Abstract
Functional MRI (fMRI) most commonly employs 2D echo-planar imaging (EPI). The advantages for fMRI brought about by the increasingly popular ultra-high field strengths are best exploited in high-resolution acquisitions, but here 2D EPI becomes unpractical for several reasons, including the very long volume acquisitions times. In this study at 7 T, a 3D EPI sequence with full parallel and partial Fourier imaging capability along both phase encoding axes was implemented and used to evaluate the sensitivity of 3D and corresponding 2D EPI acquisitions at four different spatial resolutions ranging from small to typical voxel sizes (1.5–3.0mm isotropic). Whole-brain resting state measurements (N=4) revealed a better, or at least comparable sensitivity of the 3D method for gray and white matter. The larger vulnerability of 3D to physiological effects was outweighed by the much shorter volume TR, which moreover allows whole brain coverage at high resolution within fully acceptable limits for event-related fMRI: TR was only 3.07s for 1.5 mm, 1.88 s for 2.0mm, 1.38 s for 2.5 mm and 1.07s for 3.0 mm isotropic resolution. In order to investigate the ability to detect and spatially resolve BOLD activation in the visual cortex, functional 3D EPI experiments (N=8) were performed at 1mm isotropic resolution with parallel imaging acceleration of 3×3, resulting in a TR of only 3.2s for whole-brain coverage.
From our results, and several other practical advantages of 3D over 2D EPI found in the present study, we conclude that 3D EPI provides a useful alternative for whole-brain fMRI at 7 Tesla, not only when high-resolution data are required.
Introduction
Since the discovery of the BOLD effect, 2D gradient-echo echo planar imaging (EPI) has been the workhorse of functional MRI, largely because of its high sampling speed and excellent sensitivity to signal changes related to brain activation, at both sufficient temporal (~2–3 s) and spatial resolution (~3–4 mm isotropic) with whole brain coverage (Norris, 2006). Recently, it has been shown that higher spatial resolution would be beneficial to reduce the influence of physiological fluctuations (Triantafyllou et al., 2005; Triantafyllou et al., 2006). However, sampling at higher (isotropic) spatial resolution in conventional multi-slice 2D EPI considerably increases measurement times as the echo train length (ETL) becomes larger, and more slices have to be acquired to obtain the same covered volume. Methods such as partial Fourier (PF) sampling (Jesmanowicz et al., 1998) or partial parallel imaging (PPI) (Griswold et al., 2002; Pruessmann et al., 1999) provide an efficient means for shortening the ETL, and are hence well suited for obtaining high-resolution functional images, images with reduced EPI distortion artifacts (de Zwart et al., 2006; de Zwart et al., 2002; Preibisch et al., 2003; Schmidt et al., 2005), or single-shot acquisition of multiple echoes to improve BOLD sensitivity (Poser and Norris, 2009; Poser et al., 2006). Considerable effort has gone into the optimization of 2D EPI protocols and the practical challenges associated with fMRI at high field (Speck et al., 2008).
In the conventional 2D EPI sequence, slice selective excitation is performed and the signal subsequently acquired under an oscillating read and blipped phase encode gradient, so as to acquire the data for a full 2D image in a single shot within TRslice. After repetition time TRslice·Nslices, all Nslices slices have been acquired, and the process is repeated. The effectiveness of both the PF and PPI methods for the reduction of acquisition time is hence rather limited in the case of 2D EPI, as the acceleration can only be performed along the (one and only) phase encode (PE) direction. The small potential time gain from under-sampling in 2D EPI is primarily due to the fact that TE should be kept constant to obtain the same functional contrast: For example, if the ETL is shortened by an acceleration factor of 2, the effective reduction in sampling time per slice is only TRslice – ETL/4, which for a typical protocol is a time saving of only about 10% – 15%.
Alternatively, a 3D sampling scheme can be employed whereby the same thick slab of tissue corresponding to the volume of interest is repeatedly excited, and a kx-ky plane of k-space acquired each time with a different kz increment. For application to fMRI, this concept has attracted rapidly growing interest in the recent years, and 3D implementations with Cartesian (Poser and Norris, 2005; van der Zwaag et al., 2009) and spiral (Hu and Glover, 2007a; Hu and Glover, 2007b; Lai and Glover, 1998) k-space trajectory have been presented. For EPI-like sequences a similar scheme was first suggested by (Song et al., 1994) and (Mansfield et al., 1995) who proposed to acquire even multiple kz planes in a single shot. All these methods have in common that slice selection is replaced by a secondary phase encoding direction, and the complete 3D image then obtained by 3D Fourier transform upon acquisition of all kz planes, again after time TRslice·Nslices. The important potential of 3D EPI here lies in the fact that entire kz encoding steps (kx-ky planes) can be ‘skipped’ by the use of PF or PPI along the secondary phase direction, permitting considerable reductions in the volume sampling time TRvolume: For both these undersampling strategies, or their combination, the fractional time saving is directly given by the fraction of non-acquired secondary phase encodings. A 3D acquisition scheme where undersampling can be performed along the secondary PE direction will therefore be substantially more time efficient.
