Abstract
We sought to computationally design model collagen peptides that specifically associate as heterotrimers. Computational design has been successfully applied to the creation of new protein folds and functions. Despite the high abundance of collagen and its key role in numerous biological processes, fibrous proteins have received little attention as computational design targets. Collagens are composed of three polypeptide chains that wind into triple-helices. We developed a discrete computational model to design heterotrimer forming collagen-like peptides. Stability and specificity of oligomerization were concurrently targeted using a combined positive and negative design approach. The sequences of three 30-residue peptides, A, B and C were optimized to favor charge-pair interactions in an ABC heterotrimer, while disfavoring the twenty-six competing oligomers (i.e. AAA, ABB, BCA, etc.). Peptides were synthesized and characterized for thermal stability and triple-helical structure by circular dichroism and NMR. A unique A:B:C-type species was not achieved. Negative design was partially successful, with only A+B and B+C competing mixtures formed. Analysis of computed versus experimental stabilities helps clarify the role of electrostatics and secondary-structure propensities determining collagen stability, and provide important insight into how subsequent designs can be improved.
Introduction
Collagen is the most abundant protein in higher animals, accounting for approximately one-third of total protein mass. Individual collagen chains trimerize into triple-helices, which further assemble into higher order structures such as long fibers and mesh-like networks (1–3). These structures provide tensile strength and flexibility to tissues. Collagens also play important functional roles in mediating cell polarity, initiating thrombosis and modulating tumor metastasis (1). There are twenty-eight known collagen types in humans (2), many with multiple subtypes. Subtypes are often co-expressed to assemble in heterotrimeric triple-helices. The most abundant collagen, Type I, is composed of two α1(I) and one α2(I) chains. Type IV collagen, a primary component of basement membranes, can exist as 2α1(IV):α2(IV), α3(IV):α4(IV):α5(IV) or α5(IV):2α6(IV) heterotrimers (3). Stoichiometry is controlled at multiple levels, from gene expression to protein-protein interactions. Natural collagens are over a thousand amino acids in length and contain both globular and triple-helical domains. As such, model peptide systems have been essential tools in exploring the molecular basis for stability, specificity and stoichiometry of collagen assembly. We use computational protein design to study this important class of proteins.
At one extreme, the design of collagens is quite easy; fibril forming domains are defined by a canonical Gly-X-Y triplet repeat. These repeats can extend for a thousand amino acids, resulting in triple-helices around 300 nm long. The X and Y positions are frequently proline and (4R)-hydroxyproline (abbreviated as Hyp or O) respectively. At the other extreme, understanding the thermodynamics of collagen assembly towards atomic resolution computational design applications is a challenging task. Unlike globular proteins, the fibrillar regions of collagens do not have a hydrophobic core. Instead, the triple-helix structure is mediated by a network of inter-chain backbone hydrogen bonds (4, 5). Sidechains of non-glycine positions project into solvent, where the energetic contributions of these groups to folding and stability are highly dependent on interactions with water (6, 7). High-resolution crystal structures of collagen peptides show an extended, structured hydration network surrounding the triple-helix. Modeling solvent contributions to protein folding and structure has always been a major challenge in computational methods, trying to strike a balance between the efficiency of continuum solvation and the accuracy of explicit water models (8, 9). It is also unclear to what extent hydration serves to stabilize the collagen triple-helix. Raines and colleagues have demonstrated that unique stereoelectronic effects such as a predisposition of the Cγ-exo ring pucker in hydroxyproline favors main-chain bond angles associated with the triple-helix (10, 11). Such forces are essential for collagen folding but not commonly included in molecular mechanics or design potentials. Additionally, the folding of collagen is slow, and does not appear to be two-state (12, 13), complicating the estimation of sequence contributions to the energies of native and unfolded states. Computational design of collagen peptides will provide a powerful approach for exploring the complexities of fibrous protein folding.
Current understanding of collagen structure and folding has come in large part from extensive biophysical characterization of model peptide systems. Natural collagens have a higher than normal frequency of acidic and basic residues in triple-helical domains, suggesting that electrostatic interactions play an important role in stability and the specificity of chain-chain recognition during folding (14–16). Seminal host-guest model collagen peptide studies by Brodsky and colleagues established that favorable interchain charge-pair interactions can increase thermal stability (17–19). Most model peptide studies have focused on homotrimers, although the introduction of disulfides has proved useful in stabilizing engineered heterotrimers (20, 21). Complementary pairing of electrostatic interactions can also drive the formation of highly stable heterotrimeric triple-helices (22–24).
Designing hetero-oligomeric assemblies provide unique challenges to protein design. An effective design will optimize the stability of the target fold, while disfavoring competing states. These two components are referred to as ‘positive’ and ‘negative’ design (25). Characterizing the heterotrimer energy landscape with peptides A, B and C, requires consideration of the target state, ABC, and the twenty-six competing states: AAA, BBB, CCC, AAB, BCA, etc. Positive design targets the ABC state, by maximizing its stability based on charge pair interactions between the three peptides. Additionally, an explicit negative design component selects for sequences of A, B and C with unfavorable interaction energies when computed in alternate, competing oligomerization states. Since each round of sequence selection requires calculations over multiple states rather than one, a simple, discrete model of collagen heterotrimer stability is developed that allows rapid computation.
In this study, we present a computational design protocol that concurrently optimizes target stability and specificity over competing states, towards the design of three peptides that assemble as an ABC heterotrimer. This project is novel in scope with the selection of collagen as a target and the explicit incorporation of positive and negative design. We characterize the first-generation of peptides using this approach and analyze design successes and failures toward improving the design protocol and understanding the role of electrostatics in collagen folding.
