Abstract
Bacterial type I polyketide synthases assemble structurally diverse natural products of significant clinical value from simple metabolic building blocks. The synthesis of these compounds occurs in a processive fashion along a large multi-protein complex. Transfer of the acyl intermediate across inter-polypeptide junctions is mediated, at least in large part, by N- and C-terminal docking domains. We report here a comprehensive analysis of the binding affinity and selectivity for the complete set of discrete docking domain pairs in the pikromycin and erythromycin PKS systems. Despite disconnection from their parent module, each cognate pair of docking domains retained exquisite binding selectivity. Further insights were obtained by X-ray crystallographic analysis of the PikAIII/PikAIV docking domain interface. This new information revealed a series of key interacting residues that enabled development of a structural model for the recently proposed H2–T2 class of polypeptides involved in PKS intermodular molecular recognition.
INTRODUCTION
Although naturally occurring polyketides have a wide variety of chemical structures, they are produced by three broad classes of polyketide synthases (PKSs) that share a common mechanism. This involves sequential decarboxylative condensation reactions to form carbon – carbon bonds between simple carboxylic acid extender units (1, 2). Type I modular PKSs are the large, multi-functional enzymes responsible for the production of a diverse family of structurally-rich and often biologically-active natural products (e.g. antimicrobial, antifungal, antiviral, anticancer and immunosuppressant compounds) (1, 3). Recently, structural studies have provided important new insights relating to the architecture and mechanism of type I PKSs and the related fatty acid synthases (4–7). Found in a variety of bacteria, modular PKSs direct biosynthesis via covalently-linked catalytic domains that are organized into linear modules where each module houses the requisite catalytic domains to perform a single elongation step in the building of the polyketide chain (Figure 1a–b). Each elongation module receives the nascent chain from the previous module, extends the polyketide by two carbons, and (typically) modifies this portion before passing the intermediate to the downstream PKS protein (5, 8). The final chemical structure is determined by the number of modules in the pathway, their catalytic domain composition, and arrangement in the biochemical assembly line (Figure 1a–b). Extensive research has identified signature amino acid sequences within the catalytic domains that guide substrate specificity (5, 9). However, details about the protein-protein interactions that govern acyl transfer between modules have only recently been explored (10–12).
Figure 1.
a–b) Arrangement of the PKS portions of the pikromycin and erythromycin biosynthetic pathways and their macrolactone products. c–d) Two examples of the intermodular transfer and elongation assays featuring the erythromycin PKS system (19–22). Abbreviations: ACP, acyl carrier protein, AT – acyltransferase, DH – dehydratase, ER – enoyl reductase, KR – ketoreductase, KS – ketosynthase, KSQ – decarboxylative ketosynthase, TE – thioesterase. Docking domains are colored by proposed subclass; H1–T1 are red, and H2–T2 is gold. Domain sizes are not drawn to scale.
The modular nature of type I PKSs has led many to envision rational “mix and match” bioengineering for the generation of novel polyketide products. As such, metabolic engineering or combinatorial biosynthesis has emerged as one potential route to create novel polyketide agents (13–16). Specific changes can be introduced to the final polyketide core in a controlled fashion by manipulating the genes that encode modular PKSs. Modifications at the level of the modules or the individual catalytic domains within a PKS module have been used to generate hundreds of novel polyketide structures, thereby establishing the potential of these applications (13–17). However, engineered PKS modules often fail to produce significant quantities of the desired product (17). Fundamental studies to establish the mechanistic basis for efficient molecular interactions between PKS multifunctional proteins will likely facilitate effective design and assembly of productive bioengineered pathways. The importance of this new information motivated the studies described in this report.
The fidelity and efficiency of acyl transfer at the interfaces of the individual PKS proteins is thought to be governed by helical regions, termed docking domains (dd), located at the C-terminus of the upstream and N-terminus of the downstream polypeptide chains (Figure 1a–b) (18). Two main strategies have been employed to study the specificity determinants for inter-polypeptide (e.g. module→module) communication. In the first strategy, modules (or excised domains) from the erythromycin PKS system were used to create a variety of in vitro intermodular transfer and elongation assays (see Figure 1c–d) (19–22). Typically, a variety of chimeric proteins were generated to investigate the effect of matched or mismatched docking domains in combination with a series of ACP/KS pairings. Detection of triketide lactones resulting from the transfer and elongation of diketide intermediates established that complementary docking domain pairs are required for efficient transfer of polyketide intermediates between polypeptides (11, 23). In some cases, formation of the cognate ACP and KS pairs also appears to impart a catalytic advantage, although tolerance for mispairing at this junction is also evident (19, 20).
The second strategy for analysis of PKS module-module molecular recognition has been to structurally characterize the docking interface. A docking domain complex model for the DEBS 2/DEBS 3 interface (Figure 1a) was developed via protein NMR spectroscopy (18). The structure established that the docking domains are helical and revealed two roles for the C-terminal PKS docking domain (ACP-side docking domains, ACPdd). First, this region appears important for stabilizing the PKS homodimer. Second, ACPdd is poised to interact with the downstream KS polypeptide through its terminal helix. The N-terminal PKS docking domain (KS-side docking domain, KSdd) exhibits a coiled-coil motif that has been observed both in the solution structure of the fused DEBS 2/DEBS 3 construct (18) and subsequently in the X-ray crystal structure of the DEBS 3 KS-AT didomain (6). The KSdd dimer presents a small hydrophobic patch, sometimes flanked by charged residues, as a narrow binding groove where the ACPdd terminal helix can bind.
Extension of the current DEBS 2/DEBS 3 structural model to the full range of docking domains across modular PKSs has not been possible due to low sequence similarity for a large subset of sequences. However, in a recent report based on the DEBS 2/DEBS 3 structural model and computational analysis of docking domain sequences from 42 characterized PKS systems, Thattai et al. proposed a new organization of PKS docking domains into distinct subclasses (24). Based on this classification system, the majority of docking domains (including the structurally characterized DEBS2/DEBS3 pair) fall into a single group termed H1–T1 (for head 1 and tail 1). Until this report, there was no structural information available for the proposed H2–T2 group of PKS docking domains.
