Abstract
To gain insight into the structural and functional properties of the vesicular stomatitis virus nucleocapsid-RNA complex (vN-RNA), we analyzed it by treatment with proteolytic enzymes. Chymotrypsin treatment to the vN-RNA results in complete digestion of the C-terminal 86 amino acids of the N protein. The residual chymotrypsin resistant vN-RNA complex (vΔN-RNA) carrying N-terminal 336 amino acids of the N protein (ΔN) was inactive in transcription. The ΔN protein retained its capability to protect the genomic RNA from nuclease digestion but failed to interact to the P protein. Interestingly, addition of excess amount of P protein rendered the vN-RNA complex resistant to the chymotrypsin digestion. Finally, our data revealed that the recombinant N-RNA complex purified from bacteria (bN-RNA) is resistant to chymotrypsin digestion, suggesting that the C-terminal unstructured domain (C-loop) remains inaccessible to protease digestion. Detail comparative analyses of the vN-RNA and vΔN-RNA are discussed.
Keywords: VSV Nucleocapsid-RNA complex, chymotrypsin digestion, biochemical analyses, structural properties
Introduction
Vesicular stomatitis virus (VSV), a prototype of non-segmented negative stranded RNA virus belonging to the rhabdovirus family, contains a single stranded RNA genome of negative polarity consisting of 11,161 nucleotides (Banerjee, 1987a; Banerjee et al., 1991; Banerjee and Barik, 1992; Rose and Schubert, 1987). The viral genomic RNA is encapsidated by ~1200 molecules of nucleocapsid (N) protein (47 kDa) to form the nuclease resistant viral N-RNA complex (vN-RNA), which serves as the template for mRNA transcription as well as genome replication (Banerjee, 1987a; Thomas et al., 1985). The vN-RNA complex associates with ~50 copies of L protein (242 kDa), the RNA-dependent RNA polymerase (RdRp) and ~500 copies of the P protein (30 kDa), a cofactor for the L protein, to form the transcribing viral ribonucleoprotein complex (vRNP) (Emerson and Yu, 1975; Thomas et al.,1985; De and Banerjee, 1985). The P protein, which interacts with both N and L proteins, mediates the contact between the vN-RNA template and the L protein (Mellon and Emerson, 1978). However, the interesting question as to how the RdRp gains access to the promoter for the initiation of transcription and carries out subsequent RNA chain elongation, while the N protein molecules remain associated with the genome RNA, still remains an enigma. Due to the high molecular weight and structural complexity, the crystal structure of the vRNP or the vN-RNA complexes remained unsolved to date. Similarly, effective packaging of the full-length genome RNA in vitro using recombinant N protein has also been unsuccessful. Only a short leader RNA (47 bases) was encapsidated in vitro with the N protein, albeit inefficiently although it remained protected against ribonuclease digestion (Das and Banerjee, 1992; Das et al., 1999).
Recently, Green et al. (2000, 2006a) have been able to purify recombinant N-RNA complex from E. coli and solved the crystal structure at 2.9Ǻ. This N-RNA complex (bN-RNA) consists of 10 molecules of recombinant VSV N protein and 90 nucleotide of bacterial RNA. Each N molecule in the ring like bN-RNA oligomer has a bilobed structure with the RNA tightly bound in the cavity at the interface between the N- and C-terminal helical lobes (Green et al., 2006a). Importantly, the VSV N protein was able to protect the bacterial RNA from ribonuclease action presumably by tight contacts with several key amino acid residues of the N protein residing in the middle of the protein (Green et al., 2006a). Concurrently, elucidation of crystal structure of the rabies virus recombinant N-RNA complex, purified from the insect cells also showed similar ring like and bilobed structure (Albertini et al., 2006). Recently, electron micrographic analyses of the respiratory syncytial virus (RSV) and Mumps virus (Mu V) recombinant N-RNA complexes, purified from insect cells and E coli, respectively, also revealed similar ring-like morphology (MacLellan et al., 2007; Cox et al., 2009). However, since the recombinant bN-RNA complexes lacked the viral genomic RNA it was not possible to test if they can serve as template for the viral RdRp holoenzyme complexes in a transcription/reconstitution system.
Unavailability of the crystal structure of the vN-RNA provided an impetus to probe into its structure using biochemical techniques particularly by treatment with several proteases. Here, we demonstrate that chymotrypsin treatment of the vN-RNA complex removes precisely 86 amino acids from the C-terminal end of the N protein. A detail comparative biochemical analyses of the vN-RNA and the chymotrypsin-treated vΔN-RNA provided a deeper insight into the structure of the vN-RNA complex.
Results
Protease digestion of the virion N-RNA complex
The virion N-RNA complex (vN-RNA), purified from mature VSV particles through CsCl equilibrium density gradient centrifugation was resolved in SDS-PAGE and detected by coomassie brilliant blue (CBB) staining and Western blot analysis using anti-N antibody. As shown in Fig. 1A a single band of the N protein was discernible confirming the purity of the vN-RNA preparation. This purified vN-RNA was used in all of the subsequent experiments. To probe into the spatial arrangement of the N protein bound to the genome RNA, we initiated studies by treatment of vN-RNA complex with several proteases, such as trypsin, pepsin and chymotrypsin. As shown in Fig 1B, we confirmed our earlier studies that controlled proteolysis by chymotrypsin digested a portion of N-protein (~10 kDa) from vN-RNA complex leaving a ~37 kDa N-protein intact (Banerjee et al., 1987b). Treatment with trypsin or pepsin failed to digest the N protein from the vN-RNA complex (data not shown). To obtain quantitative estimation of the extent of proteolysis, vN-RNA was incubated with chymotrypsin with different (w/w) ratios (vN-RNA: chymotrypsin = 4.25:1 or 8.5:1 or 17:1) in a buffer containing 10 mM Tris-HCl, pH 8.0 at 37°C for 20 min. As shown in Fig 1B, chymotrypsin treatment, at ratios 17:1 or 8.5:1 (lane 4 and 5) resulted in the production of ~37 kDa N-protein (ΔN protein), although some residual, uncleaved N protein was also observed. At higher concentration of the chymotrypsin (vN-RNA: chymotrypsin = 4.25:1), the N protein was completely digested to produce the ΔN protein only (Fig. 1B, lane 6). As shown in Fig. 1C, the chymotrypsin digestion for 5 min at 37°C resulted in virtually 100% conversion of N to ΔN and the ΔN remains intact till 20 min of incubation. Thus it seems that chymotrypsin treatment specifically cleaves and digests a portion of the N protein from the complex (~10 kDa). In subsequent experiments we maintained the above mentioned concentration of chrymotrypsin (vN-RNA: chymotrypsin = 4.25:1) and incubation time (20 min).
FIG. 1.
Chymotrypsin digestion of virion N-RNA. (A) Purified vN-RNA complex was mixed with SDS-PAGE loading buffer, heated to 100°C and resolved in SDS-PAGE. The N protein was visualized either by CBB stain (lane 2) or WB with anti-VSV N polyclonal antibody (lane 3). Purified VSV was used as positive control (lane 1) (B) Chymotrypsin digestion profile of vN-RNA complex at three different (w:w) ratios. (C) Time kinetics profile of chymotrypsin digestion of vN-RNA complex at 4.25:1 (w:w) (vN-RNA:Chymotrypsin) ratio from 0–20 min.
