Abstract
Biomedical applications of carbon nanotubes have attracted much attention in recent years. Here, we summarize our previously developed protocols for functionalization and bioconjugation of single-walled carbon nanotubes (SWNTs) for various biomedical applications including biological imaging; using nanotubes as Raman, photoluminescence and photoacoustic labels; sensing using nanotubes as Raman tags and drug delivery. Sonication of SWNTs in solutions of phospholipid-polyethylene glycol (PL-PEG) is our most commonly used protocol of SWNT functionalization. Compared with other frequently used covalent strategies, our non-covalent functionalization protocol largely retains the intrinsic optical properties of SWNTs, which are useful in various biological imaging and sensing applications. Functionalized SWNTs are conjugated with targeting ligands, including peptides and antibodies for specific cell labeling in vitro or tumor targeting in vivo. Radio labels are introduced for tracking and imaging of SWNTs in real time in vivo. Moreover, SWNTs can be conjugated with small interfering RNA (siRNA) or loaded with chemotherapy drugs for drug delivery. These procedures take various times ranging from 1 to 5 d.
INTRODUCTION
Carbon nanotubes with various unique physical and chemical properties have shown interesting applications in many fields, including biomedicine1,2. Functionalized carbon nanotubes with water solubility and biocompatibility are able to cross cell membranes, shuttling a wide range of biologically active molecules including drugs, proteins, DNA and RNA into cells3–7. The cytotoxicity of carbon nanotubes is largely dependent on their surface functionalization, with minimal toxic effects for well-functionalized, serum-stable nanotubes1,8. We have shown that after intravenous injection into mice, well-functionalized single-walled carbon nanotubes (SWNTs) accumulated in reticuloendothelial systems (RES) are slowly excreted, mainly through the biliary pathway, without exhibiting obvious side effects to the treated mice9,10. Recently, in vivo cancer treatment in an animal model has been realized by carbon nanotube-based drug delivery11.
Carbon nanotubes are classified as single-walled carbon nanotubes (SWNTs) and multi-walled carbon nanotubes (MWNTs), depending on the number of graphene layers from which a nanotube is composed. SWNTs are quasi-one-dimensional quantum wires with sharp densities of electronic states (electronic DOS) at the van Hove singularities and generally have more attractive unique intrinsic optical properties than MWNTS. SWNTs can be used as optical tags for biomedical detection and imaging12–18. Ultra-sensitive ex vivo protein sensing with a detection limit as low as 1 fM using SWNT Raman tags has been achieved using the resonance Raman scattering property of SWNTs and surface-enhanced Raman scattering (SERS)14. The Raman scattering, near-infrared (NIR) photoluminescence and high optical absorbance of SWNTs have all been used for biomedical molecular imaging in vitro and in vivo15–18. Thus, carbon nanotube bioconjugates are promising nanomaterials for biomedical applications.
SWNTs may have various potential advantages over other nanomaterials in different areas of nanobiotechnology. As an optical tag in biological imaging, SWNTs can be used in Raman, NIR fluorescence and photoacoustic imaging of cells and animals12,13,15–18. Multimodality optical imaging could thus be achieved using SWNTs as the contrast agent. Quantum dots or surface-enhanced Raman scattering gold/silver nanoparticles normally only have a single imaging functionality19,20. In contrast to widely used fluorescent quantum dots, carbon nanotubes contain no heavy metals and thus have a safer chemical composition. In the area of drug delivery, SWNT-based siRNA delivery works for a large range of cells, including notoriously ‘hard-to-transfect’ human T cells4, which are inert to conventional liposomal transfection agents. SWNTs can be efficiently loaded with aromatic chemotherapy drugs such as doxorubicin (DOX) through supramolecular π–π stacking21, obtaining an ultra-high loading capacity superior to other drug carriers including liposomes and micelles. Moreover, the high optical absorbance of SWNTs can also be used in photothermal therapy22,23, which may potentially be combined with chemotherapy11,21 and gene therapy4,24, both delivered by SWNTs to treat cancer in a more efficient manner.
For biomedical applications, raw hydrophobic SWNTs must be functionalized to afford water solubility and biocompatibility. Our previous studies have uncovered that the behaviors of SWNTs in biological systems in vitro (such as cellular uptake) and in vivo (such as blood circulation time and biodistribution) are highly dependent on their surface chemistry4,9,25. Developing proper surface functionalization on SWNTs is thus the most critical step to produce nanotube bioconjugates for a desired application. There are two major types of functionalization protocols for SWNTs: covalent reactions or non-covalent coating by amphiphilic molecules on nanotubes. Various covalent functionalization reactions, such as oxidation26,27 of nanotubes and 1,3-dipolar cycloaddition28 on the nanotube sidewalls, have been developed to produce water-soluble nanotubes useful in certain biomedical applications such as drug delivery2. Although covalent chemical reactions often allow stable functionalization on carbon nanotubes, the properties of SWNTs are degraded when the nanotube sidewall is damaged, dramatically decreasing the Raman scattering and NIR fluorescence signals of SWNTs1. Therefore, covalently functionalized carbon nanotubes have been widely used in drug and gene delivery2,29, but are usually not ideal for sensing and imaging applications1. In contrast, the structure and optical properties of SWNTs are largely maintained when non-covalent functionalization is used. However, the stability and biocompatibility of many non-covalently functionalized SWNTs are not satisfactory. For example, SWNTs solubilized in small-molecule surfactants (e.g., sodium dodecyl sulfate, SDS) will aggregate and precipitate if excess coating molecules are removed. An ideal functionalization should impart SWNTs with high water solubility, biocompatibility, minimal damage of nanotube structure and functional groups available for further bioconjugation.
Our group has developed systematic protocols for SWNT functionalization and bioconjugation in the past few years. Raw SWNTs are non-covalently functionalized by amphiphilic polymers, such as phospholipid-poly(ethylene glycol) (PL-PEG)6,22. Functionalized SWNTs have excellent stability in the aqueous phase and are highly biocompatible. Targeting ligands including antibodies and peptides can be conjugated to SWNTs to recognize specific cell receptors, yielding targeted SWNT bioconjugates useful for biological sensing14 and imaging15–18. We have also developed a protocol to label SWNTs with radioactive isotopes to track and image nanotubes in vivo by positron emission tomography (PET). In addition, SWNT-based siRNA transfection can be achieved by conjugating siRNA to SWNTs through a cleavable disulfide bond4,6. Furthermore, aromatic drug molecules can be non-covalently loaded onto SWNTs by simple mixing for drug delivery21.
Here, we systematically summarize the nanotube functionalization and bioconjugation protocols developed and used in our previous studies. Although our bioconjugation strategies apply for a wide range of biomolecules, only a few model systems are chosen to illustrate those protocols. Arg–Gly–Asp (RGD) peptide and Herceptin anti-Her2 antibody are used as targeting ligands. 64Cu is reported as an example of radiolabeling SWNTs. Anti-CXCR4 siRNA is chosen for siRNA conjugation and delivery. Finally, DOX is shown as an aromatic drug, loaded onto SWNTs for drug delivery. These detailed protocols should be beneficial to scientists interested in further developing biological applications of novel nanomaterials.
Experimental design
Non-covalent functionalization of SWNTs by PL-PEG
SWNTs are non-covalently functionalized by sonication of raw, hydrophobic nanotubes in water solutions of amphiphilic polymers (e.g., PL-PEG)6,22. The hydrophobic lipid chains of PL-PEG are strongly anchored onto the nanotube surface, whereas the hydrophilic PEG chain affords SWNT water solubility and biocompatibility. After removal of excess PL-PEG molecules, functionalized SWNTs show excellent stability in various aqueous phases including water, physiological buffers (e.g., phosphate buffered saline, PBS), cell medium and whole serum. The concentration of a SWNT solution can be determined by its optical density at 808 nm measured by a UV–VIS–NIR spectrometer with a weight extinction coefficient of 0.0465 mg l–1 cm–1 (dividing the optical density at 808 nm by the extinction coefficient gives the concentration)22. The length distribution of functionalized SWNTs can be determined by an atomic force microscope (AFM). Those non-covalently functionalized SWNTs retain their Raman and NIR fluorescence properties and are useful in biological detection and imaging applications. The functional group (e.g., amine) on the PEG terminal is available for further bioconjugation (Fig. 1).