To develop further our understanding of docking domain interactions in modular PKSs, and to expand fundamental information about docking domain protein structure, we pursued both biochemical and structural characterization of docking domains from two well-studied PKS biosynthetic systems. First, we report an analysis of the binding affinities of discrete docking domain pairs excised from the erythromycin (DEBS) and pikromycin (Pik) PKSs, using surface plasmon resonance and fluorescence polarization methods. In addition, we report the first X-ray crystal structure of a member of the recently proposed H2–T2 class of PKS docking domains, derived from the interface between PikAIII (module 5) and PikAIV (module 6) proteins from the Pik PKS system (Figure 1b)(25). Combining structural characterization of the PikAIII/PikAIV interface with discrete docking domain affinity measurements, we provide evidence in support of the prevailing model wherein the binding specificity that determines the linear arrangement of proteins in the biosynthetic assembly line is encoded in these small, terminal docking domains. Finally, we present a model for the observed docking domain specificity across a matrix of interacting pairs from the pikromycin and erythromycin pathways.
RESULTS AND DISCUSSION
Binding affinities of discrete docking domains via surface plasmon resonance
To test the capacity of discrete docking domains to discriminate between possible partners within a single biosynthetic pathway and/or between related pathways, we produced peptides corresponding to each ACPdd and KSdd region of the pikromycin and erythromycin PKS pathways (Figure 1a–b). Peptides were overexpressed in E.coli and purified using a His6-affinity handle followed by removal of the His-tag via TEV protease cleavage where necessary. While each of the docking domain constructs resulted in stable, soluble protein, the yields of the PikAII KSdd and PikAIII KSdd were low. Hence, for these two peptides, chemical synthesis was employed to produce larger quantities. The ability of KSdds to bind native immobilized ACPdd partners was evaluated using surface plasmon resonance (SPR). Biosensors based on SPR technology have been used to measure binding interactions across a wide range of affinities between partners (including discrete docking domains from a related mixed PKS/NRPS megasynthase system) varying from small molecules to large protein complexes (26–28). In modular PKSs, individual docking domains are identifiable by considering sequences directly downstream from the ends of the C-terminal ACP domain or directly upstream from the conserved start sites of the N-terminal KS domain. Using multiple sequence alignments of a number of characterized type I PKS systems, we designed, overexpressed, and purified (and in two cases synthesized) a complete set of discrete ACPdds and KSdds from the erythromycin and pikromycin system (Supplementary Figure 1). In these studies, we used a noncovalent method to immobilize the N-terminally His-tagged ACPdds to a nickel-loaded NTA sensor chip (Figure 2a) (29). The measured affinity (KD) of His-tagged PikAIII ACPdd to the nickel-NTA surface was 4.0 ± 0.04 nM (Supplementary Figure 2). This binding was sufficiently tight to enable measurement of the desired ACPdd-KSdd interactions when paired with tagless KSdds in solution.
Figure 2.
Binding analysis of discrete ACPdds to matched and mismatched KSdds measured by SPR. a) SPR assay design: after immobilizing the ACPdds via their His-tags, varying concentrations of discrete KSdds were injected over the ACPdd and control surfaces. b–c) KDs were calculated using a one site binding model, Y = Bmax*X / (KD + X). Dose response curves were performed in triplicate, and the error bars are SEM.
After immobilization of ACPdd, equilibrium analysis of a variety of matched or mismatched docking domains pairs was performed using sequential injections of KSdd at varying concentrations. Using docking domains from the erythromycin and pikromycin PKSs, we measured KDs for the matched docking domain pairs between 70 – 130 μM (Figure 2). Additionally, we were able to calculate individual kinetic parameters for the PikAIII/PikAIV binding pair (kon = 3000 ± 1800 M−1s−1, koff = 0.21 ± 0.03 s−1, KD = 73 ± 43 μM) (Supplementary Figure 3) that were in good agreement with the equilibrium analysis. As a negative control, a PikAIII ACPdd construct lacking the final nine amino acids was unable to bind to its partner KSdd (PiKAIV) or any other KSdds. A similar C-terminal deletion of the PikAIII ACPdd was recently shown to be incompetent for production of narbonolide in an in vitro PikAIII/PikAIV chemoenzymatic system (30). Furthermore, studies with mismatched docking domains clearly demonstrate that the ability to discriminate between potential PKS protein partners is encoded within the docking domains themselves (Supplementary Figure 4).
Ultimately, docking domains function not as discrete peptides, but as small appendages on much larger proteins (Figure 1). In addition to testing the complete library of discrete ACPdds and KSdds from the pikromycin and erythromycin PKS systems, we extended our analysis of the PikAIII/PikAIV docking interface to the neighboring domains. Assigning affinity and specificity determinants to 1) the docking domains, 2) the neighboring catalytic domains, 3) the phosphopantethiene arm, and 4) the growing polyketide chain will begin to separate the importance of the correct protein-protein interaction from the questions of substrate specificity at the catalytic centers. Although binding of a larger KSdd-containing protein (PikAIV KSdd-KS-AT) to the His-tag immobilized PikAIII ACPdd via SPR was observed, we were unable to calculate affinity values due to the high background refractive index change exhibited. We thus sought an alternative method to address this question.