In terms of MR imaging, the implication of using 3D vs. 2D EPI is twofold. First, the time interval between subsequent excitations of the same tissue is much reduced, and as a result the steady state magnetization that is available MR signal formation is lower; this is given by Eq. 1 and maximal when the flip angle α is equal to the Ernst angle (Eq. 2) which should be calculated for the T1 relaxation time of gray matter tissue. Second, the fact that Nslices more data points contribute to each data point in the 3D Fourier reconstruction leads to an intrinsic signal-to-noise (SNR) advantage of √Nslices which hence increases with increasing slice count.
| [Eq. 1] |
| [Eq. 2] |
In this study we propose the use of 3D EPI and to exploit the greater flexibility given by the second phase encoding along the third spatial dimension to either obtain higher spatial resolution within the same TRvolume, or to sample at the same spatial resolution within a much shorter time as compared to conventional 2D EPI. The proposed 3D EPI and conventional 2D EPI are compared by in vivo application with and without functional stimulation, and the various implications of using 3D EPI are discussed.
Methods
MRI measurements
A 3D EPI sequence with full partial Fourier and parallel imaging capability and flexible z-encoding order was implemented for the Siemens Magnetom scanner family (Siemens Healthcare, Erlangen, Germany). Image reconstruction was performed entirely through the vendor-provided software which uses GRAPPA parallel imaging reconstruction, including EPI specific functionality for removing Nyquist ghosting and a zeroth order phase correction to minimize B0 fluctuation from scanner drift and subject breathing. Both RF and gradient spoiling were used in the sequence. Measurements were performed on a total of 12 subjects. The experiments of this study consisted of two parts which were conducted separately on two different Magnetom 7T scanners; each part had been approved by the respective the local ethics committee.
(a) Temporal SNR
Corresponding 2D and 3D protocols with different spatial resolutions were used on four subjects using an 8-channel Tx/Rx head array (RAPID Biomedical, Germany). The following resolutions (isotropic voxel size) were used: 1.5mm, 2.0 mm, 2.5 mm, and 3.0 mm and the number of slices was adapted to cover the same volume at each resolution. An in-plane acceleration factor (AF) of 2 was used for each protocol. For the 3D protocols, an additional factor of 2 was used in the slice encoding direction, resulting in a total AF of 4. For parallel undersampling along the secondary phase encoding direction, the circular geometric arrangement of the elements in the head coil required sagittal slice orientation which was used for all protocols in part (a). The corresponding volume acquisition times at the four resolutions where TRvolume, 2D = [5.96 s, 3.65 s, 2.7 s 2.1 s], and TRvolume, 3D = [3.07 s, 1.88 s, 1.38 s, 1.07 s], respectively. These protocols are summarized in Table 1. In each scan, TE was 23 ms, and the flip angle set to the Ernst angle (see Eq. 2 above). In case that SAR limit was exceeded, the largest possible flip angle was used as allowed by the online SAR monitor. A time series of 50 volumes was acquired without the subjects performing a specific task.
Table 1.
Summary of the measurement protocols used for the sensitivity comparison.
| 2D | 3D | |||
|---|---|---|---|---|
| resolution | TR | AF | TR | AF |
| 1.5 mm | 5.96 s | 2 | 3.07 s | 2 × 2 |
| 2.0 mm | 3.65 s | 2 | 1.88 s | 2 × 2 |
| 2.5 mm | 2.70 s | 2 | 1.38 s | 2 × 2 |
| 3.0 mm | 2.10 s | 2 | 1.07 s | 2 × 2 |
(b) Visual task
3D EPI fMRI experiments with visual stimulation were performed on 8 subjects, using the following settings: 1 mm isotropic resolution using a 32-channel head coil (Wiggins et al., 2006), TE = 23 ms, TRslice = 50 ms, TRvolume, 3D = 3.2 s, AF = 3×3, BW= 2000 Hz/px, matrix size 180 × 180, 104 slices in an axial orientation with slice oversampling of 25% to prevent signal fold-over and intensity variation across the slab due to pulse profile imperfection. The fMRI task consisted of 3 minutes visual stimulation in blocks of 20/20 s on/off.
