Summary
Elucidating how chromatin organization influences gene expression patterns and ultimately cell fate is fundamental to understanding development and disease. Here, we investigate the role of the histone variant H2AZ in embryonic stem (ES) cells because it plays an essential, but poorly understood function during development. Genome-wide analysis reveals that H2AZ is enriched at a large class of developmentally important genes whose promoters harbor bivalent histone modifications in a manner that is remarkably similar to the Polycomb group (PcG) protein Suz12. By using RNAi, we demonstrate a role for H2AZ in regulating target gene expression, find that H2AZ and PcG protein occupancy is interdependent at promoters, and further show that H2AZ is necessary for ES cell differentiation. Notably, H2AZ occupies a different subset of genes in lineage-committed cells suggesting that its dynamic redistribution is necessary for cell fate transitions. These results indicate that H2AZ, together with PcG proteins, may establish chromatin states necessary for the proper execution of developmental gene expression programs.
Keywords: histone, variant, H2AZ, Polycomb group proteins, stem cells, gene regulation
Introduction
A major challenge in biology is to understand how the pluripotent cells of the mammalian embryo and their in vitro derivatives, namely embryonic stem (ES) cells, execute the diverse gene expression programs that lead to cellular specification. The regulation of chromatin structure facilitates the establishment and maintenance of heritable gene expression patterns during development. ES cells are a valuable model system to study changes in chromatin state as a function of cell state because of their unique ability to differentiate into multiple lineages (Keller, 2005; Jaenisch and Young, 2008). Recent studies have begun to reveal unique chromatin states in pluripotent and lineage-committed cells (Meshorer and Misteli, 2006; Mendenhall and Bernstein, 2008). Therefore, knowledge of how chromatin influences gene expression patterns in ES cells is expected to provide key insights into the process of cell fate specification and for understanding the progression from normal to disease states.
Chromatin structure is highly regulated by a variety of complex processes that are not well understood. These include nucleosome remodeling and post-translational modification of histone proteins (Dunn and Kingston, 2007; Kouzarides, 2007; Surani et al., 2007; Workman, 2006). An additional mechanism for chromatin regulation is the replacement of conventional histones with specific variants. Histone variants are structural components of chromatin and play important roles in all eukaryotes by influencing a wide range of DNA-mediated processes such as genome integrity, X-inactivation, DNA repair, and gene regulation (Henikoff and Ahmad, 2005; Guillemette and Gaudreau, 2006; Hake and Allis, 2006; Jin and Felsenfeld, 2007; Li et al., 2007; Raisner and Madhani, 2006). The genes that code for them are evolutionarily conserved, non-allelic variants of the major histone genes whose expression is not linked to the cell cycle and whose non-random incorporation into chromatin is independent of DNA replication. Variants also differ from canonical histones in their primary sequence and their incorporation can have functional consequences on the biophysical properties of the nucleosome core particle. These data indicate that histone variants perform specialized functions and suggest an important role for histone replacement in the regulation of chromatin states.
The histone H2A variant H2AZ is of particular interest because it is essential in multi-cellular organisms (Faast et al., 2001; Liu et al., 1996; Ridgway et al., 2004; van Daal and Elgin, 1992). H2AZ has been implicated from yeast to human in many DNA-mediated processes including gene regulation. Interestingly, H2AZ has been linked to both gene activation and repression. Genome-wide studies in a range of organisms have shown that the distribution of H2AZ across the genome appears to be largely confined to small regions flanking transcription start sites, although enrichment has also been reported at larger regions proximal to telomeres or centric heterochromatin (Albert et al., 2007; Barski et al., 2007; Guillemette et al., 2005; Li et al., 2005; Meneghini et al., 2003; Raisner et al., 2005; Rangasamy et al., 2003; Zhang et al., 2005). Moreover, studies have shown that H2AZ incorporation can affect local histone modification patterns, the activity of chromatin remodeling enzymes, and chromatin conformation (Barski et al., 2007; Fan et al., 2002; Li et al., 2005; Millar et al., 2006; Raisner et al., 2005; Swaminathan et al., 2005; Zhang et al., 2005). Thus, the function of H2AZ appears to be highly context-dependent in a manner that influences transcriptional output.
To investigate the essential role of H2AZ during mammalian development, we have generated genome-wide maps in ES cells and find that H2AZ is enriched at a large set of developmental genes in a manner that is highly similar to the Polycomb group (PcG) protein Suz12 (Boyer et al., 2006; Lee et al., 2006). PcG proteins are transcriptional repressors that play important roles in regulating developmental gene expression patterns by epigenetic modification of chromatin structure (Ringrose and Paro, 2007; Schuettengruber et al., 2007; Schwartz and Pirrotta, 2007). We also show that H2AZ is required for the regulation of target gene expression in ES cells and further demonstrate an important role for H2AZ in mediating cell fate transitions upon induction of differentiation. Conversely, H2AZ enrichment was detected at active genes that are not Polycomb targets in multi-potent neural precursors suggesting that its dynamic redistribution is necessary for lineage specification. Together, these findings establish a critical role for H2AZ in regulating developmental gene expression programs in ES cells and provide important insights into the essential nature of this variant during mammalian development.
Results
H2AZ occupies promoter regions in ES cells
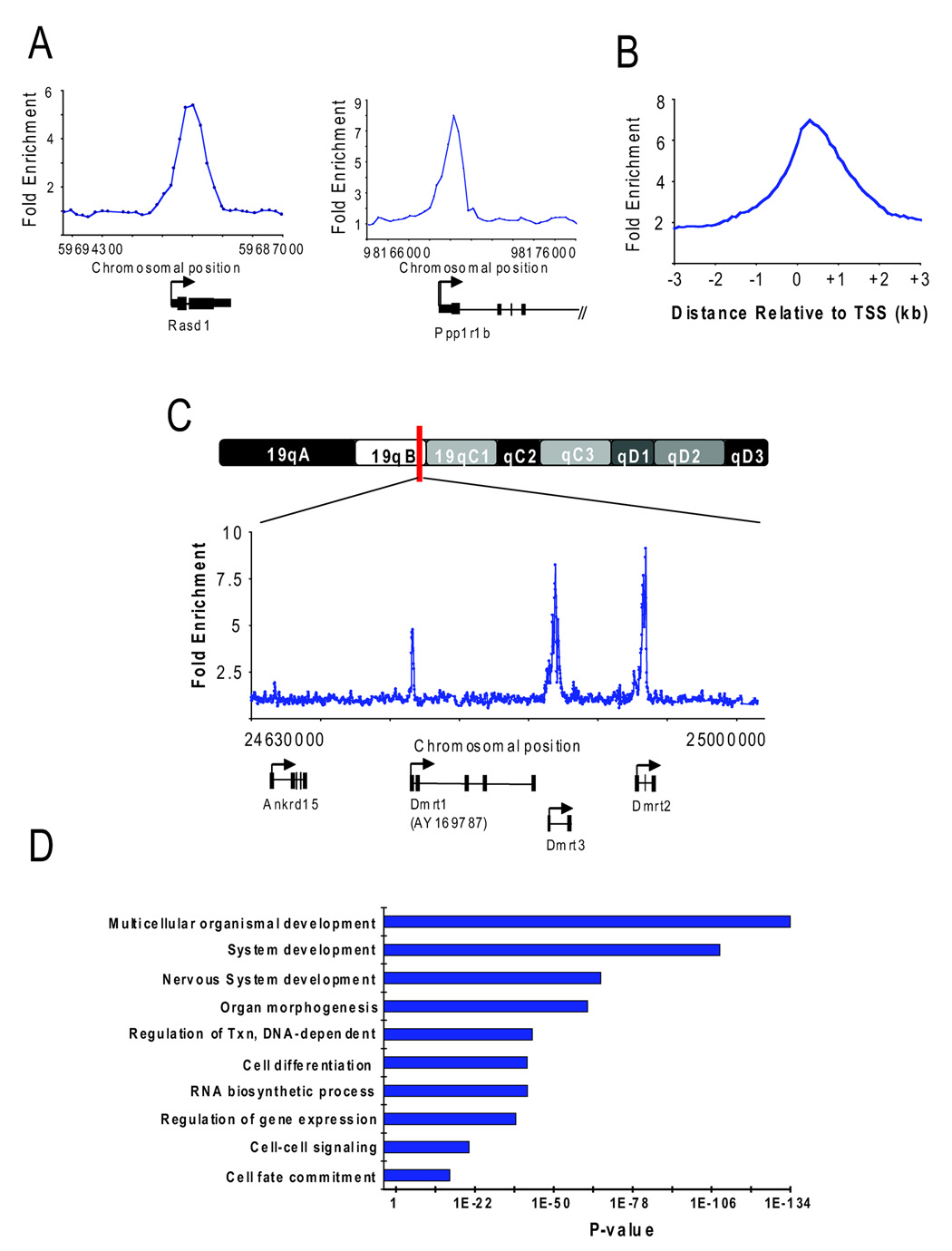
Because the localization of H2AZ might reveal its essential role during early mammalian development, we mapped this histone variant throughout the genome in murine ES cells. For this purpose, DNA sequences occupied by H2AZ were identified by chromatin immunoprecipitation (ChIP) with specific antibodies followed by hybridization to DNA microarrays (chip) as previously described (Boyer et al., 2005; Boyer et al., 2006). DNA microarrays were designed to contain 60-mer oligonucleotide probes that span the region from −4 kilobase pair (kb) to +4 kb relative to the transcription start sites (TSS) of ~20,000 annotated mouse genes (see Supplemental Data). The genomic sites occupied by H2AZ were identified as peaks of ChIP-enriched DNA using a previously validated algorithm (Figure 1A; Table S1). Notably, the majority (92%) of enriched regions were detected within 1kb of a TSS (Figure 1B; Table S2). Control ChIP experiments using antibodies against histone H3 or IgG did not yield similar enrichment (data not shown). Thus, our genome-wide analysis found that H2AZ occupied nucleosomes at the promoter regions of 1655 (8%) protein-coding genes in ES cells with high confidence.