Binding affinities of discrete docking domains via fluorescence polarization
To assess the effect of larger protein complexes on docking domain binding affinity, a fluorescence polarization (FP) assay was employed (31). We empirically determined the best fluorophore placement through the addition of a cysteine residue at each of the four possible termini (N-terminus and C-terminus of PikAIII ACPdd and PikAIV KSdd). Inclusion of a single cysteine residue enabled site-specific labeling with iodoacetamide-BODIPY-FL. Titration of increasing concentrations of the unlabeled matched docking domain identified the C-terminus of PikAIII ACPdd (termed PikAIII ACPdd-FL) as the optimal fluorophore placement, as this tracer exhibited the largest change in FP upon protein binding. The binding affinity of the PikAIV KSdd for PikAIII ACPdd-FL measured using this method provided an independent confirmation of the discrete docking domain binding affinities generated using SPR (Figure 3). When the larger KSdd-containing PikAIV proteins were titrated against ACPdd-FL, a 2-10-fold increase in affinity was observed (Figure 3). However, a construct consisting of only the KS domain of PikAIV (without its docking domain) did not bind to PikAIII ACPdd-FL (data not shown). Most likely, the presence of downstream domains in these longer constructs stabilizes the productive binding conformation of the PikAIV KSdd. Furthermore, it is possible that additional protein-protein contacts exist between the upstream ACPdd and the downstream KS-AT region of the module, although these regions have yet to be identified (10, 22).
Figure 3.
Binding analysis of fluorescent PikAIII ACPdd to PikAIV KSdd-containing proteins measured by FP. a) FP assay design. 50 nM PikAIII ACPdd-FL was mixed with varying concentrations of KSdd, KSdd-KS-AT, or full module PikAIV, and allowed to equilibrate at room temperature before reading. b–c) KDs were calculated using a one site binding model, Y = Bmax*X / (KD + X). Dose response curves were performed in triplicate, and the error bars are SEM.
Our in vitro binding affinities for these canonical modular PKS docking domains are similar to those measured by SPR and ITC for the orthogonal discrete TubB/TubC docking elements (KD ~50 μM), domains found in some mixed-PKS-NRPS synthetases, whose novel structure was reported recently (28). Additionally, the affinity of the PikAIV full module for PikAIII ACPdd as assessed by fluorescence polarization (5 ± 1 μM) is comparable to that estimated for the DEBS 1 / DEBS module 3+TE obtained by monitoring rates of tri- and tetraketide lactone synthesis (2.6 μM) in vitro (32). Thus, correct pairing of large multi-domain modules in both PKS and mixed PKS-NRPS biosynthetic assembly lines appears to result, at least in part, from specificity determinants with rather weak affinities. Despite these weak affinities, discrete docking domains from the related phoslactomycin (Plm) biosynthetic cluster have been used to separate the trimodular PikAI PKS (Figure 1b) into monomodular proteins in a Streptomycesvenezuelae strain lacking pikAI (J. Yan, S. Gupta, DHS, KAR, unpublished results). Remarkably, generation of the final macrolide products (methymycin and pikromycin) were within two-fold of the total yield compared with production when using native PikAI. How the bacteria achieve such exquisite selectivity albeit with only modest protein-protein affinities remains poorly understood. However, one clue might come from the analysis of the PksX megacomplex, a mixed PKS-NRPS responsible for producing bacillaene. In this system, the proteins of the biosynthetic machinery have been visualized via fluorescence microscopy to reside at a single organelle-like complex in the bacteria, perhaps suggesting that higher order multivalent interactions are available to further increase the affinities if needed (33).
Structure of the PikAIII/PikAIV docking interface
We next explored the structural basis for the observed binding specificity between the discrete pikromycin and erythromycin docking domain pairs. Given the low sequence similarity between the structurally characterized H1–T1 class and the uncharacterized H2–T2 class of PKS docking domains, we targeted the recently proposed H2–T2 class for structure determination (24). A direct fusion strategy had been used successfully to generate a construct to solve the DEBS 2/DEBS 3 solution structure (18). To characterize the low-affinity PikAIII/PikAIV docking domain complex, we generated constructs where the C-terminus of PikAIII ACPdd was either directly fused to the N-terminus of PikAIV KSdd or separated by one or two Gly-Gly-Gly-Ser spacers. In the PikAIII/PikAIV system, this docking domain fusion strategy yielded proteins that were highly soluble, and purification yielded 25–75 mg protein/L of culture (data not shown). Docking domain constructs derived from PikAIII/PikAIV containing all four predicted helices eluted as two oligomeric species on size exclusion chromatography, but these proteins failed to form crystals. This is likely due to the existence of mobile linker regions, as were found in the DEBS 2/DEBS 3 docking complex (18). We then targeted a smaller construct focused only on the putative inter-polypeptide docking helices (amino acids 1534–1562 of PikAIII ACPdd fused to amino acids 1–37 of the KSdd of PikAIV, together termed P3P4dock) (Figure 4). The P3P4dock crystal structure was solved by single wavelength anomalous diffraction using selenomethionyl protein. The 1.75 Å crystal structure of P3P4dock includes residues 1544–1562 PikAIII ACPdd and 1–37 of PikAIV KSdd, whereas residues 1534–1543 were disordered and remain unresolved.
Figure 4.
a–b) Packing of the PikAIII ACPdd/PikAIV KSdd crystal structure (PDB code, 3F5H). Three P3P4dock dimers are shown. The docking interaction formed by neighboring dimers is highlighted. c) In the NMR structure of the DEBS 2 ACPdd – DEBS 3 KSdd (pdb code, 1pzr), the docking interaction is intramolecular. b–c) Polypeptide chains are colored blue to red from the N-terminus to the C-terminus of the construct. d) P3P4dock sequence; top line is residual purification tag (SN) followed by PikAIII (residues 1534–1562), bottom line is PikAIV (residues 1–37). ACPdd are indicated below the sequence.