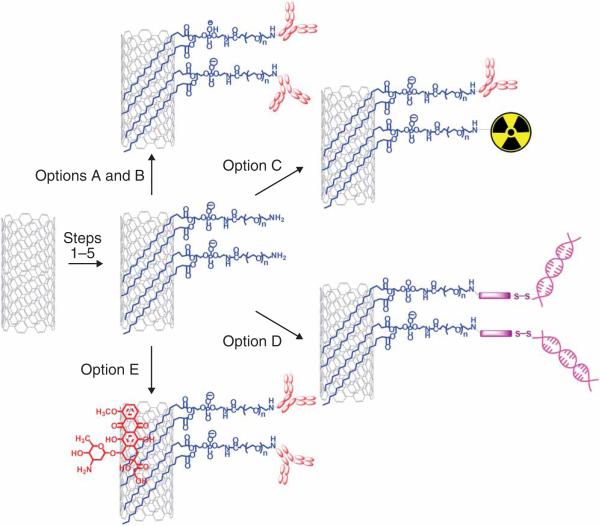
Figure 1.
Overview of the protocol. The protocol presented here contains five sub-protocols. Steps 1–5 are the functionalization of SWNTs. Step 6A and B are conjugation of targeting ligands to SWNTs. Step 6C is radiolabeling of SWNTs. Step 6D is siRNA conjugation to SWNTs. Step 6E is doxorubicin loading onto functionalized SWNTs.
Conjugation with targeting ligands
Targeting ligands including antibodies and peptides can be conjugated to SWNTs to recognize specific cell receptors (Fig. 2). Herceptin is a monoclonal antibody that binds specifically to the Her2/neu receptor over-expressed on a wide range of human breast cancer cells30. The RGD peptide targets integrin αvβ3 receptors that are upregulated on fast-growing tumor vasculature cells and many types of cancer cells31. The antibody Herceptin is first thiolated by Traut's reagent following standard protocols15,16 and used immediately after purification. The Traut's reagent reacts with amino groups on the antibody and produces active thiol groups useful for bioconjugation. Thiolated RGD peptide synthesized following a previously published protocol32 is used directly. The thiolated antibody or peptide should be protected from oxidation by adding EDTA to prevent heavy metal-catalyzed oxidization, or Tris-(2-carboxyethyl)phosphine hydrochloride (TCEP) as a reducing agent, during the conjugation with nanotubes. Maleimide groups are introduced onto SWNTs by reacting PL-PEG-amine-functionalized SWNTs with a sulfosuccinimidyl 4-N-maleimidomethyl cyclohexane-1-carboxylate (Sulfo-SMCC) bifunctional linker. The activated SWNTs are then reacted with thiolated antibodies or peptides, obtaining targeted SWNT bioconjugates, which can be used in multiplexed Raman spectroscopic imaging15,18, ultra-sensitive Raman detection of proteins14, NIR fluorescence imaging16, photoacoustic imaging17 and targeted photothermal therapy22,23. The targeting ability of SWNT bioconjugates (e.g., SWNT-RGD) can be characterized by in vitro Raman spectroscopic imaging experiments, in order to examine the staining of integrin αvβ3-positive U87MG cells and αvβ3-negative MCF-7 cells incubated with the SWNT-RGD conjugate.
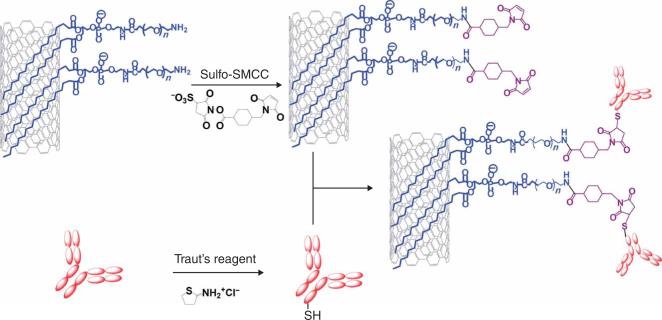
Figure 2.
A scheme showing conjugation of targeting ligands to SWNTs. PL-PEG5000-Amine-functionalized SWNTs are first activated by Sulfo-SMCC, yielding maleimide groups on SWNTs available for conjugation to thiolated antibodies or peptides. Thiolation of antibodies is carried out by treating them with Traut's reagent. Thiolated RGD peptide is synthesized following a previous protocol and used directly32.
Radiolabeling of SWNTs
To image and track SWNTs in vivo by PET, SWNTs are labeled with a radioactive isotope (Fig. 3)25. PET imaging provides three-dimensional distribution information of radiolabeled nanotubes in live mice in real time. To obtain RGD-conjugated radiolabeled SWNTs, SWNTs are first reacted with a mixture of sulfo-SMCC and N-hydroxysuccinimide (NHS) activated 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) and then conjugated to RGD-SH. After removal of excess reagents, 64Cu radioactive isotope can then be complexed to the DOTA rings on the SWNTs to achieve radiolabeling. The radiolabeled, targeted SWNT bioconjugate can then be used for in vivo PET imaging of mice bearing integrin αvβ3-positive, e.g., U87MG human glioblastomas tumors25. A total of 5–10 million of U87MG cells should be injected subcutaneously on the shoulder of a nude mouse. The mice can be used 2–3 weeks after tumor inoculation. PET imaging should be carried out at 0.5, 2, 4, 6 and 24 h post injection (p.i.). Mice may be killed at 24 h p.i. when the blood circulation of nanotubes has ended.
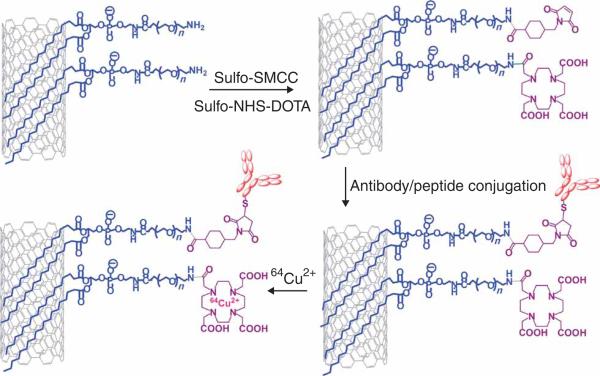
Figure 3.
A scheme showing radiolabeling of targeting SWNT bioconjugates. PL-PEG5000-Amine-functionalized SWNTs are reacted with a mixture of Sulfo-SMCC and Sulfo-NHS activated DOTA. Thiolated peptides or antibodies are then conjugated to maleimide groups on SWNTs. The targeting SWNT bioconjugate is then labeled by radioisotopes such as 64Cu through DOTA capture of metal ions.
siRNA conjugation to SWNTs through cleavable disulfide bond
The intracellular molecular delivery ability of SWNTs can be used for siRNA transfection4,6. In this example we chose to use CXCR4, a chemokine receptor that has an important role in the entry of HIV virus into human T cells33. SWNTs are first reacted with a bifunctional linker, Sulfosuccinimidyl 6-(3’-[2-pyridyldithio]-propionamido)hexanoate (Sulfo-LC-SPDP) and then conjugated with thiolated siRNA through a cleavable disulfide bond (Fig. 4). Two CXCR4 siRNAs with different sequences and a control luciferase siRNA are used. Dithiothreitol (DTT) is used to cleave any disulfide bond formed during storage of thiolated siRNA and removed by a NAP-5 column before the conjugation of siRNA with SWNTs. The SWNT–siRNA conjugates should be sterilized by centrifugation before cell incubation. Once transported into cells through endocytosis, siRNA is released from SWNTs by sulfide cleavage and then binds to CXCR4 mRNA to induce gene silencing. The CXCR4 receptor expression on CEM.NKR cells, a human T-cell line inert to commercial cationic liposome transfection agents34, is knocked down after cells are incubated with the SWNT–siRNAanti-CXCR4 conjugate for 3 d. The CXCR4 expression levels of cells can be determined by labeling cells with Phycoerythrin (PE)-anti-CXCR4 antibody and flow cytometry (FACS) measurement with dead cells excluded by propidium iodide (PI) staining. Control experiments using commercial cationic liposome-based transfection agents to transfect CEM.NKR with CXCR4 siRNA showed no obvious gene silencing effect. The mis-matched siRNA sequence (luciferase siRNA) also did not affect the CXCR4 expression.
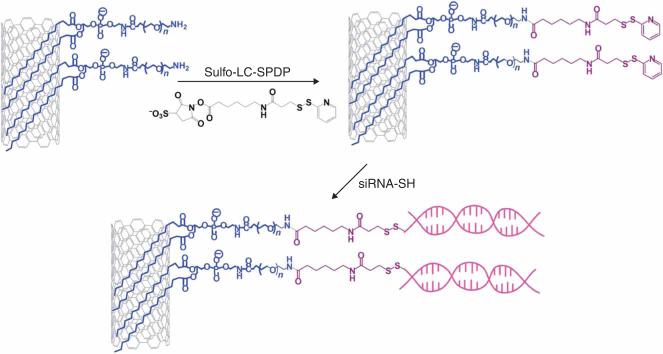
Figure 4.
