Summary
Regulatory divergence is likely a major driving force in evolution. Comparative transcriptomics provides a new glimpse into the evolution of gene regulation. Ascomycota fungi are uniquely suited among eukaryotes for studies of regulatory evolution, due to broad phylogenetic scope, many sequenced genomes, and facility of genomic analysis. Here we review the substantial divergence in gene expression in Ascomycota and how this is reconciled with the modular organization of transcriptional networks. We show that flexibility and redundancy in both cis- and trans-regulation can lead to changes from altered expression of single genes to wholesale re-wiring of regulatory modules. Redundancy thus emerges as a major driving force facilitating expression divergence while preserving the coherent functional organization of a transcriptional response.
Introduction
The incredible diversity of living creatures defies their similarity in protein sequence and gene content. King and Wilson [1] first proposed that organismal diversity is likely driven by regulatory differences controlling when, where, and how genetic material is expressed. Nearly 35 years later, although examples of regulatory divergence are known in a wide range of species [2] including bacteria [3], fungi [4], flies [5], and mammals [6], the mechanisms through which regulatory systems evolve are still poorly understood. In recent years, comparative genomics approaches have allowed us to identify the functional components of genomes and to trace evolutionary events at different time scales [7,8]. These approaches are also being used to infer the evolution of gene-expression regulation through two general approaches: characterization of cis-regulatory elements in orthologous promoter sequences, and comparative analysis of mRNA profiles across organisms. While studies relying on sequence data are more prevalent, functional studies of comparative gene regulation are now starting to shed light on how genome evolution is linked to functional changes.
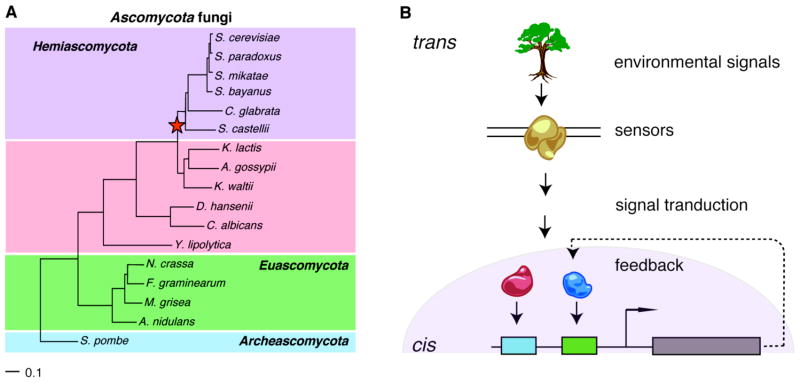
Among eukaryotes, the Ascomycota fungi (Figure 1A) are particularly suitable for studies of eukaryotic regulatory evolution. Over 100 genome sequences exist, for individuals within Saccharomyces species and across fungi spanning hundreds of millions of years of evolution. They include model organisms (S. cerevisiae, S. pombe) and important human and agricultural pathogens (e.g. C. albicans, Aspergilli, F. graminarium). Furthermore, sequenced genomes include species that diverged before and after a whole genome duplication (WGD, Figure 1A), which occurred ~150 million years ago (mya) [9,10]. This provides a unique opportunity to explore the effect of gene duplication on regulatory divergence. Finally, the relatively small (ca. 9 – 100 Mb) genomes are computationally tractable, but still display many of the hallmarks of eukaryotic gene regulation.
Figure 1. A. Ascomycota fungi.
A phylogenetic tree showing sequenced species from the major clades of the Ascomycota fungi. Red star – WGD. Tree is drawn to scale and adapted from [31]. B. Factors affecting regulatory evolution. trans factors, including differential interpretation of environmental signals by sensory and signaling proteins and transcription factors, as well as cis regulatory elements (boxes) affect gene expression and drive regulatory module evolution.
In this review, we focus on recent advances made in understanding the evolution of gene regulation in Ascomycota, from micro-evolutionary scales (<5 million years and typically within species) to macro- evolutionary time frames over tens of millions of years involving extensive speciation. (It is estimated that S. cerevisiae can undergo ~2900 generations per year [11] We examine conservation and divergence from two different perspectives: the regulation of mRNA expression at the level of single genes, and the coordinated expression of genes in regulons or ‘modules’ (i.e. sets of co-regulated target genes) within a network. As we show, flexibility and apparent redundancy in gene copies, functional elements and molecular interactions may play a major role at driving the emergence of regulatory divergence, while conserving the functional backbone of transcriptional responses.
Functional and mechanistic corollaries of expression conservation and divergence
Available compendia of genome-wide mRNA profiles have allowed direct comparison of orthologous mRNA expression patterns across species. Large datasets exist for model Ascomycota (S. cerevisiae, S. pombe and C. albicans), and smaller datasets are available for other fungi in the phylum (e.g. other sensu stricto Saccharomyces [12] C. glabrata [13], K. lactis [14], and some Euascomycota [15,16]) as well as different strains within S. cerevisiae [12,17–20]. Together, these afford a global assessment of expression divergence (ED) [12]– a quantitative measure of the differences in the expression of a pair of orthologs between two species.