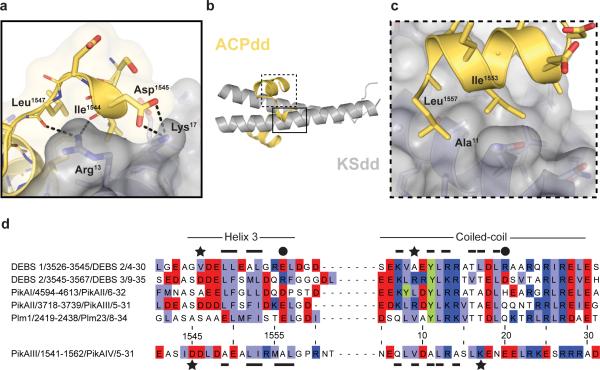
The P3P4dock protein structure consists of a short helix bound to a parallel coiled-coil (Figure 4b and 4c) (18). The relevant docking interface is made up of a coiled-coil of a single homodimer flanked by two individual ACPdd helices from neighboring protein molecules in the crystal lattice (Figure 4b and Supplementary Figure 5). The coiled-coil packing exhibits the familiar heptad repeat architecture with the “a” and “d” amino acids forming the core of the coiled-coil, and the “e” and “g” positions providing the majority of the residues for contacting the upstream PiKAIII ACPdd helix (Supplementary Figure 5d–f). The dominant interaction of the PikAIII ACPdd helix occurs in a hydrophobic patch on the PikAIV KSdd coiled-coil (Supplementary Figure 5b–c). The interacting hydrophobic surfaces display exquisite shape complementarity (Figure 5). Additional inter-domain interactions are found where residues 1544–1547 of PikAIII ACPdd fold back to interact further downstream on the KSdd dimer (Figure 5a–b). This positioning of residues 1544–1547 in PikAIII ACPdd is mediated by a charge-charge interaction between Asp1545 of PikAIII and Lys17 of PikAIV, as well as hydrogen bonds between main chain carbonyls from Ile1544 and Leu1547 of PikAIII and Arg13 of PikAIV (Figure 5a). No other charge-charge interactions are seen at the PikAIII/PikAIV docking interface. These electrostatic interactions and remarkable shape complementarity represent a potential selectivity filter (Figure 4d).
Figure 5.
Shape complementarity in PKS docking domains. a–c) Docking interface of PikAIIIPikAIV; KSdd of PikAIV is colored grey and the terminal helix of PikAIII ACPdd is light yellow. d) Multiple sequence alignment of the docking domains tested (or highlighted) in this study generated in JalView. Residue numbering is shown for the PikAIII/PikAIV pairing. Basic residues are colored dark blue, acidic residues are red, hydrophobic amino acids are light blue, and tyrosine is highlighted in green. Regions of interaction for each subclass are denoted by matching symbols.
The PikAIII/PikAIV docking domain structure revealed an overall architecture similar to that of the DEBS 2/DEBS 3 docking domain model obtained via NMR spectroscopy (Figure 4b and 4c) (18). In both the PikAIII/PikAIV and DEBS 2/DEBS 3 docking domain structures, the ACPdd helix binds to the KSdd coiled-coil approximately 30 Å (Fig. 4a) from the downstream KS catalytic domain (not present in either structure). However, many details of the structures differ. The most apparent structural difference between the PikAIII/PikAIV and DEBS 2/DEBS 3 docking domains is the length of the terminal ACPdd helix (Figures 4–5). The 9-residue PikAIII ACPdd helix (residues 1549–1557) is considerably shorter than its 15-residue DEBS 2 ACPdd counterpart. Although both the PikAIII/PikAIV and DEBS 2/DEBS 3 docking domain interfaces display well-defined shape complementarity between matched pairs, the mis-matched pairs appear highly non-complementary. Polar interactions also differ between the two interfaces.
The classification of PKS docking domains proposed by Thattai et al. is generally consistent with the two experimental structures of paired docking domains in which DEBS 2/DEBS 3 is type H1–T1 and PikAIII/PikAIV is type H2–T2 (Figure 5d). For example, the residues analogous to PikAIII Asp1545 and PikAIV Lys17 of the H2–T2 subclass are most frequently an Asp/Lys pair. In contrast, small or hydrophobic residues occupy those positions in the H1–T1 subclass of PKS docking domains (Figure 5d, Supplementary Figure 6). In addition, the key residues involved in the hydrophobic interface are shifted between the proposed H1–T1 and H2–T2 subclasses of PKS docking domains (see bars above the sequence alignments in Figure 4d). Furthermore, residue 11 is an alanine in PikAIV and all other H2–T2 KSdds, whereas in over 90% of H1–T1 KSdds the analogous residue is a tyrosine (24) (Figure 5 and Supplementary Figure 6). On the ACPdd helix, the large, hydrophobic residues Ile1553 and Leu1557 are across the interface from Ala11 of PikAIV KSdd (Figure 5c). Due to the size of these residues on the ACPdd helix, accommodating the tyrosine side chain of an H1–T1 KSdd at the position analogous to Ala11 of PikAIV KSdd appears unfavorable and likely represents another selectivity filter between the H1–T1 and H2–T2 subclasses. Altogether, these data lend support to the hypothesis that H1–T1 and H2–T2 are structurally distinct subclasses of PKS docking domains.
Specificity within the H1–T1 class appears to be driven by three distinct interaction zones. The hydrophobic core of the protein-protein interface is symbolized (Figure 5d) by solid bars above both the ACPdd and the KSdd in the multiple sequence alignment. On either side of the hydrophobic core are positions of potential charge-charge interaction (Figure 5d, stars & circles). Mismatching at any of the three zones appears to be sufficient to inhibit non-cognate docking domains from binding productively to one another in vitro. For example, at the “star” position, the DEBS 2/DEBS 3 pair and the PikAII/PikAIII pair both contain the same attractive charge-charge pair (Asp/Arg). However, at the “circle” position, a mismatched PikAII/DEBS 3 pairing would bring two negatively charged residues (Glu/Asp) in close proximity. A report of productive association and transfer between PiKAII and DEBS 3 in vivo suggests that within the H1–T1 class, inter-polypeptide interactions beyond the docking domains may also come into play (16), although further structural work is needed to identify additional contact regions.
An increased understanding of the key steps involved in PKS-mediated intermodular acyl group transfer provides a number of exciting opportunities. As the number of orphan non-collinear biosynthetic clusters rises with the completion of microbial genome sequencing projects, the ability to sequentially order the polypeptides via prediction of docking domain compatibility could enable more facile prediction of core polyketide structures. Combined with the predictive tools already in place for PKS catalytic domains, this enhanced analytical power should enable more accurate assignment of individual pathway metabolic products. The ultimate goal is to design and build hybrid PKS systems utilizing heterologous module pairs in a combinatorial fashion. To achieve more efficient polyketide production and the generation of novel drug-like products, we will be required to combine the lessons learned for optimizing key protein-protein interactions at the inter-polypeptide interface and those related to identification of catalytic domains capable of processing non-native substrates.