Mass spectrometric analysis of the ΔN protein
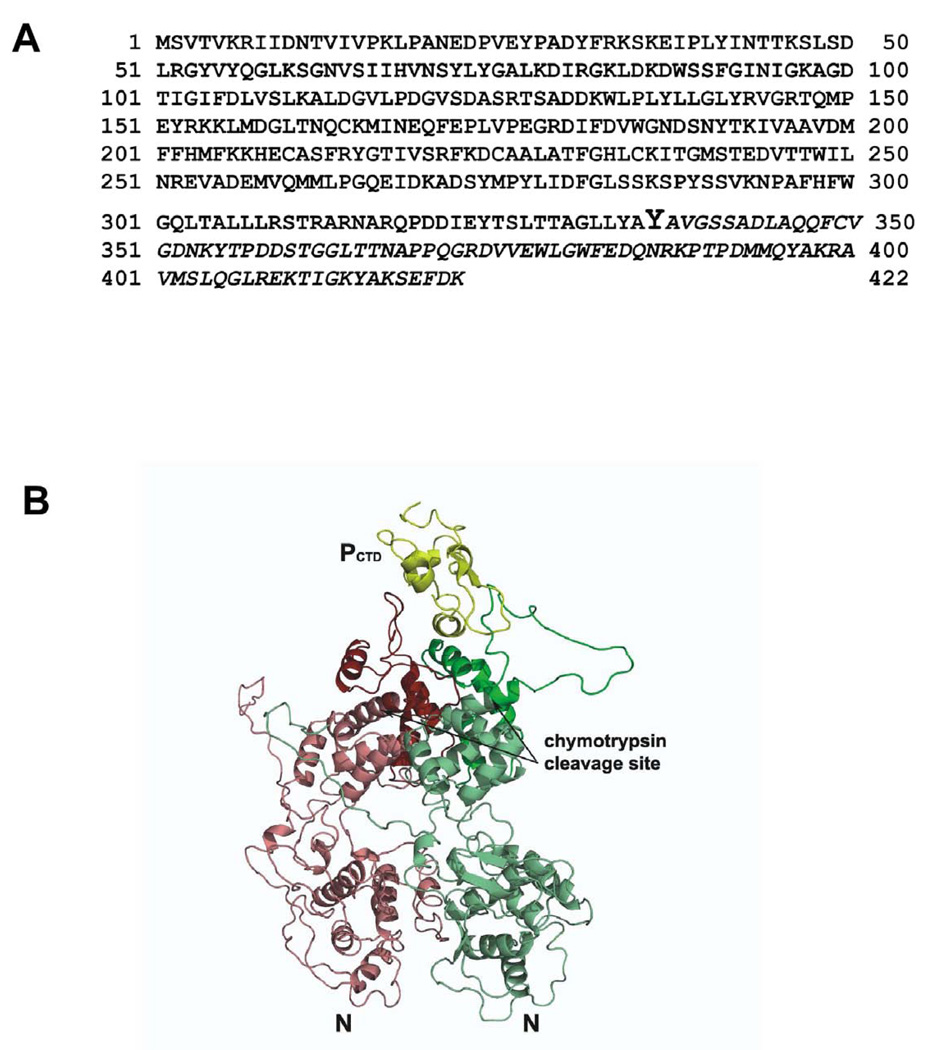
To determine the exact chymotrypsin cleavage site, the full-length N (47 kDa) and the ΔN (~37kDa) proteins were subjected to mass spectrometric analysis as described in Materials and Methods. A comparative analysis of the protein sequences identified in the N and the ΔN bands showed that the chymotryptic truncation occurred at the C-terminal portion of the N protein (Fig. 2) at a tyrosine (Y336) residue (shown in larger font). These results clearly indicate that the C-terminal 86 amino acids (designated as NC-86) of calculated molecular mass 9.5 kDa was specifically digested by chymotrypsin (shown in italics), indicating that only this domain of the N protein in the vN-RNA complex remains accessible to the chymotrypsin action. The chymotrypsin cleavage site in the N protein is shown in Fig 2B.
FIG. 2.
(A) Amino acid sequence of the VSV N protein. The ΔN portion of the protein, as determined by LC-MS analysis, is shown in regular front and the NC-86 protein (the C-terminal 86 amino acid) is shown in italics. The chymotrypsin cleavage site Y336 is shown in larger font. (B) The structure of the C-terminal nucleocapsid binding domain of the phosphoprotein (P), PCTD, in complex with two nucleocapsid protein (N) subunits is presented as a ribbon drawing. PCTD (yellow) corresponds to residues 193–265. The two N subunits (red and green) are oriented so that the cavity that encapsidates RNA is facing away. The chymotrypsin cleavage site in each subunit is marked by an arrow. The cleavage will remove the C-terminal portion of the N subunits (dark red and dark green) that makes contact with PCTD. The coordinates used for preparing this figure are derived from PDB 3HHZ. The figure was prepared with program PyMOL (DeLano, 2002).
Biochemical analyses of the vΔN-RNA
To address the structural and functional alterations that occurred to the vN-RNA complex following chymotrypsin digestion, we studied the following biochemical properties of the vΔN-RNA complex.
(a) structural integrity of the vΔN-RNA
The purified vN-RNA template was digested with chymotrypsin (vN-RNA: chymotrypsin = 4.25:1) for 20 min and banded in CsCl equilibrium density gradient ultracentrifugation as described in Materials and Methods. As a control, purified vN-RNA was similarly banded in CsCl and collected fractions were analyzed by 15% SDS-PAGE. As shown in Fig. 3A the vN-RNA (lanes 2 and 3) banded in the same fractions with similar density in the gradient as the vΔN -RNA (lanes 4 and 5) suggesting that the overall structural integrity of both complexes were similar.
FIG. 3.
Biochemical analyses of vN-RNA and vΔN-RNA. (A) vN-RNA was digested with chymotrypsin to obtain vΔN-RNA and vΔN-RNA was further subjected to CsCl equilibrium density gradient ultracentrifugation. Purified proteins were resolved in SDS-PAGE and visualized by CBB stain. Banding profile of vN-RNA (lane 2 and 3) and vΔN-RNA (lane 4 and 5) in CsCl equilibrium density gradient centrifugation is shown in figure. (B) In vitro transcription reconstitution reaction performed with vL-P and vN-RNA or vΔN-RNA complexes. 32P-labeled mRNA transcripts were analyzed in 5% urea-PAGE followed by autoradiography. All of the detected transcripts are indicated. (C) 32P-labeled VSV was prepared and vN-RNA was purified from the 32P-labeled VSV. The vN-RNA, containing 32P-labeled genome RNA, was digested with chymotrypsin and the digested products were further purified through CsCl equilibrium density gradient ultracentrifugation to obtain vΔN-RNA. vN-RNA and vΔN-RNA, containing 32P-labeled genome RNA, were digested with Micrococcal nuclease and resolved in 5% urea-PAGE. The 32P-labeled genomic RNA present within vN-RNA and vΔN-RNA were detected by autoradiography.
(b) transcription activity of vΔN-RNA
In vitro transcription-reconstitution reaction was performed with the vL-P complex using vN-RNA or vΔN-RNA complexes as templates as described in Materials and Methods. As shown in Fig. 3B, the vN-RNA supported in vitro transcription efficiently and synthesized full length mRNAs (lane 3), whereas vΔN-RNA was completely inactive in transcription (lane 5). These results confirmed our previous findings (Banerjee et al., 1987b) and indicated that removal of NC-86 portion of the N protein rendered the vN-RNA template non-functional in transcription.
(c) nuclease sensitivity of the genome RNA in vΔN-RNA
Next, we wanted to test if the truncated nucleoprotein complex retains its ability to protect the genome RNA from nuclease digestion. The genome RNA was labeled with 32P-orthophosphate and the vN-RNA and vΔN-RNA complexes containing 32P-labeled genome RNA were purified as described in the Materials and Methods. The complexes were incubated with 0, 0.06 and 0.12 (U/µg vN-RNA) of micrococcal nuclease in separate reaction mixtures containing 50 mM Tris-HCl, pH 8.0, 4 mM CaCl2 at 30°C for 1 hr. The reactions were terminated with 10 mM EGTA and SDS was added to a final concentration of 0.5%. The 32P-labeled genomic RNA was resolved in 5% PAGE containing 7 M urea and detected by autoradiography. As shown in Fig. 3C, the genome RNA from both the vN-RNA (lanes 2 and 3) and the vΔN-RNA (lanes 5 and 6) complexes were resistant to micrococcal nuclease digestion indicating that the vΔN-RNA retained the ability to protect the genome RNA from nuclease action and the NC-86 does not play a direct role in protecting the genome RNA from digestion by nuclease.