A scheme showing siRNA conjugation to SWNTs through a disulfide bond. PL-PEG2000-Amine-functionalized SWNTs are activated by the Sulfo-LC-SPDP bifuncitonal linker. The pyridyl disulfide group can then be coupled to thiolated siRNA to create a disulfide linkage through a thiol exchange reaction.
DOX loading on functionalized SWNTs
SWNTs, with all atoms exposed on their surface, have an ultra-high surface area available for binding of aromatic molecules through supramolecular π–π stacking21. Functionalized SWNTs with or without targeting ligands can be loaded with DOX, an aromatic chemotherapy drug used for various types of cancers, by simply mixing the two solutions at a slightly basic pH. Excess unloaded DOX can be removed by filtration. The optical absorption of SWNT–DOX at 490 nm after subtraction of the SWNT absorption (at the same nanotube concentration) can be used to calculate the DOX concentration and loading in the SWNT–DOX complex21. On the basis of the UV–VIS–NIR absorption spectra, up to 4 g of DOX can be loaded on 1 g of SWNTs. The toxicity of SWNT–DOX is lower than that of free DOX but can be enhanced when conjugated with a targeting ligand such as RGD peptide for targeted drug delivery. Toxicity assays can be carried out by testing cell viabilities after incubating cells with free DOX, SWNT–DOX and SWNT–RGD–DOX at series of DOX concentrations using a CellTiter 96 kit (Promega). Samples should be measured in triplicate and the cell viability should be determined compared with that of the untreated control, defined as 100% viable.
MATERIALS
REAGENTS
As Produced HIPCO® single-walled carbon nanotubes (Unidym) ! CAUTION Hipco SWNTs are very light and could become airborne. Inhalation of SWNTs should be avoided. Wear goggles, lab coat and face mask during experiments.
PL-PEG5000-Amine (NOF Corporation, cat. no. DSPE-050PA)
PL-PEG2000-Amine (NOF Corporation, cat. no. DSPE-020PA)
RGD peptide (c(RGDyK); Peptides International)
Thiolated RGD peptide (see REAGENT SETUP)
Herceptin (Genentech)
Sulfo-SMCC (sulfosuccinimidyl 4-N-maleimidomethyl cyclohexane-1-carboxylate) (Pierce, cat. no. 22322)
Sulfo-NHS (N-hydroxysulfosuccinimide) (Pierce, cat. no. 24510)
Sulfo-LC-SPDP (N-Succinimidyl 3-(2-pyridyldithio)-propionate) (Pierce, cat. no. 21650)
Traut's Reagent (2-iminothiolane●HCl) (Pierce, cat. no. 26101)
EDC (1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride) (Pierce, cat. no. 22981)
DOTA (1,4,7,10-Tetraazacyclododecane-1,4,7,10-tetraacetic acid) (Sigma-Aldrich, cat. no. 86734)
DTT (d,l-dithiothreitol) (Sigma-Aldrich, cat. no. 43815)
Sodium bicarbonate (Sigma-Aldrich, cat. no. S6297)
Sodium hydroxide (Sigma-Aldrich, cat. no. 221465) ! CAUTION It is a corrosive strong base. Wear goggles, lab coat and face mask during experiments.
DMSO (dimethyl sulfoxide), 99.9%, anhydrous (Sigma-Aldrich, cat. no. 276855)
EDTA (ethylenediaminetetraacetic acid) solution (Sigma-Aldrich, cat. no. 03690)
TCEP (Tris(2-carboxyethyl)phosphine hydrochloride) (Sigma-Aldrich, cat. no. C4706)
Sodium acetate (Sigma-Aldrich, cat. no. S8750)
Acetic acid (Sigma-Aldrich, cat. no. 242853) ! CAUTION It is evaporative and corrosive. Wear goggles, lab coat and face mask during experiments. Handle acetic acid inside a hood.
Chelex 100 sodium form (Sigma-Aldrich, cat. no. C7901)
DOX (Doxorubicin HCl, Tecoland)
U87MG human glioblastoma cells (ATCC, cat. no. HTB-14; see REAGENT SETUP)
MCF-7 human breast cancer cells (ATCC, cat. no. HTB-22; see REAGENT SETUP)
BT-474 human breast cancer cells (ATCC, cat. no. HTB-20; see REAGENT SETUP)
CEM.NKR human T-lymphoblastoid cells (NIH Aids Reagents Program, cat. no. 795081; see REAGENT SETUP)
Low-glucose DMEM medium (Invitrogen, cat. no. 11885-084)
High-glucose DMEM medium (Invitrogen, cat. no. 21063-045)
RMPI-1640 medium (Invitrogen, cat. no. 11875-093)
Fetal bovine serum (FBS) (Invitrogen, cat. no. 10437-028)
Phosphate-buffered saline (PBS), 10×, pH 7.4 (Invitrogen, cat. no. 70011-069)
Phosphate-buffered saline (PBS), 1×, pH 7.4 (Invitrogen, cat. no. 10010-049)
Penicillin–streptomycin, liquid (10,000 U penicillin;10,000 μg streptomycin) (Invitrogen, cat. no. 15140-163)
CXCR4 siRNA: sequence-a: 5′-Thiol-GCGGCAGCAGGUAGCAAAGdTdT-3′; sequence-b: 5′-Thiol-AUGGAGGGGAUCAGUAUAUdTdT-3′. (Dharmacon RNAi Technology)
Luciferase siRNA (control): 5′-Thiol-CUUACGCUGAGUACUUCGAdTdT-3′. (Dharmacon RNAi Technology)
PE-anti-CXCR4 antibody (Caltag Laboratories, cat. no. MHCXCR404)
Propidium iodide (PI) solution, 1.0 mg ml–1 in water (Sigma-Aldrich, cat. no. P4864) ! CAUTION PI causes eye, skin and respiratory irritation and is harmful if swallowed. Wear goggles, lab coat and face mask during experiments.
Lipofectamine2000 (Invitrogen, cat. no. 11668019)
LipofectamineRNAiMAX (Invitrogen, cat. no. 13778075)
siPORT (Ambion, cat. no. AM4510)
HiPerFect (Qiagen, cat. no. 301704)
CellTiter 96 MTS assay kit (Promega, cat. no. G3580)
Isoflurane (RxElite, cat. no. NDC60307-120-25) ! CAUTION Isoflurane is a profound respiratory depressant. Wear goggles, lab coat and face mask during experiments. Handle isoflurane inside a hood when appropriate. Closely seal the bottle after use.
64CuCl2 radioactive isotope (University of Wisconsin-Madison) ! CAUTION Please obtain appropriate training for handling radioactive materials. Wear goggles, lab coat, mask, radiation dosimeter badge and rings during experiment. Handling of radioactive isotopes should only be performed in designated rooms for those experiments. Check any possible radioactive contamination after experiments.
U87MG tumor-bearing athymic nude mice (Harlan; see REAGENT SETUP) ! CAUTION Please obtain appropriate training regarding handling animals. Animal protocols must be in place before performing animal studies.
EQUIPMENT
Bath Sonicator (Cole-Parmer, cat. no. 08849-00)
Accu Spin 400 centrifuge (Fisher Scientific, cat. no. 75005194)
Sorvall Legend Mach 1.6R centrifuge (Kendro Laboratory Products, cat. no. 75004337)
Amicon centrifugal filter device 4 ml, 10,000 MWCO (Millipore, cat. no. UFC801024)
Amicon centrifugal filter device 4 ml, 100,000 MWCO (Millipore, cat. no. UFC810024)
Microcon Ultracel YM-100 filter device 0.5 ml, 100,000 MWCO (Millipore, cat. no. 42412)
Illustra NAP-5 columns, Sephadex G-25 DNA Grade (GE Healthcare, cat. no. 17-0853-01)
Flow cytometer (FACScan, Becton Dickinson)
Cary-6000i UV–VIS–NIR Spectrometer (Varian)
Tecan Spectrafluor Plus microplate reader (Tecan Group)
Confocal Raman microscope (Horiba Jobin Yvon)
Micro-PET R4 rodent model scanner (Concorde Microsystems)
REAGENT SETUP
Thiolated RGD (RGD-SH) peptide
Thiolated RGD (RGD-SH) peptide is prepared following a previously reported protocol32.
10 mM RGD-SH solution
Dissolve 1.2 mg of RGD-SH in 200 μl of water (~10 mM). Store the RGD-SH solution at –20 °C in small aliquots to avoid too many freeze–thaw cycles. The solution can be stable for up to 6 months if used and stored properly.
100 μM siRNA solution
Dissolve siRNA purchased from Dharmacon in the desired amount of RNase-free water to reach a siRNA concentration of 100 μM. Store siRNA solution at 20 °C in small aliquots to avoid too many freeze–thaw cycles. The solution can be stable for up to 6 months if used and stored properly.
