Abstract
Bacteria adhere to a surface and, through cell division and coordinated expression of gene products, to develop into a structurally-complex population of adherent cells. This process, known as biofilm formation, requires that intrinsic and extrinsic signals are transduced into appropriate gene expression patterns as biofilms mature. Mutational analysis has begun to reveal the complexity of systems used by Streptococcus mutans to ensure proper biofilm formation. These studies have revealed new and unique targets for the design of broadly-effective anti-caries strategies.
Introduction
While the bulk of dental caries microbiology in the last century was focused on the pathogen Streptococcus mutans, the view of the etiology of caries has changed over the last 10 years. In particular, it is now widely-accepted that S. mutans can be absent from carious lesions and that the microbial populations associated with caries display greater “aciduricity”, or ability to tolerate low pH, than populations from healthy sites (Beighton, 2005; Brailsford et al., 2001; Corby et al., 2005; Fejerskov, 2004; Marchant et al., 2001; van Houte et al., 1994; van Ruyven et al., 2000). These advances have ramifications for development of anti-caries strategies; the former suggesting that therapies directed at S. mutans alone may not be sufficient to control caries, and the latter highlighting a general characteristic of caries pathogens that could provide promising targets for novel caries control methods.
Biofilms, the environment and ecological succession
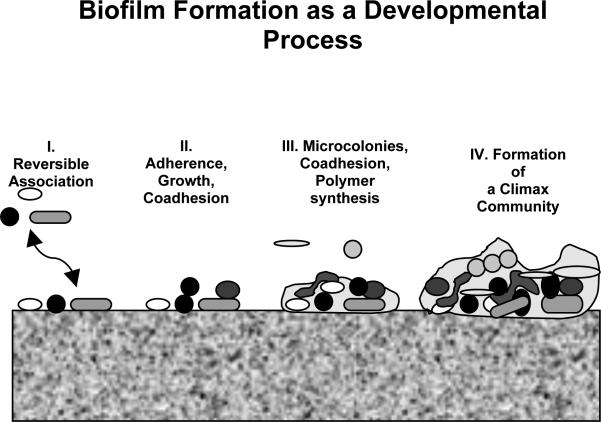
Biofilm formation by bacteria can be viewed as a developmental process (Fig. 1). A single cell, or small cluster of cells, initially colonizes a surface. If the local environment provides the necessary nutrients and environmental conditions are favorable, the cells can begin to divide. As cell division occurs and new species are recruited to the growing biofilms, the populations themselves alter the microenvironment in ways that support additional community diversity. Increases in species diversity enhance the range of substrates that can be utilized by the developing degradative community.
Figure 1.
Schematic representation of biofilm formation. Pioneer species (early colonizers) adhere to the pellicle-coated substratum and begin to divide and condition the surface (I & II). New organisms are recruited to the surface through co-adhesion and exposure of new binding sites, and polymers can be produced by the organisms or deposit from the planktonic phase (III). A mature biofilm with high community diversity becomes established (IV) and is maintained through metabolic cooperation and a balance of antagonistic and cooperative interactions of the individual organisms with the community and environment.
Importantly, if biofilms are to undergo proper maturation, it is essential that the organisms respond at the genetic and physiologic levels to the changes in the environment brought on by mass transport (diffusion) limitation and the increase in the spectrum of substrates and end products. Once formed, the complex populations on oral surfaces are fairly stable and, in most cases, compatible with health. However, the environment to which oral biofilms are exposed influences the growth characteristics and behavior of the organisms (Burne, 1998; Lemos et al., 2005). Certain environmental influences promote changes in the biofilms that favor development of cariogenic dental plaque, whereas other environmental conditions favor the stabilization of a flora that is compatible with health. We know that the sensing pathways that bacteria utilize to integrate environmental signals play majors roles in biofilm formation and virulence expression.