Interaction of vN-RNA and vΔN-RNA with the P protein
Since vΔN-RNA was found to be inactive in transcription, although retaining its ability to protect the genomic RNA from nuclease action, we wanted to test if the lack of template function was primarily due to its inability to interact with the P protein. To test this contention, the interaction of P protein with the vN-RNA and vΔN-RNA was carried out in vitro in the following manner. A plasmid expressing Myc tagged P protein (Myc-P) was transfected in HeLa cells and after 20 hour of post-transfection cell lysates were prepared and incubated with purified vN-RNA or vΔN-RNA for 1 hr at 30°C as detailed in Materials and Methods. The reaction mixtures were diluted and centrifuged at 120,000 × g for 1 hr and the pelleted protein complexes were analyzed by Western blot using anti-N and anti-Myc antibodies. As shown in Fig 4, both N (lane 2 and 3, lower panel) and ΔN (lane 4 and 5, lower panel) proteins were detected in the pellet. As expected, the vN-RNA complex was able to pellet Myc-P (lane 3, upper panel) whereas vΔN-RNA failed to do so (lane 5, upper panel). The Myc-P when incubated alone in a control experiment and centrifuged in the same manner did not pellet (lane 6, upper panel). These data indicate that vΔN-RNA lost its ability to interact with P protein due to the removal of C-terminal 86 amino acids.
FIG. 4.
In vitro interaction between the P protein and vN-RNA or vΔN-RNA. HeLa cells were transfected with a mock plasmid or a plasmid expressing Myc-P as detailed in Materials and Methods. At 20 hr post transfection lysates were incubated with the vN-RNA or the vΔN-RNA complexes at 30°C for 1 hr. The reaction mixtures were then centrifuged at 120,000 × g for 1 hr using a Sorvall TH660 rotor. Protein complexes present in the pellet were detected by Western blot analysis using anti-N or anti-Myc polyclonal antibodies.
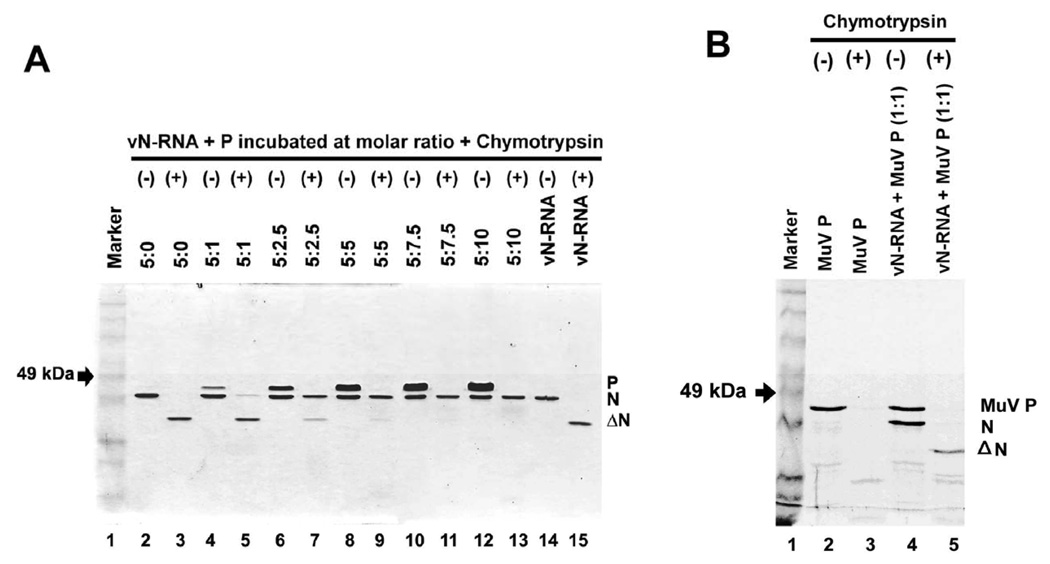
We, next, wanted to test if binding to the P protein leads to any conformational alteration of the vN-RNA complex which is active for transcription. To test this contention, the vN-RNA was incubated at 30°C for 1 hr with P protein at different molar ratios and the complexes were subjected to chymotrypsin treatment at 4.25:1 (w/w) (vN-RNA: chymotrypsin) ratio at 37°C for 20 min. The digested proteins were then resolved in 10% SDS-PAGE and detected by CBB stain. As expected, vN-RNA was digested by the chymotrypsin to form the ΔN (Fig 5A, lane 3) in the absence of P protein prior to digestion. However, when the P protein was added in increasing amounts followed by chymotrypsin treatment, a gradual decrease in the appearance of ΔN was observed (lane 4–13). Only 80% and 30% ΔN were produced compared to control at the molar ratios of vN-RNA:P 5:1 and 5:2.5, respectively (Fig. 5A, lanes 5 and 7). At vN-RNA:P molar ratios 5:5, 5:7.5 and 5:10), vN-RNA became totally resistant to the chymotrypsin digestion (Fig. 5A, lanes 9, 11 and 13). It is important to note that at all levels of added P protein, it was completely digested by the chymotrypsin (lanes 5, 7, 9, 11 and 13) and the N protein remained intact. Since, the P protein interacts with the vN-RNA complex through a small region (residues 251–265 of P; helix 4) (Green et al, 2009 and Fig 2B), it is possible that this region (which is not detectable in SDS-PAGE after chymotrypsin digestion) is sufficient to protect the vN-RNA from chymotrypsin digestion. Alternatively, the P protein, upon binding to the C-terminal domain of the N protein is capable of altering its conformation to a highly compact form rendering the protein inaccessible to the chymotrypsin action. In a control experiment when the bacterially expressed purified P protein of mumps virus was used instead of the VSV P protein the vN-RNA was effectively converted to ΔN following digestion with chymotrypsin (Fig.5B) under these conditions. Thus, it seems that P protein plays a critical role by specifically interacting with the vN-RNA and protects the vN-RNA from the chymotrypsin action.
FIG. 5.
Protease protection assay of vN-RNA complex. (A) vN-RNA complex was incubated with the P protein at 30°C for 1 hr complex formation. Chymotrypsin digestion profile of N protein within the vN-RNA complex when complexed with P protein formed at different molar ratios (vN-RNA:P = 5:0, 5:1; 5:2.5, 5:5, 5:7.5 and 5:10). (B) vN-RNA complex and purified P protein of mumps virus (MuV) incubated at 1:1 molar ratio at 30°C temperature for 1 hr and then digested with chymotrypsin as described in materials and methods.