Computational and Experimental Methods
Computing stability
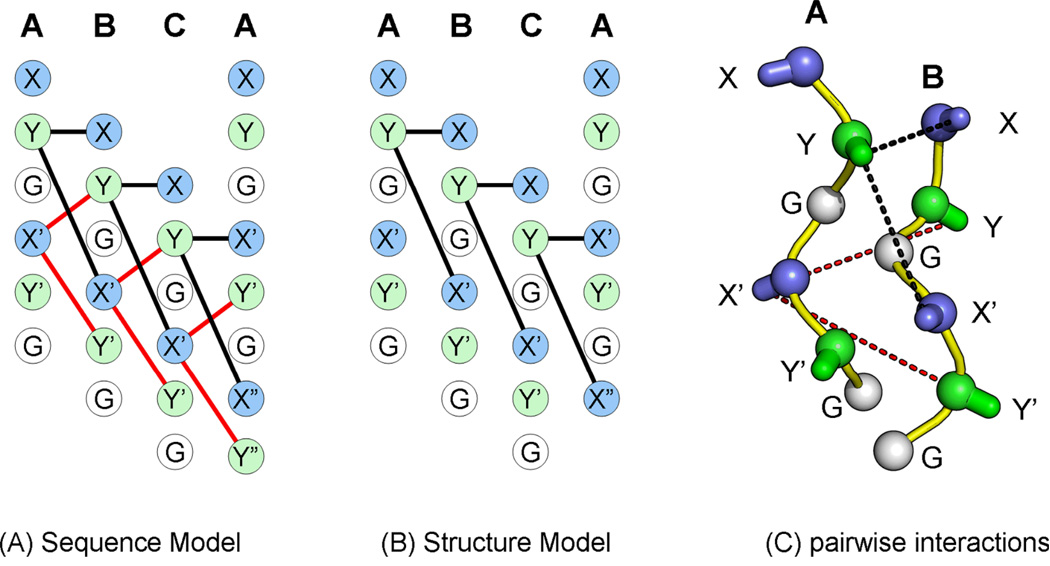
The interaction energy is computed with sequence- and structure-based models (Figure 1) shown below as:
Figure 1.
Ion pairs of (A) the sequence-based model; Y-X and Y-X’ interactions in black lines, X’-Y and X’-Y’ interactions in red lines. (B) The structure-based model. The ion pairs are connected with dashed lines. (C) Structural model of inter-chain interactions between chains A and B.
Sequenced-based model
| Eq. 1 |
Inspection of crystal structures of collagen peptides shows the pairs X’A-YB and X’A-Y’B are not within interaction distance. Based on this, we formulated the structure-based model (Eq. 2).
Structure-based model
| Eq. 2 |
An additional −1.5 kcals/mole was included for each POG triplet in the three peptides to account for imino acid contributions to triple-helix stability. The sequence-based model was used in all design calculations. The structure-based model was only used during post hoc analysis of experimental outcomes.
Computing Specificity
There are twenty-seven structurally unique triple-helical species possible with a mixture of peptides A, B and C. The specificity, PABC, of the target over competing species is defined using a Boltzmann distribution.
| Eq. 3 |
where i ∈ (AAA, AAB, AAC ... CCC) and β is a system temperature set to 1.0. In cases where all peptides in solution are predicted to assemble as ABC, PABC approaches one. If ABC is not formed, PABC approaches zero.
Sequence Optimization
To concurrently optimize the stability and specificity of the target, a combined score, S, incorporates both elements:
| Eq. 4 |
where c is a scaling factor controlling the relative contributions of stability and specificity and was assigned a value of one hundred in the current simulations.
A Monte Carlo Simulated Annealing (MCSA) protocol was used to optimize the sequence of peptides A, B and C (26–28). Simulations were run for 105 cycles where each iteration involved a triplet sequence modification to one of the three peptides. Available triplets were POG, PRG, ROG, PEG and EOG. Thus, for three thirty-amino acid peptides, there were 530 (~1021) possible sequences. Changes were accepted or rejected using the Metropolis criterion (28) with a probability a, where
| Eq. 5 |
A linear temperature cooling schedule was used with T0 = 40 and Tfinal = 10−3. Ten thousand simulations were run. The best set of A, B and C sequences were chosen from these for synthesis and characterization.
Peptides
The three peptides, A, B and C were synthesized using solid phase FMOC chemistry at the Tufts University Core Facility (http://tucf.org). N and C termini were uncapped. Although lack of acetylation or amidation was anticipated to incur repulsive interactions between amines at the N-terminus and carboxylates at the C-terminus, it has been shown that such end effects have modest effects on stability at neutral pH in model collagen peptides (17). Peptides were purified to 90% purity by reverse phase HPLC and products were verified by mass spectrometry. Concentrations were determined by monitoring absorbance at 214 nm using ε214 = 2200 M−1 cm−1 per peptide bond. Ten peptide mixtures were prepared with ratios: A, B, C, A:2B, 2A:B, A:2C, 2A:C, B:2C, 2B:C and A:B:C.* Total peptide concentrations of were 0.2 mM in 10 mM phosphate buffer at pH 7.0. Low pH studies were carried out in 0.1 mM HCl adjusted to pH 1.1.
Peptides were split into two groups prior to incubation at 4°C. In the first group, mixtures were prepared at room temperature, preheated to 40°C (more than 15°C above observed melting temperatures) for 15 minutes and then stored at 4°C for 48 to 72 hours. In the second, mixtures were prepared at room temperature and directly stored at 4°C for the same duration. Incubation at low temperatures for periods ranging from overnight to several days is a typical protocol for folding collagen peptides due to the slow kinetics of triple-helix formation.
To explore binding stoichoimetry, the Method of Continuous Variation (Job Plot) was applied (29); 0.2 mM total peptide solutions were prepared in 10 mM phosphate buffer at pH 7.0 as A:nB, nB:C, and A:nB:C, where the ratio n was 0.0, 0.5, 1.0, 1.5, 2.0, 2.5, 3.0 and 3.5. Mixtures were incubated overnight at 4°C prior to measurement. Each experiment was repeated three times with newly prepared samples.
Circular Dichroism
CD experiments were performed on an AVIV Model 400 Spectrophotometer. Optically matched 0.1 cm path length quartz cuvettes (Model 110-OS, Hellma USA) were used. Wavelength scans were conducted from 190 to 260 nm at 0°C (number of scans 1, averaging 1.0 second). Values are reported as molar ellipticity, correcting for concentration, sequence length and cell path length.
Thermal denaturation CD measurements were performed on the same instrument. Ellipticity was monitored at 223 nm. Temperatures were sampled from 0 to 40°C at 0.33°C/step, 2 minutes equilibration time. In order to calculate an apparent melting temperature, Tm, we estimated the fraction folded using:
| Eq. 6 |
where θ(T) is the observed ellipticity and θF(T) and θU(T) are estimated ellipticities derived from linear fits to the folded and unfolded baselines. The melting temperature is estimated as T where F(T) = 0.5.
NMR
The three basis peptides A, B, C were mixed to get six different combinations: A, B, C, A:2B, 2B:C, and A:B:C. Final concentrations were 1.0 mM in D2O with 25 mM NaDPO4, pD 7.0. 1H-13C single-bond correlation spectra were recorded at 4°C on a Varian Inova 600 MHz spectrometer using sensitivity-enhanced constant-time HSQC as described earlier (30). Spectral widths were 7000 Hz for 1H and 12065 Hz for 13C.