Historically, anti-caries strategies have focused on biofilm removal or targeting of specific virulence attributes of S. mutans, e.g. adhesins. However, in light of what has been learned about biofilm formation and caries in recent years, a reasonable approach for development of anticaries therapies could be to focus on those signaling pathways that allow bacteria to respond appropriately to environmental input. In the case of dental caries, those factors are pH, the amount and type of carbohydrate, and oxygen (Lemos et al., 2005). Therapies directed at adaptive strategies for coping with these environmental fluctuations could prevent the development of cariogenic biofilms and foster stabilization of healthy plaque communities, and thus be broadly effective at preventing dental caries irrespective of the etiologic agents.
Acid Tolerance
The pH of supragingival dental plaque may be the most important factor influencing plaque ecology and the development of caries. In fact, the most consistently-reported chemical difference in biofilms in caries versus health is that resting plaque pH is lower, and the depth and duration of acidification following carbohydrate intake is substantially greater, in caries-active sites. Given the importance of acid production in the pathogenesis of caries, it is not surprising that aciduricity is a major characteristic of organisms associated with caries. Aciduricity is a complex phenotype that manifests as an enhanced capacity to grow and to produce acids at pH values that are considered extreme in dental plaque, which, in terms of caries, translates to a greater depth and duration of acidification of dental biofilms. Sustained exposure of oral biofilms to low pH during caries development selects for organisms that gain a competitive advantage by virtue of their aciduricity (Marsh, 2000; 2003). Eventually, these organisms accumulate in proportions that are sufficient to cause detectable damage to tooth enamel.
For some time, it has been known that the inherent resistance of S. mutans to low pH is considerably greater than dental plaque bacteria that are not generally associated with caries, such as Streptococcus sanguinis. Early work by Marquis and coworkers associated the elevated aciduricity of S. mutans with the F1FO H+-translocating ATPase enzyme (Bender et al., 1986; Bender and Marquis, 1987). The F-ATPase of oral streptococci is the primary engine for extrusion of protons, which maintains the cytoplasmic pH in a range where metabolic enzymes and transporters can function. S. mutans was shown to display higher levels of activity of the ATPase enzyme and the enzyme functioned at lower pH values compared with the less-aciduric species associated with dental health (Bender et al., 1986).
In addition to constitutional acid resistance, many bacteria, including S. mutans, are able to mount an adaptive acid tolerance response (ATR) (Lemos, 2005). The existence of an ATR can be revealed experimentally by showing that cells become far more resistant to killing at a lethal pH (e.g. 3) and have a greater capacity to produce acid at low pH, after exposure to a sublethal acidic pH (e.g. 5.5) than cells that were maintained at neutral pH (Belli and Marquis, 1991). Extrapolation of this experimental system to life in dental plaque would suggest that repeated exposure of S. mutans to low pH in vivo could actually alter the phenotype of the organisms in a way that would enhance its cariogenicity.
Genomic science has helped to advance our understanding of the molecular basis for acid resistance and the adaptive ATR in S. mutans, revealing an array of genetic determinants that affect the ability of the organisms to cope with low pH. Proteomics has shown that the synthesis of a large number of proteins is altered when cells sense low pH for short periods of time (acid shock), or when they become adapted to growth at low pH (Len et al., 2003; Len et al., 2004; Svensater et al., 2000). Gene products ranging from molecular chaperones (Jayaraman et al., 1997), to global regulatory proteins, to products governing transport, to surface proteins and autolytic enzymes, all impact the manifestation of aciduricity in vitro (Lemos et al., 2005). Quorum sensing of peptides involved in the development of genetic competence, which is the ability to take up exogenous DNA, and the universal bacterial communication molecule produced by the LuxS enzyme, autoinducer-2 (AI-2), also impact on acid resistance by S. mutans (Li et al., 2001; Wen and Burne, 2004). Interestingly, the underlying mechanisms of acid adaptation by mutans streptococci are not universally conserved. Even the close relative of S. mutans, Streptococcus sobrinus, uses a different strategy to adapt to growth at low pH. Rather than up-regulating the expression of the ATPase enzyme, like S. mutans, mounting of an ATR by S. sobrinus involves up-regulation of glucose phosphoenolpyruvate:sugar phosphotransferase system (PTS) (Nascimento et al., 2004), which presumably allows the cells to generate more ATP to move more protons out of the cells. Collectively, these data indicate that the regulation of acid resistance and adaptation are control points governing the growth and persistence of cariogenic organisms. Thus, short-circuiting the underlying mechanisms controlling the ability to sense and regulate bacterial growth and metabolism at low pH may be an approach that is more widely effective at controlling caries than targeting of specific enzymatic activities. In fact, such approaches have been used already to target the quorum sensing competence peptide system of S. mutans (Eckert et al., 2006).