An emerging theme is that different classes of genes have different propensities for ED. [20–22]. Most notably, genes with high- versus low- ED are distinguished by a broad functional dichotomy: genes with conserved expression are typically involved in growth and general metabolism, whereas those with divergent expression are often subtelomeric, responsive to external and internal signals (e.g. stress) and are nonessential. This dichotomy in divergence between species [22,23] is also mirrored when measuring variation between isogenic cells in a yeast culture [24] or between genetic variants of S. cerevisiae [18,20,25].
Expression divergence is also correlated to distinct promoter architectures. The promoters of genes with conserved expression (low-ED) lack upstream TATA elements, contain well-positioned nucleosomes, and display a substantial nucleosome-depleted region where most regulatory elements reside. Conversely, promoters of high-ED genes contain TATA elements and display distributed nucleosomes, which are regulated through dynamic chromatin remodeling [12,26,27]. Promoters of high-ED genes are also associated with the presence of unstable tandem repeats proposed to drive changes in nucleosome organization and gene expression [28].
That specific genes tend to show large versus small expression differences both within clonal populations and across species suggests a higher tolerance for expression differences at particular gene classes, and thus a higher prevalence of evolved expression. Indeed, ED levels are correlated with the level of constraint measured on steady-state expression across S. cerevisiae strains [KH Eng, et al., unpublished]. It is intriguing that genes belonging to particular functional classes contain promoters linked to high or low ED. Indeed, variable expression of certain genes within a population (e.g. stress defense genes) may have been directly selected for through modification of promoter structure [26].
Gene duplication facilitates regulatory neo-functionalization
Gene duplication may provide a unique opportunity for ED of at least one of the two paralogs [29–31]. In a comprehensive study of S. cerevisiae paralogs whose origins range to the last common ancestor with S. pombe (Figure 1A, root) we found surprisingly little divergence of the molecular function of paralogs, but substantial (~70%) divergence in gene regulation, as reflected in the cis-regulatory elements, the transcription factors (TFs) bound to the genes’ promoters, and the gene regulon to which they belong [31]. This is consistent with the idea that regulatory divergence occurs at an elevated rate compared to divergence of coding sequence of paralogs.
Two likely scenarios can underlie diverged expression of paralogs. Regulatory sub-functionalization occurs when multiple, distinct regulatory elements controlling expression of the single ancestral gene are ‘split’ between its two descendants, such that each paralog retains only some of the regulatory inputs. In contrast, regulatory neo-functionalization occurs when one paralog evolves a new control not used by the ancestral gene. A recent study [32] proposed neo-functionalization as the dominant mode of diverged expression of S. cerevisiae paralogs, since often the expression pattern of only one paralog was distinct from that of the single pre-duplication ortholog in C. albicans.
Expression divergence of paralogs may have an important adaptive role. First, regulatory divergence can effectively lead to sub-functionalization of paralogs, even if paralogs have the same biochemical function (but serve different roles due to expression differences). This may have occurred in the post-WGD regulatory divergence of glycolysis/gluconeogenesis enzymes [33], such as the hexose kinases HXK2 and HXK1 that share hexokinase activity but display distinct expression patterns. Notably, sub-functionalization may be partial and remnants of the shared (joint) ancestral control may still allow paralogs to ‘backup’ each other [34]. Finally, constraints on expression levels could in turn influence the evolution of gene copy number [29,31]}, since low-ED genes also exhibit few duplication and loss events, whereas high-ED genes have substantial variation in copy number between species.
Flexibility in regulatory mechanisms can drive expression divergence
Genetic changes in both cis and trans elements can contribute to expression divergence (Figure 1B). A genetic change can affect expression in cis, either directly by altering regulatory sequences controlling gene expression, or indirectly by modifying the activity of the gene’s product and consequently affecting expression through feedback [35]. Polymorphisms in cis appear to contribute most to ED in phylogenetically close species, independent of environmental factors [22]. Alternatively, a polymorphism distant to the affected gene can affect its expression in trans, by affecting its regulators directly (e.g. a polymorphism in a regulatory protein), or indirectly by altering physiology that provokes an expression response despite perfect conservation of the direct regulatory system [36,37].
Gain and loss of cis-regulatory motifs, and the potential for corresponding changes in transcription factor binding, can occur on relatively short time scales (<5 – 20 million years), both within [38] and between species [22,39–41]. Of the lineage-specific binding-site losses within sensu stricto Saccharomyces, over half may correspond to newly emerged binding sites in the same regulatory regions [39]. Such binding site turnover – where the appearance of a new binding site allows loss of the other – explains how expression can be maintained despite plasticity in regulatory sequences.