Results
A Discrete Model of Collagen Stability
The goal of this project was to develop a computational scheme that optimized the stability of a target species, an ABC collagen-like heterotrimeric triple-helix, while disfavoring the formation of competing states. The interaction energies of all species, the target ABC heterotrimer and the other 26 undesired states, were evaluated with a discrete model, where the ion pairs of charged residues were counted. Interactions scores were adapted from a similar approach to α-helical coiled-coil design in Summa et. al. (31) (Eq. 7). The energy of an Arg–Arg repulsion was weighted less than Glu-Glu due to the expectation that the longer, flexible sidechain of Arg would more easily avoid unfavorable interactions. Interactions of any residue with proline or hydroxyproline were assigned a score of zero. A single-body energy of −1.5 kcals/mole was added for each POG triplet to approximate the favorable backbone stability provided by this neutral motif.
| Eq. 7 |
Intermolecular neighboring pairs were included using the sequenced-based model (Eq. 1).
Surveying the Energy Landscape
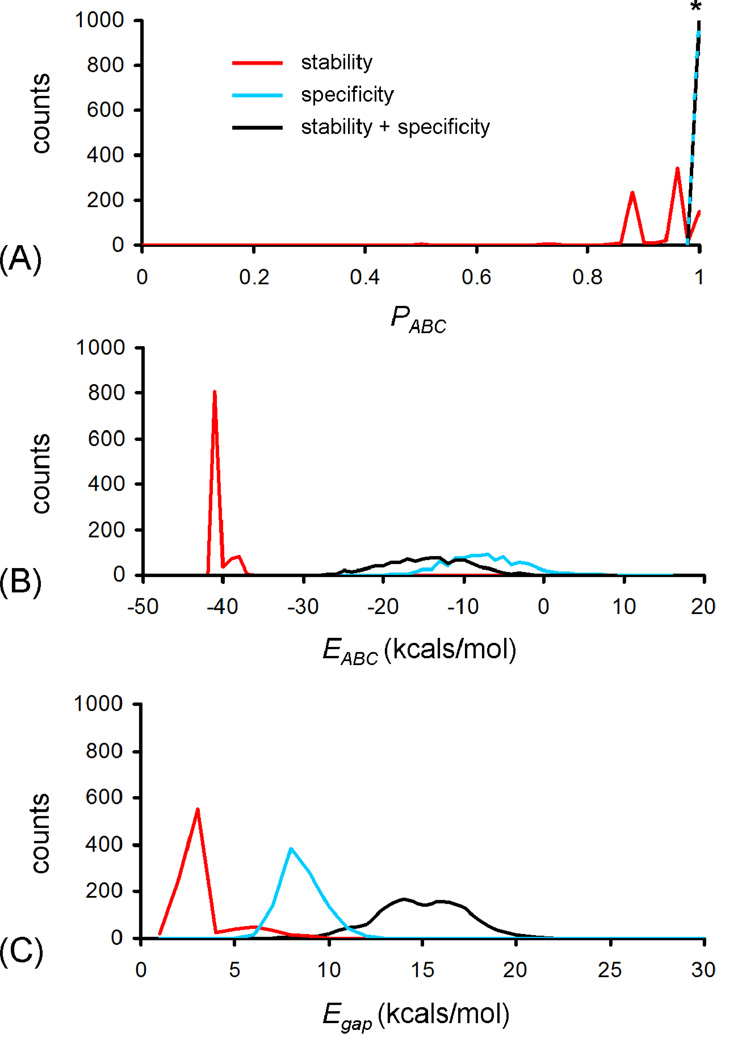
Stability and specificity were simultaneously optimized to separate the computed target heterotrimer ABC stability from other competing hetero- and homotrimers (Figure S1). The stability of the ABC species was optimized using positive design, maximizing the number of favorable interactions in a discrete, sequence-based interaction model. Specificity was targeted using negative design, applying the same discrete interaction model to competing species, and optimizing a Boltzmann probability based specificity score (Eq. 3). To evaluate the relative contributions of positive and negative design to energy and specificity distributions, one thousand simulations were performed for each scenario: positive design alone for stability using Eq. 1, negative design alone for specificity using Eq. 3, or concurrent optimization of both stability and specificity using Eq. 4.
In simulations where only stability of ABC was optimized, final EABC values were favorable, between −38 and −40 kcals/mol (Figure 2). Only six out of one thousand simulations resulted in PABC < 0.5, and over one third gave PABC values around 0.95. This observation is consistent with common wisdom in protein design that focusing on stability can often also result in good target specificity.
Figure 2.
(A) PABC (Eq.3), (B) EABC (Eq. 1) and (C) energy gap (Eq. 8) distributions for 1000 simulations optimizing stability alone (red), 1000 simulations optimizing specificity alone (black) or 1000 simulations optimizing both elements simultaneously (Eq. 4) (blue). *Both specificity and stability + specificity simulations consistently resulted in final PABC values between 0.98 and 1.0.
Optimizing PABC alone resulted in a significantly weaker and broader distribution of stabilities for ABC, with a mean of −5 kcals/mole. Not surprisingly, specificities were very tightly clustered around 1.0. Such tradeoffs between stability and specificity had been observed previously in lattice chain model systems employing charge-pair interactions (32, 33). Combining positive and negative design elements resulted in intermediate distributions. The mean stability for ABC was around −15 kcals/mol while PABC still clustered around one.
A statistical mechanic metric that correlates with fold specificity is the energy gap between the native target and the lowest energy of an unwanted competing state (34–37).