Management of catabolic pathways
For extended periods of time, bacteria in oral biofilms exist only on the nutrients derived from salivary molecules, and the remains of dead bacteria and sloughed epithelial cells. Organisms like S. mutans depend on carbohydrates to generate ATP, so a major contributor to the ability of cariogenic organisms to survive is the PTS. The PTS is a high-affinitymulti-substrate enzyme complex (Fig. 2) that internalizes a variety of mono- and disaccharides; operating when sugars are present in concentrations as low as 10 μM. Recently, the importance of the PTS and other systems that play roles in genetic and biochemical regulation of carbohydrate catabolism in S. mutans have been highlighted. Biofilm maturation, regulation of acid tolerance, global control of gene expression, and regulation of catabolic systems are all under the influence of a subset of sugar permeases of the PTS (Abranches et al., 2006; Abranches et al., 2003; Zeng et al., 2006). For example, we have shown that the Enzyme II for mannose (EIIman) affects the expression of over 60 genes and modulates catabolite repression, which is characterized by down-regulation of genes for utilization of a non-preferred energy source when a preferred source is present (Abranches et al., 2006). We have also shown that at least three additional sugar permeases modulate the expression of genes for catabolism of inulin, an extracellular storage compound produced from sucrose by multiple oral bacteria (Wen et al., 2001; Zeng et al., 2006).
Figure 2.
A diagram of the bacterial sugar:phosphotransferase system (PTS) Phosphoenolpyruvate (PEP) provided by glycolysis is cleaved by Enzyme I (EI) and the phosphate group is transferred to general phosphcarrier protein HPr, which in turn acts as a phosphate donor to a variety of membrane-bound Enzyme II (EII) complexes that concomitantly phosphorylate and internalize a spectrum of mono- and di-saccharides. Specific EII enzymes, HPr and EI can also be involved in regulation of gene expression and enzyme activity through direct interaction with other proteins or by phospho-relay.
An interesting example of the evolution of control of catabolic pathways in S. mutans is the production of tetra- and penta-phosphorylated guanine nucleotides (p)ppGpp, which function as intracellular stress signal molecules. (p)ppGpp accumulates during amino acid starvation (Cashel and Rudd, 1987) and triggers a “stringent response”, which is characterized by the down regulation of genes for ribosomal RNAs and certain anabolic processes, and up-regulation of genes that allow for synthesis of essential nutrients and stress tolerance. Recently, it was shown that other Gram-positive bacteria, including Bacillus subtilis and Staphylococcus aureus, had a single (p)ppGpp synthase/hydrolase enzyme, RelA, but S. mutans possesses RelA and two additional gene products that can make (p)ppGpp (Lemos et al., 2007). Notably, one of the (p)ppGpp synthases is co-transcribed with, and regulated by, a two-component signal transduction system (TCS). TCSs are widely disseminated in bacteria and regulate gene expression by relaying external signals to the transcriptional machinery (Lemos et al., 2007). Why this finding is particularly important is that it shows that S. mutans is up-regulating (p)ppGpp synthesis, which would in turn down-regulate growth and shift metabolism to storage and survival (Nascimento et al., 2008), in response to exogenous signals. In fact, cells lacking RelP the RelRS TCS grow faster and to a higher density in culture (Lemos et al., 2007). We believe the RelPRS system is germane to the survival and persistence of S. mutans in the mouth because bacteria have to balance growth against acquiring sufficient storage compounds during dietary intake by the host in order to remain competitive during fasting periods. The RelRS TCS and novel (p)ppGpp synthases represent attractive potential targets to upset gene regulation in S. mutans in a way that should reduce competitive fitness.