Analysis
(a) Temporal SNR
The data sets of each subject were first motion corrected and high-pass filtered (> 0.01 Hz) using Feat (FMRI Expert Analysis Tool, version5.98, part of FSL (FMRIB’s Software Library, www.fmrib.ox.ac.uk/fsl, Oxford, UK). The 2D data sets were then linearly coregistered to the 1.5mm resolution 2D data set using an affine 9 parameter transformation and normalized correlation as a cost function (Flirt, FSL, Oxford). The 3D data sets were coregistered to the 2D data sets with the corresponding resolution using only a 6 parameter rigid body transformation, since the in-plane geometric distortions of the 2D and 3D protocols are identical. In ROIs for the different brain tissues, i.e. occipital gray matter (GM), parietal white matter (WM) and cerebrospinal fluid in the lateral ventricle (CSF), the sensitivity of the protocols was compared as follows: first, temporal SNR (tSNR) was obtained pixelwise in the conventional manner as the mean signal over imaging volumes divided by standard deviation along the timecourse. To facilitate comparison of protocols with different TRvolume (but the same TE), the measure for ‘sensitivity’ was taken as tSNR/√TRvolume.
(b) Visual task
fMRI data processing was carried out using FEAT. First, an analysis with a relatively large spatial smoothing kernel (3mm) was performed mainly for visualization purposes and to analyze the functional data in a commonly used way. The following pre-statistics processing was applied; motion correction using MCFLIRT (Jenkinson et al., 2002) non-brain removal using BET (Smith, 2002); spatial smoothing using a Gaussian kernel of 3mm FWHM; grand-mean intensity normalization of the entire 4D dataset by a single multiplicative factor; high-pass temporal filtering (Gaussian-weighted least-squares straight line fitting, with sigma= 20.0 s). Time series statistical analysis was carried out using FILM (Woolrich et al., 2001). Z-statistic images (Gaussianized T/F) were thresholded using clusters determined by Z> 5.3 and a (corrected) cluster significance threshold of P= 0.00005. In one subject no significantly activated cluster was found. The largest cluster of activated voxels in the visual cortex was then used to assess the number of activated pixels, average and maximum z-values, and relative signal change. In addition, noise was estimated from the residuals of the GLM analysis and used to calculate CNR as signal change divided by noise and tSNR as mean signal divided by noise. Subsequently, for visualization purposes and to show the potential of the high spatial resolution acquisition, the analysis was repeated by using only very little (0.75mm kernel) or no spatial smoothing, while leaving the other parameters except the significance threshold (Z > 4.3 for 0.75 mm smoothing; Z > 3.3 for no smoothing) unchanged.
Results
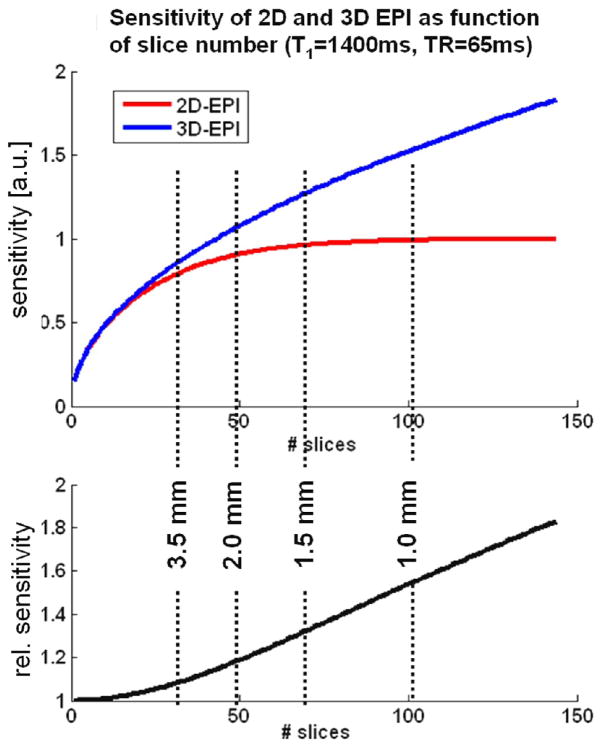
Using Eqs. 1 and 2, the sensitivity of 2D vs. 3D EPI as a function of slice number were simulated under the assumption of Gaussian noise, a gray matter T1 at 7T of 1400 ms, a constant TRslice of 65ms, and no use of parallel acceleration along the slice direction. Note that since the resulting volume TR are identical for both methods, their sensitivities are directly given by their respective MR signal; this is shown in the top panel of Fig. 1. The bottom panel of the figure indicates performance gain of 3D EPI over 2D EPI. The vertical dashed lines indicate the approximate number of slices required for whole brain coverage (taken here as a 100mm axial slab) at the different slice thicknesses. The plot also clearly illustrates the increasing benefit of 3D with increasing resolution along the slice direction.