0.5 M sodium bicarbonate (NaHCO3) buffer
Dissolve 8.4 g of sodium bicarbonate in 200 ml water. The solution can be stored at room temperature (~22 °C) in a plastic bottle and stable for 6 months.
0.1 M sodium hydroxide (NaOH) solution
Dissolve 1.0 g of sodium hydroxide in 250 ml water. The solution can be stored at room temperature in a plastic bottle and stable for 6 months. ! CAUTION Sodium hydroxide is a corrosive strong base. Wear goggles, lab coat and face mask during experiments.
0.1 M sodium acetate buffer (NaAcO, pH 6.5)
Dissolve 1.64 g of sodium acetate in 200 ml of water. Add 22 μl of acetic acid. Add 1 g of Chelex 100 beads into the buffer to avoid heavy metal ion contamination. Sodium acetate buffer can be stored for 6 months at room temperature. ! CAUTION Acetic acid is evaporative and corrosive. Wear goggles, lab coat and face mask during experiments. Handle acetic acid inside a hood.
Cell culture
Culture U87MG cells in DMEM (low glucose) supplemented with 10% (vol/vol) FBS and 1% (vol/vol) penicillin–streptomycin at 37 °C. Culture MCF-7 cells in DMEM (high glucose) supplemented with 10% (vol/vol) FBS and 1% (vol/vol) penicillin–streptomycin at 37 °C. Culture BT474 cells in DMEM (high glucose) supplemented with 5 g l–1 of glucose, 10% (vol/vol) FBS and 1% (vol/vol) penicillin–streptomycin at 37 °C. Culture CEM.NKR cells in RPMI-1640 supplemented with 10% (vol/vol) FBS at 37 °C. All cells are cultured in 5% CO2 atmosphere.
U87MG tumor model
Inject 5 × 106 U87MG cells subcutaneously into athymic nude mice (on their shoulders). Wait for 3–4 weeks before imaging, until tumor sizes reach 300–500 mm3. ! CAUTION Please obtain appropriate training regarding animal handling and have animal protocols in place before performing animal studies.
EQUIPMENT SETUP
Raman microscope
Confocal Raman spectroscopic imaging is carried out using a Horiba Jobin Yvon Raman confocal microscope with a 785-nm laser (80 mW) as the excitation light source. A × 50 objective was used for imaging with ~1 μm laser spot size. A 1-mm pin pole was applied to restrict the spatial resolution in z-axis to ~1 μm. Each Raman spectroscopic map contains at least 100 × 100 spectra with a 0.5-s integration time for each spectrum.
FACS machine
FACS measurement is carried out using a Becton Dickinson FACScan. A 488-nm laser is used as the excitation light source. Channels 1, 2 and 3 collect green, yellow and red fluorescence, respectively. A flow rate of 200–400 cells per s is used in the measurement.
PROCEDURE
Functionalization of SWNTs ● TIMING 1–2 d
-
1|
Weigh 1 mg of Hipco SWNTs and 5 mg of PL-PEG5000-Amine or PL-PEG2000-Amine into a 20-ml glass scintillation vial. Add 5 ml of water. Dissolve DSPE-PEG completely by shaking.
! CAUTION Hipco SWNTs are very light and could become airborne. Inhalation of SWNTs should be avoided. Wear goggles, lab coat and face mask during experiments.
-
2|
Sonicate the vial in a bath sonicator for 60 min at room temperature (~22 °C). Change the water in the water bath every 20 min to avoid overheating.
▲ CRITICAL STEP Make sure the vial is at the best position in the bath sonicator to ensure the most efficient sonication.
? TROUBLESHOOTING
-
3|
Centrifuge the SWNT suspension for 6 h, at 24,000g, room temperature. Collect the supernatant solution.
-
4|
Record the UV–VIS–NIR absorption spectrum of the obtained SWNT solution. The final SWNT concentration normally ranges from 40 to 70 mg l–1. Store the SWNT solution at 4 °C.
■ PAUSE POINT The PL-PEG-functionalized SWNT solution can be stored at 4 °C for 1–2 months before bioconjugation. It is highly recommended to store SWNT solutions in the presence of excess PL-PEG (e.g., before the removal of excess PL-PEG in Step 5).
-
5|
Add 1 ml of SWNT solution from the stock prepared in Step 3 into a 4-ml Amicon centrifugal filter device with a molecular weight cutoff (MWCO) of 100 kDa. Add 3 ml of water and centrifuge the device for 10 min, at 4,000g, room temperature. The leftover volume in the filter should be <0.5 ml. Fill the filter device with water to 4 ml. Wash 5–6 times by repeating the centrifuge/water adding steps, in order to completely remove excess PL-PEG in the SWNT solution. After the final wash step, measure the concentration of the SWNT solution by UV–VIS–NIR spectrometer with a weight extinction coefficient of 0.0465 l mg–1 cm–1 at 808 nm. Adjust the concentration of the SWNTs to ~50 mg l–1 by adding the required amount of water.
? TROUBLESHOOTING
-
6|
SWNTs can be functionalized with option A (targeting antibodies), option B (radiolabeling), option C (siRNA) and option D (DOX).
(A) Conjugation with RGD peptide ● TIMING 3 d
Dissolve 0.5 mg of Sulfo-SMCC in 50 μl of DMSO. Add 0.5 ml of 50 mg l–1 SWNT functionalized by PL-PEG5000-Amine solution from Step 5. Add 60 μl of PBS (10×). Let the reaction occur at room temperature for 2 h.
- Remove excess Sulfo-SMCC using an Amicon centrifugal filter device (MWCO = 100 kDa). Wash 5–6 times as described in Step 5. Add water to a volume of 0.5 ml.
- ▲ CRITICAL STEP The final SWNT solution should be immediately mixed with thiolated RGD.
- Dissolve ~2 mg of TCEP in 20–25 μl of 0.5 M sodium bicarbonate. Add sodium bicarbonate buffer slowly until the solution reaches pH 6. Measure the pH using pH test paper. Add the required volume (calculated by the exact weight of TCEP and the exact volume of sodium bicarbonate buffer added) of water so that the final TCEP concentration is 0.2 M.
- ! CAUTION Always prepare the TCEP solution immediately before use. TCEP-HCl solution in water has a very acidic pH. Wear goggles, lab coat and face mask during experiments.
Mix 0.5 ml of Sulfo-SMCC-activated SWNT solution from Step 6A(ii) with 50 μl of 10× PBS, 10 μl of 100 μM RGD-SH solution and 25 μl of the TCEP solution from Step 6A(iii). The final reaction solution (based on the volumes and initial concentrations of various solutions added into the reaction mixture) should have a SWNT concentration of ~40 mg l–1, RGD-SH concentration of ~0.2 mM and TCEP concentration of ~10 mM. Allow the reaction to proceed for 24 h at 4 °C.
- Remove excess RGD and TCEP in the above solution using an Amicon centrifugal filter device (MWCO = 100 kDa). Wash 5–6 times as described in Step 5. Store the SWNT–RGD conjugate at ~50 mg l–1 in water at 4 °C.
- ■ PAUSE POINT The SWNT–RGD conjugate can be stored at 4 °C for 2–3 weeks without losing targeting activity.
The SWNT–RGD conjugation can be verified by cell staining. Collect U87MG cells and MCF-7 cells by trypsinization from separate culture flasks (T25), aliquot, and gently centrifuge for 7 min in 1.5 ml centrifuge tubes, at 300g, room temperature. Resuspend the pellet in RMPI-1640 cell medium with 10% (vol/vol) FBS. Count the number of cells to ensure a density of ~1 million cells per ml.
Add 50 μl of SWNT-RGD from Step 6A(v) to 200 μl of U87MG cells (positive) and 200 μl of MCF-7 cells (negative). Incubate for 1 h at 4 °C.
- Centrifuge the cells for 7 min at 300g, room temperature and remove the supernatant. Wash the cells with 200 μl of 1× PBS and centrifuge as before, repeat the wash step three times.
- ■ PAUSE POINT The cells may be stored in 1× PBS at 4 °C for a few hours before Raman imaging without changing the Raman imaging results.
For Raman imaging, seal a drop of the cell suspension between two thin plastic coverslips.
Carry out Raman spectroscopic imaging of the labeled SWNTs using a Horiba Jobin Yvon Raman confocal microscope15.
(B) Conjugation with targeting antibodies ● TIMING 2 d
Dissolve 0.5 mg of Sulfo-SMCC in 50 μl of DMSO. Add 0.5 ml of SWNT functionalized by PL-PEG5000-Amine solution from Step 5. Add 60 μl of PBS (10×). Let the reaction occur at room temperature for 2 h.
- Remove excess Sulfo-SMCC using an Amicon centrifugal filter device (MWCO = 100 kDa). Wash 5–6 times as described in Step 5. Add water to a final volume of 0.5 ml.