| Eq. 8 |
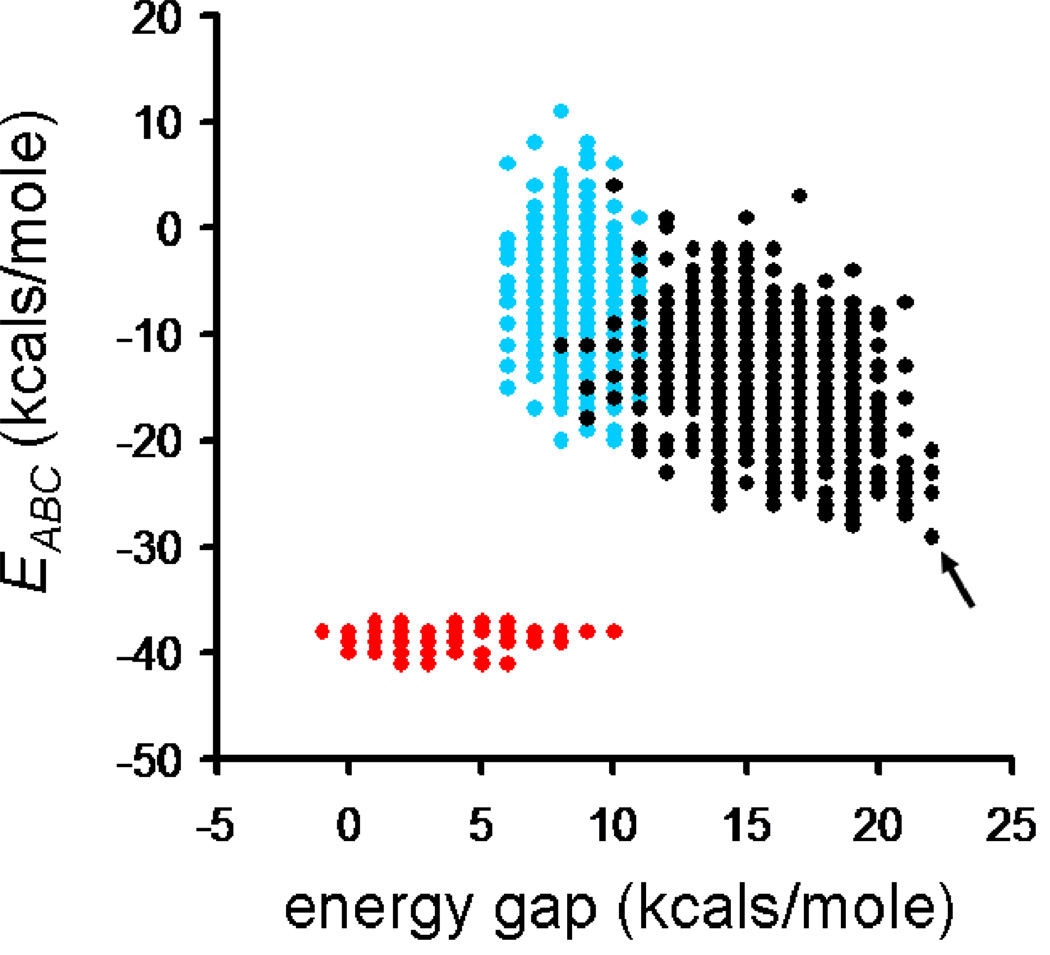
In computing the Egap distribution for the three simulation scenarios, combining positive and negative design raised the mean gap energy over either component alone (Figure 2C). Based on these observations, we ran ten-thousand MCSA simulations combining positive and negative design elements. From these, a final set of A, B and C peptide sequences were selected that had an optimal stability of ABC and a maximal energy gap (Figure 3).
Figure 3.
Stability versus gap for three optimization conditions: stability alone (red), specificity alone (blue) and both terms combined (black). The solution which combined the best stability and a high energy gap was selected for synthesis and characterization (arrow).
Optimized Peptide Sequences
Using the sequence-based model, a set of peptide sequences was chosen where EABC = −29 kcals/mole and Egap = 22 kcals/mole. The next most stable states were AAB and BAB with computed stabilities of −7 kcals/mol (Table 1).
peptide A: EOGROGPEGROGPEGPEGEOGEOGPEGROG
peptide B: PEGEOGROGROGPRGPRGPRGROGROGPRG
peptide C: PRGROGROGROGPRGEOGPEGPEGEOGEOG
Table 1.
Predicted interaction scores of all 27 possible combinations of peptides A, B, C. The lowest scores with the same ratio of peptides are highlighted in bold.
| Species | Interaction score | |
|---|---|---|
| Sequence- based |
Structure- based |
|
| AAA | 20 | 9 |
| BBB | 30 | 18 |
| CCC | 29 | 20 |
| ABB | −1 | −3 |
| BAB | −7 | 6 |
| BBA | 0 | 4 |
| AAB | −7 | −7 |
| ABA | −5 | 5 |
| BAA | −6 | 0 |
| BCC | −5 | −2 |
| CBC | −3 | 4 |
| CCB | −1 | −4 |
| BBC | −3 | −1 |
| CBB | 1 | −3 |
| BCB | −6 | 0 |
| ACC | 2 | 17 |
| CAC | 22 | 32 |
| CCA | 32 | 19 |
| AAC | −2 | 14 |
| ACA | 21 | 27 |
| CAA | 32 | 16 |
| ABC | −29 | −4 |
| ACB | −4 | 1 |
| BAC | −7 | 9 |
| BCA | −7 | 4 |
| CAB | −7 | 3 |
| CBA | 5 | 2 |
Five triplets, PRG, ROG, EOG, PEG, and POG were used as motif elements in the sequence optimization based on their thermal stability measured with host-guest methods (38). Other triplets, such as REG and ERG with two charged residues that could provide additional charge pair stability (17), were avoided. In test simulations where RRG, EEG, REG and ERG were initially included, no imino acid containing triplets were found in the final sequences. Limiting triplets to those containing a Hyp or Pro was a ‘compositional constraint’, a form of negative design used to destabilize the unfolded state (39).
Although the propensity for POG to stabilize the triple-helix backbone was accommodated by adding −1.5 kcals/mol per POG, final sequences only used charged residue containing triplets. The three peptides combined had a 2+ charge, with A being net positive, B net negative and C neutral. In top scoring sequences, the frequencies of acidic and basic residues were close to equal (Table S1). The same behavior was observed when scoring on stability alone.
Eliminated Competing States
Although there were twenty-seven possible triple-helix combinations of A, B and C, due to experimental challenges with determining collagen structure, only stoichiometry of association was characterized. As such, ten mixtures were studied: A, B, C, A:2B, 2A:B, A:2C, 2A:C, B:2C, 2B:C and A:B:C (Table 2).
Table 2.