In other cases the apparent loss of a binding site, without concomitant turnover, corresponds to a change in gene expression pattern, and ultimately loss of TF control (Figures 2B and 3) [39,40]. For example, ChIP-chip studies of two TFs, Ste12p and Tec1p, in three sensu stricto Saccaromyces species (~20 mya) [7,40] show that only 20% of TF binding events were conserved across all three species. Similar conservation (15%) was observed for the binding of Mcm1 between more distant species (S. cerevisiae, K. lactis, and C. albicans) [41]. In many cases the loss of TF binding correlated with loss of the binding site, but in some cases TF binding occurred despite absence of a discernible DNA motif [41]. Such ‘promiscuous’ binding to non-canonical sites followed by selection for optimal binding may facilitate the acquisition of new targets [42].
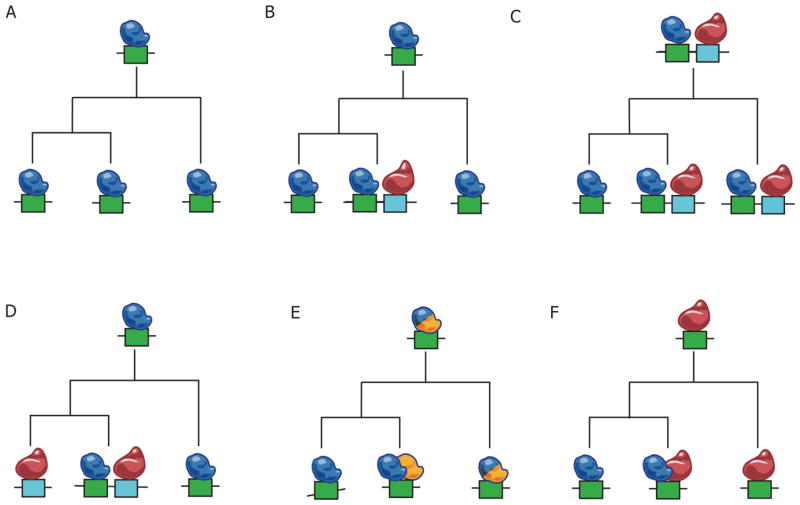
Figure 2. Alternative modes for the regulatory evolution of transcriptional modules.
Each panel shows a distinct scenario of the inferred evolution of an ancestral program (at tree root) into programs observed in extant species (leaves). A. Conservation of both cis-elements (boxes) and TFs (ovoids). B. Elaboration of a program by the emergence of a new cis-element in addition to the ancestral one. C. Divergence of a program by loss of cis-element. D–F. Divergence of a program through cis- or trans-redundancy. D. In cis, a switch from one cis-element to another through an intermediate that has both sites, bound by distinct factors. E. In trans, two paralogous TFs bind the same site following gene duplication. After sub-functionalization, only one of the factors controls the module. F. In trans, switching of one TF for another that does not share ancestry, but can bind a similar site.
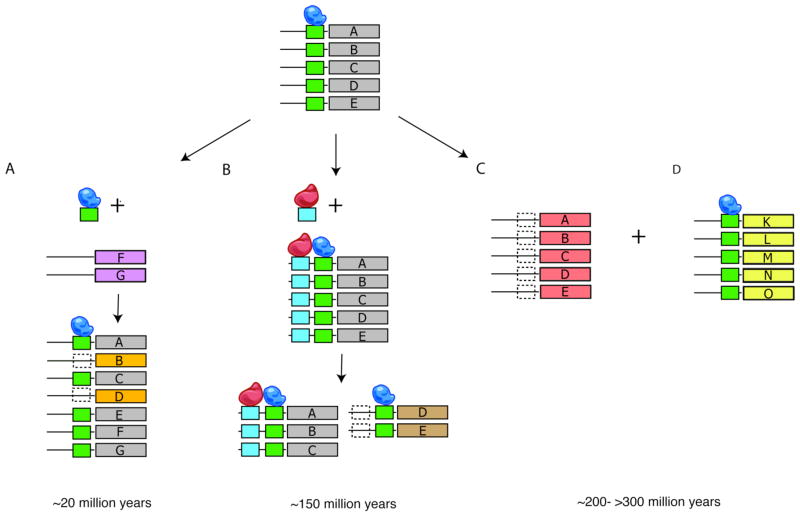
Figure 3. Different mechanisms of evolution in transcriptional regulatory systems at distinct time scales.
An ancestral module (genes A–E) controlled by one TF (blue ovoid) and site (green box) is shown on top. A. At shorter time scales (~20 million years), loss and gain of the cis-regulatory site allows fine-tuning of module membership. B. Gain of a new control through a new cis-regulatory element (blue box) and TF (red ovoid) can lead to elaboration. Subsequent loss of one of the elements from some of the module’s genes can lead to splitting (bottom), at longer time scales (~150 million years). C,D. Wholesale re-wiring is observed at longer times (~200->300 million years), by full switching of the targets of the regulatory system (trans factor and cis elements) from one set of genes (A–E, red) to another (K–O, yellow).