- ▲ CRITICAL STEP The final SWNT solution should be immediately mixed with thiolated Herceptin.
Immediately after the Sulfo-SMCC reaction with SWNTs is started (Step 6B(ii)), weigh 1–2 mg of Traut's reagent into a 2-ml plastic tube. On the basis of weight, add the desired volume of water so that the final concentration of Traut's reagent is 5 mM. In a plastic tube, add 6 μl of 140 μM Herceptin, 10 μl of 1× PBS, 1.7 μl of Traut's reagent solution and 1 μl of 0.5 M of EDTA solution. The molar ratio of Traut's reagent/antibody is about 10:1. Incubate the reaction solution for 1.5–2 h at 4 °C.
- Add 300 μl of 1× PBS into the solution prepared in Step 6B(iii), remove the excess Traut's reagent by filtration using a Microcon Ultracel YM-100 filter device. Centrifuge for 6–8 min at 10,000g, room temperature until the leftover volume is <10 μl.
- ▲ CRITICAL STEP Use the thiolated Herceptin immediately.
- Mix the thiolated Herceptin from Step 6B(iv) with 0.5 ml of Sulfo-SMCC-modified SWNTs from Step 6B(ii). Add 50 μl of 10× PBS and 2 μl of 0.5 M EDTA. Incubate the reaction solution for 24 h at 4 °C. The conjugate can be used directly without further purification.
- ■ PAUSE POINT The SWNT–Herceptin conjugate can be stored at 4 °C for 2–3 weeks without losing targeting activity.
(C) Radiolabeling of SWNTs ● TIMING 5 d
- Dissolve 2 mg of DOTA in 50 μl of 0.1 M NaOH. Dissolve 2 mg of Sulfo-NHS and 1.5 mg of EDC in 50 μl of water. Add 30 μl of the Sulfo-NHS/EDC solution to the 50 μl of DOTA solution. Incubate for 15 min at room temperature. Check the solution pH using pH test paper. The pH in the incubation solution should be pH 5–6. Slowly add more 0.1 M of sodium hydroxide solution if the pH is below 5. Molar ratio: DOTA/EDC/Sulfo-NHS of ~1:1:1.2.
- ! CAUTION Sodium hydroxide is a corrosive strong base. Wear goggles, lab coat and face mask during experiments.
- ▲ CRITICAL STEP Use EDC and Sulfo-NHS immediately after they are dissolved in water.
Dissolve 0.5 mg of Sulfo-SMCC in 20 μl of DMSO and mix with 10 μl of the DOTA/EDC/Sulfo-NHS solution from Step 6C(i). Add 500 μl of PL-PEG5000-Amine-functionalized SWNT solution from Step 5. Add 60 μl of 10× PBS. Incubate the reaction solution for 2 h at room temperature.
Remove excess Sulfo-SMCC, DOTA, EDC and Sulfo-NHS using an Amicon centrifugal filter device (MWCO = 100 kDa). Wash 5–6 times as described in Step 5. Add water to a final volume of 0.5 ml.
Mix 0.5 ml of the SWNT solution from Step 6C(iii) with 50 μl of 10× PBS, 10 μl of 100 μM RGD-SH solution and 10 μl of 0.2 mM TCEP solution prepared in Step 6A(iii). The final reaction solution has a SWNT concentration of ~40 mg l–1, RGD-SH concentration of ~0.2 mM and TCEP concentration of ~10 mM. Let the reaction proceed for 24 h at 4 °C.
- Remove excess RGD and TCEP from the solution using an Amicon centrifugal filter device (MWCO = 100 kDa). Wash 5–6 times as described in Step 5. Measure the concentration of final SWNT–RGD solution by UV–VIS–NIR spectroscopy. Store the SWNT–RGD conjugate at ~50 mg l–1 in water at 4 °C.
- ■ PAUSE POINT The SWNT–RGD conjugate can be stored at 4 °C for 2–3 weeks without losing targeting activity.
- Dilute 10–20 mCi of 64CuCl2 in 0.5 ml of 0.1 M sodium acetate buffer (pH 6.5). Add a fraction of this solution (2 mCi activity) to 100 μl of RGD/DOTA co-conjugated SWNT solution prepared in Step 6C(v). Add 300 μl of 0.1 M sodium acetate buffer (pH 6.5). Incubate the reaction mixture for 1 h at 40 °C with constant shaking.
- ! CAUTION Obtain appropriate training for handling radioactive materials. Wear goggles, lab coat, mask, radiation dosimeter badge and rings during experiment. Handling of radioactive isotopes should only be performed in designated rooms for those experiments. Check for any possible radioactive contamination after experiments.
- Remove excess 64Cu by filtration using a Microcon Ultracel YM-100 filter device. Centrifuge for 6–8 min at 10,000g until the leftover volume is <10 μl. Wash 3–4 times by adding 200–300 μl of water and centrifuge for 6–8 min at 10,000g each time. Resuspend the final labeled SWNTs in 500 μl of 1× PBS (SWNT concentration ~10 mg l–1). Determine the radiolabeling yield by g-counting.
- ! CAUTION Radio isotope waste including contaminated devices such as filters should be collected and disposed in designated containers shielded with lead.
- ? TROUBLESHOOTING
- Inject 150 μl of ~10 mg l–1 radiolabeled SWNTs (Step 6C(vii)) into U87MG tumor bearing mice through the tail vein.
- ! CAUTION Obtain appropriate training regarding animal handling. Animal protocols must be in place before performing animal studies.
Image the mice under a micro-PET scanner at different time points p.i., e.g., 0.5, 2, 4, 6, 24 h. Process PET data according to a previously established protocol25.
- Kill mice 24 h p.i.
- ! CAUTION The animal bodies should be collected in a designated freezer for radioactive contaminated biohazardous waste. Label all animal cages clearly with radioactive marks. Leave contaminated cages in the radioactive work designated animal room for a week until a complete decay of 64Cu (half-life ~12.7 h) before cleaning. Check for any possible radioactive contamination after experiments.
(D) siRNA conjugation to SWNTs through cleavable disulfide bond ● TIMING 5–6 d
Mix 500 μl of PL-PEG2000-amine-functionalized SWNTs from Step 5 with 0.5 mg of Sulfo-LC-SPDP. Add 50 μl of 10× PBS. Incubate for 2 h at room temperature.
Immediately after Step 6D(i) is initiated, prepare 10 mM of DTT solution by dissolving 1.54 mg of DTT in 1 ml of water. Mix 15 μl of 100 μM siRNA (siRNACXCR4 or siRNAluc) with 1.5 μl of DTT solution. Allow the reaction to proceed for 1.5 h at room temperature.
- After Step 6D(i), remove the excess Sulfo-LC-SPDP from the SWNT solution using an Amicon centrifugal filter device (MWCO = 100 kDa). Wash 5–6 times by adding 3–4 ml DNase/RNase-free water and centrifuge for 6–8 min at 10,000g each time. The leftover volume in the filter should be <0.5 ml.
- ▲ CRITICAL STEP The obtained activated SWNTs should be used immediately for siRNA conjugation.
After Step 6D(ii), and in parallel with Step 6D(iii), purify DTT treated with the siRNA using a NAP-5 column following the manufacturer's protocol. Elute the siRNA from the column with 500 μl of DNase/RNase free 1× PBS.
- Resuspend the activated SWNTs from Step 6D(iii) with the 500 μl of purified siRNA solution from Step 6D(iv). Allow the conjugation to proceed for 24 h at 4 °C. The final SWNT and siRNA concentrations are ~40 mg l–1 and ~2.5 μM, respectively.
- ▲ CRITICAL STEP Use the SWNT–siRNA conjugate immediately after synthesis. This will reduce the chance of bacteria contamination and siRNA degradation.
For the siRNA transfection, plate CEM.NKR cells in a 24-well plate with 500 μl of cells per well. The cell density should be ~1 × 105 cells per ml.
To remove aggregates, centrifuge the SWNT–siRNA solutions prepared in Step 6D(v) for 10 min, at 10,000g, 4 °C. Collect the supernatant containing either SWNT–siRNACXCR4 or SWNT–siRNAluc conjugates and add 100 μl to each well of cells. The final SWNT and siRNA concentrations are ~10 mg l–1 and ~500 nM, respectively, in the cell medium.
Transfect CEM.NKR cells with 500 nM siRNACXCR4 using commercial transfection agents (e.g., Lipofectamine2000 (Invitrogen), LipofectamineRNAiMAX (Invitrogen), siPORT (Ambion) and HiPerFect (Qiagen)) following the manufacturers’ protocols.
- Incubate the cells at 37 °C, 5% CO2 for 3 d before analysis.