Predicted interaction scores and experimentally measured stabilities of triple-helix formation for mixtures of peptides A, B, C at pH 1.0 and 7.0, respectively. Scores and apparent Tm of species showing cooperative unfolding are highlighted in bold.
| Mixture | Scorea |
Apparent Tm |
|||
|---|---|---|---|---|---|
| pH 7.0 |
pH 1.1 |
pH 7.0d |
|||
| Eq. 1 | Eq. 2 | Eq. 2 | preheatedb | not preheated |
|
| A | 20 | 9 | 0 | N/A | N/A |
| B | 30 | 18 | 10 | N/A | N/A |
| C | 29 | 20 | 8 | N/A | N/A |
| A:2B | −7 | −3 | 2 | 20 | 20 |
| 2A: B | −7 | −7 | 0 | 20 | 19 |
| B:2C | −5 | −4 | 2 | N/Ac | N/Ac |
| 2B:C | −6 | −3 | 2 | 22 | 20 |
| A:2C | 2 | 17 | 4 | N/A | N/A |
| 2A:C | −2 | 14 | 2 | N/A | N/A |
| A:B:C | −29 | −4 | 0 | 20 | 19 |
the lowest scores among the scores of the species with the same mixing ratio of peptides but different staggering phases such as AAB, ABA, and BAA are shown.
before incubation at 0 °C for 2–3 days, the mixtures are incubated at 40 °C for 15 min.
Thermal melting performed after elimination of white precipitates by centrifugation.
No triple-helical structure observed for any mixture at pH 1
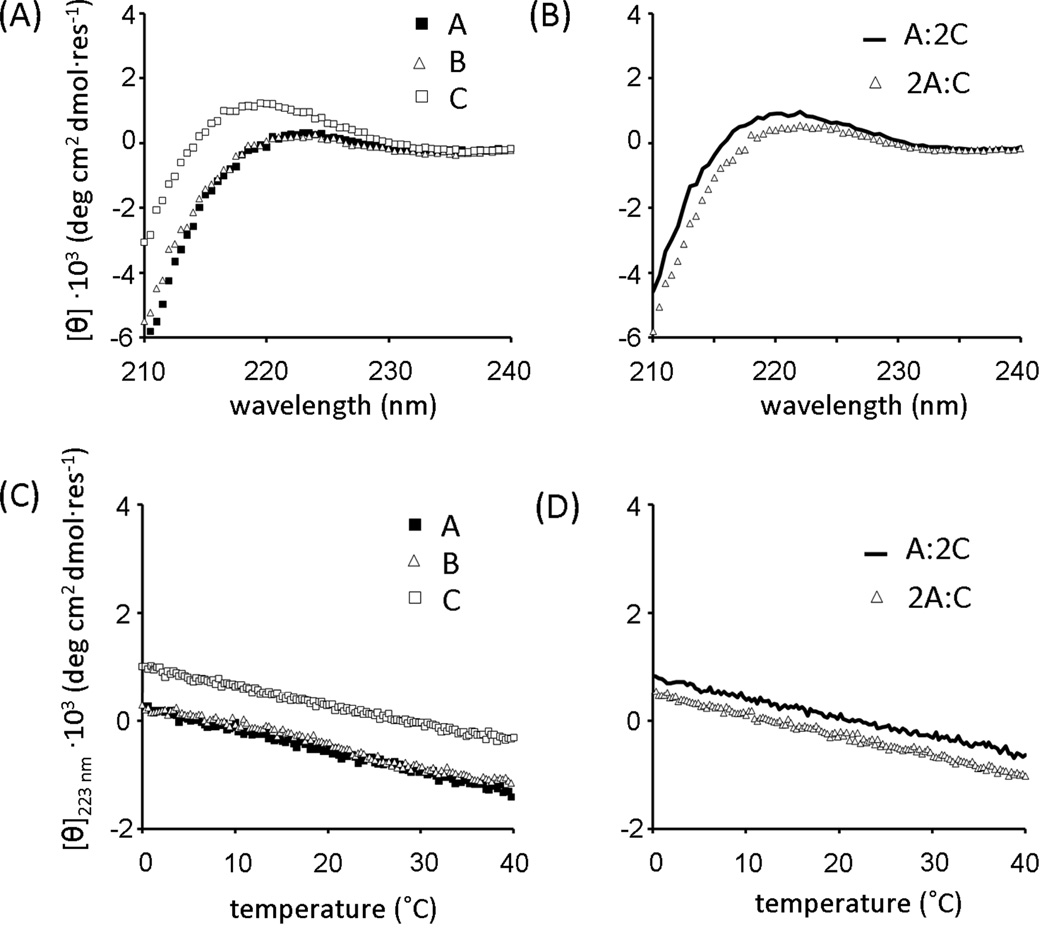
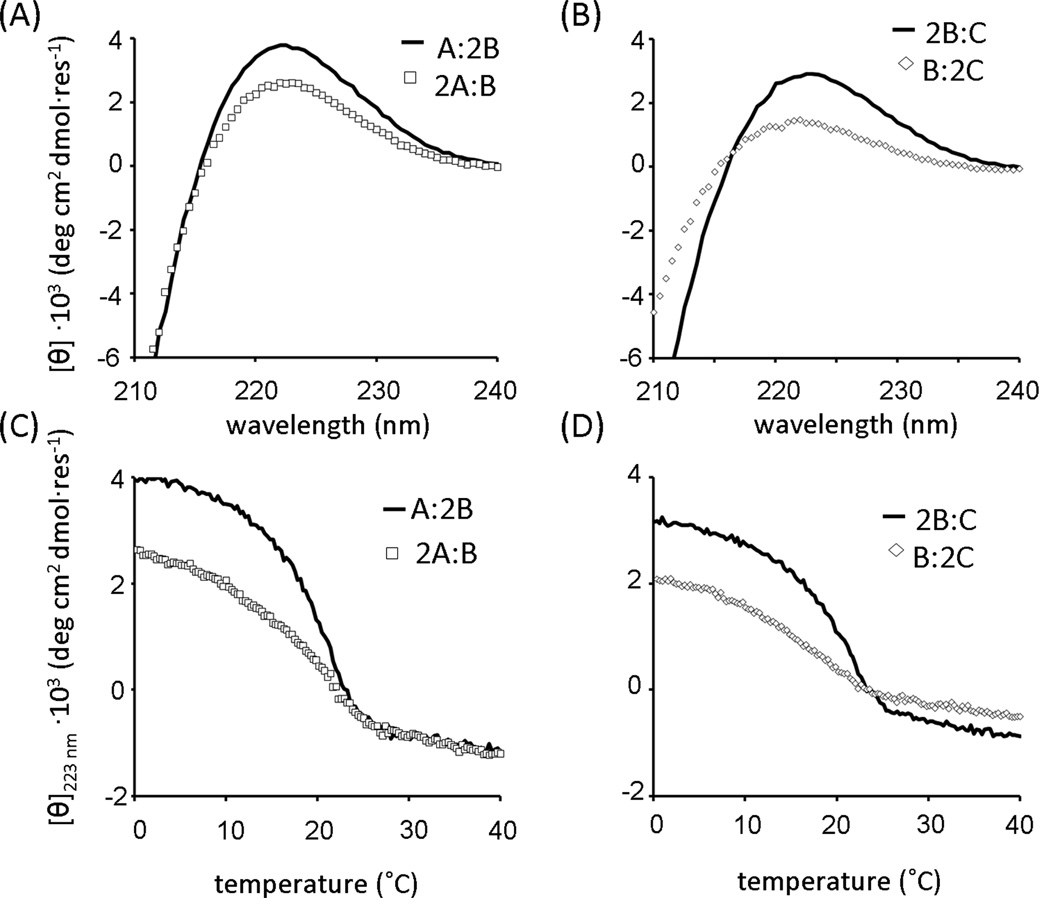
Through analysis of the far-UV CD spectrum and thermal denaturation experiments, the possible existence of homotrimers of A, B, C and heterotrimers A:2C and 2A:C was successfully excluded (Figure 4). Solutions A, B, A:2C, and 2A:C showed no significant peak at 223 nm. None of these mixtures exhibited a cooperative change in secondary structure as a function of temperature between zero and 40°C. In peptide C, a positive band at 218–220 nm was observed, which was an unusually short wavelength for a triple-helix. No cooperative unfolding of peptide C alone was observed, supporting the assertion that this peak was not associated with triple-helix structure.