Structural properties of N-RNA complex purified from the infected cells
Since in vitro interaction of the P protein and the vN-RNA complex protected the latter from chymotrypsin digestion, it was of interest to test if the RNP complex purified from VSV infected BHK cells (iRNP) and its corresponding N-RNA (iN-RNA) prepared from it can be digested with chymotrypsin. The vRNP and iRNP complexes were purified as described in the Materials and Methods and subjected to chymotrypsin treatment at a ratio of 4.25:1 (w/w) (vRNP/iRNP:chymotrypsin) in the buffer containing 10 mM Tris-HCl, pH 8.0 at 37°C for 20 min. As shown in Fig 6A, as expected, the chymotrypsin treatment of vRNP resulted in the formation of 37 kDa ΔN product; the L and P proteins, present in vRNP, were totally digested by the chymotrypsin (lane 4). In contrast, the purified iRNP, when treated similarly with chymotrypsin, no 37 kDa ΔN product was produced and the N protein remained resistant to the chymotrypsin digestion (lane 6). As expected, the L and P proteins present in the iRNP were completely digested by the chymotrypsin (lane 6). Thus, it seems that similar to the P protein interaction with vN-RNA described above, the association of excess P proteins to the N protein present in the iRNP (Hsu et al, 1979; Patton et al., 1983), protected the N protein from the chymotrypsin digestion. The vRNP, on the other hand, probably due to the presence of low quantities of P proteins is digestible by chymotrypsin.
FIG. 6.
Chymotrypsin digestion of virion RNP (vRNP), infected cell RNP (iRNP) and infected cell N-RNA (iN-RNA) complexes. (A) Comparative analysis of the Chymotrypsin digestion profile of vRNP and infected cell RNP (iRNP) (B) Chymotrypsin digestion profile of the purified vN-RNA and infected cell N-RNA (iN-RNA), obtained from iRNP.
Next, the vN-RNA and iN-RNA (Fig 6B, lane 2 and 4) complexes purified from the corresponding RNPs were subjected to chymotrypsin treatment at a ratio of 4.25:1 (w/w) (N-RNA: chymotrypsin). As shown in Fig. 6B, lane 5 the iN-RNA complex, upon chymotrypsin treatment, produced the same ΔN fragment suggesting that removal of the L and P proteins produced a complex which is similar to vN-RNA complex, again indicating that excess of intracellular L and P proteins bound to iRNP rendered the structure different from vRNP.
Structural properties of the N-RNA complex purified from E. coli
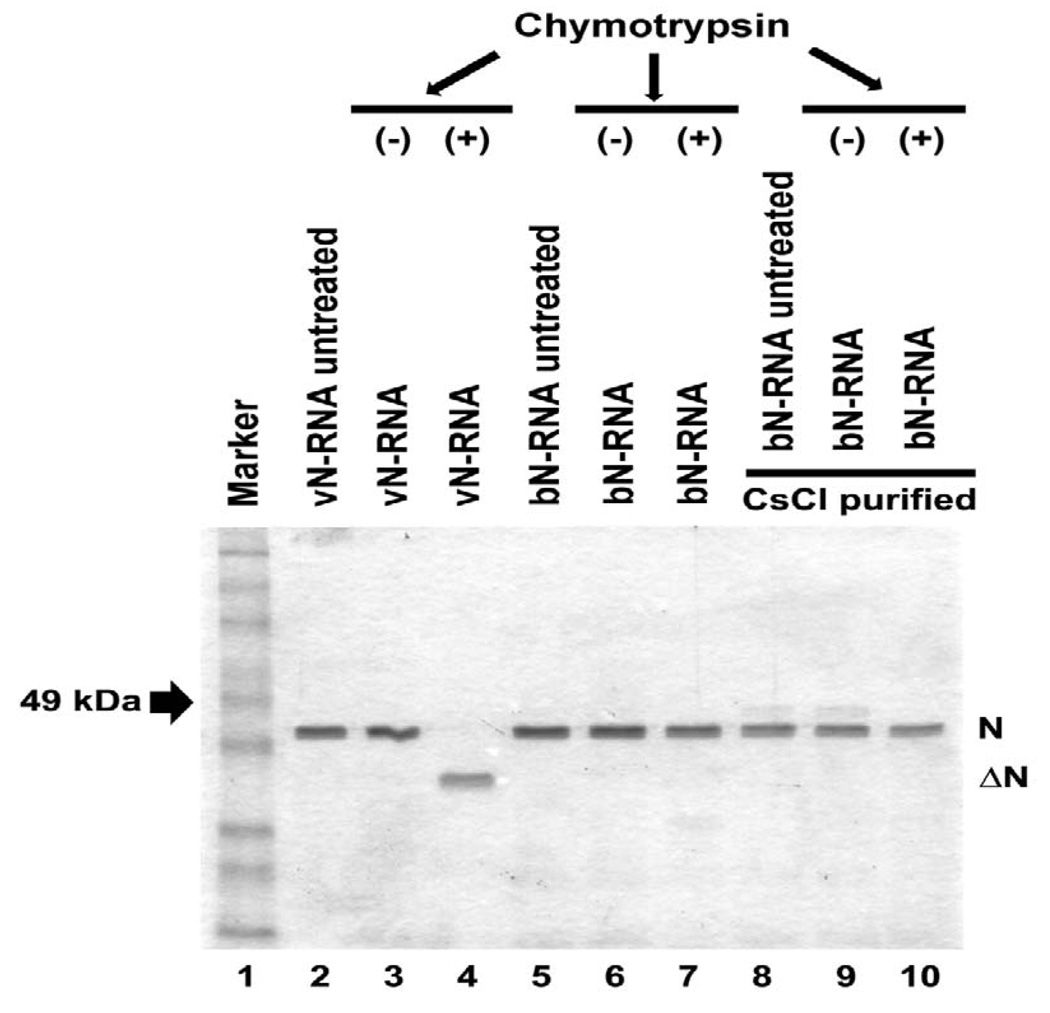
Finally, we wanted to study the structure of the N-RNA obtained from E. coli (bN-RNA), the crystal structure of which was recently solved (Green et al., 2006a). The complex was purified as previously described (Green et. al., 2006a). The purity of the bN-RNA protein complex was verified by resolving it in 10% SDS-PAGE followed by CBB staining and also by Western blot analyses using anti-N and anti-P antibodies (data not shown). The bN-RNA complex was then digested with the chymotrypsin under the same reaction conditions as described from vRNA. In contrast to vN-RNA, the bN-RNA complex remained totally resistant to the chymotrypsin digestion (Fig 7, lane 7). Further purification of bN-RNA through the CsCl equilibrium density gradient ultracentrifugation (bN-RNACsCl) to remove any contaminating P protein, also failed to produce ΔN fragment upon treatment with chymotrypsin (Fig. 7, lane 10). These results strongly suggest that the quaternary structures of the vN-RNA and the bN-RNA are significantly different.
FIG. 7.
Chymotrypsin digestion profile of bacterial N-RNA complex. SDS-PAGE analysis of the purified bacterial N-RNA complex (bN-RNA) from bacteria and CsCl equilibrium density gradient purified bacterial N-RNA complex (bN-RNACsCl) with respect to vN-RNA complex when digested with and without chymotrypsin at 4.25:1 (w:w) ratio.
Discussion
The N protein of VSV encapsidates the genomic RNA to form vN-RNA complex and the genomic RNA inside the vN-RNA remains resistant to RNAase digestion (Banerjee, 1987a; Chanda and Banerjee, 1979). The vN-RNA complex within the virion also packages L and P proteins that constitute the vRNP which is trancriptionally active in vitro and in vivo. There are ~1200 N protein monomers that encapsidates the genome RNA (~11kb), thus, approximately 1 N monomer covers 9 bases. Due to the large size and complexity the crystal structure of the vRNP or the vN-RNA complexes remained unsolved. Recently, however, the recombinant N protein expressed in E. coli was found to contain 10 molecules of VSV N protein bound to 90 nucleotides of bacterial RNA which was subsequently crystallized and its structure solved (Green et al., 2000; Green et al., 2006a). The crystal structure clearly showed that the N molecules in the complex have a bilobed structure with the bacterial RNA tightly bound in the cavity at the interface between the N- and C-terminal lobes. The RNA within the complex was resistant to nuclease. The RNA binding cavity consisted of side chains of Arg143, Arg146 and Lys155 from N-terminal lobe and side chains of Lys286, Arg317 and Arg408 from C-terminal lobe seem to be involved in the binding of the phosphate groups of the RNA (Green et al., 2006). Each monomer has an extended N-terminal arm and a C-terminal extended loop. The network of contacts among the monomers possibly brings the orderly assembly of the N protein on the nascent RNA during replication. Recent structural analyses of recombinant N-RNA complexes of several viruses belonging to the mononegavirales order revealed similar bilobed, ring-like structures (Albertini et al., 2006; Luo et al, 2007; Green et al., 2006a; Green et al., 2006b; MacLellan et al., 2007; Cox et al., 2009; Tawar et al., 2009). However, the ability of the recombinant N-RNA complexes to act as template for viral RdRp heterocomplexes can not be tested due to the lack of viral RNA in the complexes.