Figure 1. H2AZ occupies promoter regions in ES cells.
(A) Representative examples of DNA sequences occupied by H2AZ isolated using chromatin immunoprecipitation (ChIP) and promoter microarrays. ChIP with core histone H3 was hybridized together with H2AZ to control for nucleosome density. Note that hybridization of H2AZ ChIP-DNA with bulk chromatin as input yielded highly similar results in a replicate set of experiments. The plots show unprocessed enrichment ratios (blue) for all probes within a genomic region. Chromosomal positions are from NCBI build 34 (mm6) of the mouse genome. Genes are shown to scale below plots. The start and direction of transcription are both indicated by an arrow. (B) Distribution of the distance between bound probes and the closest transcription start site (TSS). Data are the average unprocessed enrichment ratios for each oligonucleotide probe within the –4 kb to +4 kb genomic region for all enriched genes. (C) Representative example of DNA sequences occupied by H2AZ using ChIP and tiled chromosome 19 microarrays. The plots show unprocessed enrichment ratios (blue) for all probes within a genomic region (ChIP versus histone H3). Genes are shown to scale below plots as in (A). (D) Gene ontology analysis for biological process of H2AZ-enriched genes. Ontology terms are represented on the y-axis and the p-value for enrichment of bound genes relative to all genes represented on the microarray is shown for each category on the x-axis.
Because promoter arrays were used to interrogate enrichment, it was possible that H2AZ occupied other genomic sequences in ES cells. To test this, DNA microarrays were designed to contain 60-mer probes that span the entire non-repeat portion of chromosome 19 in the mouse genome (see Supplemental Data). H2AZ ChIP-enriched DNA was hybridized to chromosome arrays and binding events were identified using the above criteria (Table S3). Similar to the promoter array data, H2AZ occupied genomic regions in close proximity to known or predicted TSSs and additional sites of enrichment were not observed along the chromosome (Figure 1C). These data indicate that H2AZ predominantly occupies promoter regions at a defined set of genes in ES cells.
H2AZ is enriched at key developmental regulators in ES cells
To gain insights into the biological roles of the genes occupied by H2AZ in ES cells, we determined which gene ontology (GO) terms were over-represented in this set relative to all genes with representative probes on the microarray. This analysis revealed a striking enrichment for genes connected to the developmental and transcription hierarchies including those with roles in organ and system development, cell differentiation and cell fate commitment, as well as in the regulation of gene expression (Figure 1D; Table S4). Further analysis indicated that many of these genes are transcription factors with important roles in a variety of developmental processes.
H2AZ was enriched at the majority of homeodomain genes including members of the Pou, Pax, Six, Dlx, Irx, and Lhx gene families and all of the homeotic genes found in Hox gene clusters (Table S2 and Table S4). Homeodomain-containing transcription factors are evolutionarily conserved regulators that specify cell fate during embryonic development through transcriptional control of other developmental regulators (Deschamps, 2007; Pearson et al., 2005). H2AZ also occupied promoters of members of the Fox, Sox, Gata and Tbx transcription factor families that have important roles in development and disease (Burch, 2005; Lehmann et al., 2003; Schepers et al., 2002; Showell et al., 2004). Interestingly, the targets were also enriched for genes that encode components of signaling pathways such as members of the Wnt family (Clevers, 2006). These data suggest that H2AZ predominantly occupies the promoters of genes in ES cells that when expressed, would promote developmental progression and differentiation.
H2AZ and Suz12 occupy a highly similar set of genes in ES cells
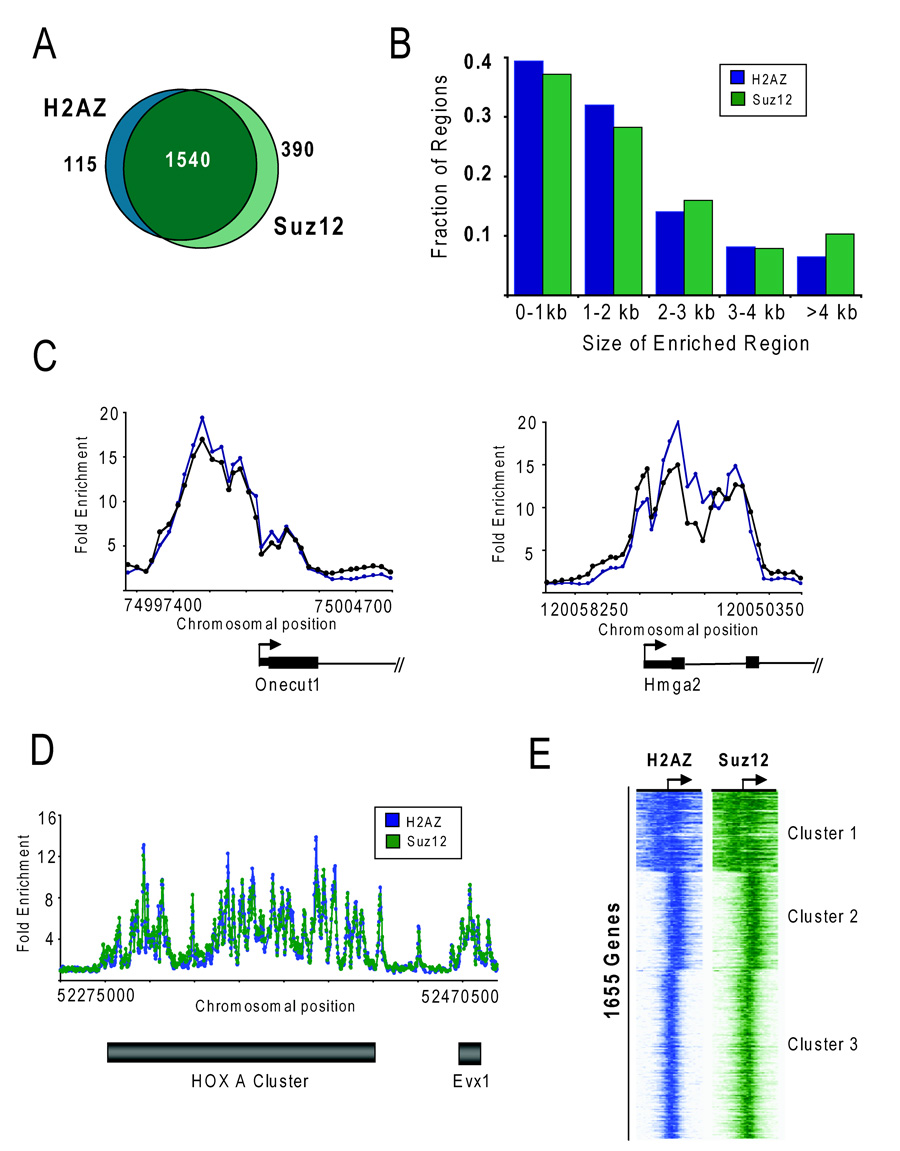
It was particularly intriguing that H2AZ occupied the promoter regions of developmental genes in ES cells, including many that code for transcription factors and signaling components. Previous studies in both human and mouse ES cells have revealed that Polycomb group (PcG) proteins occupy the promoters of developmental genes (Boyer et al., 2006; Lee et al., 2006). This led us to compare the set of H2AZ-enriched genes with PcG protein binding targets. In this study, Suz12 was used as a proxy for PcG protein occupancy because core components of PRC2 (Suz12 and Eed as well as the modification catalyzed by PRC2, H3K27me3) and PRC1 (Rnf2 and Phc1) were found to significantly overlap in ES cells (Boyer et al., 2006) and because Suz12 has been shown to be critical for the function of the PRC2 complex (Cao and Zhang, 2004; Pasini et al., 2004). To control for potential differences in the probes used in the prior genome-wide analysis, binding data for Suz12 (Table S5) were generated on the same promoter microarray design platform as described above. As expected, the Suz12 target genes were highly similar to those previously reported (Table S6) (Boyer et al., 2006). Further analysis found that most (93%) of the H2AZ enriched regions, were also occupied by Suz12 (Figure 2A). PcG proteins have been shown to globally maintain their target genes in a silent state (Boyer et al., 2006; Bracken et al., 2006; Lee et al., 2006). Therefore, H2AZ occupies the promoter regions of a defined set of genes in ES cells that are targets of Polycomb-mediated repression.