Figure 4.
Circular dichroism wavelength scans at 0°C for (A) peptides A, B or C alone, and (B) A+C mixtures. Thermal melting profiles from 0 to 40 °C monitored at 223 nm for (C) peptides A, B or C alone, and (D) A+C mixtures.
Formation of Competing Heterotrimers in Binary Mixtures
Despite the inclusion of a negative design component in the computational protocol, undesired heterotrimers involving A + B and B + C were formed (Figure 5). Each of these species also showed a cooperative unfolding transition upon thermal denaturation with Tm values between 19 and 22°C. Relative [θ]223 nm intensities of A:2B versus 2A:B suggesting that A:2B was the primary species in A + B mixtures.
Figure 5.
Circular dichroism wavelength scans at 0°C for (A) peptides A+B mixtures and (B) B+C mixtures. Thermal melting profiles from 0 to 40 °C monitored at 223 nm for (C) peptides A+B mixtures, and (D) B+C mixtures.
B + C also formed heterotrimers, but determining the ratio (B:2C versus 2B:C) was complicated by the significant ellipticity of free C at 223 nm. Additionally, it was found that 0.2 mM B:2C formed aggregates upon two to three days of incubation at 0°C. No aggregation was observed in 2B:C mixtures. Scattering of B:2C showed high turbidity (O.D. at 313 nm = 0.75 versus an average value of around 0.09 for other peptide mixtures). Absorbance at 214 nm after centrifugation of precipitates showed approximately 40% of the peptide was in the aggregate after three days of incubation. The ratio of B to C in aggregates was approximately 1:4 as determined by HPLC of aggregates once the supernatant was removed. C alone could also form aggregates, but over a much longer time (several weeks at 0°C).
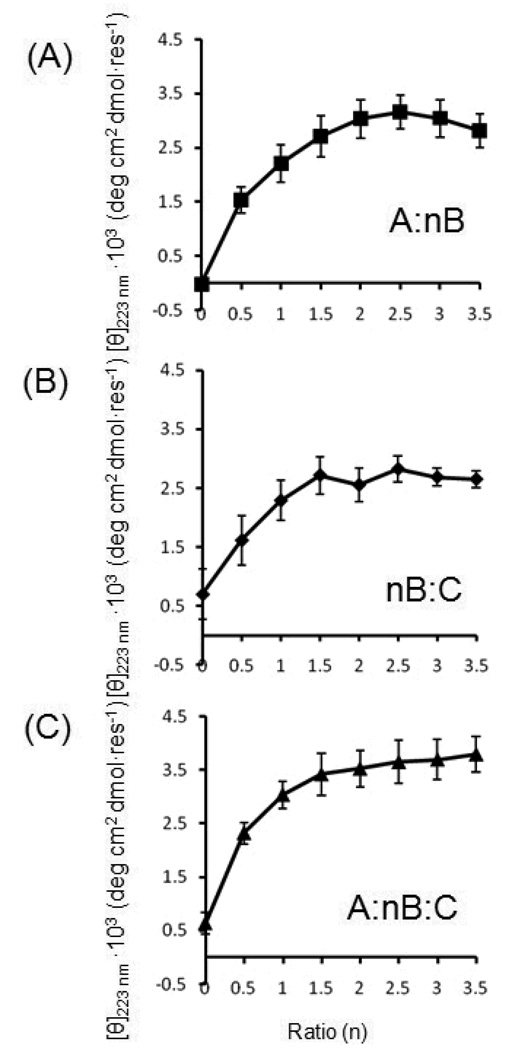
Job plots with varying ratios of A:nB and nB:C were utilized to further probe the stoichiometry of heterotrimers. Ellipticity at 223 nm for A:nB peaks at n = 2.5, consistent with a weakly associated heterotrimer A:2B (Figure 6A). The decreasing ellipticity at n = 3 and 3.5 indicated the amount of A became a limiting factor in complex formation.
Figure 6.
Job plots of three mixtures A:nB, nB:C and A:nB:C, where n is the mixing ratio. Total molar peptide concentration in all mixtures is 0.2 mM. Experiments were conducted in triplicate with standard error shown.
Interpreting the outcome nB:C (Figure 6B) was complicated by competing signal changes from triple-helix complex formation and loss of monomer C signal. Separation of the nB:C titration into component spectra of triple-helix, free B and free C indicated that maximal triple-helix was observed at B:2C (Figure S2). However, aggregation at low ratios of B to C (n < 2) made it challenging to definitively assign stoichiometry.