Fig 1.
Theoretical sensitivity of 2D and 3D EPI as a function of the number of acquired slices (top) the relative performance of 3D vs. 2D EPI (bottom) in the absence of physiological noise as derived from the signal equations (Eqs1 and 2) for the case without parallel undersampling. Since the resulting volume TR are identical for both methods, their relative sensitivities scale with the MR signal. The dashed lines indicate the approximate number of slices needed for whole-brain coverage (≥ 100mm axial slab) at the different spatial resolutions.
Table 2 summarizes the results of the tSNR and sensitivity analysis of 2D and 3D EPI protocols: shown are the tSNR and sensitivity values for the two methods in the gray matter, white matter and CSF ROIs, at each of the four spatial resolutions. As expected, and in accordance with Eq.1, tSNR is higher for 2D EPI due to the longer TR which passes between successive excitations of the same slice. Correcting for the different TR by factor 1/√TR yields the sensitivity which for 3D is significantly higher than for 2D at the highest resolution (1.5 mm, p< 0.05) and comparable at the lower resolutions for gray matter; for white matter the sensitivity is consistently higher. In contrast, the CSF sensitivity is lower in 3D than in 2D EPI. For illustration purposes the results are plotted in Fig. 2, showing the values for gray matter, white matter and CSF in the top, middle an bottom panel, respectively.
Table 2.
Temporal SNR (tSNR) and corresponding sensitivity of the different EPI protocols used in part(a) of the study. The effective functional sensitivity for fMRI is calculated as tSNR/√TRvolume. The error terms give the standard error over subjects (N= 4).
| (a) tSNR of 2D and 3D EPI | ||||
|---|---|---|---|---|
| 2D | 1.5 mm | 2 mm | 2.5 mm | 3 mm |
| GM | 31.4 ± 2.3 | 42.5 ± 4.1 | 52.2 ± 4.0 | 50.7 ± 6.8 |
| WM | 30.7 ± 2.8 | 46.5 ± 2.2 | 71.1 ± 1.7 | 81.5 ± 3.0 |
| CSF | 59.1 ± 7.2 | 64.7 ± 7.9 | 77.4 ± 9.6 | 75.7 ± 11.5 |
| TRvolume | 5960 ms | 3650 ms | 2700 ms | 2090 ms |
| 3D | ||||
| GM | 29.9 ± 1.3 | 32.4 ± 3.8 | 35.8 ± 4.9 | 45.5 ± 4.0 |
| WM | 31.4 ± 1.6 | 39.9 ± 1.8 | 54.6 ± 3.4 | 65.3 ± 1.9 |
| CSF | 30.5 ± 3.6 | 40.5 ± 4.5 | 38.0 ± 3.4 | 53.0 ± 6.8 |
| TRvolume | 3068 ms | 1880 ms | 1376 ms | 1066 ms |
| (b) Corresponding sensitivity of 2D and 3D EPI | ||||
| 2D | 1.5 mm | 2 mm | 2.5 mm | 3 mm |
| GM | 12.9 ± 1.0 | 22.3 ± 2.2 | 31.8 ± 2.5 | 35.1 ± 4.7 |
| WM | 12.6 ± 1.1 | 24.4 ± 1.2 | 43.3 ± 1.1 | 56.4 ± 2.1 |
| CSF | 24.2 ± 2.9 | 33.9 ± 4.1 | 47.1 ± 5.8 | 52.4 ± 8.0 |
| TRvolume | 5960 ms | 3650 ms | 2700 ms | 2090 ms |
| 3D | ||||
| GM | 17.1 ± 0.7 | 23.7 ± 2.7 | 30.6 ± 4.2 | 44.1 ± 3.8 |
| WM | 17.9 ± 0.9 | 29.1 ± 1.3 | 46.5 ± 2.9 | 63.3 ± 1.8 |
| CSF | 17.4 ± 2.0 | 29.6 ± 3.3 | 32.4 ± 2.9 | 51.3 ± 6.6 |
| TRvolume | 3068 ms | 1880 ms | 1376 ms | 1066 ms |
Fig 2.
Plots of sensitivity against voxel volume for the 2D (dashed lines) versus 3D EPI protocols (straight lines), shown separately for GM (top), WM (middle) and CSF (bottom). The errors bars indicate standard error over subjects (N= 4).