- ? TROUBLESHOOTING
- To analyze the effect of the RNAi, remove the medium and wash each well of the cells with 200 μl of 1× PBS twice by centrifuging for 7 min, at 300g, room temperature. Resuspend the cells in 200 μl of 1× PBS. Add 2 μl of PE-anti-CXCR4 antibody solution. Incubate for 1 h at 4 °C.
- The samples need to be centrifuged (7 min, 300g, room temperature) and collected before the wash step.
- Centrifuge the cells for 7 min at 300g, room temperature. Wash the cells with 200 μl of 1× PBS and centrifuge for 7 min, at 300g, room temperature to collect cells; wash the cells a total of three times. Dilute 5 μl of 1 mg ml–1 PI solution in 5 ml of 1× PBS. Resuspend the cells in 200 μl of the PBS containing 1 μg ml–1 PI.
- ! CAUTION PI causes eye, skin and respiratory irritation and is harmful if swallowed. Wear goggles, lab coat and face mask during experiments.
- ■ PAUSE POINT The stained cells can be stored at 4 °C for up to 4 h without losing substantial viability.
To analyze the cells by flow cytometry, measure PE and PI fluorescence by channel 2 (yellow) and 3 (red), respectively. Exclude PI-positive cells (dead cells) in the data analysis. Determine the CXCR4 expression levels on CEM.NKR cells after various treatments by calculating the mean fluorescence intensity of each cell sample.
(E) DOX loading on functionalized SWNTs ● TIMING 5–6 d
- Mix 0.5 ml of PL-PEG5000-Amine-functionalized SWNTs from Step 5 or RGD-conjugated SWNTs from Step 6A(v) with 30 μl of 10 mM DOX solution. Add 50 μl of 10× PBS and 2 μl of 0.5 M sodium bicarbonate buffer. The final pH is around 8. The final SWNT and DOX concentrations are ~40 mg l–1 and ~0.5 mM, respectively. Incubate the solution at 4 °C for 24 h.
- ▲ CRITICAL STEP Ensure that the pH is not too basic. The SWNT–RGD–DOX will have reduced stability if the pH is more than 9.
- Remove excess DOX using an Amicon centrifugal filter device (MWCO = 10 kDa). Wash at least six times using 3–4 ml of water each time until the filtrate solution appears to be almost colorless. Re-suspend SWNT–DOX in 0.5 ml of water. Centrifuge the SWNT–DOX solution for 10 min, at 10,000g, 4 °C to remove aggregates.
- ? TROUBLESHOOTING
- Record UV–VIS–NIR absorption spectra of SWNT (SWNT–RGD) and SWNT–DOX (SWNT–RGD–DOX) conjugates21. To determine DOX loading onto SWNTs, normalize the spectra by the absorption at 808 nm. DOX has a molar extinction coefficient of 1.05 × 104 M–1 cm–1. The final DOX loading should be 600–800 DOX molecules per SWNT.
- ■ PAUSE POINT The SWNT–DOX conjugate can be stored at 4 °C for 3–5 d without obvious loss of activity.
To determine the cell toxicity of the DOX-conjugated SWNTs, plate U87MG and MCF-7 cells in two 96-well plates with ~10,000 cells in 100 μl medium per well. Culture the cells overnight at 37 °C, 5% CO2. Prepare triplicate wells for each sample to be tested, plus triplicate wells for untreated control cells.
Centrifuge the SWNT–DOX solutions prepared in Step 6E(iii) for 10 min at 10,000g, 4 °C to remove any aggregates. Add a range of concentrations (1–40 μM) of free DOX, SWNT–DOX and SWNT–RGD–DOX into different wells of cells. Untreated cells are used as the control. Add each sample to triplicate wells. Incubate the plates for 1–2 h at 37 °C, 5% CO2.
Gently wash the cells with 200 μl of 1× PBS twice for 7 min at 300g, room temperature to remove the excess drug. Add 100 μl of fresh cell medium and incubate at 37 °C, 5% CO2 for 48 h.
Measure the cell viability in each well using a CellTiter 96 kit following the manufacturer's instructions. Measure the absorbance of the cells at 490 nm and determine the relative cell viabilities21.
● TIMING
Steps 1–5, Functionalization of SWNTs: 1–2 d
Step 6A, Conjugation of SWNTs with RGD peptide: 3 d
Step 6B, Conjugation of SWNTs with antibody (Herceptin): 2 d
Step 6C, Radiolabeling of SWNTs: 5 d
Step 6D, siRNA conjugation on SWNTs: 5–6 d
Step 6E, DOX loading on functionalized SWNTs: 5–6 d
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
TABLE 1.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 2 | SWNTs are not well suspended by PL-PEG | Sonication is not efficient because of inappropriate vial position in the sonicator | Adjust the position and angle of the vial in the sonicator. Ensure the water bath is at the recommended level |
| 5 | Nanotubes are very sticky on the filter device during filtration | A certain amount of stickiness is normal for PL-PEG2000-Amine functionalization but not for PL-PEG5000-Amine. This may be due to poor coating of PL-PEG molecules on nanotubes. | |
| The PL-PEG/SWNT ratio is not sufficient when making nanotube suspensions. | Reduce the amount of SWNTs or increase PL-PEG concentration during sonication | ||
| Water in bath is too hot during sonication | Change water in the water bath sonicator more frequently during sonication | ||
| 6C (vii) | The radiolabeling yield is very low (e.g., <30%) | This may be due to metal ion contamination in the SWNT solution | Dialyze the SWNT solution against deionized distilled water with a 10-kDa MWCO membrane for 2 d before radiolabeling |
| 6D (ix) | Cells are contaminated by bacteria after 3 d incubation | SWNT solution is not well sterilized before adding into the cell culture | Normally centrifuging SWNT-siRNA solution for 10 min at 10,000g will resolve this problem. Repeating the centrifuge step 2–3 times will be helpful. Carefully take out supernatant and discard any aggregates after centrifugation. Use sterilized containers during experiments |
| 6E(ii) | A large amount of SWNT–DOX complex precipitates during centrifugation | The DOX loading on nanotubes is too high in this case. This stability issue is more frequent when making SWNT–RGD–DOX | Reduce pH in the loading solution to 7.8–8.0 by adding less sodium bicarbonate buffer Add 10–20 μM of PL-PEG into the loading solution. This will increase the stability of SWNT–RGD–DOX and not affect DOX loading and RGD targeting |
ANTICIPATED RESULTS
Functionalization of SWNTs
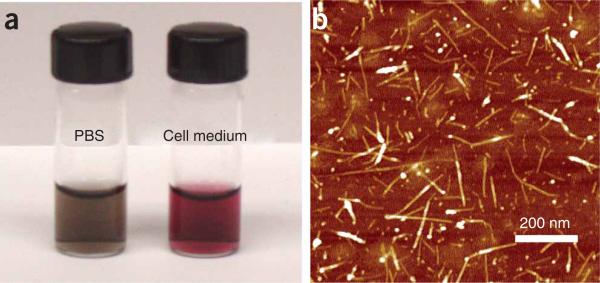
After Step 5, PL-PEG-functionalized SWNTs should have excellent water solubility and are stable in various biological solutions without any visible aggregation after a long period of incubation time (weeks) (Fig. 5). Atomic force microscope (AFM) images of SWNTs show that the lengths of SWNTs range from 50 to 300 nm, with an average of ~100 nm.
Figure 5.
Functionalization of SWNTs by PL-PEG. A photo showing PL-PEG2000-Amine-functionalized SWNTs in PBS and RPMI-1640 cell medium (10% (vol/vol) FBS). The non-covalently functionalized SWNTs exhibit excellent stability in various biological solutions even after removal of excess PL-PEG molecules from the solution (a). An Atomic Force Microscopy image of SWNTs on a silicon substrate. These SWNTs are mostly single nanotubes with a few small bundles, showing lengths of 50–300 nm (b). This figure is adapted from our previously published work and reproduced with permission from Wiley InterScience4.