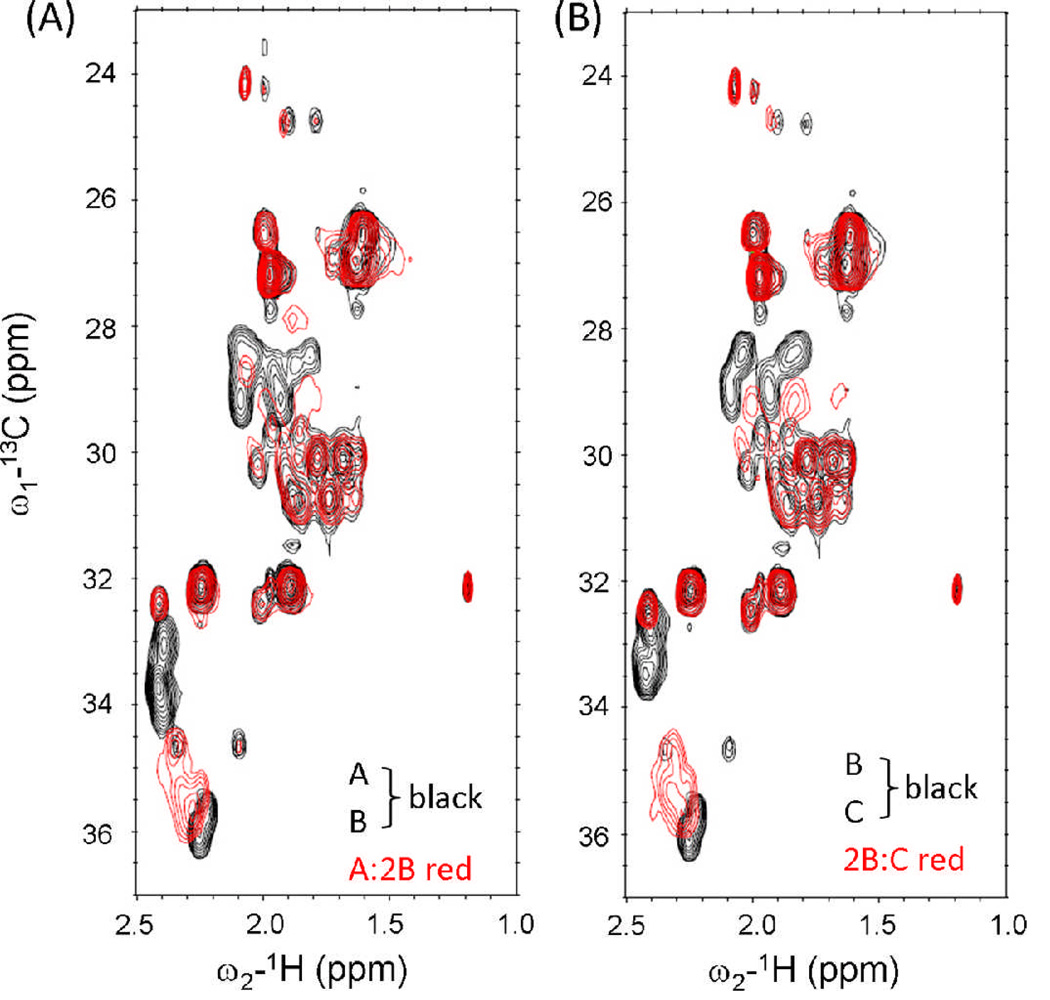
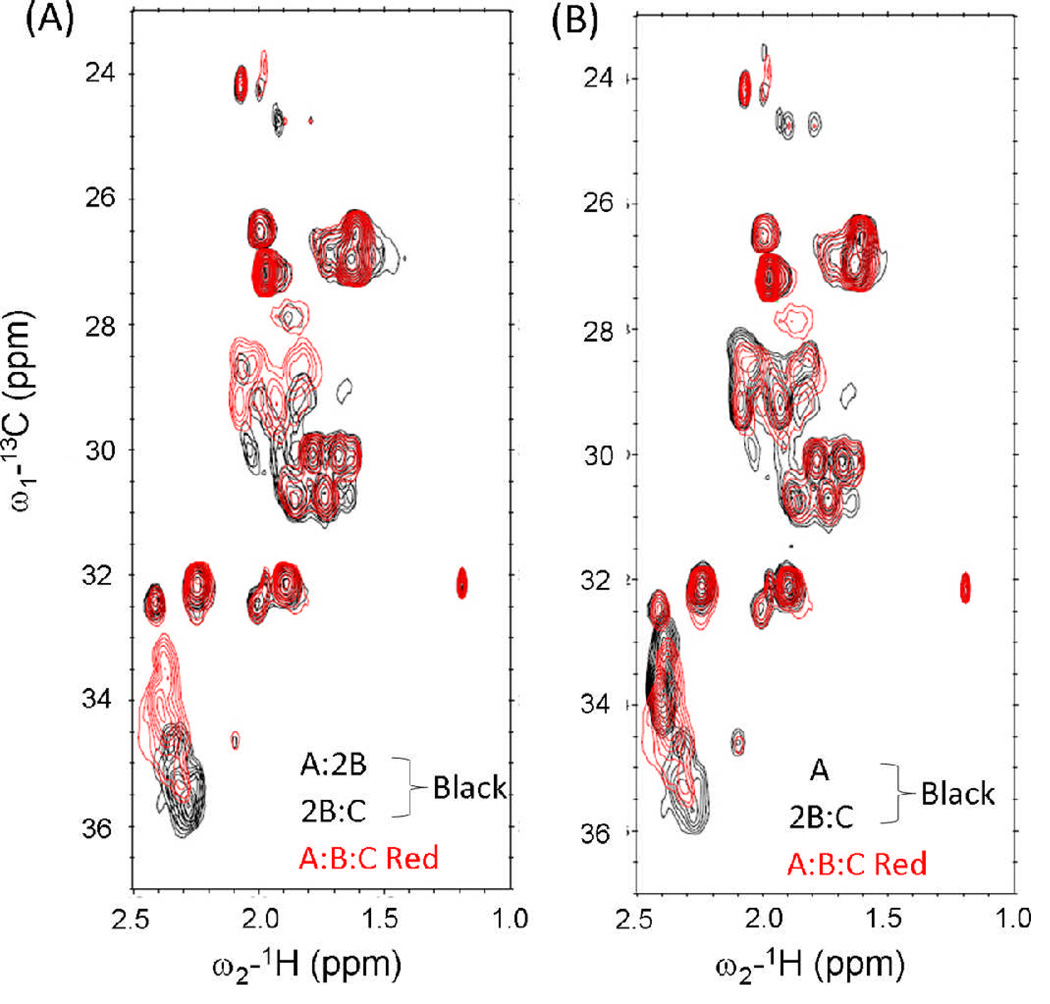
The formation of competing A + B and B + C heterotrimers were corroborated by NMR HSQC measurements. Residues in different species would experience dissimilar chemical environments, giving rise to distinct resonances for each species. New resonances in A:2B which were not observed in the spectra of A or B, were consistent with complex formation (Figure 7). Similarly, the new resonances in 2B:C spectra indicated complex formation between B and C.
Figure 7.