Regulation of gene expression and cell surface composition by oxygen
Bacteria on a freshly cleaned tooth surface are exposed to concentrations of oxygen close to that of air-saturated water (Marquis, 1995). The redox potential of early dental biofilms is high, approximately +300 mV, and drops significantly as plaque matures to values of -140 mV and below (Mettraux et al., 1984). Although molecular oxygen (O2) is not directly toxic to bacteria, chemically- and enzymatically-mediated single-electron reductions of oxygen produce reactive oxygen species (ROS) that create problems for bacteria lacking sufficient quantities of the enzymes used to detoxify ROS, e.g. superoxide dismutase and peroxidases (Marquis, 1995). A major effort has been invested in dissecting how eubacteria cope with oxygen, although relatively little work has been done specifically with oral streptococci (Marquis, 1995). Recently, we began to explore how growth in the presence of air affects S. mutans. We discovered that the presence of oxygen, at levels that do not inhibit the growth of the bacteria, is strongly inhibitory to formation of biofilms (Ahn and Burne, 2007). The underlying molecular basis for this behavior appears to involve changes in the transcriptome of the organisms when oxygen is present, coupled with post-transcriptional modification of the surface of the bacterial cell in a way that alters known virulence properties of the organisms (Ahn et al., 2007).
Importantly, we have identified a number of pathways in S. mutans that are responsible for modulating the genetic and phenotypic properties of the cells in response to growth in the presence of air. One is a two-component system (TCS) that appears to regulate gene expression in response to redox. The Vic TCS is integral to the formation of biofilms, expression of virulence attributes and modulation of acid tolerance (Ahn and Burne, 2007; Senadheera et al., 2005). The Vic TCS modulates the maturation of an important autolysin, known as AtlA, which is a surface-associated protein that is required for biofilm formation by S. mutans (Brown et al., 2005). Loss of AtlA dramatically affects the ability of the cells to generate a normal cell surface, as does growth in the presence of oxygen; affecting hydrophobicity and altering GTF localization (Ahn et al., 2007). Thus, there is a complex regulatory network in S. mutans for dealing with oxygen that may be targeted to diminish establishment or virulence.
Summary
For many years, researchers focused on specific virulence attributes of S. mutans with the idea that highly targeted therapeutics that disrupted adherence or persistence of this pathogen would be the most effective way to control caries. Detailed studies of the genetic regulatory pathways that control the expression of genes contributing to the virulence of S. mutans were often viewed as laboratory exercises that were of interest to a handful of molecular biologists, but not particularly useful for the design of novel anti-caries strategies. Over the last few years, since the genomes of S. mutans and many related bacteria has become available, and technologies like whole genome profiling have become commonplace in oral microbial pathogenesis studies, evidence has accumulated that supports that a re-examination of how to approach control of cariogenic biofilms or to stabilize “healthy” biofilms is needed. We suggest that directing therapies at regulatory pathways used by S. mutans to coordinate gene expression in a way that allows the organisms i) to properly sequence biofilm maturation, ii) to balance growth at the expense of ensuring persistence and survival, and iii) to cope with environmental stresses are likely to be the most broadly effective at controlling the biofilm-forming, aciduric species associated with caries development.
Acknowledgements
This work was supported by NIDCR DE12236, DE10362 and DE13239.