To gain insight into the arrangement of N proteins on full length VSV genomic RNA, we characterized the vN-RNA complex biochemically by digesting it with proteases, such as trypsin, pepsin and chymotrypsin. Our analyses revealed that chymotrypsin, on limited proteolysis, can produce a 37 kDa (ΔN) fragment from the 47 kDa full length N protein present in the vN-RNA complex (Fig. 1), while trypsin and pepsin could not digest the vN-RNA (data not shown). LC-MS studies of the N and ΔN proteins, confirmed that the C-terminal 86 amino acids (NC-86) of the N protein were completely digested by the chymotrypsin (Fig 2). It is important to note that the proteolytic cleavage site (Y336) falls in the α helical region, α-12, just prior to the extended loop region of the previously solved crystal structure of bN-RNA, which, interestingly, contains a characteristic unstructured region (Green et al., 2006a and Fig 2). Thus, it seems that this extended loop may be the target for the chymotrypsin action (Fig 2). Further analysis showed that both vN-RNA and vΔN-RNA banded at the same position in the CsCl density gradient ultracentrifugation during the purification, suggesting that the overall structural integrity of the vΔN-RNA was maintained (Fig 3). However, the vΔN-RNA failed to produce mRNA transcripts in trancripstion reaction when reconstituted with the vL-P complex in vitro (Fig. 3), but micrococcal nuclease treatment to the 32P labeled vN-RNA and vΔN-RNA confirmed that the genomic RNA of VSV in 32P-vΔN-RNA complexes was resistant to the nuclease treatment (Fig. 3). Coupled with the fact that recent analyses revealed that point mutations in the C-loop region of the N protein, although resulted in alerted transcription/replication, whereas it did not affect RNA encapsidation in vivo (Harouaka et al, 2009), it seems that the NC-86, although indispensable for transcription, is not directly involved in protecting the genomic RNA from nuclease digestion. This region possibly remains protruded out of the vN-RNA complex, and thus, digestable by chymotrypsin; following proteolysis the residual N molecules are sufficient to protect the genome RNA. Consistent with this observation, mutation of a single amino acid (Ser290→Trp), which lies within the ΔN region, leads to loss of the capability of the bacterially expressed N protein to encapsidate RNA (Zhang et al., 2008). Moreover, since the C-terminal 5 amino acids of the N protein have been shown to play a critical role in encapsidation of the leader RNA in vitro (Das et al., 1999) it is, thus, possible that the extreme C-terminus of the NC-86 might be involved in the initiation of the encapsidation process.
Subsequent biochemical analyses revealed that the vΔN-RNA, unlike vN-RNA, failed to interact to the P protein (Fig 4). Interestingly, trypsin digestion of rabies virus N-RNA also removes a C-terminal region, which results in loss of P binding (Schoehn et al., 2001). Consistent with this observation, the recent structural analyses revealed that each monomeric PCTD (amino acids 178–265 of P protein; consists of an antiparallel β-turn and 5 α-helices) of VSV interacts with two adjacent N monomers by binding to the C-loop of the N monomers (Green and Luo, 2009; Ribeiro et al, 2008; also shown in Fig 2B). The 17 amino acids of two adjacent N monomers (11 and 6 residues from each monomer) that contribute to this specific interaction are present in a contiguous stretch, between 354 and 386 (Green and Luo, 2009). Therefore, our data strongly suggest that the ΔN-RNA failed to act as template in transcription primarily due to its inability to interact to the P protein indicating that the vN-RNA complex must interact with the P protein (and the RdRp) through the NC-86 region (Green and Luo, 2009; Ribeiro et al, 2009). These interactions, which are mostly H bonds and hydrophobic interactions are, as suggested, transient and a ‘dynamic make and break’ of these interactions, are necessary to facilitate the movement of the RdRp to the next N molecule and hence the next 9 nucleotides of the genome RNA (Green and Luo, 2009; Longhi, 2009).
Our further biochemical analyses revealed that excess amount of P protein interact with the vN-RNA complex and renders the latter inaccessible to the chymotrypsin action (Fig 5). It appears that the small vN-RNA binding domain of P protein (Green and Luo, 2009) is sufficient to protect the complex from chymotrypsin digestion. Alternatively, the P protein, due to its chaperone action (Chen et al., 2007; Davis et al., 1986; Masters and Banerjee, 1988a; Masters and Banerjee, 1988b, Majumder et al., 2001; Majumder et al., 2004), might possess intrinsic capability to alter the conformation of the N protein even when the N protein is present in vN-RNA complex. These observations are consistent with the fact that the interaction between N and P proteins causes structural changes in both the proteins and the significant change occurs in the C-loop of the N monomers, where the P protein interacts to the N protein (Green and Luo, 2009). This contention was further borne out by our findings that the structure of nucleocapsid isolated from the infected cells (iRNP) which contain excess of intracellular L and P proteins were similarly resistant to chymotrypsin action (Fig 6). In contrast, the vRNP which contains a limited quantity of the P protein release the ΔN protein upon chymotrypsin digestion. Interestingly, the iN-RNA that was purified from the iRNP, when digested with chymotrypsin, produced the same ΔN fragment as that of vN-RNA, suggesting that core N-RNA structures of both complexes are similar (Fig. 6).
Finally, our observation that recombinant bN-RNA was resistant to such proteolytic digestion strongly suggest that the quaternary structure of the complex is different from both vN-RNA and iN-RNA complexes. It is possible that despite containing the unstructured C-terminal loop the structure of smaller 10 subunit ring of bN-RNA complex, due to the close contacts between the N and C-lobes, is relatively more compact and hence is resistant to proteolytic digestion. Further studies, along these lines may shed light on the detail structural aspect of the N-RNA complex.
Material and Methods
Cells and viruses
BHK-21 and HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). VSVIND Mudd-summers strain was propagated in BHK-21 monolayer cells and purified as described previously (Barik and Banerjee, 1991). Recombinant vaccinia virus (vTF7-3) carrying the bacteriophage T7 RNA polymerase gene was propagated in HeLa cells.
Purification of viral L-P and N-RNA complexes
Purified virus were then subjected to further purification steps for the isolation of purified vRNP, vL-P and vN-RNA complexes following the previously described earlier (Ogino and Banerjee, 2007).