Expression divergence in transcriptional regulatory networks
It is challenging to reconcile the substantial evolutionary diversity in the expression of individual genes with the functional organization of regulatory networks. In particular, it is well established that transcriptional modules (regulons of co-regulated genes), play a central role in regulatory networks [43–45]. Various modules are conserved across organisms from E. coli to humans [46,47], including fungi [21]. However, if the regulation of individual genes is highly evolvable, how are multiple evolutionary events coordinated to sustain conservation of co-expression in regulons? One possibility is that changes occur predominantly in trans (e.g. divergent TF responsiveness but conserved cis-regulatory elements), thus conserving co-expression (module identity) while diverging module expression. However, several cases demonstrate dramatic differences in module composition or its associated cis-elements. As we show below, multiple forms of functional redundancy allow for complex divergence of regulatory mechanism while maintaining co-regulation within modules.
Conservation of regulatory modules over long evolutionary timeframes
Many regulatory modules are conserved across Ascomycota, often associated with conserved cis-regulatory elements and transcription factors (Figure 2A). Conserved modules can be identified by a statistically significant overlap in orthologous genes between modules of genes with correlated expression within each species [21,48]. Alternatively, we can identify conserved regulation in gene modules [4] by comparing cis-regulatory elements enriched in the promoters of sets of orthologous co-regulated genes. Three-quarters of the cis-regulatory elements associated with recognizable modules are conserved by the latter criterion between S. cerevisiae and species that emerged around the WGD (~150 mya). Nearly a third of these modules are conserved out to C. albicans (~230 mya), and several are conserved in Sz. pombe (>300 mya) [4,21].
These computational results have been confirmed experimentally, showing conserved regulatory connections for a TF, its cognate cis-element, and a large fraction of orthologous targets. Examples include the Gcn4/Cpc1 TFs and amino-acid responsive genes [16,49], AP-1 factors and oxidative-stress defense genes [13,50,51] and several TFs and their cell-cycle regulated targets [41,52,53]. Features of binding site organization within the promoter are often conserved as well [4,21,41,54], suggesting that regulatory mechanisms and combinatorial regulation can also be under purifying selection.
Re-shaping modules by gain and loss of gene targets
That orthologous regulatory modules can be detected over long time frames does not preclude substantial regulatory evolution. First, co-expression of genes in a module can collectively evolve simply by altering TF activation in response to different environmental cues and upstream signals. Secondly, cis-regulatory changes can drive gain and loss of targets to affect module identity, as well as regulatory patterns. Although the underlying mechanism is the same, the effects of target gain and loss can appear very different depending on the time scale. Over short timescales (5 – 20 million years), this can appear as fine-tuning of module members (due to neutral or selective forces), as suggested for Ste12/Tec1 targets [39,40] (Figure 3A). Selection over longer time may promote the gain or loss of entire functional sub-modules, thus splitting or merging modules to create higher-order coordination [55] (Figure 3B). This may have occurred in the evolution of mitochondrial and cytoplasmic ribosomal protein (mRP or cRP) gene expression [55]. mRPs and cRPs were proposed to be ancestrally regulated as a single module through the Rapid Growth Elements (RGE, AATTTT), but the module apparently split in post-WGD species when mRPs lost the site, leading to de-coupling of the mRP and cRP expression in species that can decouple fermentation from respiration.
Conversely, the gain of new targets by an existing transcription factor, whether by drift or selection, can lead to the co-option of an existing regulatory module under more elaborate control, to the ‘birth’ of a new module of co-regulated genes, or to a wholesale change in a target regulon, such that there is little or no overlap in the targets of orthologous regulators (Figure 3C,D). Such dramatic changes in regulatory systems are typically observed over longer time scales (~150 to over 300 million years). For example, in C. albicans, Mcm1 control, along with combinatorial regulation by Wor1, emerged in a novel host-adaptation module in C. albicans [41], whereas the Sko1 TF was co-opted to regulate a cell-wall biogenesis module, in addition to ancestral targets involved in stress [56]. Such co-option may lead to higher order coordination between modules, and is likely related to adaptation of C. albicans to its human host. In more extreme cases, drift in TF targets can lead to a full switch in regulated genes, along with co-evolved differences in environmental responsiveness. For example, sometime after the divergence of the lineages leading to C. albicans and S. cerevisiae, the Gal4 TF acquired galactose metabolism (GAL) genes under its control, lost the ancestral glycolysis and subtelomeric targets still seen in C. albicans, and acquired responsiveness to galactose [49,57–59], resulting in a complete functional and mechanistic switch.
The role of redundancy in mediating regulatory re-wiring of conserved modules
The formation of ‘seemingly redundant’ regulatory mechanisms may facilitate the dramatic rewiring of regulatory mechanisms while maintaining gene co-regulation. In several cases, wholesale rewiring is observed between species diverged 200 – 300 mya, and appears to have been preceded by a period of redundant regulation on the order of 100 – 150 mya.