Figure 2. H2AZ and Suz12 occupy a highly similar set of genes in ES cells.
(A) Venn diagram showing the overlap of genes enriched with H2AZ and occupied by Suz12 at high confidence in ES cells as determined by ChIP combined with promoter arrays. 93% of H2AZ-enriched genes were also occupied by Suz12 using high-confidence threshold criteria. (B) H2AZ and Suz12 enriched regions were binned according to length of the bound region as determined by promoter microarrays. The majority of bound regions are less then 2 kb whereas a small fraction (~15%) exhibit an extended domain of binding > 4 kb. (C) H2AZ and Suz12 display the same spatial patterning at target genes. The plots show unprocessed enrichment ratios for H2AZ (blue) and Suz12 (green) for all probes within a genomic region as in Figure 1A. (D) H2AZ and Suz12 occupy large domains (> 100kb) that span multiple contiguous genes encompassing the HOX gene clusters. Data are derived from the promoter microarrays that were also designed to contain probes tiling the entire HoxA locus. Unprocessed enrichment ratios for H2AZ (blue) and Suz12 (green) for all probes within the region are shown. The grey bars represent the approximate location of the gene cluster. (E) H2AZ and Suz12 display a highly similar defined spatial pattering across target genes in ES cells. K-means clustering was performed using the Cluster algorithm (http://rana.standord.edu/software) and the set of 1655 H2AZ-enriched genes. Each horizontal line represents an individual gene and the enrichment values for H2AZ (blue) and Suz12 (green) for each probe within the region –3.5 kb to + 3.5 kb relative to the TSS are represented by color intensity (darker color represents higher enrichment ratio).
H2AZ and Suz12 display highly similar spatial patterning in the ES cell genome
Genome-wide studies in yeast, Drosophila, and human T cells have shown that H2AZ maps to a limited number of nucleosomes around TSSs (Albert et al., 2007; Barski et al., 2007; Guillemette et al., 2005; Mavrich et al., 2008; Raisner et al., 2005). In agreement with these studies, ~75% of H2AZ-enriched regions in ES cells are localized within discrete intervals of less than 2 kb around the TSS (Figure 2B). Surprisingly, ~25% of the promoter regions showed extended regions of H2AZ occupancy of greater than 2 kb. Analysis of Suz12 revealed a similar length distribution in ES cells. These data indicate that although the majority of enriched regions are found within small domains at promoters in ES cells, H2AZ also extends across larger regions.
A previously observed feature of Suz12 binding in ES cells at the subset of genes encoding developmental transcription factors was the extensive span over which the regulator occupied the locus. For example, Suz12 occupancy was found to encompass large domains extending from the promoter into the gene (Lee et al., 2006). A similar genomic distribution has also been observed for H3K27me3, the histone modification catalyzed by PRC2 (Boyer et al., 2006; Mikkelsen et al., 2007). Strikingly, we found that the distribution pattern at target genes were comparable between H2AZ and Suz12 (Figure 2C). Thus, H2AZ also exhibits an unusual tendency to occupy extended regions at development genes.
An additional unique feature of PcG protein occupancy was the finding that Suz12 encompassed multiple contiguous genes that extended across the entire HoxA, HoxB, HoxC and HoxD clusters but that it did not occupy adjacent genomic sequences (Lee et al., 2006). This led us to compare H2AZ and Suz12 enrichment across the HoxA gene cluster as well as flanking genomic regions (Table S1 and Table S5). Surprisingly, H2AZ extended over contiguous genes that encompassed the HoxA cluster yielding a highly defined spatial pattern nearly identical to Suz12 (Figure 2D). Because a similar spatial patterning of H2AZ and Suz12 suggests an important functional interaction between these two chromatin regulatory pathways in ES cells, their genomic distribution was examined at all target loci. To this end, the pattern of H2AZ and Suz12 enrichment was compared for the set of 1655 H2AZ occupied regions. The probe enrichment scores across a 7 kb region centered at the TSS were clustered using K-means clustering selecting three nodes (Figure 2E). Higher node numbers were tested but additional nodes showed similar binding behavior based on composite profiles (data not shown). This analysis revealed a remarkably similar distribution between H2AZ and Suz12 occupancy across all target genes in ES cells (Table S8 lists genes in each cluster). These data reveal that the genomic distribution of H2AZ in ES cells is remarkably similar to Suz12.