METHODS
Design of expression constructs
Plasmids for the expression of the discrete docking domain fragments, PikAIV KSdd-KS-AT, and the full module of PikAIV were generated by amplification using PCR with LIC overhangs and inserted into the vector pMCSG7 (34). DEBS 1, DEBS 2, and DEBS 3 docking domains were amplified from cosmid pDHS-9746. PikAI ACPdd and KSdd were amplified from plasmid pDHS-0030. PikAII ACPdd and KSdd were amplified from plasmid pDHS-0805. PikAIII ACPdd, PikAIII ACPdd-C-FL and KSdd were amplified from plasmid pDHS-8011. PikAIV KSdd, KSdd-KS-AT, and the full module were amplified from plasmid pDHS-0137. All primers are listed in Supplementary Table 1. All PCR fragments were inserted in the vector pMCSG7 via ligation independent cloning. Similarly, a construct lacking the N-terminal docking domain, termed PikAIV KS, was amplified from plasmid pDHS0137 and inserted into pMocr (35). The C-terminus of the PikAIV discrete ketosynthase construct terminates at a position near that of a recently reported soluble DEBS module 3 KS (36).
A plasmid encoding the full PikAIII ACPdd fused to the PikAIV KSdd (pDHS-9672) was generated via sequential PCR amplification of 1) individual dd's PikAIII and PikAIV containing appropriate overlapping DNA at the ends using plasmid DNA for PikAIII (pDHS-8011) and PikAIV (pDHS-0137) and 2) the fused construct from PCR amplification of the combined fragments using outside primers. The plasmid pDHS-9570 (encoding P3P4dock) was generated by PCR amplification of a fragment of pDHS-9672 followed by insertion into the vector pMCSG7. All DNA sequences were confirmed by sequencing.
Expression and purification of docking domain proteins
Plasmids encoding TEV protease-cleavable N-terminal His6-fusion proteins were transformed into E. coli BL21(DE3) and grown at 37 °C in TB medium to an OD600 of ~1.0 in 2L flasks. The cultures were cooled to 18 °C, and isopropyl β-D-thiogalactopyranoside was added to a final concentration of 0.2 mM and grown 12–16 h with shaking. The cells were harvested by centrifugation and frozen at either −20 or −80 °C. Selenomethionyl protein was produced in a similar fashion using selenomethionine minimal medium (37). Cell pellets were thawed to 4 °C and resuspended in 5× volume of lysis buffer (20 mM HEPES, pH 7.8, 300 mM NaCl, 20 mM imidazole, 1 mM MgCl2, and ~100 mg CelLytic Express (Sigma-Aldrich)) before lysis via sonication. For discrete KSdds, Complete Protease Inhibitor Cocktail tablets (Roche) were added to the lysis buffer. Centrifugation at 25,000×g for 30 min provided clarified lysates. Proteins were purified using Ni-Sepharose affinity chromatography on an Akta FPLC. Briefly, after filtration of the supernatant through 0.45 μm membrane, the solution was loaded onto a 5-mL HisTrap nickel-nitrilotriacetic acid column. The column was washed with 10 column volumes of buffer A (20 mM HEPES, pH 7.8, 300 mM NaCl, 20 mM imidazole) and eluted with a linear gradient of buffer B (20 mM HEPES, pH 7.8, 300 mM NaCl, 400 mM imidazole). His-tag removal was achieved by TEV protease incubation overnight at 4 °C in HEPES buffered saline (20 mM HEPES, pH 7.8, 150 mM NaCl, HBS) or buffer A containing 1 mM TCEP. His-tagged peptides and TEV protease were removed by repassaging the solution over the HisTrap column. Flow-through fractions were pooled, concentrated, and loaded onto a HiLoad 16/60 Superdex 75 (GE Healthcare) column equilibrated with HBS. Fractions were combined, concentrated, frozen, and stored at −80 °C. Because many of the small peptides lack amino acids with appreciable absorbance at 280 nm, protein concentration was determined using the bicinchoninic acid (BCA) method using BSA as a standard. Protein yields varied from 1 – 75 mg/L of cell culture. PikAII and PikAIII KSdd were chemically synthesized by Genscript corporation. Proteins were further purified by size exclustion chromatography on the HiLoad 16/60 Superdex 75 as above to remove residual HPLC purification contaminants before using the peptides in binding assays. To assure that no undesired cleavage products were formed during TEV protease incubation, the PikAIV, DEBS2 and DEBS3 KSdds were subjected to high resolution mass spectrometry (data not shown). For each peptide, the observed molecular weight was consistent with cleavage exclusively at the predicted TEV protease site (Supplementary Figure 1).
Expression and purification of PikAIV KS, PikAIV KSdd-KS-AT and PikAIV full module
Proteins were expressed as described for the docking domains above except that the PikAIV full module construct was grown in BAP1 E.coli cells (38) to provide post-translational modification of its ACP domain. Proteins were purified as above using the following buffers. For cell lysis, lysis buffer with reductant (20 mM HEPES, pH 7.8, 300 mM NaCl, 20 mM imidazole, 0.5 mM TCEP 1 mM MgCl2, and ~100 mg CelLytic Express) was used. During FPLC purification, wash buffer was buffer C (20 mM HEPES, pH 7.8, 300 mM NaCl, 20 mM imidazole, 10% glycerol, 0.5 mM TCEP) and the elution buffer used was buffer D (20 mM HEPES, pH 7.8, 300 mM NaCl, 400 mM imidazole, 10% glycerol, 0.5 mM TCEP). Size exlclustion chromatography was performed on a a HiLoad 16/60 Superdex 200 (GE Healthcare) column equilibrated with storage buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol, 0.5 mM TCEP). Protein concentrations were determined using absorbance at 280 nm and calculated extinction coefficients (PikAIV KS, 1 A280 = 1.0 mg/ml; PikAIV KSdd-KS-AT, 1 A280 = 0.91 mg/ml; PikAIV full module, 1 A280 = 0.94 mg/ml) (Supplementary Figure 1).