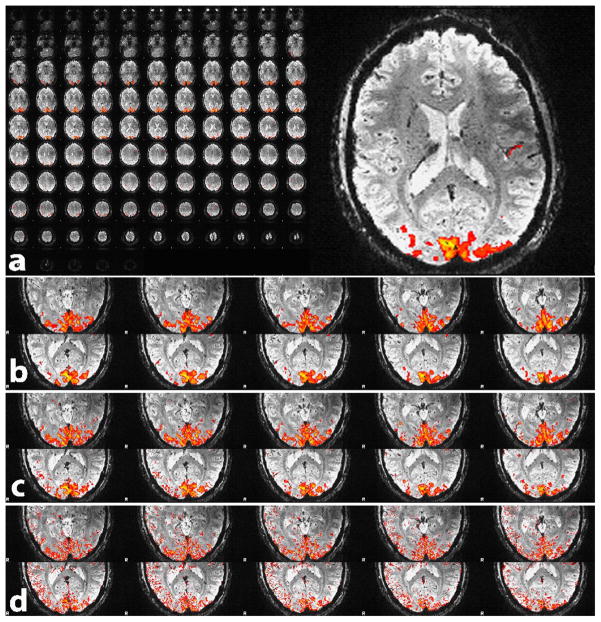
The results from the functional experiments (part ‘b’) are shown in Table 3, which gives the number of activated voxels, average and maximum Z-score, relative BOLD signal changes, CNR and tSNR for each subject. In Fig. 3 the temporal mean of all slices of the 1 mm resolution data set from a single subject is shown (panel (a)). Note the excellent image quality and gray-white matter contrast, as well as the detailed depiction of veins and deep brain nuclei which is seen best in zoomed view of one selected slice (slice 49). Sections (b) - (d) give a zoomed view of ten slices covering the occipital cortex for better visibility of the activation pattern. Overlaid on these images are the significantly activated clusters after spatially smoothing with a 3 mm kernel (b) which was used for improved visualization of the activation clusters. In (c) smoothing was reduced to a 0.75 mm kernel and in (d) no spatial smoothing was used. Note how the activation nicely follows the sulci of the occipital cortex. In these experiments with whole-brain acquisition the AF of 3×3 resulted in a TR of only 3.2 s.
Table 3.
Number of activated pixels, average and maximum z-values, relative signal change, and BOLD contrast-to-noise (CNR) and tSNR in the ROI defined by the activation cluster.
| Subject | # voxels | avg. z | max z | rel dS | CNR | tSNR |
|---|---|---|---|---|---|---|
| dc001 | 23070 | 6.85 | 10.66 | 5.50% | 3.42 | 62.20 |
| dc002 | 6396 | 6.78 | 10.29 | 4.95% | 3.33 | 67.26 |
| dc004 | 872 | 6.05 | 7.92 | 5.25% | 2.79 | 53.15 |
| dc005 | 2729 | 6.60 | 9.74 | 4.81% | 3.25 | 67.54 |
| dc006 | 6355 | 6.32 | 8.90 | 5.44% | 2.94 | 54.05 |
| dc007 | 7638 | 6.47 | 9.71 | 4.78% | 3.10 | 64.96 |
| dc008 | 1226 | 6.30 | 9.35 | 5.33% | 2.96 | 55.47 |
| Mean | 6898 | 6.48 | 9.51 | 5.15% | 3.11 | 60.66 |
| SD | 2880.53 | 0.11 | 0.34 | 0.11% | 0.09 | 2.38 |
Fig 3.
Whole brain mean intensity images with 1mm3 resolution, obtained with 3D EPI at 7 T using the 32 channel coil and factor 3×3 acceleration, resulting in a TR of only 3.2 s. The top panel (a) shows all 104 slices in mosaic view as well as a zoomed view of slice49. Panel (b) - (d) give a zoomed view of ten slices covering the occipital cortex, overlaid with the visual activation obtained with a smoothing kernel of 3mm (b), 0.75 mm (c) and no smoothing (d).