Literature Cited
- 1.Abranches J, Candella MM, Wen ZT, Baker HV, Burne RA. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J Bacteriol. 2006;188(11):3748–56. doi: 10.1128/JB.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abranches JA, Chen YM, Burne RA. Characterization of Streptococcus mutans strains deficient in EIIABMan of the sugar phosphotransferase system. Appl Environ Microbiol. 2003;69:4760–4769. doi: 10.1128/AEM.69.8.4760-4769.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn SJ, Burne RA. Effects of oxygen on biofilm formation and the AtlA autolysin of Streptococcus mutans. J Bacteriol. 2007;189:6293–302. doi: 10.1128/JB.00546-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn SJ, Wen ZT, Burne RA. Effects of oxygen on virulence traits of Streptococcus mutans. J Bacteriol. 2007;189(23):8519–27. doi: 10.1128/JB.01180-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beighton D. The complex oral microflora of high-risk individuals and groups and its role in the caries process. Community Dent Oral Epidemiol. 2005;33(4):248–55. doi: 10.1111/j.1600-0528.2005.00232.x. [DOI] [PubMed] [Google Scholar]
- 6.Belli WA, Marquis RE. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl Environ Microbiol. 1991;57:1134–1138. doi: 10.1128/aem.57.4.1134-1138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bender GR, Sutton SVW, Marquis RE. Acid tolerance, proton permeabilities, membrane ATPases of oral streptococci. Infection and Immunity. 1986;53(331-338) doi: 10.1128/iai.53.2.331-338.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bender GR, Marquis RE. Membrane ATPases and acid tolerance of Actinomyces viscosus and Lactobacillus casei. Appl Environ Microbiol. 1987;53:2124–2128. doi: 10.1128/aem.53.9.2124-2128.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brailsford SR, Shah B, Simons D, Gilbert S, Clark D, Ines I, et al. The predominant aciduric microflora of root-caries lesions. J Dent Res. 2001;80:1828–33. doi: 10.1177/00220345010800091101. [DOI] [PubMed] [Google Scholar]
- 10.Brown TA, Jr., Ahn SJ, Frank RN, Chen YY, Lemos JA, Burne RA. A hypothetical protein of Streptococcus mutans is critical for biofilm formation. Infect Immun. 2005;73:3147–51. doi: 10.1128/IAI.73.5.3147-3151.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burne RA. Oral streptococci: products of their environment. J Dental Research. 1998;77:445–452. doi: 10.1177/00220345980770030301. [DOI] [PubMed] [Google Scholar]
- 12.Cashel M, Rudd KE. In Escherichia coli and Salmonells typhimurium : Cellular and Molecular Biology. American Society for Microbiology; Washington, D.C.: 1987. The stringent response. [Google Scholar]
- 13.Corby PM, Lyons-Weiler J, Bretz WA, Hart TC, Aas JA, Boumenna T, et al. Microbial risk indicators of early childhood caries. J Clin Microbiol. 2005;43:5753–9. doi: 10.1128/JCM.43.11.5753-5759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckert R, He J, Yarbrough DK, Qi F, Anderson MH, Shi W. Targeted killing of Streptococcus mutans by a pheromone-guided “smart” antimicrobial peptide. Antimicrob Agents Chemother. 2006;50:3651–7. doi: 10.1128/AAC.00622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fejerskov O. Changing paradigms in concepts on dental caries: consequences for oral health care. Caries Res. 2004;38:182–91. doi: 10.1159/000077753. [DOI] [PubMed] [Google Scholar]
- 16.Jayaraman GC, Penders JE, Burne RA. Transcriptional analysis of the Streptococcus mutans hrcA, grpE, and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol Microbiol. 1997;25:329–341. doi: 10.1046/j.1365-2958.1997.4671835.x. [DOI] [PubMed] [Google Scholar]
- 17.Lemos JA, Abranches J, Burne RA. Responses of cariogenic streptococci to environmental stresses. Curr Issues Mol Biol. 2005;7:95–107. [PubMed] [Google Scholar]