Surface plasmon resonance assays
Sensor chips (NTA) and HBS-P buffer were purchased from GE Healthcare Life Sciences. SPR experiments were performed on a BIAcore 3000 instrument. Running buffer for SPR was HBS-P+E (10 mM Hepes, pH 7.4, 0.15 M NaCl, 0.005% surfactant P20, 50 μM EDTA). The surface was prepared for immobilization of ACPdd by activating with 12 μL of 500 μM NiCl2 in HBS-P. Both the loading concentration and contact time were empirically determined for each ACPdd so that the maximum amount of protein was immobilized on the chip and that this protein was stably bound for the course of the experiment. ACPdd concentrations used for loading varied depending on the protein between 50 nM – 1 μM. Typically, 700 – 1500 RU of ACPdd was bound to the Ni-NTA sensor chip for each experiment. To measure binding to ACPdd by SPR, solutions of KSdd in HBS-P+E were injected over the prepared surface as well as a nickel only flow cell at a flow rate of 10 μL/min. After multiple injections (8 – 10 concentrations), the surface was regenerated using 30 μL of 175 mM EDTA in HBS-P, pH 8.3. Maximum testable concentrations for the KSdds were limited by either the solubility of the peptide or its level of nonspecific binding to the nickel-only control lane. Kinetic data analysis was carried out using Scrubber2 (BioLogic Software) and BIAevaluation (GE Healthcare Life Sciences). Nonlinear curve fitting of the equilibrium binding response was carried out using GraphPad Prism software.
Fluorescence polarization assays
Labeled ACPdds were generated by reaction of BODIPY® FL C1-IA (N-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-yl)methyl)iodoacetamide) (Invitrogen) with cysteine-containing ACPdds. Briefly, 4 μL of 100 mM TCEP in water and 40 μL of 10 mM BODIPY® FL C1-IA in DMSO was added to 360 μL of 500 μM ACPdd in HBS. Reactions were protected from light and proceeded for 2 hours at room tempetature. Unreacted BODIPY® FL C1-IA was removed from the labeled protein by passing the mixture over a preequilibrated Zeba spin desalting column (Pierce) and dialyzing into HBS. FP assays were performed at 20 μL total volume in a low volume black opaque polystyrene plate (Matrix Technologies). Proteins (50 nM PikAIII ACPdd-FL tracer and varying concentrations of unlabeled KSdd-containing PikAIV constructs) were allowed to incubate together for 10 minutes at room temperature in HBS-P (10 mM Hepes, pH 7.4, 0.15 M NaCl, 0.005% surfactant P20). Fluorescence polarization measurements were made at high sensitivity setting on a SpectraMax M5 (Molecular Devices) using 485 nm excitation, 538 nm emission, and 530 nm cutoff filter. The G factor was determined experimentally by setting a standard of 50 nM fluorescein in 0.1 N NaOH to 20 mP. Nonlinear curve fitting of the equilibrium binding response was carried out using GraphPad Prism software. Control experiments using up to 1 mg/ml BSA confirmed that the polarization increase upon incubation of PikAIII ACPdd-FL with unlabeled PikAIV KSdd was due to a specific protein-protein interaction (data not shown).
Crystallization, data collection and structure determination
Initial screening with P3P4dock produced small crystals of needle morphology under a variety of conditions containing high concentrations of organic solvents such as dioxane and MPD. The best-diffracting native crystals grew in 4–8 weeks at 4 °C using hanging-drop vapor diffusion techniques. Similarly, selenomethionyl P3P4dock crystals grew in 1–2 weeks at 4 °C after microseeding with native crystals. For crystal growth, an equal volume of protein solution (2.5 –5 mg/ml) in HBS (20 mM HEPES, pH 7.8, 150 mM NaCl) was mixed with mother liquor containing 55% 2-methyl-2,4-pentanediol (MPD), 150 – 200 mM sodium acetate, pH 5.0. The crystals were harvested in loops and frozen in liquid N2. Diffraction data were collected at 100 K on GM/CA-CAT beamlines 23ID-B and 23ID-D at the Advanced Photon Source in the Argonne National Laboratory (Argonne, IL). The data were processed using the HKL2000 suite (39). Initial phasing by the single-wavelength anomalous diffraction (SAD) method was performed using data collected at the wavelength with strongest anomalous signal from a single selenomethionyl-labeled protein crystal (Table 1). To minimize radiation damage, the dataset was assembled from 45° wedges of data collected from multiple points along a single crystal using a 10-micron X-ray beam (40). The PHENIX software package located five of the six selenium atoms and approximately two-thirds of the structure was automatically built from the 3.0 Ǻ SAD-phased map (41). Two molecules were present in the asymmetric unit (Vm = 2.40, 49% solvent). Modeling was completed manually using COOT (42). The model was refined against the 1.75 Ǻ native dataset using REFMAC5 of the CCP4 suite (43–45). (Tables 1–2).
Table 1.
Diffraction Data
| Parameter | Native | SeMet |
|---|---|---|
| Space Group | C2221 | C2221 |
| Dimensions (Å) a,b,c | 59.0, 117.9, 41.8 | 59.7, 118.5, 41.9 |
| X-ray source | 23ID-D | 23ID-D |
| Wavelength λ (Å) | 0.97934 | 0.97940 |
| dmin(Å)a | 1.75 (1.81–1.75) | 2.80 (2.90–2.80) |
| Unique observations | 15,084 | 3,917 |
| Rmerge (%)a,b | 6.9 (50.7) | 12.3 (29.7) |
| 〈I/σ〉a | 15.6 (2.1) | 11.5 (4.3) |
| Completeness (%)a | 99.2 (98.0) | 100 (100) |
| Avg. redundancya | 3.6 (3.1) | 5.1 (5.1) |
Values in parenthesis are for outer shell
Rmerge = Σ|Ii−〈I〉|/ΣIi, where Ii is the intensity of the ith observation and 〈I〉 is the mean intensity
Table 2.
