mOrange, TagRFP, and TagRFP-T photobleaching suggests that chromophore isomerization and Glu215 decarboxylation may play only a minor role for such proteins under normoxic conditions. Additionally, we found no evidence by mass spectrometry that photobleaching using the solar simulator led to any detectable decarboxylation of Glu215 in mOrange (data not shown). Under some conditions mOrange2 shows an initial photoactivation of about 5% (Fig. 1a, d) before bleaching takes over. At present we have no molecular explanation for this minor effect or the reversible photoswitching shared by most fluorescent proteins (see Results and Supplementary Note 3 online).
For mRFP1 variants, we have clearly observed the importance of residue 163 in influencing photostability (see Supplementary Note 1 online), but have also seen somewhat context-specific effects of 163 and surrounding residues on different wavelength-shifted variants. This region, composed of residues 64, 97, 99, and 163, appears to be important in determining photostability. However, of these, only residue 163 is in direct contact with the chromophore. It may be that the mutations Q64H and F99Y together lead to a rearrangement of the other side chains in the vicinity of the chromophore so as to hinder a critical oxidation that leads to loss of fluorescence.
Discrepancies in tubulin and connexin localization (see Supplementary Note 2 online) when fused to mOrange2 versus mKO or tdTomato can probably be attributed to the three-dimensional structure of the fluorescent protein and potential steric hindrance in the fusions. mOrange2 contains extended N- and C- termini derived from EGFP to improve performance in fusions, whereas the much shorter protein, mKO (236 vs. 218 amino acids, respectively), may experience steric interferences that lead to poorer performance in similar fusions. The fused dimeric character of tdTomato effectively doubles its size compared to the monomeric orange fluorescent proteins, so steric hindrance is the most likely culprit in preventing tubulin localization. For most fusions, however, we observed little or no difference in performance between mOrange2 and mKO, suggesting that many proteins are more tolerant of fusion partners than tubulin or connexins.
We have shown that our photostability selection method may be applied to TagRFP, which, though it already possesses reasonably good photostability, was still amenable to improvements. From a saturation-mutagenesis library of two chromophore-proximal residues (consisting of 400 independent clones), we selected a single clone with substantially enhanced photostability. The selected mutant, TagRFP-T, should prove to be a very useful addition to the fluorescent protein arsenal, as it is the most photostable monomeric fluorescent protein of any color yet described under both arc lamp and confocal laser illumination.
As the applications of genetically encoded fluorescent markers continue to diversify and become more complex, the demand for greater photostability than is available in current fluorescent proteins has likewise continued to grow. We have expanded existing directed evolution approaches by utilizing medium-throughput photostability selection. We expect this novel screening method to be applicable to any of the large number of existing fluorescent proteins, and, with modifications, to be useful in selecting for more efficient photoconvertible and photoswitchable fluorescent proteins as well3, 5, 10, 20, 27-31. Possible enhancements to this selection technique could include time-lapse imaging of bacterial plates during bleaching to enable direct selection for kinetics (independent of absolute brightness) and the use of higher intensity illumination from other light sources (such as lasers) during screening to select for or against non-linear photobleaching behavior. Ideally, a selection scheme that allows true simulation of microscopic imaging light intensities while maintaining a medium- to high-throughput should allow selection of fluorescent proteins with the most beneficial properties for imaging applications.
Supplementary Material
Supplementary Information
Supplementary Figure 1 Excitation, emission, and absorbance spectra of novel fluorescent protein variants.
Supplementary Note 1 Directed evolution and characterization of mApple.
Supplementary Note 2 Directed evolution and characterization of mOrange2.
Supplementary Note 3 Summary of reversible photoswitching data with representative examples.
Supplementary Methods
Acknowledgements
L. A. Gross performed mass spectroscopy. S. R. Adams performed gel filtration experiments. We thank R. E. Campbell and C. T. Dooley for helpful discussion. Sequencing services were provided by the University of California, San Diego Cancer Center shared sequencing resource and the Florida State University Bioanalytical and Molecular Cloning DNA Sequencing Laboratory. N.C.S. was a Howard Hughes Medical Institute Predoctoral fellow during this work. This work was additionally supported by NIH (NS27177 and GM72033) and the Howard Hughes Medical Institute.