Targeting SWNT bioconjugates for Raman imaging and sensing
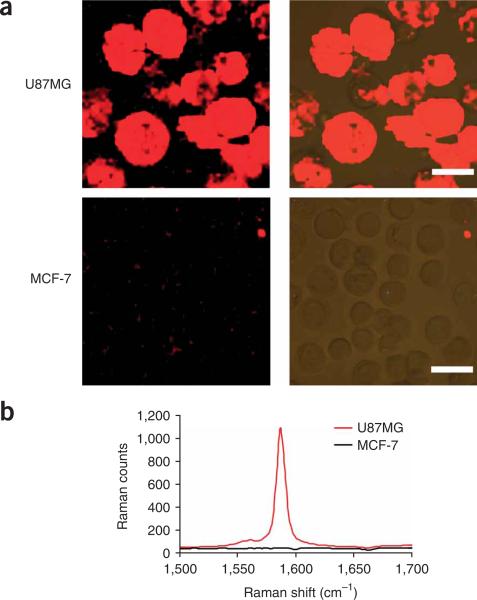
In 6A(vii), confocal Raman spectroscopic images of cells stained by SWNT–RGD should exhibit strong SWNT Raman signals on integrin αvβ3-positive U87MG cells but not on αvβ3-negative MCF-7 cells (Fig. 6a). Nonspecific binding of SWNTs to MCF-7 cells should be minimal. The ratio of Raman signals on positive versus negative cells is >40 (Fig. 6b). Raman imaging of BT474 (Her2-positive) and MCF-7 (Her2-negative) cells after staining with SWNT-Herceptin (Step 6B) should give very similar results (data not shown). Those targeted SWNT bioconjugates may also be used in ultra-sensitive protein microarray detection14.
Figure 6.
Targeting SWNT bioconjugates for cell labeling and Raman imaging. Confocal Raman images of U87MG (integrin αvβ3-positive) and MCF-7 (integrin αvβ3-negative) cells (left column) after incubation with SWNT–RGD for 1 h at 4 °C (a). Optical images overlaid with Raman images are shown in the right column. Scale bar = 20 mm. (b) Averaged spectra of two Raman images in panel (a). Very strong Raman G-band signals are observed on U87MG cells. Negative MCF-7 cells exhibit minimal nonspecific absorption signals of SWNT-RGD. This figure is adapted from previously published work14.
Radiolabeled SWNTs for in vivo PET imaging and tumor targeting
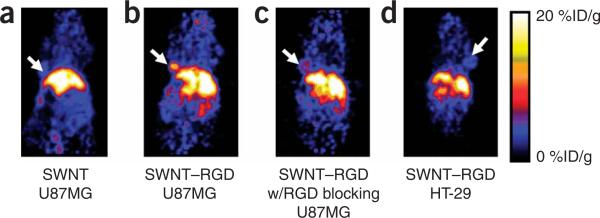
In Step 6C(ix), PET images (Fig. 7) at 6 h p.i. should show high tumor uptake (10–15%ID/g) in U87MG tumor bearing mice injected with SWNT–RGD (functionalized by PL-PEG5000)25. Radiolabeled SWNTs without RGD conjugation should show a reduced uptake in the tumor (3–5%ID/g) in comparison with RGD-conjugated SWNTs. Tumor uptake will be significantly reduced if mice are pre-injected with a high dose of free RGD peptide before injection of SWNT–RGD (4–6%ID/g). Control integrin αvβ3-negative HT29 tumors should have a lower uptake of SWNT–RGD (3–5%ID/g) (Fig. 7d).
Figure 7.
Radiolabeled SWNTs for in vivo PET imaging and tumor targeting radiolabeled nanotubes. Images are obtained at 6 h post injection of radiolabeled nanotubes. (a) Mice carrying U87MG tumors were injected with control radiolabeled SWNTs without RGD conjugation show low uptake in the tumor. (b) Mice carrying U87MG tumors were injected with radiolabeled SWNT–RGD, high tumor SWNT uptake is observed. (c) A control experiment showing mice carrying U87MG tumors injected with radiolabeled SWNT–RGD and co-injection of free c(RGDyK). (d) Mice carrying a tumor formed from HT-29 cells (integrin αvβ3-negative) injected with radiolabeled SWNT–RGD, showing low uptake of SWNT–RGD. Efficient tumor targeting is achieved by conjugating SWNTs with RGD peptide, which binds specifically to integrin αvβ3 expressed on tumor cells and tumor vasculature. The arrow points to the tumor. Animal experiments were conducted under the protocols of Administrative Panel on Laboratory Animal Care (APLAC) at Stanford University. This figure is adapted from our previously published work25.
SWNT-based siRNA transfection and RNAi effect
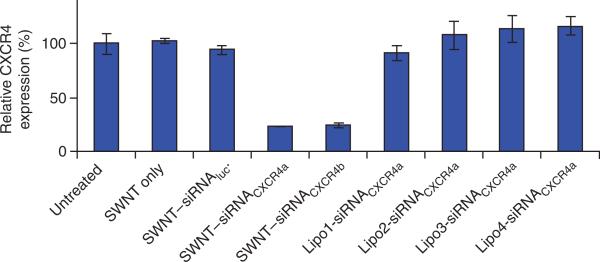
In Step 6D(vii), as shown in Figure 8, SWNT only and SWNT–siRNAluc mis-matched control-treated CEM.NKR cells show normal CXCR4 expressions4. The CXCR4 expression should be significantly reduced after treatment with SWNT–siRNACXCR4 (two sequences) compared with untreated cells. The RNAi effect should range from 70 to 90%. Other types of commercial cationic liposome-based siRNA transfection agents do not show significant siRNA transfection effects to CEM.NKR cells because human T cells are well known to be difficult to transfect.
Figure 8.
CXCR4 expression levels on CEM.NKR cells after various treatments. CEM.NKR cells were left untreated, treated with SWNTs only or SWNTS conjugated to a luciferase (Luc) siRNA control (SWNT–siRNAluc). Cells were also treated with SWNTs conjugated to two different CXCR4 siRNAs: SWNT–siRNACXCR4a or SWNT–siRNACXCR4b. Both siRNACXCR4a and siRNACXCR4b target CXCR4 mRNA but have different sequences. Cells were also treated with SWNT–siRNACXCR4a in combination with four types of commercial cationic liposome agents (Lipo1–4). The siRNA concentration used was 500 nM in the incubation solution, SWNTs were used at a concentration of 10 mg l–1. Cells were incubated for 3 d before the analysis of CXCR4 expression levels using FACS analysis. Human T cells are not transfected by liposome-based transfection agents. In contrast, the SWNT-conjugated siRNA transfection method was highly efficient at targeting CXCR4. The error bars represent the standard deviation (s.d.) of three samples. This figure is adapted from our previously published work and reproduced with permission from Wiley InterScience4.
DOX loading on functionalized SWNTs
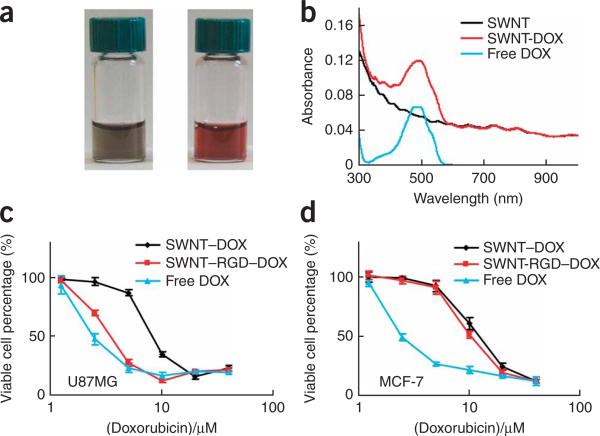
In Step 6E, the SWNT–DOX solution is reddish in color because of the UV–VIS absorption of DOX stacked onto SWNTs (Fig. 9a). The DOX absorption peak at 490 nm after subtraction of SWNT absorption at this wavelength is used to determine DOX concentration (Fig. 9b). In Step 6E(iv), SWNT–DOX has a lower toxicity than free DOX. Conjugation of RGD enhances the toxicity of DOX-loaded SWNTs to integrin αvβ3-positive U87MG cells but not to αvβ3-negative MCF-7 cells (Fig. 9c,d)21.
Figure 9.
Doxorubicin (DOX) on functionalized SWNTs for drug delivery. (a) Photos of PL-PEG-functionalized SWNT solutions with (right image) and without (left image) bound DOX. (b) UV–VIS–NIR absorbance spectra of solutions of free DOX (blue), SWNTs with PL-PEG functionalization (black), and PL-PEG SWNTs complexed with DOX (red) after incubation in a DOX solution at pH 8–9. The absorption peak at 490 nm is due to DOX π-stacked on SWNTs, and used for analyzing the amount of molecule loaded onto nanotubes. Concentration-dependent survival curves of U87MG cells (c) and MCF-7 cells (d) treated with the various samples indicated. The viable cell percentages are measured by the MTS assay (CellTiter 96 kit). SWNT–DOX has relatively lower toxic effect than free DOX to both types of cells, whereas SWNT–RGD–DOX exhibits increased toxicity to U87MG cells but not to MCF-7 cells. Cell viability was calculated by normalizing to the untreated control sample, which is determined as 100%. Error bars are based on standard deviations of triplicate samples. This figure is adapted from our previously published work and reproduced with permission from ACS Publications21.