Refinement Statistics
| P3P4dock | |
|---|---|
| Date range | 50–1.75 |
| R/Rfreea,b | 0.201/0.250 |
| RMSD bond length (Å) | 0.011 |
| RMSD bond angle (Å) | 1.216 |
| Avg. Protein B-factor (Å2) | 24.4 |
| Avg. Solvent B-factor (Å2) | 39.5 |
| Wilson B (Å2) | 20.3 |
| Ramachandran plotc | |
| Favored | 100 |
| Allowed | 0.0 |
| Disallowed | 0.0 |
| Protein atoms | 919 |
| Water molecule | 151 |
| Other atoms | 1 |
R = Σ|Fo−|Fc||/Σ|Fo| where Fo is the observed structure factor and Fc is the calculated structure factor used in the refinement
Rfree = Σ|Fo−|Fc||/Σ|Fo| where Fo is the observed structure factor and Fc is the calculated structure factor from 5% of reflections not used in the refinement
From output of MOLProbity (45)
Sequence and structure analysis
Multiple sequence alignments were performed using the CustalX method within JalView software (46). Structural figures were generated with PyMOL (DeLano Scientific).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank C. Rath for assistance with mass spectrometry analysis of KSdds. BAP1 E. coli cells were a generous gift from C. Khosla (Stanford University). This work was funded by NIH grant R01 GM076477 and the H. W. Vahlteich Professorship (to D.H.S.) and by NIH grant R37 DK042303 (to J.L.S.). GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357.
Footnotes
Accession Code The atomic coordinates of the PikAIII / PikAIV complex have been made publicly available through the Protein Data Bank (www.rcsb.org/pdb) with the PDB ID 3F5H.
Supporting Information Available: This material is available free of charge via the Internet.
REFERENCES
- 1.Hill AM. The biosynthesis, molecular genetics and enzymology of the polyketide-derived metabolites. Nat. Prod. Rep. 2006;23:256–320. doi: 10.1039/b301028g. [DOI] [PubMed] [Google Scholar]
- 2.Shen B. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr. Opin. Chem. Biol. 2003;7:285–295. doi: 10.1016/s1367-5931(03)00020-6. [DOI] [PubMed] [Google Scholar]
- 3.Newman DJ, Cragg GM. Natural Products as Sources of New Drugs over the Last 25 Years. J. Nat. Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 4.Maier T, Jenni S, Ban N. Architecture of mammalian fatty acid synthase at 4.5 Å resolution. Science. 2006;311:1258–1262. doi: 10.1126/science.1123248. [DOI] [PubMed] [Google Scholar]
- 5.Smith S, Tsai SC. The type I fatty acid and polyketide synthases: a tale of two megasynthases. Nat. Prod. Rep. 2007;24:1041–1072. doi: 10.1039/b603600g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang Y, Kim C-Y, Mathews II, Cane DE, Khosla C. The 2.7-Angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11124–11129. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khosla C, Tang Y, Chen AY, Schnarr NA, Cane DE. Structure and mechanism of the 6-deoxyerythronolide B synthase. Annu. Rev. Biochem. 2007;76:11.11–11.27. doi: 10.1146/annurev.biochem.76.053105.093515. [DOI] [PubMed] [Google Scholar]
- 8.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 9.Reeves CD, Murli S, Ashley GW, Piagentini M, Hutchinson CR, McDaniel R. Alteration of the substrate specificity of a modular polyketide synthase acyltransferase domain through site-specific mutations. Biochemistry. 2001;40:15464–15470. doi: 10.1021/bi015864r. [DOI] [PubMed] [Google Scholar]
- 10.Weissman KJ, Müller R. Protein-protein interactions in multienzyme megasynthetases. ChemBioChem. 2008;9:826–848. doi: 10.1002/cbic.200700751. [DOI] [PubMed] [Google Scholar]
- 11.Tsuji SY, Cane DE, Khosla C. Selective protein-protein interactions direct channeling of intermediates between polyketide synthase modules. Biochemistry. 2001;40:2326–2331. doi: 10.1021/bi002463n. [DOI] [PubMed] [Google Scholar]
- 12.Worthington AS, Hur GH, Meier JL, Cheng Q, Moore BS, Burkart MD. Probing the compatibility of type II ketosynthase-carrier protein partners. ChemBioChem. 2008;9:2096–2103. doi: 10.1002/cbic.200800198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menzella HG, Reeves CD. Combinatorial biosynthesis for drug development. Curr. Opin. Microbiol. 2007;10:238–245. doi: 10.1016/j.mib.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Menzella HG, Carney JR, Santi DV. Rational design and assemby of synthetic trimodular polyketide synthases. Chem. Biol. 2007;14:143–151. doi: 10.1016/j.chembiol.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Kittendorf JD, Sherman DH. Developing tools for engineering hybrid polyketide synthetic pathways. Curr. Opin. Biotechnol. 2006;17:597–605. doi: 10.1016/j.copbio.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 16.Tang L, Fu H, McDaniel R. Formation of functional heterologous complexes using subunits from the picromycin, erythromycin and oleandomycin polyketide synthases. Chem. Biol. (Cambridge, MA, U. S.) 2000;7:77–84. doi: 10.1016/s1074-5521(00)00073-9. [DOI] [PubMed] [Google Scholar]
- 17.Floss HG. Combinatorial biosynthesis - potential and problems. J. Biotechnol. 2006;124:242–257. doi: 10.1016/j.jbiotec.2005.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broadhurst RW, Nietlispach D, Wheatcroft MF, Leadlay PF, Weissman KJ. The structure of docking domains in modular polyketide synthases. Chem. Biol. 2003;10:723–731. doi: 10.1016/s1074-5521(03)00156-x. [DOI] [PubMed] [Google Scholar]
- 19.Wu N, Tsuji SY, Cane DE, Khosla C. Assessing the balance between protein-protein interactions and enzyme-substrate interactions in the channeling of intermediates between polyketide synthase modules. J. Am. Chem. Soc. 2001;123:6465–6474. doi: 10.1021/ja010219t. [DOI] [PubMed] [Google Scholar]