One prominent mode of redundancy is the formation of a module under the control of multiple regulatory systems, through distinct cis-regulatory sites (Figure 2D). In this ‘regulatory switching’ scenario, an ancient regulatory program is augmented by the appearance of a new set of distinct cis-regulatory elements, followed by loss of one system’s sites from the target gene promoters. For example, the switch in the regulation of the RP genes from the Homol-D system in S. pombe to the Rap1 system in S. cerevisiae, may have occurred through the formation of an intermediate, where both sites appear in close proximity in RP promoters, as is still observed in several extant species from A. gossyipii to C. glabrata (100–150 mya) [4,21,60]. A similar redundancy is observed in the regulation of the GAL module [58] and may have contributed to a switch from one cis-regulatory element to another in the control of the mating module [54]. Notably, in species whose module harbors both regulatory controls, they may have distinct roles, e.g. in regulating distinct expression levels, while being redundant for co-expression.
From the trans side, the presence of multiple TFs capable of binding similar sites can lead to re-wiring. For example, redundant binding by two paralogous TFs can result in sub-functionalization of one paralog and its subsequent loss of control of a regulatory system (Figure 2E). This has occurred for the paralogous TFs Mcm1 and Arg80, each of which regulates a subset of the ancestral protein’s targets [41,61]. More generally, many TFs are derived from a few broad families, resulting in overlapping specificities and the propensity for ‘redundant’ regulation through the same elements (Figure 2F). This may have contributed to the replacement of Gcn4 by another bZIP family member as the regulator of the amino acid metabolism module in S. pombe [21]. Finally, relative promiscuity in binding similar sites can lead to switching from one TF to another even when they do not share ancestry, as has been suggested for the switch from the ancestral a2 activator (seen in C. albicans) to the derived alpha2 repressor (in S. cerevisiae) in the control of the mating module [62].
Flexibility and redundancy in cooperative and combinatorial interactions between distinct transcription factors can also facilitate re-wiring [63]. For example, flexibility in the required components for a multi-TF complex may underlie the co-option of Hap4 TF to regulate iron uptake in S. pombe [64]. In certain cases, this flexibility involves an elaborate interplay of trans- and cis- factors. The transition from a2 to alpha2 regulation in the mating module may have been facilitated through interactions of both factors with the conserved cofactor Mcm1 [54,62]. Interestingly, evolved Mcm1-DNA interactions at elements with high flanking AT content in S. cerevisiae may have alleviated the need for a2-dependent activation, allowing for loss of a2 regulation at these genes [54]. Since regulation through combinatorial and cooperative control by a larger number of TFs may be associated with greater binding flexibility, combined cis-trans redundancy may be a major driving force for regulatory evolution.
Future prospects
While great advances have been made in the phenomenology and mechanistic understanding of the evolution of gene regulation, the relative roles of neutral drift and selective forces in promoting divergence remain largely unknown. For individual genes, purifying selection can be effectively invoked for the conservation of sites in closely related species, and for the low-ED of genes involved in growth processes. However, it is unclear how much of the increased divergence of high-ED genes is due to direct positive selection vs. neutral drift and relaxed constraint. For gene modules, purifying selection may act to conserve co-expression and co-regulation of certain regulons. However, the repeated divergence of the regulatory mechanisms controlling other conserved modules (e.g., RPs) suggests the potential adaptive importance of conserving co-expression while diverging how a module is coupled to its inputs. Identifying these selective forces is highly challenging. Understanding how changes in the direct control of modules are related to upstream changes in signal sensing [65] may shed light on this question.
The major promise of Ascomycota for studies of evolution of gene expression is in the facility of experiments in both model- and non-model organisms in this phylogeny. In S. cerevisiae, experimental evolution and direct engineering [66,67] can highlight the effect of well-defined selective pressures on regulatory evolution. At a broader phylogenetic scope, recent studies comparing expression or in vivo TF-binding profiles in two or three organisms have shown early promise. A critical next step is to expand the experimental scope to cover a broader phylogenetic range and density, thus allowing us to directly study the divergence of expression in individual genes and gene modules.
Acknowledgments
The authors thank Sigrid Hart for graphics included in the figures. DJW was supported by an NLM training grant 5T15LM007359. DAT was supported by a Human Frontiers Science Program Research Grant. APG was supported by an NSF CAREER award (#0447887) and NIGMS R01GM083989-01. AR was supported by HHMI, by a Career Award at the Scientific Interface from the Burroughs Wellcome Fund, by an NIH Pioneer Award and by the Sloan Foundation.