Chymotrypsin treatment of N-RNA complex and purification of ΔN-RNA complex
The vN-RNA template was treated with chymotrypsin A (Boehringer Mannheim) at a ratio of 4.25:1, 8.5:1 and 17:1 (w/w) (vN-RNA: chymotrypsin) in the buffer containing 10 mM Tris-HCl, pH 8.0 at 37°C for 20 min. Reactions were stopped by adding Laemmli buffer and analyzed immediately by SDS-PAGE. A time kinetics was also performed with the ratio of 4.25:1 (w/w) (vN-RNA: chymotrypsin) in 10mM Tris-HCl, pH 8.0 at 37°C from 0 to 20 min at different time intervals. 100 µg of vN-RNA template was digested with chymotrypsin at a ratio of 4.25:1 to generate vΔN-RNA complex and then was diluted in 4 ml of TE buffer containing 0.1% (w/v) Triton X-100 and CsCl to give an initial density of 1.29 g/cc, and centrifuged to equilibrium in a Sorvall TH-660 rotor at 120,000 × g for 16 hr at 20°C. The fractions containing the vΔN-RNA were diluted eightfold with the high salt buffer, HSB (1600 mM NaCl, 40 mM Tris-HCl [pH 8.0], 2 mM DTT) and then then centrifuged at 120,000 × g for 1 hr in Sorvall TH-660 rotor. The resulting pellet of the vΔN-RNA was resuspended in TE buffer containing 10% glycerol and was dialyzed against the same buffer and then concentrated using Amicon Ultra-15 centrifugal filter device (Millipore) and stored at −80°C freezer.
Mass spectrometric analysis
The purified vN-RNA and vΔN-RNA protein bands were cut out from the gel, divided into smaller pieces, destained and washed with water and dehydrated in acetonitrile. The protein bands were then alkylated with iodoacetamide and digested in-gel by adding 5 µl of 20 ng/µl trypsin in 50 mM (NH4)2CO3 and incubated overnight at room temperature. The resulting peptides were extracted from the polyacrylamide in two aliquots of 30 µl 50% acetonitrile with 5% formic acid. These extracts were combined and evaporated to <10 µl in a speedvac, resuspended in 30µl of 1% acetic acid and analyzed by Liquid Chromatography Mass Spectrometry (LC-MS) using a Finnigan LCQ ion trap mass spectrometer system. A detail LC-tandem MS and the CID spectra generated multiple fragments of N protein sequence (data not shown). These analyses were carried out in the mass spectrometry core facility of Lerner Research Institute, Cleveland Clinic.
In vitro transcription reaction
In vitro reconstituted transcription reaction was performed with the vN-RNA or vΔN-RNA complex and vL-P complex as described earlier (Ogino and Banerjee, 2007). The synthesized mRNAs were resolved in 5% PAGE containing 7M urea visualized by autoradiography.
Micrococcal nuclease treatment of 32P labeled viral N-RNA and ΔN-RNA complexes
For labeling the VSV genome, BHK-21 monolayer cells were grown in confluent manner and infected with VSV at 10 pfu/cell in presence of 70µCi/ml of 32P-orthophosphorus (Perkin-Elmer) and labeled for 24 hr following the previous protocol (Chanda and Banerjee, 1979). 32P-labeled virus was recovered and subsequently 32P-labeled virion nucleocapsid (32P-vN-RNA) was purified as described above. 100µg of 32P-vN-RNA template was digested with chymotrypsin A at a ratio of 4.25:1 (w/w) (32P-vN-RNA: chymotrypsin) in the buffer containing 10 mM Tris-HCl, pH 8.0 at 37°C for 20 min and then was diluted in 4 ml of TE buffer containing 0.1% (w/v) Triton X-100 and CsCl to develop an initial density of 1.29 g/cc, and centrifuged to equilibrium in a Sorvall TH-660 rotor at 120,000 × g for 16 hr at 20°C. The fractions containing the 32P-vΔN-RNA were then purified following the above mentioned protocol. 3 µg of 32P-vN-RNA or 32P-vΔN-RNA were digested in separate reaction mixtures containing 0, 0.06 and 0.12 (U/µg vN-RNA) of micrococcal nuclease containing 50 mM Tris-HCl, pH 8.0, 4 mM CaCl2 and digested at 30°C for 1 hr. The reactions were stopped with 10 mM ethyleneglycol-bis-(2-amino ehtylether) N, N’ tetractetic acid (EGTA) to inactivate the nuclease and SDS was added to a final concentration of 0.5%. The micrococcal nuclease digested 32P labeled RNA samples were analyzed by 5% PAGE containing 7M urea, followed by auto radiography.
In vitro complex formation between viral N-RNA/ΔN-RNA and P protein
HeLa cells were infected with recombinant vaccinia virus (vTF7-3) expressing T7 RNA polymerase and at 2 hour of post infection cells were transfected with 1 µg of plasmid expressing Myc-P (Chen et al., 2006) using lipofectin (Invitrogen) using manufacturer’s protocol. At 20 hr. post transfection cells were lysed and 216 µg of either Myc-P or mock cell lysates were incubated with either 5 µg of vN-RNA or vΔN-RNA at 30°C for 1 hour in a buffer containing 50 mM Tris (pH 8.0), 100 mM NaCl, 5 mM MgCl2 and 1 mM DTT. These reaction mixtures were then diluted 40 times in the same buffer and centrifuged at 120,000 × g for 1 hr using a Sorvall TH660 rotor. The protein complexes pelleted at the bottom of the tube were suspended in SDS-PAGE loading buffer, boiled and resolved in 10% SDS-PAGE. The resolved proteins were analyzed by Western blot using anti-N and anti-Myc rabbit polyclonal antibodies (Chen et al., 2007).
Purification of the bacterially expressed VSV P protein
The plasmid pET-3a-VSVIND-PHis containing P gene of Indiana serotype with histidine tag (Chen et al., 1997) was transformed into E. coli BL21 (DE3) and the his-tagged P protein was purified as described previously with slight modifications in the protocol (Barik and Banerjee, 1991). In brief, bacterial cells, induced with IPTG, were harvested by centrifugation and suspended in 15ml of buffer P-1 (50mM Tris-HCl [pH 7.5], 10% glycerol, 1 mM DTT, 0.1mM EDTA) and protease-inhibitor cocktail (Roche Applied Sciences). Then 1.5 ml of 50 mM EDTA, 10% Triton X-100, 2.5 mg/ml of lysozyme was added and the suspension was incubated in ice for 45 min with occasional mixing. The lysed cell mass was briefly sonicated to break DNA and centrifuged at 100,000 × g for 45 min in Sorvall TH-641 rotor. The pellet containing nearly all of the expressed His-P protein was dissolved in 20 ml of buffer P-2 containing 100 mM NaH2PO4, 10 mM Tris-HCl [pH 8.0] containing 6M guanidine hydrochloride with the aid of a homogenizer in the cold and incubated in ice for another 20 min. This soluble His-P protein solution was loaded on a pre equilibrated Ni-NTA column. The bound protein in column matrix was washed with the washing buffer (100 mM NaH2PO4, 10 mM Tris-HCl [pH 6.3], 8 M urea) and finally His-P protein was eluted with elution buffer (100 mM NaH2PO4, 10 mM Tris-HCl [pH 4.5], 8 M urea). The eluted protein sample was dialyzed against P-3 buffer 50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM DTT, 10% glycerol with three changes and then concentrated using Amicon Ultra-15 centrifugal filter device (Millipore). Protein concentration was determined using Bradford reagent (BioRad) and the purity of the P protein was checked by SDS-PAGE (SDS-polyacrylamide gel electrophoresis) and Western blot analyses.