- 20.Wu N, Cane DE, Khosla C. Quantitative analysis of the relative contributions of donor acyl carrieer proteins, acceptor ketosynthases, and linker regions to intermodular transfer of intermediates in hybrid polyketide synthases. Biochemistry. 2002;41:5056–5066. doi: 10.1021/bi012086u. [DOI] [PubMed] [Google Scholar]
- 21.Kumar P, Li Q, Cane DE, Khosla C. Intermodular communication in modular polyketide synthases: structural and mutational analysis of linker mediated protein-protein interactions. J. Am. Chem. Soc. 2003;125:4097–4102. doi: 10.1021/ja0297537. [DOI] [PubMed] [Google Scholar]
- 22.Weissman KJ. Single amino acid substitutions alter the efficiency of docking in modular polyketide biosynthesis. ChemBioChem. 2006;7:1334–1342. doi: 10.1002/cbic.200600185. [DOI] [PubMed] [Google Scholar]
- 23.Weissman KJ. The structural basis for docking in modular polyketide biosynthesis. ChemBioChem. 2006;7:485–494. doi: 10.1002/cbic.200500435. [DOI] [PubMed] [Google Scholar]
- 24.Thattai M, Burak Y, Shraiman BI. The origins of specificity in polyketide protein interactions. PLoS Comput. Biol. 2007;3:1827–1835. doi: 10.1371/journal.pcbi.0030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue Y, Zhao L, Liu HW, Sherman DH. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: architecture of metabolic diversity. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12111–12116. doi: 10.1073/pnas.95.21.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myszka DG. Kinetic, equilibrium, and thermodynamic analysis of macromolecular interactions with BIACORE. Methods Enzymol. 2000;323:325–340. doi: 10.1016/s0076-6879(00)23372-7. [DOI] [PubMed] [Google Scholar]
- 27.Papalia GA, Leavitt S, Bynum MA, Katsamba PS, Wilton R, Qiu H, Steukers M, Wang S, Bindu L, Phogat S, Giannetti AM, Ryan TE, Pudlak VA, Matusiewicz K, Michelson KM, Nowakowski A, Pham-Baginski A, Brooks J, Tieman BC, Bruce BD, Vaughn M, Baksh M, Cho YH, Wit MD, Smets A, Vandersmissen J, Michiels L, Myszka DG. Comparative analysis of 10 small molecules binding to carbonic anhydrase II by different investigators using Biacore technology. Anal. Biochem. 2006;359:94–105. doi: 10.1016/j.ab.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 28.Richter CD, Nietlispach D, Broadhurst RW, Weissman KJ. Multienzyme docking in hybrid megasynthetases. Nat. Chem. Biol. 2008;4:75–81. doi: 10.1038/nchembio.2007.61. [DOI] [PubMed] [Google Scholar]
- 29.Nieba L, Nieba-Axmann SE, Persson A, Hämäläinen M, Edebratt F, Hansson A, Lidholm J, Magnusson K, Karlsson ÅF, Plückthun A. BIACORE analysis of histidine-tagged proteins using a chelating NTA sensor chip. Anal. Biochem. 1997;252:217–228. doi: 10.1006/abio.1997.2326. [DOI] [PubMed] [Google Scholar]
- 30.Kittendorf JD, Beck BJ, Buchholz TJ, Seufert W, Sherman DH. Interrogating the molecular basis for multiple macrolactone ring formation by the pikromycin polyketide synthase. Chem. Biol. 2007;14:944–954. doi: 10.1016/j.chembiol.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owicki JC. Fluorescence polarization and anisotropy in high throughput screening: perspectives and primer. J. Biomol. Screen. 2000;5:297–306. doi: 10.1177/108705710000500501. [DOI] [PubMed] [Google Scholar]
- 32.Gokhale RS, Hunziker D, Cane DE, Khosla C. Mechanism and specificity of the terminal thioesterase domain from the erythromycin polyketide synthase. Chem. Biol. 1999;6:117–125. doi: 10.1016/S1074-5521(99)80008-8. [DOI] [PubMed] [Google Scholar]
- 33.Butcher RA, Schroeder FC, Fischbach MA, Straight PD, Kolter R, Walsh CT, Clardy J. The identification of bacillaene, the product of the PksX megacomplex in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1506–1509. doi: 10.1073/pnas.0610503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expression Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 35.Delproposto J, Majmudar CY, Smith JL, Brown WC. Mocr: A novel fusion tag for enhancing solubility that is compatible with structural biology applications. Protein Expression Purif. 2009;63:40–49. doi: 10.1016/j.pep.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen AY, Cane DE, Khosla C. Structure-based dissociation of a type I polyketide synthase module. Chem. Biol. (Cambridge, MA, U. S.) 2007;14:784–792. doi: 10.1016/j.chembiol.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guerrero SA, Hecht J-J, Hofmann B, Biebl H, Singh M. Production of selenomethionine-labelled proteins using simplified culture conditions and generally applicable host/vector systems. Appl. Microbiol. Biotechnol. 2001;56:718–723. doi: 10.1007/s002530100690. [DOI] [PubMed] [Google Scholar]
- 38.Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C. Biosynthesis of complex polyketides in a metabolically engineered strain of E.coli. Science. 2001;291:1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolec. Crystallogr., Pt. A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.Sanishvili R, Nagarajan V, Yoder D, Becker M, Xu S, Corcoran S, Akey DL, Smith JL, Fischetti RF. A 7 micron mini-beam improves diffraction data from small or imperfect crystals of macromolecules. Acta Crystallogr. Sect. D Biol. Crystallogr. 2008;64:425–435. doi: 10.1107/S0907444908001741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 44.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 45.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.