References
- 1.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 2.Jensen LJ, Jensen TS, de Lichtenberg U, Brunak S, Bork P. Co-evolution of transcriptional and post-translational cell-cycle regulation. Nature. 2006;443:594–597. doi: 10.1038/nature05186. [DOI] [PubMed] [Google Scholar]
- 3.McAdams HH, Srinivasan B, Arkin AP. The evolution of genetic regulatory systems in bacteria. Nat Rev Genet. 2004;5:169–178. doi: 10.1038/nrg1292. [DOI] [PubMed] [Google Scholar]
- 4.Gasch AP, Moses AM, Chiang DY, Fraser HB, Berardini M, Eisen MB. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol. 2004;2:e398. doi: 10.1371/journal.pbio.0020398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prud’homme B, Gompel N, Carroll SB. Emerging principles of regulatory evolution. Proc Natl Acad Sci USA. 2007;104(Suppl 1):8605–8612. doi: 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khaitovich P, Enard W, Lachmann M, Pääbo S. Evolution of primate gene expression. Nat Rev Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- 7.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 8.Cliften P. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 9.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 10.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 11.Fay J, Benavides J. Evidence for Domesticated and Wild Populations of Saccharomyces cerevisiae. PLoS Genetics. 2005;1:e5. doi: 10.1371/journal.pgen.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tirosh I, Weinberger A, Carmi M, Barkai N. A genetic signature of interspecies variations in gene expression. Nat Genet. 2006;38:830–834. doi: 10.1038/ng1819. [DOI] [PubMed] [Google Scholar]
- 13.Lelandais G, Tanty V, Geneix C, Etchebest C, Jacq C, Devaux F. Genome adaptation to chemical stress: clues from comparative transcriptomics in Saccharomyces cerevisiae and Candida glabrata. Genome Biol. 2008;9:R164. doi: 10.1186/gb-2008-9-11-r164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suleau A, Gourdon P, Reitz-Ausseur J, Casaregola S. Transcriptomic analysis of extensive changes in metabolic regulation in Kluyveromyces lactis strains. Eukaryot Cell. 2006;5:1360–1370. doi: 10.1128/EC.00087-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen MR, Vongsangnak W, Panagiotou G, Salazar MP, Lehmann L, Nielsen J. A trispecies Aspergillus microarray: comparative transcriptomics of three Aspergillus species. Proc Natl Acad Sci USA. 2008;105:4387–4392. doi: 10.1073/pnas.0709964105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian C, Kasuga T, Sachs MS, Glass NL. Transcriptional profiling of cross pathway control in Neurospora crassa and comparative analysis of the Gcn4 and CPC1 regulons. Eukaryot Cell. 2007;6:1018–1029. doi: 10.1128/EC.00078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 18.Kvitek DJ, Will JL, Gasch AP. Variations in stress sensitivity and genomic expression in diverse S. cerevisiae isolates. PLoS Genet. 2008;4:e1000223. doi: 10.1371/journal.pgen.1000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL. Genetic properties influencing the evolvability of gene expression. Science. 2007;317:118–121. doi: 10.1126/science.1140247. [DOI] [PubMed] [Google Scholar]
- 20.Landry CR, Oh J, Hartl DL, Cavalieri D. Genome-wide scan reveals that genetic variation for transcriptional plasticity in yeast is biased towards multi-copy and dispensable genes. Gene. 2006;366:343–351. doi: 10.1016/j.gene.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 21.Tanay A, Regev A, Shamir R. Conservation and evolvability in regulatory networks: The evolution of ribosomal regulation in yeast. Proc Natl Acad Sci USA. 2005;102:7203–7208. doi: 10.1073/pnas.0502521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22**.Tirosh I, Reikhav S, Levy AA, Barkai N. A yeast hybrid provides insight into the evolution of gene expression regulation. Science. 2009;324:659–662. doi: 10.1126/science.1169766. Using allele-specific gene expression profiling in two yeast species and their hybrid, the authors confirm that the majority of ED between species is due to cis changes, but that the majority of hybrid-specific expression occurs through trans effects. Additionally, they show that parental genomes accumulate compensating cis-trans mutations, suggesting a mechanism for emergence of novel hybrid phenotypes. [DOI] [PubMed] [Google Scholar]
- 23.Tirosh I, Weinberger A, Bezalel D, Kaganovich M, Barkai N. On the relation between promoter divergence and gene expression evolution. Mol Syst Biol. 2008;4:159. doi: 10.1038/msb4100198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raser J. Control of sochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang D, Sung HM, Wang TY, Huang CJ, Yang P, Chang T, Wang YC, Tseng DL, Wu JP, Lee TC, et al. Expression evolution in yeast genes of single-input modules is mainly due to changes in trans-acting factors. Genome Res. 2007;17:1161–1169. doi: 10.1101/gr.6328907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. By examining genome-wide data for nucleosome positioning in yeast, Tirosh and Barkai identify two distinct classes of genes based upon chromatin organization: one group with high ED, and the other with low ED. Based on these data, the authors suggest that two distinct mechanisms for gene regulation by chromatin are employed, depending on the gene set. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Field Y, Fondufe-Mittendorf Y, Moore IK, Mieczkowski P, Kaplan N, Lubling Y, Lieb JD, Widom J, Segal E. Gene expression divergence in yeast is coupled to evolution of DNA-encoded nucleosome organization. Nat Genet. 2009;41:438–445. doi: 10.1038/ng.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vinces M, Legendre M, Caldara M, Hagihara M, Verstrepen K. Unstable tandem repeats in promoters confer transcriptional evolvability. Science. 2009;324:1213–1216. doi: 10.1126/science.1170097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu Z, Rifkin S, White K, Li W. Duplicate genes increase gene expression diversity within and between species. Nat Genet. 2004;36:577–579. doi: 10.1038/ng1355. [DOI] [PubMed] [Google Scholar]
- 30.Guan Y, Dunham MJ, Troyanskaya OG. Functional analysis of gene duplications in Saccharomyces cerevisiae. Genetics. 2007;175:933–943. doi: 10.1534/genetics.106.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31*.Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. Using a novel approach to assigning gene orthology, the authors trace the evolutionary history of gene duplications and losses across 17 Ascomycota fungi. The study reveals new insights into the consequences of gene amplification and loss, demonstrating that the propensity for dynamic gene copy number changes varies by gene functional group, essentiality, and transcription regulation. The authors also discuss the inferred consequences of gene duplication on the evolution of biochemical function and gene-expression regulation. [DOI] [PubMed] [Google Scholar]
- 32.Tirosh I, Barkai N. Comparative analysis indicates regulatory neofunctionalization of yeast duplicates. Genome Biol. 2007;8:R50. doi: 10.1186/gb-2007-8-4-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conant G, Wolfe K. Increased glycolytic flux as an outcome of whole-genome duplication in yeast. Mol Syst Biol. 2007;3:12. doi: 10.1038/msb4100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kafri R, Bar-Even A, Pilpel Y. Transcription control reprogramming in genetic backup circuits. Nat Genet. 2005;37:295–299. doi: 10.1038/ng1523. [DOI] [PubMed] [Google Scholar]
- 35.Ronald J, Brem RB, Whittle J, Kruglyak L. Local regulatory variation in Saccharomyces cerevisiae. PLoS Genet. 2005;1:e25. doi: 10.1371/journal.pgen.0010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith EN, Kruglyak L. Gene-environment interaction in yeast gene expression. PLoS Biol. 2008;6:e83. doi: 10.1371/journal.pbio.0060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yvert G, Brem RB, Whittle J, Akey JM, Foss E, Smith EN, Mackelprang R, Kruglyak L. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- 38.Doniger SW, Kim HS, Swain D, Corcuera D, Williams M, Yang SP, Fay JC. A catalog of neutral and deleterious polymorphism in yeast. PLoS Genet. 2008;4:e1000183. doi: 10.1371/journal.pgen.1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doniger SW, Fay JC. Frequent gain and loss of functional transcription factor binding sites. PLoS Comput Biol. 2007;3:e99. doi: 10.1371/journal.pcbi.0030099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40**.Borneman AR, Gianoulis TA, Zhang ZD, Yu H, Rozowsky J, Seringhaus MR, Wang LY, Gerstein M, Snyder M. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317:815–819. doi: 10.1126/science.1140748. Using ChIP-chip, the authors examined the binding of two transcription factors in three sensu stricto Saccharomycetes and found a surprising amount of cis-regulatory diversity between closely related species. In only about 20% of cases did a transcription factor bind to the same site in all three species, supporting a major role for cis-regulatory mutations in evolution. [DOI] [PubMed] [Google Scholar]
- 41**.Tuch BB, Galgoczy DJ, Hernday AD, Li H, Johnson AD. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 2008;6:e38. doi: 10.1371/journal.pbio.0060038. Using ChIP-chip and fungal comparative genomics, the authors identify gene expression modules dependent on Mcm1 and its transcriptional cofactors in three hemiascomycetes. The work characterizes lineage-specific rates of target gene gain and loss and demonstrates species-specific differences in binding-site architecture. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanay A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 2006;16:962–972. doi: 10.1101/gr.5113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ihmels J, Friedlander G, Bergmann S, Sarig O, Ziv Y, Barkai N. Revealing modular organization in the yeast transcriptional network. Nat Genet. 2002;31:370–377. doi: 10.1038/ng941. [DOI] [PubMed] [Google Scholar]