H2AZ and H3K27me3 are enriched at a distinct set of genes in neural precursors
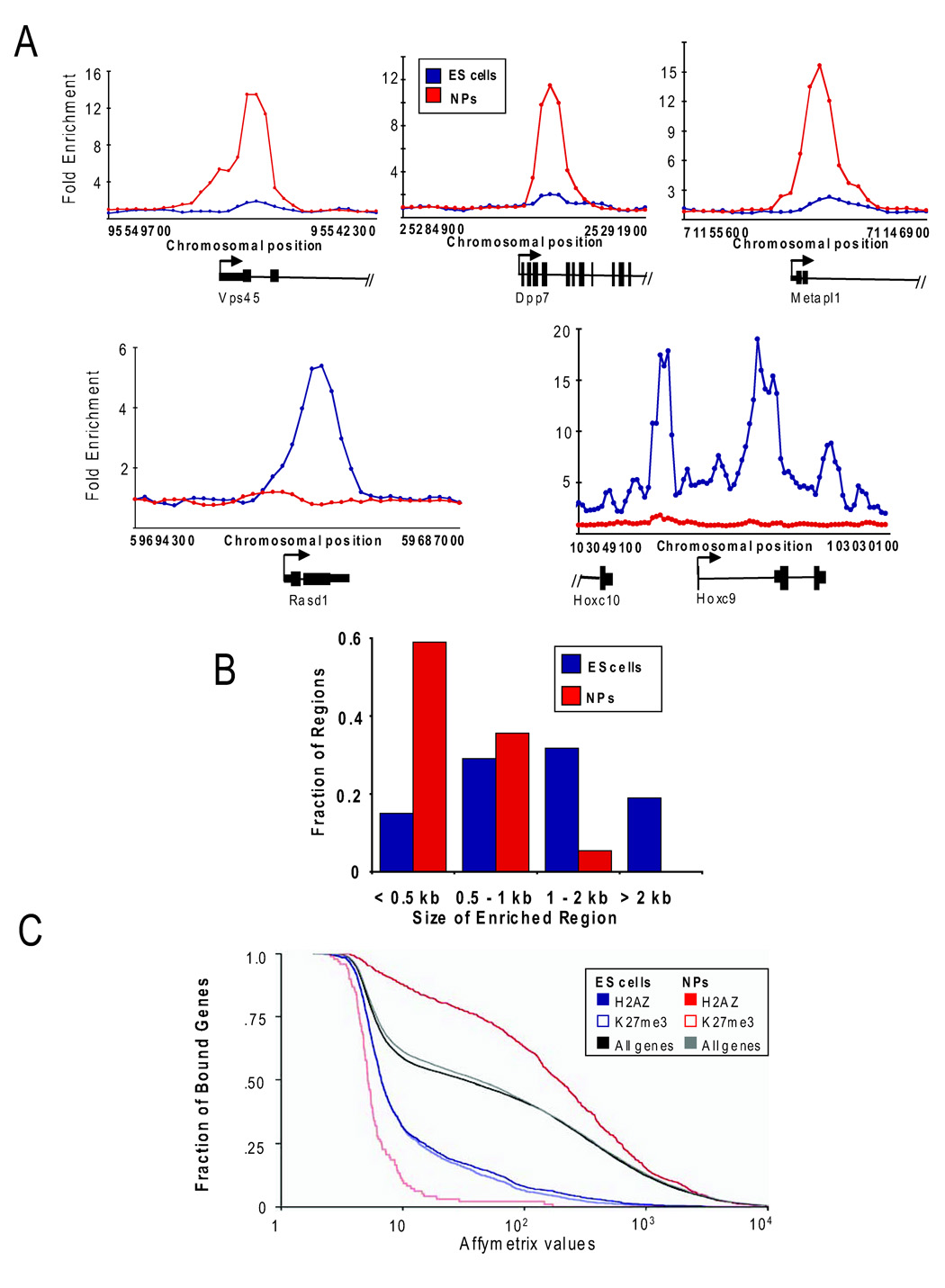
The process of lineage commitment is characterized by a dramatic reorganization of chromatin structure and recent evidence suggests that every cell type may harbor a unique chromatin signature (Mendenhall and Bernstein, 2008; Meshorer and Misteli, 2006). Given the unexpected similarity between H2AZ and Suz12 enrichment in ES cells, we next asked if this pattern was maintained in differentiated cells. H2AZ occupancy was examined in neural precursors (NPs) because these cells can be directly derived from ES cells and because many of the target genes have known roles in neural development (see Figure 1D). Similarly to its distribution in ES cells, H2AZ was significantly enriched at the promoter regions of a large set of genes (1206) in NPs and 94% of the bound regions were within 1 kb of a TSS (Figure 3A; Table S9 and Table S10). In contrast, the set of H2AZ-enriched genes was remarkably different in NPs as compared to ES cells (Table S10). Moreover, analysis of the spatial distribution in NPs found that H2AZ localized primarily to discrete intervals of less than 2 kb and that the more extended regions of enrichment were unique to ES cells (Figure 3B). These data indicate that H2AZ occupies the promoters of a unique set of genes in ES cells and neural precursors.
Figure 3. H2AZ is enriched at a distinct set of genes in neural precursors.
(A) Representative examples of DNA sequences occupied by H2AZ from a replicate set of experiments isolated using ChIP and promoter microarrays. ChIP with core histone H3 was hybridized together with H2AZ to control for nucleosome density. The plots show unprocessed enrichment ratios for H2AZ in ES cells (blue) and NPs (red) for all probes within a genomic region as shown in Figure 1A. (B) The spatial distribution of H2AZ differs in NPs as compared to ES cells. H2AZ enriched regions in ES cells (blue) and NPs (red) were binned according to length of the bound region based on data from the promoter microarrays. (C) H2AZ is enriched at active genes in NPs. Cumulative distribution plot of the expression of genes associated with H2AZ or H3K27me3 enrichment in ES cells and NPs. Cumulative distribution of all genes in ES cells (black) and NPs (gray) are shown as a reference. Affymetrix expression and H3K27me3 enrichment data were derived from Mikkelsen et al., 2007.
Given that H2AZ and Suz12 co-occupy target genes that code for developmental regulators that are silent in ES cells, we performed a similar comparison in NPs. To this end, genome-wide H3K27me3 binding and expression data were analyzed in both cell types (Mikkelsen et al., 2007; data are from the same genetic background as used in this study). H3K27me3 enrichment has been shown to strongly correlate with Suz12 in ES cells (Boyer et al., 2006; Lee et al., 2006). As expected, H2AZ and H3K27me3 were enriched at genes in ES cells with lower expression levels when compared to all genes (Figure 3C). Conversely, while H3K27me3 occupancy correlated with lower expression in NPs consistent with its role as a mark of silent chromatin, H2AZ was predominantly enriched at genes that displayed high expression levels as compared to all genes in NPs. Further analysis of the set of H2AZ occupied genes in NPs revealed a significant enrichment for GO terms associated with a wide range of metabolic processes (Figure S1; Table S10). H2AZ enrichment at active gene promoters in NPs is consistent with prior studies in other differentiated cell types in mammals (Barski et al., 2007; Schones et al., 2008). These data suggest that co-occupancy of H2AZ and PcG proteins at promoters is particular to ES cells and that H2AZ localization changes dramatically during development and differentiation.
H2AZ is not required for ES cell self-renewal and pluripotency
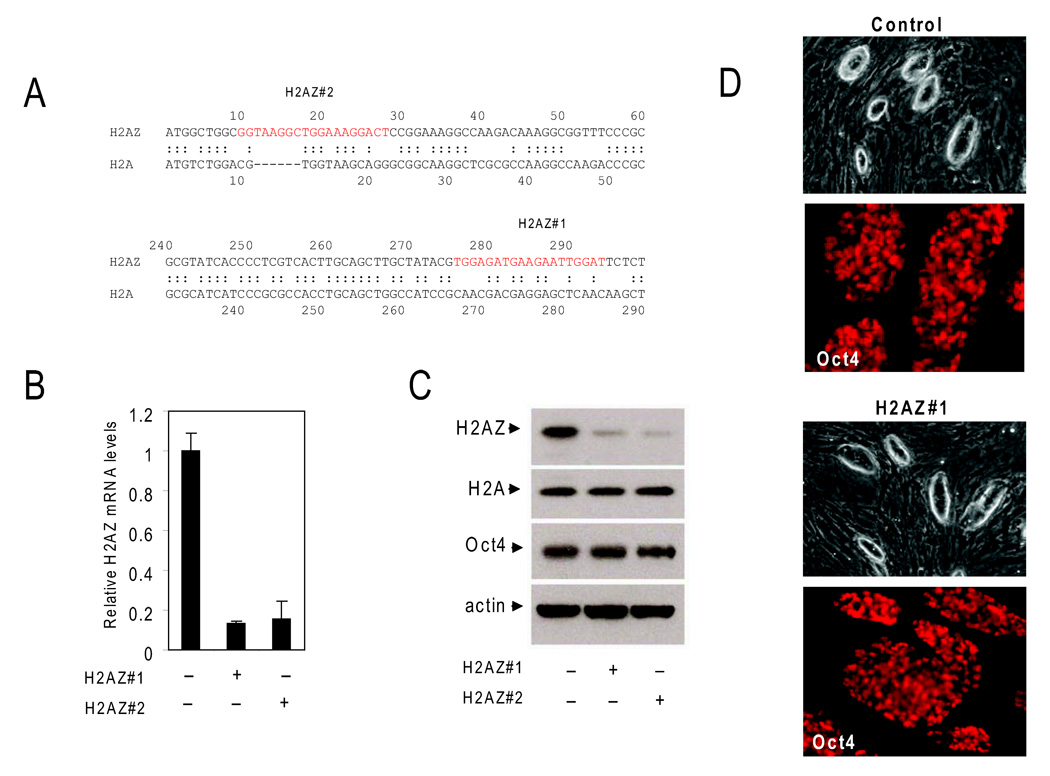
In order to investigate the role of H2AZ, ES cell lines that stably expressed sequence specific hairpins were generated by retroviral transduction. The expression of two unique hairpins (Figure 4A) resulted in efficient and specific reduction in H2AZ mRNA levels in ES cells as compared to a control cell line expressing a non-targeting GFP specific hairpin (Figure 4B; Figure S2). Notably, a reduction in mRNA levels resulted in suppression of H2AZ protein abundance, but did not alter the levels of the major histone H2A or the essential pluripotency gene Pou5f1/Oct4 as compared to the control ES cell line (Figure 4C). H2AZ-depleted ES cell lines showed typical colony morphology and Oct4 expression patterns (Figure 4D), displayed normal cell cycle kinetics, proliferated normally under ES cell growth conditions (Figures S3 and S4), and maintained a normal karyotype in pro-longed culture (Figure S5). This suggests that H2AZ-depletion per se does not affect maintenance of the ES cell state.