Discussion and Conclusion
The purpose of the present study was to investigate the usefulness of parallel accelerated 3D EPI for high-resolution fMRI at 7T. Several practical aspects, in particular the reduced volume acquisition time make 3D EPI an attractive option; it has however been far from clear that 3D would be the better choice in terms of functional sensitivity. In the presence of physiological noise as caused by breathing, cardiac pulsation, and motion, and at typical spatial resolution for fMRI, a single-shot 2D sampling strategy is usually preferable since here the detrimental effects are weaker: as a consequence of the shorter effective sampling window (TRslice vs. TRvolume) physiological fluctuations have less time to impose themselves. For voxel resolutions ranging from (1.5mm)3 to (3.0mm)3 we find in this study a larger, or at least comparable, functional sensitivity of 3D EPI for the GM and WM signal. The value of fast 3D methods for fMRI has been explored in earlier studies at 1.5T, however using the PRESTO-SENSE sequence (Golay et al., 2000; Klarhofer et al., 2003) which combines the ‘principle of echo shifting with a train of observations’ technique (Liu et al., 1993) with SENSE parallel imaging. In a comparative study at 3T (Neggers et al., 2008) the authors found a highly accelerated PRESTO-SENSE sequence (AF = 2 × 1.8, TR= 0.5 s) to be more sensitive than a corresponding 2D EPI protocol (TR= 1.9 s), which is attributed to the larger number of observations per unit time thanks to the short TR. This finding at 3T is hence consistent with our results at 7 T, and underpins the importance of rapid sampling of the BOLD signal, even at the price of image SNR, in the regime where temporal noise is dominated by physiological fluctuations. While very attractive at clinical field strengths, PRESTO is not a viable option at 7T and higher particularly when high spatial resolution is desired, as the typically short required TE will not leave enough time for acquiring the shifted echo from the previous excitation.
There are several important considerations for the choice of approach in practice: Immediately obvious is the desire, or need, for short repetition times which should ideally not exceed 3 seconds for adequate sampling of the BOLD response when using event-related stimulation paradigms (Norris, 2006). For a typical TRslice of 60 ms this would limit the possible number of 2D slices to only about 50, which only just allows whole-brain coverage with a stack of axial 2mm slices. The greatest advantage thus lies in the capacity of 3D EPI to be accelerated in the second (‘slice’) phase encoding direction, moreover, with time savings given by the nominal acceleration factor. Furthermore, as the demand for large slice numbers increases, the advantage of 3D sampling simultaneously increases due to the inherent factor √Nslice sensitivity increase as a result of Nslice k-space points contributing to each image voxel. Finally decisive for a suitable choice –if TR is not an issue and 2D hence remains an option– is the effective functional sensitivity of each available alternative. Even if the within-image SNR, or tSNR calculated along the time series, is lower for a given protocol, it may nevertheless be the better choice if the effect of shorter TR, and hence more sampling points per unit time outweighs the SNR loss. This was indeed the case for the results obtained in this study (re. Table 2), where we found a greater GM and WM sensitivity for 3D EPI for all but one protocol (2.5 mm, sensitivity 31.8 vs. 30.6).
An advantage that follows from 3D acquisition is that the sensitivity reduction (‘g’-factor noise) due to parallel undersampling is less when spread out over the two phase encoding dimensions; this is also favored by the circular or helmet design of most phased array coils. Increasingly high acceleration factors are possible as RF coil arrays with large numbers of elements become more and more readily available; it has furthermore been shown that the SNR sacrifice due to high acceleration factors is relatively reduced at 3T and above (Wiesinger et al., 2004). It is worth pointing out here that the data for the tSNR and sensitivity analysis in part (a) were acquired with only an eight-element coil, implying that substantially larger benefits of 3D vs. 2D EPI in absolute terms can be expected for the same protocols when using the 16-, 24- or 32- channel coils that are now available on most human 7T systems.
One observation that was repeatedly made throughout the study is that image artifacts related to the use parallel imaging (i.e. effects comparable to ‘residual fold-over’ in SENSE) were generally lower in 3D EPI. Especially in 2D slices near inferior brain regions relatively stronger artifacts were sometimes observed, which can likely be attributed to the low SNR signal from these regions that makes the coil weight estimation for the GRAPPA reconstruction less reliable. The consequence is a further enhanced SNR difference between inferior and superior brain regions, potentially confounding for statistical fMRI analysis. In case of 3D EPI, one set of coil weights is fitted in 3D k-space, and hence there is no such slice dependence introduced via the reconstruction algorithm.
Independent of the considerations related to functional sensitivity, there are three practical limitations of 2D EPI sometimes encountered in high resolution protocols: First, SAR safety limits are rapidly reached even in GE-EPI without fat-saturation due to the large Ernst flip angle that is needed for the typically long TR (Speck et al., 2008); the factor Nslices shorter TR in 3D requires very much lower flip angles and hence energy deposition. Second, the effective spatial resolution and SNR may be compromised by the imperfect 2D slice profile; this is not an issue with 3D phase encoding, where only a few edge slices need discarding in case of axial slice positioning (no slices need discarding in case of sagittal whole-brain acquisitions). A sharp 2D slice profile can in principle be achieved by using pulses with a high bandwidth-time product, but this further increases SAR values and/or makes the pulses too long. Third, the slice-select gradient amplitude required for thin slices (<2 mm) easily reaches system limits and/or causes nerve stimulation unless the pulse duration, and thereby the minimum possible TE, is increased; for thick (3D) slabs this is no concern.