ACKNOWLEDGMENTS
The multiple projects involved here were supported by a Stanford Graduate Fellowship, a Stanford Bio-X grant, CCNE-TR at Stanford University, NIH-NCI R01 CA135109-02 and Ensysce Biosciences Inc. Drs Nadine Wong Shi Kam, Sarunya Bangsaruntip, Xiaowu Tang, Xiaoming Sun, Xiaoyuan Chen, Weibo Cai and Ms Nozomi Nakayama have also contributed in the development of this protocol.
Footnotes
Published online at http://www.natureprotocols.com.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions.
References
- 1.Liu Z, Tabakman S, Welsher K, Dai H. Carbon nanotubes in biology and medicine: in vitro and in vivo detection, imaging and drug delivery. Nano Res. 2009;2:85–120. doi: 10.1007/s12274-009-9009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bianco A, Kostarelos K, Partidos CD, Prato M. Biomedical applications of functionalised carbon nanotubes. Chem. Commun. 2005:571–577. doi: 10.1039/b410943k. [DOI] [PubMed] [Google Scholar]
- 3.Kam NWS, Jessop TC, Wender PA, Dai HJ. Nanotube molecular transporters: internalization of carbon nanotube-protein conjugates into mammalian cells. J. Am. Chem. Soc. 2004;126:6850–6851. doi: 10.1021/ja0486059. [DOI] [PubMed] [Google Scholar]
- 4.Liu Z, Winters M, Holodniy M, Dai HJ. siRNA delivery into human T cells and primary cells with carbon-nanotube transporters. Angew. Chem. Int. Ed. Engl. 2007;46:2023–2027. doi: 10.1002/anie.200604295. [DOI] [PubMed] [Google Scholar]
- 5.Kam NWS, Liu ZA, Dai HJ. Carbon nanotubes as intracellular transporters for proteins and DNA: an investigation of the uptake mechanism and pathway. Angew. Chem. Int. Ed. Engl. 2006;45:577–581. doi: 10.1002/anie.200503389. [DOI] [PubMed] [Google Scholar]
- 6.Kam NWS, Liu Z, Dai H. Functionalization of carbon nanotubes via cleavable disulfide bonds for efficient intracellular delivery of siRNA and potent gene silencing. J. Am. Chem. Soc. 2005;127:12492–12493. doi: 10.1021/ja053962k. [DOI] [PubMed] [Google Scholar]
- 7.Kam NWS, Dai H. Carbon nanotubes as intracellular protein transporters: generality and biological functionality. J. Am. Chem. Soc. 2005;127:6021–6026. doi: 10.1021/ja050062v. [DOI] [PubMed] [Google Scholar]
- 8.Sayes CM, et al. Functionalization density dependence of single-walled carbon nanotubes cytotoxicity in vitro. Toxicol. Lett. 2006;161:135–142. doi: 10.1016/j.toxlet.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 9.Liu Z, et al. Circulation and long-term fate of functionalized, biocompatible single-walled carbon nanotubes in mice probed by Raman spectroscopy. Proc. Natl. Acad. Sci. USA. 2008;105:1410–1415. doi: 10.1073/pnas.0707654105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schipper ML, et al. A pilot toxicology study of single-walled carbon nanotubes in a small sample of mice. Nat. Nanotech. 2008;3:216–221. doi: 10.1038/nnano.2008.68. [DOI] [PubMed] [Google Scholar]
- 11.Liu Z, et al. Drug delivery with carbon nanotubes for in vivo cancer treatment. Cancer Res. 2008;68:6652–6660. doi: 10.1158/0008-5472.CAN-08-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heller DA, Baik S, Eurell TE, Strano MS. Single-walled carbon nanotube spectroscopy in live cells: towards long-term labels and optical sensors. Adv. Mater. 2005;17:2793–2799. [Google Scholar]
- 13.Leeuw TK, et al. Single-walled carbon nanotubes in the intact organism: near-IR imaging and biocompatibility studies in Drosophila. Nano Lett. 2007;7:2650–2654. doi: 10.1021/nl0710452. [DOI] [PubMed] [Google Scholar]
- 14.Chen Z, et al. Protein microarrays with carbon nanotubes as multicolor Raman labels. Nat. Biotechnol. 2008;26:1285–1292. doi: 10.1038/nbt.1501. [DOI] [PubMed] [Google Scholar]
- 15.Liu Z, et al. Multiplexed multi-color Raman imaging of live cells with isotopically modified single walled carbon nanotubes. J. Am. Chem. Soc. 2008;130:13540–13541. doi: 10.1021/ja806242t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welsher K, Liu Z, Daranciang D, Dai H. Selective probing and imaging of cells with single walled carbon nanotubes as near-infrared fluorescent molecules. Nano Lett. 2008;8:586–590. doi: 10.1021/nl072949q. [DOI] [PubMed] [Google Scholar]
- 17.de la Zerda L, et al. Photoacoustic molecular imaging in living mice utilizing targeted carbon nanotubes. Nat. Nanotechnol. 2008;3:557–562. doi: 10.1038/nnano.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zavaleta C, et al. Noninvasive Raman spectroscopy in living mice for evaluation of tumor targeting with carbon nanotubes. Nano Lett. 2008;8:2800–2805. doi: 10.1021/nl801362a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian XM, et al. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat. Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 20.Alivisatos AP, Gu WW, Larabell C. Quantum dots as cellular probes. Ann. Rev. Biomed. Eng. 2005;7:55–76. doi: 10.1146/annurev.bioeng.7.060804.100432. [DOI] [PubMed] [Google Scholar]
- 21.Liu Z, Sun X, Nakayama N, Dai H. Supramolecular chemistry on water-soluble carbon nanotubes for drug loading and delivery. ACS Nano. 2007;1:50–56. doi: 10.1021/nn700040t. [DOI] [PubMed] [Google Scholar]
- 22.Kam NWS, O'Connell M, Wisdom JA, Dai H. Carbon nanotubes as multifunctional biological transporters and near-infrared agents for selective cancer cell destruction. Proc. Natl. Acad. Sci. USA. 2005;102:11600–11605. doi: 10.1073/pnas.0502680102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakravarty P, et al. Thermal ablation of tumor cells with anti body-functionalized single-walled carbon nanotubes. Proc. Natl. Acad. Sci. USA. 2008;105:8697–8702. doi: 10.1073/pnas.0803557105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang ZH, et al. Delivery of telomerase reverse transcriptase small interfering RNA in complex with positively charged single-walled carbon nanotubes suppresses tumor growth. Clin. Cancer Res. 2006;12:4933–4939. doi: 10.1158/1078-0432.CCR-05-2831. [DOI] [PubMed] [Google Scholar]
- 25.Liu Z, et al. In vivo biodistribution and highly efficient tumour targeting of carbon nanotubes in mice. Nat. Nanotechnol. 2007;2:47–52. doi: 10.1038/nnano.2006.170. [DOI] [PubMed] [Google Scholar]
- 26.Niyogi S, et al. Chemistry of single-walled carbon nanotubes. Acc. Chem. Res. 2002;35:1105–1113. doi: 10.1021/ar010155r. [DOI] [PubMed] [Google Scholar]
- 27.Rosca ID, Watari F, Uo M, Akaska T. Oxidation of multiwalled carbon nanotubes by nitric acid. Carbon. 2005;43:3124–3131. [Google Scholar]
- 28.Tagmatarchis N, Prato M. Functionalization of carbon nanotubes via 1,3-dipolar cycloadditions. J. Mater. Chem. 2004;14:437–439. [Google Scholar]
- 29.Liu Y, et al. Polyethylenimine-grafted multiwalled carbon nanotubes for secure noncovalent immobilization and efficient delivery of DNA. Angew. Chem. Int. Ed. Engl. 2005;44:4782–4785. doi: 10.1002/anie.200500042. [DOI] [PubMed] [Google Scholar]
- 30.Ross JS, Fletcher JA. The HER-2/neu oncogene in breast cancer: prognostic factor, predictive factor, and target for therapy. Stem Cells. 1998;16:413–428. doi: 10.1002/stem.160413. [DOI] [PubMed] [Google Scholar]
- 31.Jin H, Varner J. Integrins: roles in cancer development and as treatment targets. Br. J. Cancer. 2004;90:561–565. doi: 10.1038/sj.bjc.6601576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai WB, Chen XY. Preparation of peptide-conjugated quantum dots for tumor vasculature-targeted imaging. Nat. Protoc. 2008;3:89–96. doi: 10.1038/nprot.2007.478. [DOI] [PubMed] [Google Scholar]
- 33.Anderson J, Banerjea A, Planelles V, Akkina R. Potent suppression of HIV type 1 infection by a short hairpin anti-CXCR4 siRNA. AIDS Res. Hum. Retroviruses. 2003;19:699–706. doi: 10.1089/088922203322280928. [DOI] [PubMed] [Google Scholar]
- 34.Novina CD, et al. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]