Figure 4. RNAi-mediated H2AZ depletion in ES cells.
(A) Schematic representation of H2AZ hairpin targeting sequences (red) and the homology between H2A and H2AZ. (B) mRNA levels were significantly reduced upon RNAi-mediated suppression of H2AZ as measured by quantitative real-time PCR in H2AZ-depleted ES cell lines relative to the control ES cell line. Data were normalized to Gapdh and error bars represent 2 standard deviations. (C) H2AZ protein abundance s determined by Immunoblot analysis is specifically reduced in depleted ES cell lines whereas no reduction in histone H2A or Oct4 levels is observed in the control cell line. Actin serves as a loading control. (D) ES cell colony morphology and Oct4 levels are similar between control and H2AZ-deficient ES cell lines.
H2AZ levels are necessary for regulation of target gene expression
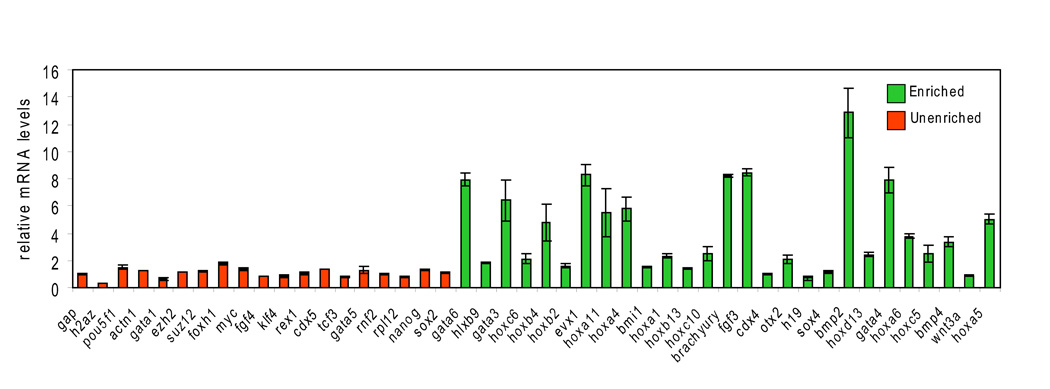
The observation that H2AZ occupied promoter regions in ES cells suggested a role in gene regulation. To examine whether H2AZ occupancy influences target gene expression, real-time polymerase chain reaction (rt-PCR) was used to compare the mRNA levels of H2AZ target genes in control and H2AZ-depleted ES cell lines. Notably, enriched genes displayed an overall tendency to become de-repressed in H2AZ-deficient ES cells whereas no significant difference was observed at a set of un-enriched genes (Figure 5; Table S12). Importantly, these data are similar to that derived from the loss of PRC2 components such as Suz12 and Eed in ES cells (Boyer et al., 2006; Chamberlain et al., 2008; Pasini et al., 2007). The finding that H2AZ directly contributes to the regulation of target genes with known roles in development may explain, at least in part, the essential requirement for H2AZ during early mammalian development.
Figure 5. H2AZ is necessary for control of target gene expression.
Target genes display a propensity to become de-repressed upon RNAi-mediated suppression of H2AZ in ES cells. Real-time PCR analysis of mRNA levels in H2AZ-depleted ES cells for genes that are enriched (green) or un-enriched (orange). Data were normalized to Gapdh and are shown relative to control ES cell lines. Reactions were performed in triplicate and error bars represent 2 standard deviations of the mean.
H2AZ and PcG protein occupancy at target promoters is interdependent in ES cells
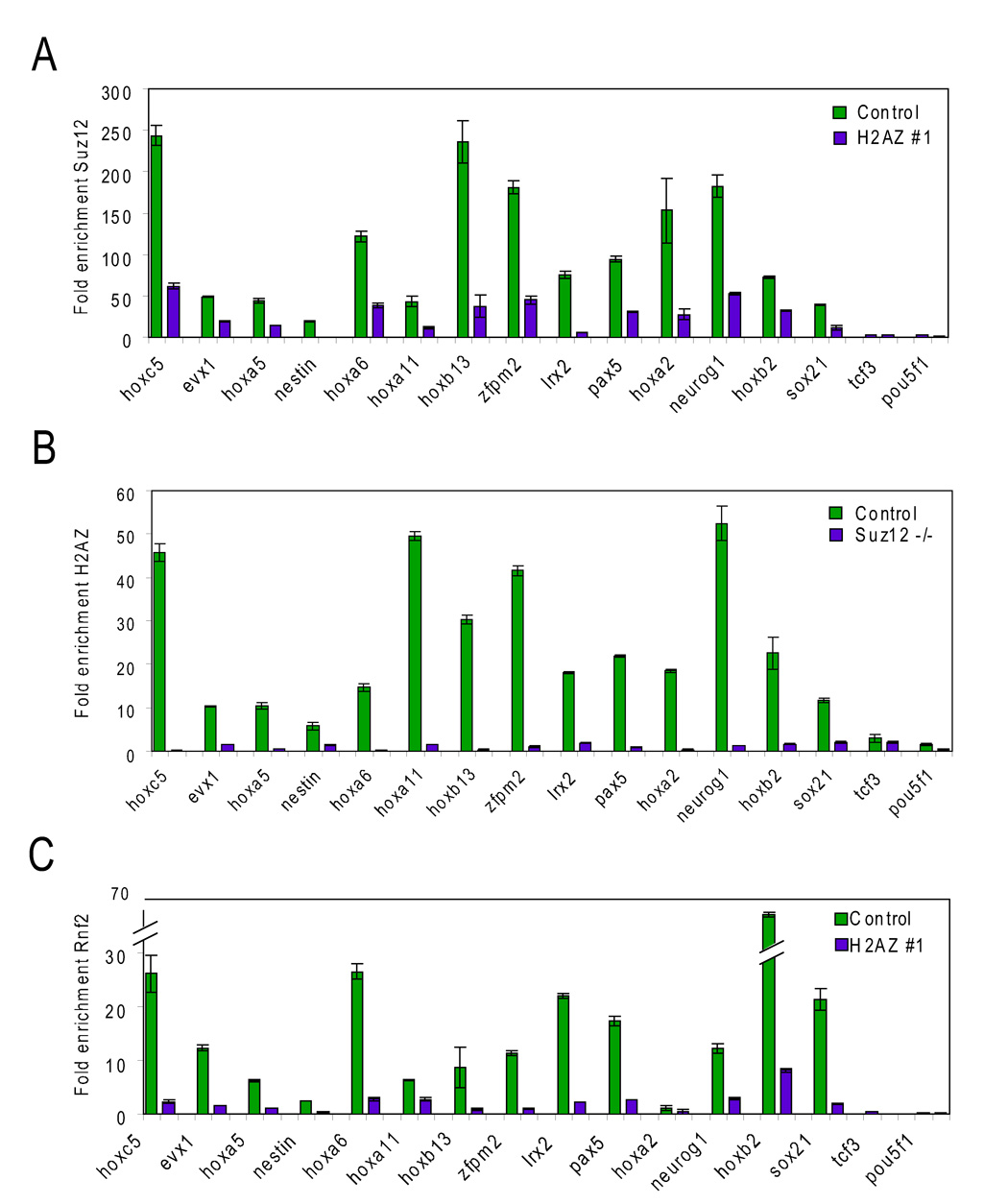
The co-occupancy of H2AZ and Suz12 at promoters in ES cells suggested that the localization of one could be dependent on the other. To investigate this, ChIP and sitespecific rt-PCR was performed for a subset of genes in control, H2AZ-deficient, and suz12 null ES cells (Figure 6; Table S13). While significant Suz12 enrichment was detected at target genes in control ES cells, this enrichment was reduced in H2AZ-deficient ES cells (Figure 6A). The low level of Suz12 detected at these genes is most likely the result of the incomplete depletion of H2AZ due to RNAi. Surprisingly, H2AZ-enrichment was also reduced to background levels (similar to the negative controls Pou5f1/Oct4 and Tcf3) in suz12 null ES cells (Figure 6B) suggesting that the localization of these regulators is mutually interdependent at target genes. Importantly, both H2AZ and Suz12 protein levels were unchanged in the reciprocal mutant ES cell line (Figure S6). Although components of PRC2 (Suz12, Eed) and PRC1 (Rnf2/Ring1b, Phc1) co-occupy a highly similar set of genes in ES cells (Boyer et al., 2006), PRC1 can be recruited independently to target regions (Pasini et al., 2007; Schoeftner et al., 2006). Therefore, enrichment for the essential component Rnf2/Ring1b was analyzed in control and H2AZ-deficient ES cells. Similarly to Suz12, Rnf2/Ring1b enrichment was significantly reduced at target genes (Figure 6C). We next tested whether Suz12 and H2AZ could directly interact by co-immunoprecipitation (Figure S7). Although an interaction was detected between Suz12 and Ezh2, an association with H2AZ was not observed in the ES cell extracts. These data suggest that H2AZ and PcG proteins do not directly interact and that their interdependence at promoters may be mediated at the level of chromatin or by an unknown factor.