The studies by Trianafyllou et al. (Triantafyllou et al., 2005; Triantafyllou et al., 2006) suggest that to increase functional sensitivity it may be beneficial to sample at a higher spatial resolution than actually required for a given study, and to retrospectively ‘smooth down’ to the desired resolution in a post-processing step. This way the relative effect of physiological noise sources can be reduced during acquisition, but the SNR loss from the smaller voxel volume is fully regained by the smoothing. While conceptionally appealing, this is only to some degree practicable in 2D EPI due to the additional required investment into TR. This is not so in 3D EPI where undersampling can be applied along the slice direction, making this strategy much more viable in practice.
In conclusion, a 3D EPI sequence with full parallel and partial Fourier imaging capability along both phase encoding axes was implemented on a Siemens platform. 3D and corresponding 2D EPI acquisitions with a resting state paradigm at four different spatial resolutions (1.5–3.0mm isotropic voxels) revealed a comparable or better sensitivity of the 3D method. The larger vulnerability of 3D to physiological effects was outweighed by the much shorter volume TR, which moreover allows whole brain coverage at high resolution (1 mm isotropic voxels) within acceptable limits for event-related fMRI. Together with the improved practical aspects as discussed above (slice profile, reduced artifacts, shorter volume TR) our results show that 3D EPI can be a superior alternative to 2D EPI at 7T and should be considered especially if (near) whole-brain coverage is required where conventional 2D acquisition would lead to impracticable long TR.
Acknowledgments
Two authors (T.W. and L.L.W.) acknowledge support from National Institutes of Health grants: NCRR P41RR14075, NIBIB R01EB006847 and research support from Siemens Healthcare. One of the authors (L.L.W.) has obtained consulting income from Siemens Healthcare.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- de Zwart JA, van Gelderen P, Golay X, Ikonomidou VN, Duyn JH. Accelerated parallel imaging for functional imaging of the human brain. NMR Biomed. 2006;19:342–351. doi: 10.1002/nbm.1043. [DOI] [PubMed] [Google Scholar]
- de Zwart JA, van Gelderen P, Kellman P, Duyn JH. Application of sensitivity-encoded echo-planar imaging for blood oxygen level-dependent functional brain imaging. Magn Reson Med. 2002;48:1011–1020. doi: 10.1002/mrm.10303. [DOI] [PubMed] [Google Scholar]
- Golay X, Pruessmann KP, Weiger M, Crelier GR, Folkers PJ, Kollias SS, Boesiger P. PRESTO-SENSE: an ultrafast whole-brain fMRI technique. Magn Reson Med. 2000;43:779–786. doi: 10.1002/1522-2594(200006)43:6<779::aid-mrm1>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA) Magn Reson Med. 2002;47:1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- Hu Y, Glover G. Increasing the sensitivity to BOLD contrast in high resolution fMRI studies by using 3D spiral technique. Proceedings of the 15th Annual Meeting of ISMRM; Berlin. 1945.2007a. [Google Scholar]
- Hu Y, Glover GH. Three-dimensional spiral technique for high-resolution functional MRI. Magn Reson Med. 2007b;58:947–951. doi: 10.1002/mrm.21328. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Jesmanowicz A, Bandettini PA, Hyde JS. Single-shot half k-space high-resolution gradient-recalled EPI for fMRI at 3 Tesla. Magn Reson Med. 1998;40:754–762. doi: 10.1002/mrm.1910400517. [DOI] [PubMed] [Google Scholar]
- Klarhofer M, Dilharreguy B, van Gelderen P, Moonen CT. A PRESTO-SENSE sequence with alternating partial-Fourier encoding for rapid susceptibility-weighted 3D MRI time series. Magn Reson Med. 2003;50:830–838. doi: 10.1002/mrm.10599. [DOI] [PubMed] [Google Scholar]
- Lai S, Glover GH. Three-dimensional spiral fMRI technique: a comparison with 2D spiral acquisition. Magn Reson Med. 1998;39:68–78. doi: 10.1002/mrm.1910390112. [DOI] [PubMed] [Google Scholar]