- 44.Segal E, Shapira M, Regev A, Pe’er D, Botstein D, Koller D, Friedman N. Module networks: Identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 45.Tanay A, Sharan R, Kupiec M, Shamir R. Revealing modularity and organization in the yeast molecular network by integrated analysis of highly heterogeneous genomewide data. Proc Natl Acad Sci USA. 2004;101:2981–2986. doi: 10.1073/pnas.0308661100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bergmann S, Ihmels J, Barkai N. Similarities and differences in genome-wide expression data of six organisms. PLoS Biol. 2004;2:E9. doi: 10.1371/journal.pbio.0020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 48.Ihmels J, Bergmann S, Berman J, Barkai N. Comparative gene expression analysis by differential clustering approach: application to the Candida albicans transcription program. PLoS Genet. 2005;1:e39. doi: 10.1371/journal.pgen.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martchenko M, Levitin A, Whiteway M. Transcriptional activation domains of the Candida albicans Gcn4p and Gal4p homologs. Eukaryot Cell. 2007;6:291–301. doi: 10.1128/EC.00183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Cao YY, Jia XM, Cao YB, Gao PH, Fu XP, Ying K, Chen WS, Jiang YY. Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans. Free Radic Biol Med. 2006;40:1201–1209. doi: 10.1016/j.freeradbiomed.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 51.Quinn J, Findlay VJ, Dawson K, Millar JB, Jones N, Morgan BA, Toone WM. Distinct regulatory proteins control the graded transcriptional response to increasing H2O2 levels in fission yeast Schizosaccharomyces pombe. Mol Biol Cell. 2002;13:805–816. doi: 10.1091/mbc.01-06-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oliva A, Rosebrock A, Ferrezuelo F, Pyne S, Chen H, Skiena S, Futcher B, Leatherwood J. The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol. 2005;3:e225. doi: 10.1371/journal.pbio.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng X, Karuturi RK, Miller LD, Lin K, Jia Y, Kondu P, Wang L, Wong LS, Liu ET, Balasubramanian MK, et al. Identification of cell cycle-regulated genes in fission yeast. Mol Biol Cell. 2005;16:1026–1042. doi: 10.1091/mbc.E04-04-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsong AE, Tuch BB, Li H, Johnson AD. Evolution of alternative transcriptional circuits with identical logic. Nature. 2006;443:415–420. doi: 10.1038/nature05099. [DOI] [PubMed] [Google Scholar]
- 55.Ihmels J, Bergmann S, Gerami-Nejad M, Yanai I, McClellan M, Berman J, Barkai N. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- 56.Rauceo J, Blankenship J, Fanning S, Hamaker J, Deneault J, Smith F, Nantel A, Mitchell A. Regulation of the Candida albicans cell wall damage response by transcription factor Sko1 and PAS kinase Psk1. Mol Biol Cell. 2008;19:2741. doi: 10.1091/mbc.E08-02-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57*.Hittinger CT, Carroll SB. Gene duplication and the adaptive evolution of a classic genetic switch. Nature. 2007;449:677–681. doi: 10.1038/nature06151. In this study, Hittinger and Carroll examined the evolution of a pair of duplicated S. cerevisiae genes, GAL1 and GAL3, by testing fitness differences resulting from differing gene regulation in K. lactis, which has not undergone the gene duplication. They find evidence supporting sub-functionalization of the ancestral gene following duplication. [DOI] [PubMed] [Google Scholar]
- 58.Martchenko M, Levitin A, Hogues H, Nantel A, Whiteway M. Transcriptional rewiring of fungal galactose-metabolism circuitry. Curr Biol. 2007;17:1007–1013. doi: 10.1016/j.cub.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rokas A, Hittinger CT. Transcriptional rewiring: the proof is in the eating. Curr Biol. 2007;17:R626–628. doi: 10.1016/j.cub.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 60**.Hogues H, Lavoie H, Sellam A, Mangos M, Roemer T, Purisima E, Nantel A, Whiteway M. Transcription factor substitution during the evolution of fungal ribosome regulation. Mol Cell. 2008;29:552–562. doi: 10.1016/j.molcel.2008.02.006. The authors examined the regulation of ribosomal-protein gene expression in C. albicans, using ChIP-chip to define the Tbf1/Cbf1 regulon and comparative genomics to chronicle the switch in RP regulation between C. albicans and other fungi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jamai A, Dubois E, Vershon A, Messenguy F. Swapping functional specificity of a MADS Box protein: residues required for Arg80 regulation of arginine metabolism. Mol Cell Biol. 2002;22:5741–5752. doi: 10.1128/MCB.22.16.5741-5752.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsong AE, Miller MG, Raisner RM, Johnson AD. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell. 2003;115:389–399. doi: 10.1016/s0092-8674(03)00885-7. [DOI] [PubMed] [Google Scholar]
- 63.Tuch BB, Li H, Johnson AD. Evolution of eukaryotic transcription circuits. Science. 2008;319:1797–1799. doi: 10.1126/science.1152398. [DOI] [PubMed] [Google Scholar]
- 64.Mercier A, Pelletier B, Labbe S. A transcription factor cascade involving Fep1 and the CCAAT-binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe. Euk Cell. 2006;5:1866–1881. doi: 10.1128/EC.00199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beltrao P, Trinidad JC, Fiedler D, Roguev A, Lim WA, Shokat KM, Burlingame AL, Krogan NJ. Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 2009;7:e1000134. doi: 10.1371/journal.pbio.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, DeSevo CG, Botstein D, Dunham MJ. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009 doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]