Purification of bacterially expressed mumps virus P protein
For the purification of mumps virus (MuV; strain 88–1961) P protein a single vector (NP-P-pET28b) co-expressing the MuV NP protein and N-terminally His-tagged P protein was used (Cox et al., 2009). The plasmid was transformed into E. coli strain BL21 (DE3) and the bacterial cells were grown at 37 °C in LB broth in the presence of kanamycin (30mg/mL). After reaching an optical density of 0.6 at 600 nm (A600), protein expression was induced with 1mM IPTG at 29 °C for six hours. The cells were then collected by centrifugation and resuspended in a buffer containing 20mM Tris (pH 7.9), 500mM NaCl and 5mM imidazole. Cells were then sonicated and centrifuged for 1hr at 14,000rpm. Soluble fractions were then collected and passed through a Ni-Affinity column (HiTrap Chelating HP, GE Healthcare) in accordance with the manufacturer’s protocol. The NP and P proteins eluted from the Ni-affinity column as a complex and were further separated by ion exchange chromatography (Hitrap Q HP, GE Healthcare). The NP protein was found to bind to the Hitrap Q HP column while the His-P protein was detected in the flow through. The flow through was then passed through the Ni affinity column and elution fractions containing the His-P protein were further purified on a Sephacryl S-300 column (GE Healthcare) using a buffer containing 50mM Tris (pH 7.5), 500mM NaCl. Fractions containing the recombinant P protein were then concentrated using a Ni affinity column (HiTrap Chelating HP, GE Healthcare) and eluted into a buffer containing 50mM Tris (pH 7.5), 500mM NaCl, 500mM imidazole and then dialyzed against a buffer containing 50 mM Tris (pH 8.0), 100 mM NaCl and 1 mM DTT containing 10 % (vol/vol) glycerol for further use.
In vitro protease protection assay of vN-RNA complexed with VSV P protein
The vN-RNA and the VSV His-P protein (expressed and purified from E. coli) were incubated in 10 mM Tris-HCl, pH 8.0 buffer for 1 hr at 30°C water bath to form complexes. The molar ratios between vN-RNA: P were varied from 5:0, 5:1, 5:2.5, 5:5, 5:7.5 and 5:10.. These reaction mixtures were then subjected to chymotrypsin treatment at 4.25:1 (w/w) (vN-RNA: chymotrypsin) ratio at 37°C for 20 min and the digestion profile was studied by 10% SDS-PAGE (acrylamide: bis-acrylamide = 30:0.4) analysis.
Purification of infected cell RNP and N-RNA from BHK-21 cells
Infected cell VSV RNP and N-RNA were purified following the previously described method with some modifications (Toneguzzo and Ghosh, 1976). In brief, confluent monolayer of BHK-21 cells were infected with VSVIND Mudd-summers strain at 10 pfu/cell and incubated at 37°C for 6 hrs. After 6hr post-infection, cells were washed twice with 1X PBS and collected by a cell scrapper in 20 ml 1X PBS. VSV infected cells were recovered by centrifugation at 800 × g for 10 min at 4°C. Cell pellets were suspended in dounce buffer (10 mM Tris-HCl, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 10% Glycerol) at a ratio of 10 ml/60 plates (150 mm) and were allowed to swell on ice for 30 min. Cells were disrupted by 60 strokes in a dounce homogenizer on ice in the presence of protease-inhibitor cocktail (Roche applied Sciences). Finally, KCl was added to this disrupted cell suspension to get a final concentration of 150 mM. Cellular debris was removed by centrifugation at 10,000 × g for 20 min at 4°C. The 10,000 × g supernatant fraction was collected and again centrifuged at 120,000 × g in Beckman SW40 Ti rotor for 2 hr at 4°C. A milky white pellet of infected cell ribonucleoprotein complex (iRNP) was pelleted at the bottom of the tube. This iRNP was further solubilized in 20 mM Tris-HCl, pH 8.0, 1mM EDTA, 10% glycerol. This iRNP solution was then layered on 30% glycerol layer in 10 mM Tris-HCl, pH 7.5, 10 mM KCl, 1.5 mM MgCl2 buffer having 100% glycerol as a cushion underneath and centrifuged at 155,000 × g for 4 hr at 4°C in Sorvall TH-641 rotor. The iRNP was collected above the glycerol cushion and diluted with 1X TE buffer to make the glycerol concentration in between 10–16%. This step was again repeated and finally the collected iRNP in TE buffer. The iRNP was treated with equal volume of 2X HSB and incubated on ice for at least 1 hr with occasional inverting to dissociate the infected cell polymerase (iL-P) complex from the infected cell nucleocapsid (iN-RNA) complex. Then the iN-RNA complex was further purified by CsCl equilibrium density gradient ultracentrifugation as discussed above.
Purification of bacterial N-RNA complex
The purified bacterial N-RNA complex (bN-RNA) (Green et al. 2006a) was either used directly for chymotrypsin digestion or purified through CsCl equilibrium density gradient centrifugation prior to chymotrypsin treatment. After CsCl density gradient ultracentrifugation for 16 hr, fractions were collected and each fractions were analyzed by 10% SDS-PAGE followed by CBB stain. The layers containing the bN-RNA were then collected, diluted eightfold with the HSB and centrifuged again at 120,000 × g for 1 hr at 4°C in Sorvall TH-660 rotor. The obtained pellet (bN-RNACsCl) was suspended in TE buffer containing 10% glycerol, was dialyzed against the same buffer and concentrated using Amicon Ultra-15 centrifugal filter device (Millipore) and stored at −80°C freezer for future use. The purified vN-RNA, bN-RNA and bN-RNACsCl were subjected to chymotrypsin digestion following the protocol described above and the digestion profile was studied in 10% SDS-PAGE (acrylamide: bis-acrylamide = 30:0.4) analysis.
Acknowledgement
This work is supported by a grant from NIH (AI26585) to AKB. We thank Michael Kinter for mass spectrometric analysis of the proteins.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albertini AA, Wernimont AK, Muziol T, Ravelli RB, Clapier CR, Schoehn G, Weissenhorn W, Ruigrok RW. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313(5785):360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- Banerjee AK. Transcription and replication of rhabdoviruses. Microbiol Rev. 1987a;51(1):66–87. doi: 10.1128/mr.51.1.66-87.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AK, Barik S. Gene expression of vesicular stomatitis virus genome RNA. Virology. 1992;188(2):417–428. doi: 10.1016/0042-6822(92)90495-b. [DOI] [PubMed] [Google Scholar]
- Banerjee AK, Barik S, De BP. Gene expression of nonsegmented negative strand RNA viruses. Pharmacol Ther. 1991;51(1):47–70. doi: 10.1016/0163-7258(91)90041-j. [DOI] [PubMed] [Google Scholar]
- Banerjee AK, Roy J, Chattopadhyay DJ. Effect of the proteolytic digestion on the function of vesicular stomatitis virus ribonucleoprotein complex. J. Biosci. 1987b;11:515–523. [Google Scholar]
- Barik S, Banerjee AK. Cloning and expression of the vesicular stomatitis virus phosphoprotein gene in Escherichia coli: analysis of phosphorylation status versus transcriptional activity. J Virol. 1991;65:1719–1726. doi: 10.1128/jvi.65.4.1719-1726.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda PK, Banerjee AK. Two distinct populations of vesicular stomatitis virus ribonucleoprotein cores with differential sensitivities to micrococcal nuclease. Biochem. Biophys. Res. Commun. 1979;91:1337–1345. doi: 10.1016/0006-291x(79)91213-0. [DOI] [PubMed] [Google Scholar]