Figure 6. Interdependent localization of H2AZ and Polycomb complexes at target promoters in ES cells.
ChIP combined with real time PCR for (A) Suz12 in control (green bars) or H2AZ-depleted ES cells (purple bars), (B) H2AZ in control (green bars) or suz12 null ES cells (purple bars) and (C) Rnf2/Ring1b in control (green bars) or H2AZ-depleted ES cells (purple bars) for the promoter regions of the indicated genes. All ChIP-qPCR reactions were performed in triplicate and error bars represent 2 standard deviations of the mean. Primers were designed to amplify a region within 1 kb of the TSS and are listed in Table S13. Tcf3 and Oct4 (Pou5f1) represent negative controls. Similar results were obtained in both H2AZ-depleted ES cell lines.
H2AZ is necessary for ES cell differentiation
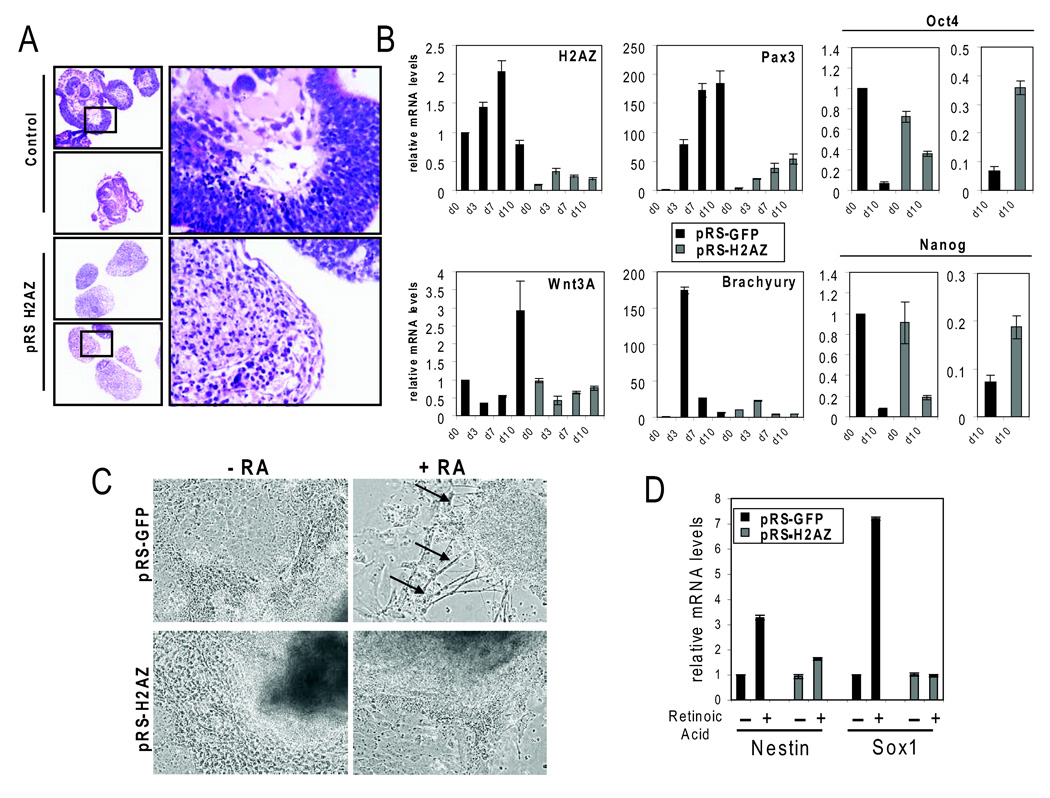
Because H2AZ is essential for early embryonic development and given that suz12 null ES cells display de-repression of target genes and an inability to initiate differentiation programs in vitro (Pasini et al., 2007), we asked whether H2AZ was also required for lineage specification. To test this, control and H2AZ-depleted ES cells were cultured under non-adherent conditions in the absence of LIF to induce aggregation, a process that results in embryoid body (EB) formation. EBs are similar to, albeit more disorganized than, egg cylinder stage embryos, and are capable of differentiation into the three germ layers. Of note is the finding that H2AZ-depleted ES cells failed to support normal development in vivo by tetraploid complementation and chimeric analysis (see Supplemental Data and Figure S8) indicating that the knockdown cells represent a good model to assess the functional role of H2AZ during ES cell differentiation. In contrast to control ES cells, H2AZ-depleted EBs failed to give rise to typical structures representing differentiated cell types (Figure 7A). Moreover, induction of early differentiation markers such as Brachyury, Pax3 and Wnt3A required normal H2AZ levels (Figure 7B). Consistent with this, higher levels of the pluripotency markers Oct4 and Nanog were observed in the H2AZ-depleted EBs. Thus, although target genes displayed a propensity to become de-repressed in H2AZ-depleted ES cell lines, these genes fail to become appropriately activated during differentiation.
Figure 7. H2AZ is necessary for ES cell differentiation.
(A) Hematoxylin and eosin staining of sectioned day 10 embryoid bodies (EBs) derived from control (upper panel) or H2AZ-depleted (bottom panel) ES cells. (B) Real-time PCR on mRNA isolated from control (black bars) or H2AZ-depleted EBs (grey bars) for the indicated genes. Relative mRNA levels are normalized to Gapdh. Days shown indicate the time EBs were maintained in culture prior to RNA isolation. Far right panel shows end-point comparison of Oct4 and Nanog levels. Reactions were performed in triplicate and error bars represent 2 standard deviations. Similar results were obtained using both H2AZ-depleted ES cells lines. (C) Panels show EB images cultured for 7 days in the absence (left panels) or presence (right panels) of retinoic acid. Upper panels display control cells while the lower panels represent EBs derived from H2AZ-depleted ES cells. Arrows indicate formation of neuron like structures typical for retinoic acid induced differentiation. (D) Real-time PCR of mRNA levels for neural differentiation markers Sox1 and Nestin in EBs as shown in (C). Reactions were performed in triplicate and error bars represent 2 standard deviations. Similar results were obtained using both H2AZ-depleted ES cells lines.
Although the EBs did not differentiate using standard conditions, it was possible that H2AZ-depleted cells could be induced to differentiate into particular lineages when given particular developmental cues and culture support. To test this, EBs derived from control and H2AZ-depleted cells were treated with retinoic acid (RA) to induce neuronal differentiation. Under these conditions the control cells undergo morphological changes consistent with neural differentiation (Glaser and Brustle, 2005; Li et al., 1998), whereas the H2AZ-depleted aggregates failed to do so (Figure 7C). Similarly, RA-treated control cells showed induction of early neuronal markers such as Sox1 and Nestin, while the H2AZ-depleted cells did not display significant expression of these markers (Figure 7D). ES cells can also be robustly differentiated into hematopoietic lineages as evidenced by the expression of classical hematopoietic markers (Kyba et al., 2002). Further experiments revealed that H2AZ-depleted cells were similarly defective when induced to differentiate into hematopoietic lineages (Figure S9). These studies argue that H2AZ plays a role with PcG proteins in arbitrating cell fate transitions and lineage specification during ES cell differentiation.