- 18.Lemos JA, Lin VK, Nascimento MM, Abranches J, Burne RA. Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol Microbiol. 2007;65:1568–81. doi: 10.1111/j.1365-2958.2007.05897.x. [DOI] [PubMed] [Google Scholar]
- 19.Len AC, Cordwell SJ, Harty DW, Jacques NA. Cellular and extracellular proteome analysis of Streptococcus mutans grown in a chemostat. Proteomics. 2003;3:627–46. doi: 10.1002/pmic.200300391. [DOI] [PubMed] [Google Scholar]
- 20.Len AC, Harty DW, Jacques NA. Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiol. 2004;150:1339–51. doi: 10.1099/mic.0.27008-0. [DOI] [PubMed] [Google Scholar]
- 21.Li YH, M NH, Svensater G, Ellen RP, Cvitkovitch DG. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J Bacteriol. 2001;183:6875–84. doi: 10.1128/JB.183.23.6875-6884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchant S, Brailsford SR, Twomey AC, Roberts GJ, Beighton D. The predominant microflora of nursing caries lesions. Caries Res. 2001;35:397–406. doi: 10.1159/000047482. [DOI] [PubMed] [Google Scholar]
- 23.Marquis RE. Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms. J Ind Microbiol. 1995;15:198–207. doi: 10.1007/BF01569826. [DOI] [PubMed] [Google Scholar]
- 24.Marsh PD. Oral ecology and its impact on oral microbial diversity Norfolk. Horizon Scientific Press; England: 2000. [Google Scholar]
- 25.Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiol. 2003;149:279–94. doi: 10.1099/mic.0.26082-0. [DOI] [PubMed] [Google Scholar]
- 26.Mettraux GR, Gusberti FA, Graf H. Oxygen tension (pO2) in untreated human periodontal pockets. J Periodontol. 1984;55(9):516–21. doi: 10.1902/jop.1984.55.9.516. [DOI] [PubMed] [Google Scholar]
- 27.Nascimento MM, Lemos JA, Abranches J, Goncalves RB, Burne RA. Adaptive acid tolerance response of Streptococcus sobrinus. J Bacteriol. 2004;186:6383–90. doi: 10.1128/JB.186.19.6383-6390.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nascimento MM, Lemos JA, Abranches J, Lin VK, Burne RA. Role of RelA of Streptococcus mutans in global control of gene expression. J Bacteriol. 2008;190:28–36. doi: 10.1128/JB.01395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Senadheera MD, Guggenheim B, Spatafora GA, Huang YC, Choi J, Hung DC, et al. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J Bacteriol. 2005;187:4064–76. doi: 10.1128/JB.187.12.4064-4076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svensater G, Sjogreen B, Hamilton IR. Multiple stress responses in Streptococcus mutans and the induction of general and stress-specific proteins. Microbiol. 2000;146:107–117. doi: 10.1099/00221287-146-1-107. [DOI] [PubMed] [Google Scholar]
- 31.van Houte J, Lopman J, Kent R. The predominant cultivable flora of sound and carious human root surfaces. J Dent Res. 1994;73:1727–34. doi: 10.1177/00220345940730110801. [DOI] [PubMed] [Google Scholar]
- 32.van Ruyven FOJ, Lingstrom P, van Houte J, Kent R. Relationship among mutans streptococci, “low-pH” bacteria, and iodophilic polysaccharide-producing bacteria in dental plaque and early enamel caries in humans. J Dent Res. 2000;79:778–784. doi: 10.1177/00220345000790021201. [DOI] [PubMed] [Google Scholar]
- 33.Wen ZT, Browngardt C, Burne RA. Characterization of two operons that encode components of fructose-specific enzyme II of the sugar:phosphotransferase system of Streptococcus mutans. FEMS Microbiol Lett. 2001;205:337–42. doi: 10.1111/j.1574-6968.2001.tb10969.x. [DOI] [PubMed] [Google Scholar]
- 34.Wen ZT, Burne RA. LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J Bacteriol. 2004;186:2682–91. doi: 10.1128/JB.186.9.2682-2691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng L, Wen ZT, Burne RA. A novel signal transduction system and feedback loop regulate fructan hydrolase gene expression in Streptococcus mutans. Mol Microbiol. 2006;62:187–200. doi: 10.1111/j.1365-2958.2006.05359.x. [DOI] [PubMed] [Google Scholar]