- Chen M, Ogino T, Banerjee AK. Mapping and functional role of the self-association domain of vesicular stomatitis virus phosphoprotein. J Virol. 2006;80(19):9511–9518. doi: 10.1128/JVI.01035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Ogino T, Banerjee AK. Interaction of vesicular stomatitis virus P and N proteins: identification of two overlapping domains at the N terminus of P that are involved in N0-P complex formation and encapsidation of viral genome RNA. J Virol. 2007;81(24):13478–13485. doi: 10.1128/JVI.01244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R, Green TJ, Qiu S, Kang J, Tsao J, Prevelige PE, He B, Luo M. Characterization of a mumps virus nucleocapsidlike particle. J Virol. 2009;83(21):11402–11406. doi: 10.1128/JVI.00504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T, Banerjee AK. Role of the phosphoprotein (P) in the encapsidation of presynthesized and de novo synthesized vesicular stomatitis virus RNA by the nucleocapsid protein (N) in vitro. Cell Mol Biol. 1992;38(1):17–26. [PubMed] [Google Scholar]
- Das T, Chakrabarti BK, Chattopadhyay D, Banerjee AK. Carboxy-terminal five amino acids of the nucleocapsid protein of vesicular stomatitis virus are required for encapsidation and replication of genome RNA. Virology. 1999;259(1):219–227. doi: 10.1006/viro.1999.9768. [DOI] [PubMed] [Google Scholar]
- Davis NL, Arnheiter H, Wertz GW. Vesicular stomatitis virus N and NS proteins form multiple complexes. J. Virol. 1986;59:751–754. doi: 10.1128/jvi.59.3.751-754.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De BP, Banerjee AK. Requirements and functions of vesicular stomatitis virus L and NS proteins in the transcription process in vitro. Biochem. Biophys. Res. Commun. 1985;126:40–49. doi: 10.1016/0006-291x(85)90568-6. [DOI] [PubMed] [Google Scholar]
- Delano WL. The PyMOL User’s Manual. San Carlos, CA: Delano Scientific; 2002. [Google Scholar]
- Emerson SU, Yu YH. Both NS and L proteins are required for the in vitro RNA synthesis by vesicular stomatitis virus. J. Virol. 1975;15:1348–1356. doi: 10.1128/jvi.15.6.1348-1356.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green TJ, Zhang X, Wertz GW, Luo M. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 2006a;313:357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- Green TJ, Luo M. Structure of the vesicular stomatitis virus nucleocapsid in complex with the nucleocapsid-binding domain of the small polymerase cofactor, P. Proc Natl Acad Sci U S A. 2009;106(28):11713–11718. doi: 10.1073/pnas.0903228106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green TJ, Luo M. Resolution improvement of X-ray diffraction data of crystals of a vesicular stomatitis virus nucleocapsid protein oligomer complexed with RNA. Acta Crystallogr. D. 2006b;62:498–504. doi: 10.1107/S0907444906006809. [DOI] [PubMed] [Google Scholar]
- Green TJ, Macpherson S, Qiu S, Lebowitz J, Wertz GW, Luo M. Study of the assembly of vesicular stomatitis virus N protein: Role of the P protein. J. Virol. 2000;74:9515–9524. doi: 10.1128/jvi.74.20.9515-9524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harouaka D, Wertz GW. Mutations in the C-terminal loop of the nucleocapsid protein affect vesicular stomatitis virus RNA replication and transcription differentially. J Virol. 2009;83(22):11429–11439. doi: 10.1128/JVI.00813-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CH, Kingsbury DW, Murti KG. Assembly of vesicular stomatitis virus nucleocapsids in vivo: a kinetic analysis. J Virol. 1979;32(1):304–313. doi: 10.1128/jvi.32.1.304-313.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhi S. Nucleocapsid structure and function. Curr Top Microbiol Immunol. 2009;329:103–128. doi: 10.1007/978-3-540-70523-9_6. [DOI] [PubMed] [Google Scholar]
- Luo M, Green TJ, Zhang X, Tsao J, Qiu S. Conserved characteristics of the rhabdovirus nucleoprotein. Virus Res. 2007;129(2):246–251. doi: 10.1016/j.virusres.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclellan K, Loney C, Yeo RP, Bhella D. The 24-angstrom structure of respiratory syncytial virus nucleocapsid protein-RNA decameric rings. J Virol. 2007;81(17):9519–9524. doi: 10.1128/JVI.00526-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A, Bhattacharya R, Basak S, Shaila MS, Chattopadhyay D, Roy S. P-protein of Chandipura virus is an N-protein-specific chaperone that acts at the nucleation stage. Biochemistry. 2004;43:2863–2870. doi: 10.1021/bi035793r. [DOI] [PubMed] [Google Scholar]
- Majumder A, Basak S, Raha T, Chowdhury SP, Chattopadhyay D, Roy S. Effect of osmolytes and chaperone-like action of P-protein on folding of nucleocapsid protein of Chandipura virus. J. Biol. Chem. 2001;276:30948–30955. doi: 10.1074/jbc.M011705200. [DOI] [PubMed] [Google Scholar]
- Masters PS, Banerjee AK. Resolution of multiple complexes of phosphoprotein NS with nucleocapsid protein N of vesicular stomatitis virus. J. Virol. 1988a;62:2651–2657. doi: 10.1128/jvi.62.8.2651-2657.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters PS, Banerjee AK. Complex formation with vesicular stomatitis virus phosphoprotein NS prevents binding of nucleocapsid protein N to nonspecific RNA. J. Virol. 1988b;62:2658–2664. doi: 10.1128/jvi.62.8.2658-2664.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon MG, Emerson SU. Rebinding of transcriptase components (L and NS proteins) to the nucleocapsid template of vesicular stomatitis virus. J Virol. 1978;27(3):560–567. doi: 10.1128/jvi.27.3.560-567.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino T, Banerjee AK. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol Cell. 2007;25:85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Patton JT, Davis NL, Wertz GW. Cell-free synthesis and assembly of vesicular stomatitis virus nucleocapsids. J Virol. 1983;45(1):155–164. doi: 10.1128/jvi.45.1.155-164.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro EA, Jr, Favier A, Gerard FC, Leyrat C, Brutscher B, Blondel D, Ruigrok RW, Blackledge M, Jamin M. Solution structure of the C-terminal nucleoprotein-RNA binding domain of the vesicular stomatitis virus phosphoprotein. J Mol Biol. 2008;382(2):525–538. doi: 10.1016/j.jmb.2008.07.028. [DOI] [PubMed] [Google Scholar]
- Ribeiro ED, Jr, Leyrat C, Gérard FC, Albertini AA, Falk C, Ruigrok RW, Jamin M. Binding of Rabies Virus Polymerase Cofactor to Recombinant Circular Nucleoprotein-RNA Complexes. J Mol Biol. 2009;394:558–575. doi: 10.1016/j.jmb.2009.09.042. [DOI] [PubMed] [Google Scholar]
- Rose JK, Schubert M. Rhabdovirus genomes and their products. In: Wagner RR, editor. The rhabdoviruses. New York, N.Y: Plenum Press; 1987. pp. 127–166. [Google Scholar]
- Schoehn G, Iseni F, Mavrakis M, Blondel D, Ruigrok RW. Structure of recombinant rabies virus nucleoprotein-RNA complex and identification of the phosphoprotein binding site. J Virol. 2001;75(1):490–498. doi: 10.1128/JVI.75.1.490-498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawar RG, Duquerroy S, Vonrhein C, Varela PF, Damier-Piolle L, Castagné N, MacLellan K, Bedouelle H, Bricogne G, Bhella D, Eléouët JF, Rey FA. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science. 2009;326(5957):1279–1283. doi: 10.1126/science.1177634. [DOI] [PubMed] [Google Scholar]
- Thomas D, Newcomb WW, Brown JC, Wall JS, Hainfeld JF, Trus BL, Steven AC. Mass and molecular composition of vesicular stomatitis virus: a scanning transmission electron microscopy analysis. J. Virol. 1985;54:598–607. doi: 10.1128/jvi.54.2.598-607.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toneguzzo F, Ghosh HP. Characterization and translation of methylated and unmethylated vesicular stomatitis virus mRNA synthesized in vitro by ribonucleoprotein particles from vesicular stomatitis virus-infected L cells. J Virol. 1976;17(2):477–491. doi: 10.1128/jvi.17.2.477-491.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Green TJ, Tsao J, Qiu S, Luo M. Role of intermolecular interactions of vesicular stomatitis virus nucleoprotein in RNA encapsidation. J Virol. 2008;82(2):674–682. doi: 10.1128/JVI.00935-07. [DOI] [PMC free article] [PubMed] [Google Scholar]