Discussion
How chromatin organization in ES cells contributes to the preservation of cell identity while maintaining the genome in a flexible state that allows for differentiation into multiple lineages remains an open question. In ES cells, the promoters of a subset of silent genes with roles in development are marked with “bivalent” histone modifications that correlate with both transcription initiation (H3K4me3) and gene silencing (H3K27me3) (Azuara et al., 2006; Bernstein et al., 2006; Mikkelsen et al., 2007). These chromatin marks are a result of the activity of Trithorax- and Polycomb-group (Trx and PcG) proteins, respectively. Core components of Polycomb Repressive Complex PRC2 (Suz12 and Eed, as well as the modification it catalyzes, H3K27me3) and PRC1 (Rnf2 and Phc1) have been found to significantly overlap in ES cells (Boyer et al., 2006; Lee et al., 2006). Our finding that H2AZ and Suz12 occupy the promoter regions of the same genes in ES cells suggests that this variant is an additional regulatory component at bivalent promoters and links H2AZ to a large class of genes with known roles in development. Consistent with a role for H2AZ in gene regulation, target genes are de-repressed and PcG proteins are lost from promoters upon RNAi-mediated H2AZ depletion in ES cells. It remains unclear, however, whether H2AZ incorporation is necessary for gene repression similarly to PcG proteins or whether it may be required for subsequent gene activation as has been observed in yeast (Li et al., 2005; Zhang et al., 2005).
Prior studies in metazoans have shown that H2AZ enrichment is directly proportional to gene activation (Barski et al., 2007; Mavrich et al., 2008; Schones et al., 2008), so it was surprising that H2AZ was enriched at a large subset of silent genes in ES cells. Recent studies have revealed that bivalent promoters experience transcription initiation but show no evidence of elongation, suggesting that they may be regulated at post-initiation steps (Guenther et al., 2007). An additional feature of these promoters is the presence of a paused RNA Polymerase II (Pol II) complex (Guenther et al., 2007; Stock et al., 2007). Interestingly, Polycomb-mediated ubiquitination of histone H2A, as catalyzed by the PRC1 component Rnf2/Ring1b (de Napoles et al., 2004), is necessary to maintain this configuration because its loss results in release of the paused polymerase and in gene de-repression (Stock et al., 2007). The observed loss of gene silencing and of Rnf2/Ring1b from target promoters in H2AZ-depleted cells is consistent with the idea that H2AZ is a target for Polycomb-mediated ubiquitination. It has been proposed that the incorporation of H2AZ favors nucleosome eviction (Mavrich et al., 2008), which would promote histone turnover and chromatin accessibility. As such, it will also be of interest to investigate whether H2AZ loss or de-ubiquitination is required for release of a paused polymerase and transcriptional elongation. Thus, H2AZ incorporation may be a key mechanism to allow developmental genes to remain silent, yet poised for activation in ES cells.
While the majority of H2AZ bound regions encompass narrow intervals (< 2 kb) at gene promoters, a small proportion of these regions extended from the promoter into the coding region and in some cases included contiguous genes such as those located in the HOX gene clusters (Lee et al., 2006). Biophysical analyses of H2AZ-containing nucleosomal arrays suggest that H2AZ inhibits the formation of highly compacted chromatin fibers (Fan et al., 2004). In this scenario, H2AZ incorporation into large regions with PcG proteins may allow these genes to remain primed for activity by contending with Polycomb-mediated chromatin compaction (Francis et al., 2004). Together, these observations suggest that H2AZ and PcG proteins together may establish a specialized, structurally distinct chromatin conformation that has important consequences on gene regulation.
The promoter configuration at developmental regulators where both H2AZ and PcG proteins are necessary to maintain proper gene control appears to be unique to pluripotent cells. We find that H2AZ and H3K27me3 are co-enriched at genes in ES cells whereas this association is not maintained upon differentiation into multi-potent neural precursors. Rather, in NPs, H2AZ occupies active genes with a wide range of roles in metabolic processes. This observation is in accordance with previous studies in human T cells in that H2AZ localized to discrete regions at the promoters of active genes (Barski et al., 2007; Schones et al., 2008). How can H2AZ mediate seemingly opposing functions? H2AZ can be post-translationally modified and its incorporation correlates with particular histone modification patterns such as acetylation (Guillemette and Gadreau, 2006). Interestingly, the acetylated form of H2AZ correlates with its localization to the 5’ regions of active genes in yeast and vertebrates (Bruce et al., 2005; Millar et al., 2006) suggesting that post-translational modification of H2AZ may underlay its differential distribution in ES cells compared to lineage-committed cells. Alternatively, H2AZ and the histone variant H3.3 can co-occupy the same nucleosome and this has been shown to impact nucleosome stability (Jin and Felsenfeld, 2007). Thus, the function of H2AZ may depend on the nucleosome into which it is incorporated. As such, it will be of tremendous interest to determine how H3.3 is distributed in pluripotent and lineage-committed cells. Together, these studies suggest that H2AZ distribution changes dramatically during development and that its developmental or stage-specific localization and function likely depends on additional modifiers.
We find that the association between H2AZ and PRC1 and PRC2 is mutually interdependent at promoters, as loss of one leads to eviction of the other. This provides strong evidence for an obligate relationship between these regulators and their effect on chromatin structure and target gene regulation in ES cells. Their highly defined distribution patterns suggest that these regulators are specifically recruited to genomic sites. While considerable evidence exists that PcG proteins are targeted by DNA binding factors in Drosophila (Ringrose and Paro, 2007), it is currently unknown how these repressors are localized to target genes in mammals. It is intriguing to speculate that H2AZ may play such a role in ES cells. However, we failed to detect an interaction between these proteins by co-immunoprecipitation assay suggesting that recruitment may occur through a different mechanism. Interestingly, a recent report suggests that long non-coding RNA may facilitate PcG protein localization to HOX gene clusters in human cells (Rinn et al., 2007), highlighting a potential role for ongoing transcription of non-coding RNA in this process. Given that H2AZ is also redistributed during ES cell differentiation, how H2AZ is conscripted at target promoters is an equally important question. H2AZ deposition is catalyzed by the ATP-dependent activity of the SRCAP/SWR1 complex in mammals (Wong et al., 2007). Thus, it is possible that H2AZ incorporation is mediated by recruitment of this complex independently of PcG proteins. Given that both H2AZ and PcG proteins have been linked to cancer progression (Hua et al., 2008; Sparmann and van Lohuizen, 2006), it will be of extreme interest to elucidate the mechanisms by which these regulators are recruited to genomic sequences.
This study demonstrates a requirement for H2AZ to initiate developmental programs, but not for maintenance of the ES cell state suggesting an important role for H2AZ in mediating cell fate transitions. This phenotype is similar to loss of Suz12 where null ES cells display de-repression of target genes and failure to differentiate into multiple lineages (Pasini et al., 2007). In contrast, although the PRC2 component Eed is essential for early development, null ES cells can contribute to tissues of all three germ layers in chimeric assays (Chamberlain et al., 2008) so it will be important to further explore its relationship between H2AZ and PRC2 in ES cells. Our findings demonstrate an important functional interaction between these two chromatin regulatory pathways in ES that is necessary for the control of developmental gene expression programs. Genetic interactions between H2AZ or its deposition complex and PcG proteins have been reported in Drosophila (Ruhf et al., 2001; Swaminathan et al., 2005). It will be important to explore this relationship in mammals to fully understand the nature of this interaction. Nonetheless, our data suggest that the association between H2AZ and PcG proteins provides an important functional switch to control the initial stages of lineage commitment. Moreover, this work provides a model for understanding the role of chromatin states in cell fate specification as well as in the progression from a normal to disease state.
Experimental Procedures
All procedures and analyses are described in detail in Supplemental Data.
Cells and Cell Culture
V6.5 (129SvJae /C57BL/6; male) ES cells were cultured as previously described (Boyer et al., 2006). H2AZ-depleted ES cell lines were similarly cultured with the addition of 1ug/ml puromycin (Sigma) to maintain selection of hairpin expression. Neural precursors were derived from V6.5 ES cell lines as described in Supplemental Data. The suz12 null ES cell lines were cultured in standard ES cell media.
Antibodies, Chromatin Immunoprecipitation (ChIP), and DNA microarray analysis
The antibodies used in this study as well as ChIP methods, array design and data analysis are described in detail in Supplemental Data.
Gene ontology
Gene ontology analysis was performed using DAVID (http://david.abcc.ncifcrf.gov/). EntrezGene IDs were used to generate enrichment statistics for the biological process category based on a background list of all represented genes on the promoter microarray design.