Methods
Mutagenesis
The cDNA for mOrange2 and TagRFP (Evrogen)13, both of which had been previously human codon-optimized, were used as the initial templates for library construction by random mutagenesis. Error-prone PCR was performed using the GeneMorph II kit (Stratagene) following the manufacturer's protocol, using primers containing BamHI and EcoRI sites for mOrange variants or BamHI and BsrGI sites for TagRFP variants. Error-prone PCR products were digested with BamHI and EcoRI or BsrGI and ligated into a modified pBAD vector (Invitrogen) or a constitutive bacterial expression vector pNCS, both of which encode an N-terminal 6xHis tag and linker identical to that found in pRSET B (Invitrogen). Site-directed mutagenesis was performed using the QuikChange II kit (Stratagene) following the manufacturer's protocol or by overlap-extension PCR. Sequences for all primers used in this study are given in Supplementary Methods online. Chemically competent or electrocompetent Escherichia coli strain LMG194 (Invitrogen) were transformed with libraries and grown overnight on LB/agar supplemented with 50 μg/mL ampicillin (Sigma) and 0.02% (wt/vol) L-arabinose (Fluka) (for pBAD-based libraries) at 37°C.
Library screening
For each round of random mutagenesis, between 20,000 and 100,000 colonies (10 to 50 plates of bacteria) were screened, a number sufficient to sample all possible single-site mutants and a limited number of double mutants. For each round of site-directed mutagenesis, a number of colonies approximately 3-fold higher than the expected library diversity (e.g. 1200 colonies for a 400-member library) were screened to ensure full coverage. Whole plates of bacteria were photobleached for 10 to 120 minutes (determined empirically for each round of directed evolution) on a Spectra-Physics 92191-1000 solar simulator with 1600 W mercury arc lamp and two Spectra-Physics SP66239-3767 dichroic mirrors to remove infrared and ultraviolet wavelengths. Remaining light was filtered through 10 cm square bandpass filters (Chroma Technology Corp.) appropriate to the fluorescent protein being bleached (540/30 nm (B540/30) for mOrange- and TagRFP-based libraries or 568/40 nm (B568/40) for mApple libraries). Final light intensities produced by the solar simulator were measured by a miniature integrating-sphere detector (SPD024 head and ILC1700 meter, International Light Corp.) to be 95 mW/cm2 for the 540/30 filter and 141 mW/cm2 for the 568/40 filter. The bacterial plates were kept at 20°C using a home-built water-cooled aluminum block. For mOrange mutant selection, plates were examined by eye as previously described32 using a 150 W xenon lamp equipped with a 540/30 nm excitation filter and fiber optic light guides to illuminate the plates and 575 nm long pass filter for visualizing emission. For TagRFP mutant selection, plates were imaged before and after bleaching on a UVP imaging system using 535/45 nm excitation and 605/70 nm emission filters. In either case, colonies maintaining bright fluorescence after photobleaching and/or those with high post- to pre-bleach fluorescence ratios were cultured for 8 h in 2ml Luria-Bertani (LB) medium supplemented with 100 μg/mL ampicillin, and then culture volume was increased to 4ml with additional LB supplemented with ampicillin and 0.2% (wt/vol) L-arabinose to induce fluorescent protein expression and were grown overnight. A fraction of each cell pellet was extracted with B-PER II (Pierce) and spectra were obtained using a Safire 96-well plate reader with monochromators (TECAN). When screening for photostable variants, spectra were obtained before and after photobleaching extracted protein on the solar simulator.
Protein production and characterization
fluorescent proteins were expressed from pBAD vectors in E. coli strain LMG194, purified on Ni-NTA agarose (Qiagen), and characterized as described2. Photobleaching measurements were performed on aqueous droplets of purified protein under oil as described2, 16. To determine if the presence of molecular oxygen influenced bleaching, we performed our standard bleaching experiment before and after equilibrating the entire bleaching apparatus under humidified N2.
GenBank accession numbers
Sequences for mOrange2, mApple, and TagRFP-T have been deposited under accession numbers DQ336159, DQ336160, and EU582019, respectively.
Footnotes
Competing financial interests
N.C.S., M.R.M., M.Z.L., and R.Y.T. are listed as inventors in a U.S. patent application assigned to the University of California, which includes these proteins as claims.
Primer list, Mass spectrometry analysis, Mammalian expression vectors, Live cell imaging, and LSCM live cell photobleaching may be found in Supplementary Methods online.