- Liu G, Sobering G, Duyn J, Moonen CT. A functional MRI technique combining principles of echo-shifting with a train of observations (PRESTO) Magn Reson Med. 1993;30:764–768. doi: 10.1002/mrm.1910300617. [DOI] [PubMed] [Google Scholar]
- Mansfield P, Coxon R, Hykin J. Echo-volumar imaging (EVI) of the brain at 3.0 T: first normal volunteer and functional imaging results. J Comput Assist Tomogr. 1995;19:847–852. doi: 10.1097/00004728-199511000-00002. [DOI] [PubMed] [Google Scholar]
- Neggers SF, Hermans EJ, Ramsey NF. Enhanced sensitivity with fast three-dimensional blood-oxygen-level-dependent functional MRI: comparison of SENSE-PRESTO and 2D-EPI at 3 T. NMR Biomed. 2008;21:663–676. doi: 10.1002/nbm.1235. [DOI] [PubMed] [Google Scholar]
- Norris DG. Principles of magnetic resonance assessment of brain function. J Magn Reson Imaging. 2006;23:794–807. doi: 10.1002/jmri.20587. [DOI] [PubMed] [Google Scholar]
- Poser A, Norris D. BOLD Contrast Sensitivity Enhancement and Artifact Reduction with Parallel-Acquired Inhomogeneity Desensitized (PAID) fMRI. Proceedings to HBM; Toronto. 2005. p. 500. [DOI] [PubMed] [Google Scholar]
- Poser BA, Norris DG. Investigating the benefits of multi-echo EPI for fMRI at 7 T. Neuroimage. 2009;45:1162–1172. doi: 10.1016/j.neuroimage.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Poser BA, Versluis MJ, Hoogduin JM, Norris DG. BOLD contrast sensitivity enhancement and artifact reduction with multiecho EPI: parallel-acquired inhomogeneity-desensitized fMRI. Magn Reson Med. 2006;55:1227–1235. doi: 10.1002/mrm.20900. [DOI] [PubMed] [Google Scholar]
- Preibisch C, Pilatus U, Bunke J, Hoogenraad F, Zanella F, Lanfermann H. Functional MRI using sensitivity-encoded echo planar imaging (SENSE-EPI) Neuroimage. 2003;19:412–421. doi: 10.1016/s1053-8119(03)00080-6. [DOI] [PubMed] [Google Scholar]
- Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999;42:952–962. [PubMed] [Google Scholar]
- Schmidt CF, Degonda N, Luechinger R, Henke K, Boesiger P. Sensitivity-encoded (SENSE) echo planar fMRI at 3T in the medial temporal lobe. Neuroimage. 2005;25:625–641. doi: 10.1016/j.neuroimage.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song AW, Wong EC, Hyde JS. Echo-volume imaging. Magn Reson Med. 1994;32:668–671. doi: 10.1002/mrm.1910320518. [DOI] [PubMed] [Google Scholar]
- Speck O, Stadler J, Zaitsev M. High resolution single-shot EPI at 7T. Magma. 2008;21:73–86. doi: 10.1007/s10334-007-0087-x. [DOI] [PubMed] [Google Scholar]
- Triantafyllou C, Hoge RD, Krueger G, Wiggins CJ, Potthast A, Wiggins GC, Wald LL. Comparison of physiological noise at 1.5 T, 3 T and 7 T and optimization of fMRI acquisition parameters. Neuroimage. 2005;26:243–250. doi: 10.1016/j.neuroimage.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Triantafyllou C, Hoge RD, Wald LL. Effect of spatial smoothing on physiological noise in high-resolution fMRI. Neuroimage. 2006;32:551–557. doi: 10.1016/j.neuroimage.2006.04.182. [DOI] [PubMed] [Google Scholar]
- van der Zwaag W, Kober T, Marques J, Glover G, Gruetter R, Krueger G. Comparison of single-shot 3D EPI and segmented EVI acquisition for fMRI at 7T. Proceedings of the 17th Annual Meeting of ISMRM; Honolulu. 2009. p. 1550. [Google Scholar]
- Wiesinger F, Boesiger P, Pruessmann KP. Electrodynamics and ultimate SNR in parallel MR imaging. Magn Reson Med. 2004;52:376–390. doi: 10.1002/mrm.20183. [DOI] [PubMed] [Google Scholar]
- Wiggins GC, Wiggins CJ, Potthast A, Alagappan V, Kraff O, Reykowski A, Wald LL. A 32 Channel Receive-only Head Coil And Detunable Transmit Birdcage Coil For 7 Tesla Brain Imaging. Proc Intl Soc Mag Reson Med. 2006:415. [Google Scholar]
- Woolrich MW, Ripley BD, Brady M, Smith SM. Temporal autocorrelation in univariate linear modeling of FMRI data. Neuroimage. 2001;14:1370–1386. doi: 10.1006/nimg.2001.0931. [DOI] [PubMed] [Google Scholar]