K-means cluster analysis of binding data
The spatial pattern of H2AZ and Suz12 enrichment was compared at the high-confidence set of 1655 H2AZ-enriched genes using K-means clustering with three nodes (Table S8). For each start site, the raw enrichment ratio for the probe closest to that start site were selected in 250 bp increments over a 7kb region (+/− 3.5 kb relative to the TSS). Promoters where >25% of probes were not represented on the array (typically due to repeat masking) were removed from the analysis. The ratios from the two experiments were used to create a single vector for each start site. The data were processed and the cluster diagram was generated using Cluster software (http://rana.standord.edu/software).
RNAi-mediated silencing of H2AZ in ES cells
19-mer hairpin oligonucleotides for H2AZ#1 (5’-TGGAGATGAAGAATTGGAT-3’) and H2AZ#2 (5’-GGTAAGGCTGGAAAGGACT-3’) and for GFP (5’- GCTGACCCTGAAGTTCATC-3’) were cloned into pRetro-Super as described previously (Brummelkamp et al., 2002). Ecotropic retroviral supernatants were generated by transfection of phoenix packaging cells and ES cells were infected overnight and allowed to recover for 48 hr in fresh ES cell medium. Stable viral integrants were selected with1 ug/ml puromycin for 72 hr. Single colonies were picked and clonally expanded under puromycin selection.
Quantitative real-time PCR
Transcript levels in ES cells were determined by reverse transcriptase quantitative real-time PCR as described in Supplemental Data. Primers specific for H2AZ enriched (E) and un-enriched (U) genes are listed in Table S12. Relative mRNA levels were measured in a triplicate set of reactions for each gene in control ES cells as well as in both H2AZ-depleted ES cell lines where relative Gapdh levels were used for normalization.
ChIP and quantitative site-specific real-time PCR analysis
Chromatin immunoprecipitation was performed in control (GFP-hairpin) and in both H2AZ-depleted ES cell lines with antibodies directed against H2AZ, Suz12, or Rnf2/Ring1b using our standard ChIP protocol (Boyer et al., 2006). Real-time quantitative PCR was performed in triplicate reactions as described in Supplemental Data. Primers were chosen to amplify a region within 1 kb upstream of the transcriptional start site and are listed in Table S13.
ES cell differentiation
Embryoid body formation and retinoic acid induction of differentiation was carried out using standard procedures and is described in Supplemental Data.
Supplementary Material
Supplemental data includes Supplemental Text and Experimental Procedures, Supplemental References, thirteen tables, and nine figures that can be found online with this article at http://www.cell.com.
Acknowledgements
We thank Patrick Shorderet and Dongdong Fu for technical assistance and members of the Boyer lab, especially Lauren Surface, Sera Thornton, and Lilly Torrey for critical review of the manuscript. We are grateful to Kristian Helin for the suz12 null ES cells. MC is supported by a grant from the Dutch Cancer Foundation (KWF). JH is supported by the Helen Hay Whitney Foundation. RAY was supported by NIH R01 HG002668 and RJ was supported by NIH grants 5 R37CA084198, 5-RO1-HDO45022. LAB was supported in part by a fellowship award from Genzyme Corp.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Raw Data and Accession Numbers
Microarray data are available at ArrayExpress under the accession designation E-TABM-556.
References
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Burch JB. Regulation of GATA gene expression during vertebrate development. Semin Cell Dev Biol. 2005;16:71–81. doi: 10.1016/j.semcdb.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Deschamps J. Ancestral and recently recruited global control of the Hox genes in development. Curr Opin Genet Dev. 2007;17:422–427. doi: 10.1016/j.gde.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Dunn RK, Kingston RE. Gene regulation in the postgenomic era: technology takes the wheel. Mol Cell. 2007;28:708–714. doi: 10.1016/j.molcel.2007.11.022. [DOI] [PubMed] [Google Scholar]
- Faast R, Thonglairoam V, Schulz TC, Beall J, Wells JR, Taylor H, Matthaei K, Rathjen PD, Tremethick DJ, Lyons I. Histone variant H2A.Z is required for early mammalian development. Curr Biol. 2001;11:1183–1187. doi: 10.1016/s0960-9822(01)00329-3. [DOI] [PubMed] [Google Scholar]
- Fan JY, Rangasamy D, Luger K, Tremethick DJ. H2A.Z alters the nucleosome surface to promote HP1alpha-mediated chromatin fiber folding. Mol Cell. 2004;16:655–661. doi: 10.1016/j.molcel.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Glaser T, Brustle O. Retinoic acid induction of ES-cell-derived neurons: the radial glia connection. Trends Neurosci. 2005;28:397–400. doi: 10.1016/j.tins.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:384–394. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette B, Gaudreau L. Reuniting the contrasting functions of H2A.Z. Biochem Cell Biol. 2006;84:528–535. doi: 10.1139/o06-077. [DOI] [PubMed] [Google Scholar]
- Hake SB, Allis CD. Histone H3 variants and their potential role in indexing mammalian genomes: the "H3 barcode hypothesis". Proc Natl Acad Sci U S A. 2006;103:6428–6435. doi: 10.1073/pnas.0600803103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K. Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol. 2005;21:133–153. doi: 10.1146/annurev.cellbio.21.012704.133518. [DOI] [PubMed] [Google Scholar]
- Hua S, Kallen CB, Dhar R, Baquero MT, Mason CE, Russell BA, Shah PK, Liu J, Khramtsov A, Tretiakova MS, Krausz TN, Olopade OI, Rimm DL, White KP. Genomic analysis of estrogen cascade reveals histone variant H2A.Z associated with breast cancer progression. Mol Syst Biol. 2008;4:188–202. doi: 10.1038/msb.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kyba M, Perlingeiro RC, Daley GQ. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Young RA. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann OJ, Sowden JC, Carlsson P, Jordan T, Bhattacharya SS. Fox's in development and disease. Trends Genet. 2003;19:339–344. doi: 10.1016/S0168-9525(03)00111-2. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci U S A. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Pevny L, Lovell-Badge R, Smith A. Generation of purified neural precursors from embryonic stem cells by lineage selection. Curr Biol. 1998;8:971–974. doi: 10.1016/s0960-9822(98)70399-9. [DOI] [PubMed] [Google Scholar]
- Liu X, Li B, Gorovsky M. Essential and nonessential histone H2A variants in Tetrahymena thermophila. Mol Cell Biol. 1996;16:4305–4311. doi: 10.1128/mcb.16.8.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Bernstein BE. Chromatin state maps: new technologies, new insights. Curr Opin Genet Dev. 2008;18:109–115. doi: 10.1016/j.gde.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5' ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Madhani HD. Patterning chromatin: form and function for H2A.Z variant nucleosomes. Curr Opin Genet Dev. 2006;16:119–124. doi: 10.1016/j.gde.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Rangasamy D, Berven L, Ridgway P, Tremethick DJ. Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J. 2003;22:1599–1607. doi: 10.1093/emboj/cdg160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway P, Brown KD, Rangasamy D, Svensson U, Tremethick DJ. Unique residues on the H2A.Z containing nucleosome surface are important for Xenopus laevis development. J Biol Chem. 2004;279:43815–43820. doi: 10.1074/jbc.M408409200. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhf ML, Braun A, Papoulas O, Tamkun JW, Randsholt N, Meister M. The domino gene of Drosophila encodes novel members of the SWI2/SNF2 family of DNA-dependent ATPases, which contribute to the silencing of homeotic genes. Development. 2001;128:1429–1441. doi: 10.1242/dev.128.8.1429. [DOI] [PubMed] [Google Scholar]
- Schepers GE, Teasdale RD, Koopman P. Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell. 2002;3:167–170. doi: 10.1016/s1534-5807(02)00223-x. [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Showell C, Binder O, Conlon FL. T-box genes in early embryogenesis. Dev Dyn. 2004;229:201–218. doi: 10.1002/dvdy.10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Swaminathan J, Baxter EM, Corces VG. The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev. 2005;19:65–76. doi: 10.1101/gad.1259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Daal A, Elgin SC. A histone variant, H2AvD, is essential in Drosophila melanogaster. Mol Biol Cell. 1992;3:593–602. doi: 10.1091/mbc.3.6.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong MM, Cox LK, Chrivia JC. The chromatin remodeling protein, SRCAP, is critical for deposition of the histone variant H2A.Z at promoters. J Biol Chem. 2007;282:26132–26139. doi: 10.1074/jbc.M703418200. [DOI] [PubMed] [Google Scholar]
- Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental data includes Supplemental Text and Experimental Procedures, Supplemental References, thirteen tables, and nine figures that can be found online with this article at http://www.cell.com.