References
- 1.Campbell RE, et al. A monomeric red fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 3.Chudakov DM, et al. Photoswitchable cyan fluorescent protein for protein tracking. Nat Biotechnol. 2004;22:1435–1439. doi: 10.1038/nbt1025. [DOI] [PubMed] [Google Scholar]
- 4.Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J Biol Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- 5.Habuchi S, et al. Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa. Proc Natl Acad Sci U S A. 2005;102:9511–9516. doi: 10.1073/pnas.0500489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karasawa S, Araki T, Nagai T, Mizuno H, Miyawaki A. Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer. Biochem J. 2004;381:307–312. doi: 10.1042/BJ20040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagai T, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen AW, Daugherty PS. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 9.Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 10.Wiedenmann J, et al. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci U S A. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zapata-Hommer O, Griesbeck O. Efficiently folding and circularly permuted variants of the Sapphire mutant of GFP. BMC Biotechnol. 2003;3:5. doi: 10.1186/1472-6750-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ai HW, Henderson JN, Remington SJ, Campbell RE. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem J. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merzlyak EM, et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 14.Matz MV, et al. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 15.Petersen J, et al. The 2.0-A crystal structure of eqFP611, a far red fluorescent protein from the sea anemone Entacmaea quadricolor. J Biol Chem. 2003;278:44626–44631. doi: 10.1074/jbc.M307896200. [DOI] [PubMed] [Google Scholar]
- 16.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 17.Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 18.Sinnecker D, Voigt P, Hellwig N, Schaefer M. Reversible photobleaching of enhanced green fluorescent proteins. Biochemistry. 2005;44:7085–7094. doi: 10.1021/bi047881x. [DOI] [PubMed] [Google Scholar]
- 19.Habuchi S, et al. Evidence for the isomerization and decarboxylation in the photoconversion of the red fluorescent protein DsRed. J Am Chem Soc. 2005;127:8977–8984. doi: 10.1021/ja047023o. [DOI] [PubMed] [Google Scholar]
- 20.Chudakov DM, Feofanov AV, Mudrik NN, Lukyanov S, Lukyanov KA. Chromophore environment provides clue to “kindling fluorescent protein” riddle. J Biol Chem. 2003;278:7215–7219. doi: 10.1074/jbc.M211988200. [DOI] [PubMed] [Google Scholar]
- 21.Andresen M, et al. Structure and mechanism of the reversible photoswitch of a fluorescent protein. Proc Natl Acad Sci U S A. 2005;102:13070–13074. doi: 10.1073/pnas.0502772102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andresen M, et al. Structural basis for reversible photoswitching in Dronpa. Proc Natl Acad Sci U S A. 2007 doi: 10.1073/pnas.0700629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henderson JN, Ai HW, Campbell RE, Remington SJ. Structural basis for reversible photobleaching of a green fluorescent protein homologue. Proc Natl Acad Sci U S A. 2007;104:6672–6677. doi: 10.1073/pnas.0700059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Thor JJ, Gensch T, Hellingwerf KJ, Johnson LN. Phototransformation of green fluorescent protein with UV and visible light leads to decarboxylation of glutamate 222. Nat Struct Biol. 2002;9:37–41. doi: 10.1038/nsb739. [DOI] [PubMed] [Google Scholar]
- 25.Bell AF, Stoner-Ma D, Wachter RM, Tonge PJ. Light-driven decarboxylation of wild-type green fluorescent protein. J Am Chem Soc. 2003;125:6919–6926. doi: 10.1021/ja034588w. [DOI] [PubMed] [Google Scholar]
- 26.van Thor JJ, Georgiev GY, Towrie M, Sage JT. Ultrafast and low barrier motions in the photoreactions of the green fluorescent protein. J Biol Chem. 2005;280:33652–33659. doi: 10.1074/jbc.M505473200. [DOI] [PubMed] [Google Scholar]
- 27.Verkhusha VV, Sorkin A. Conversion of the monomeric red fluorescent protein into a photoactivatable probe. Chem Biol. 2005;12:279–285. doi: 10.1016/j.chembiol.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukyanov KA, Chudakov DM, Lukyanov S, Verkhusha VV. Innovation: Photoactivatable fluorescent proteins. Nat Rev Mol Cell Biol. 2005 doi: 10.1038/nrm1741. [DOI] [PubMed] [Google Scholar]
- 30.Patterson GH, Lippincott-Schwartz J. Selective photolabeling of proteins using photoactivatable GFP. Methods. 2004;32:445–450. doi: 10.1016/j.ymeth.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 31.Tsutsui H, Karasawa S, Shimizu H, Nukina N, Miyawaki A. Semi-rational engineering of a coral fluorescent protein into an efficient highlighter. EMBO Rep. 2005;6:233–238. doi: 10.1038/sj.embor.7400361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc Natl Acad Sci U S A. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Figure 1 Excitation, emission, and absorbance spectra of novel fluorescent protein variants.
Supplementary Note 1 Directed evolution and characterization of mApple.
Supplementary Note 2 Directed evolution and characterization of mOrange2.
Supplementary Note 3 Summary of reversible photoswitching data with representative examples.
Supplementary Methods