NMR measurement of 1H-13C HSQC spectra. (A) Merged spectra of A and B alone versus A:2B mixture. (B) Merged spectra of B and C alone versus 2B:C mixture.
Due to the lack of homotrimer formation, it was expected that melting and re-annealing peptides was not necessary for assembly of heterotrimers as required in other systems designed using similar principles (22). To test this, two sets of peptide mixtures were prepared; one was mixed and then immediately stored at 4°C for 2–3 days, while the other was preheated at 40°C for 15 minutes before incubation at 4°C. Consistent CD spectra and apparent Tm values were obtained for both sets of peptides (Table 2), indicating that melting and annealing was not required for triple-helix assembly.
Was an A:B:C Heterotrimer Formed?
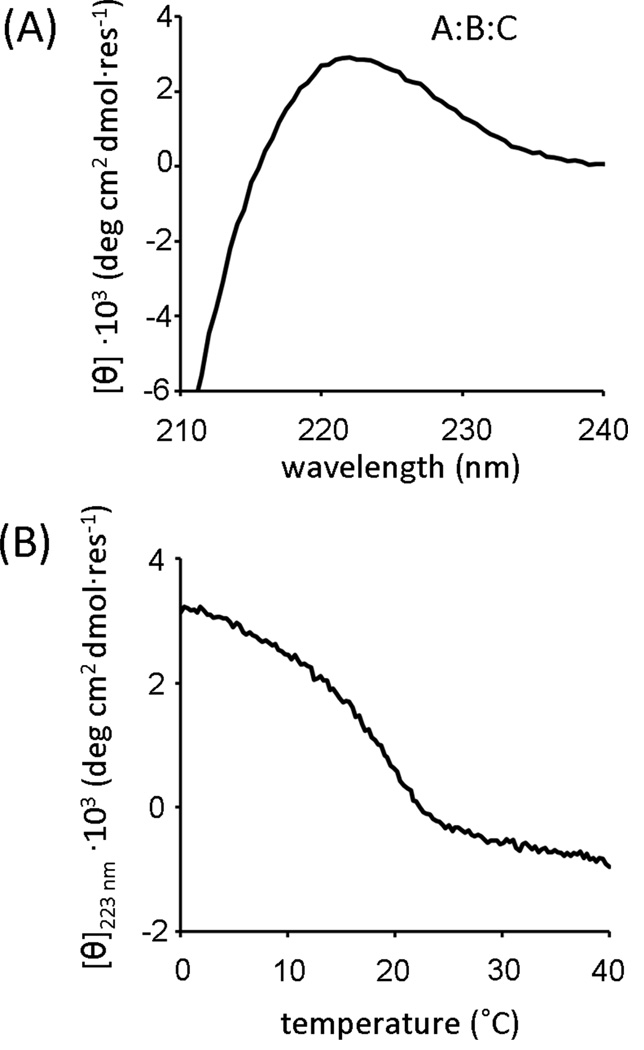
A key objective of this study, the positive design of an ABC heterotrimer, was not conclusively achieved. CD spectra and thermal melting profiles were consistent with triple-helix structure and cooperative unfolding for A:B:C mixtures (Figure 8). However, other lines of evidence indicated A:B:C mixtures were a combination of multiple species including A:2B, B:2C and free A. The ellipticity of A:nB:C stopped increasing at n=1.5 (Figure 6C), suggesting A:B:C could at least be a mixture of A:2B + xB:C. The HSQC spectrum of A:B:C was reconstructed by combining A:2B, 2B:C and free A spectra (Figure 9). Based on the existing characterization, ABC does not appear to be a major, unique species in A+B+C mixtures. Rather, the combination of the three peptides results in a complex mixture of unfolded monomers and binary heterotrimers.
Figure 8.
(A) Circular dichroism wavelength scans at 0°C and (B) thermal melting profiles from 0 to 40°C monitored at 223 nm for equimolar A+B+C mixtures.
Figure 9.
NMR measurement of 1H-13C HSQC spectra. (A) Merged spectra of A:2B and 2B:C versus A:B:C mixture. (B) Merged spectra of A and 2B:C versus the A:B:C mixture.
Assembly at Low pH
To further establish the role of electrostatic interactions in mediating triple-helix stability and specificity, assembly was measured in a low pH environment. Lowering pH prevented triple-helix formation in all combinations of A, B and C peptides. Low ellipticities at 223 nm (Table 2) and lack of cooperative melting transitions were found in CD measurements at a pH of 1.1. The pK of the glutamate carboxyl is around four and sidechains should be neutral at this acidic pH. Using the structure-based model, A, 2A:B and A:B:C had interaction scores of zero at low pH (Table 2). However, none of these mixtures show any evidence of triple helix formation, suggesting that a net balance in favor of attractive interactions is important for facilitating assembly in these model systems.
Discussion
Computational Optimization of Stability and Specificity
The design strategy implemented in this study was inspired by previous designs in α-helical coiled coil systems. Many of these explicitly included an energy gap term in the optimization scheme. For example, in the design of an A2B2 four-helix bundle metalloprotein, Summa and co-workers used an energy gap function:
| Eq. 9 |
where one competing topology was selected to represent the undesired state (31). Only interfacial charge pair interactions were considered. The interaction energies utilized in our work were adapted from this study. A similar energy gap function was used to specify protein-protein interfaces that formed homo or heterodimeric interactions. (40). Havranek and Harbury created coiled-coil homodimers and heterodimers using an energy gap fitness function that considered multiple competing species (41):
| Eq. 10 |
An explicit atomic energy function was used and aggregates and unfolded states were included as competitors. Recently, Grigoryan and colleagues developed a novel algorithmic framework that optimized target stability and the energy gap in distinct phases of the calculation to address the issue of stability/specificity tradeoff in intermolecular associations of bZIP coiled-coils (42).
In this study, we sought to simultaneously optimize stability and specificity using a single scoring function (Eq. 4). A stability/specificity tradeoff was clearly observed in comparisons of the energy landscapes for stability and specificity components in isolation to the landscape of both terms combined. However, while there was a tradeoff between EABC and PABC, Egap was largest in the combined scoring function over either term alone, even though it was not explicitly included as done previously (Eqs. 9, 10). This is a key feature of this approach, the concurrent optimization of target stability, specificity and the energy gap.
Despite the favorable predicted energy landscapes derived using the explicit positive and negative design scoring function implemented in this study, the experimental results were not consistent with the predictions. Three major issues were observed: competing species were formed instead of the target; triple-helical stabilities were poor, requiring low temperatures to observe structure; and over time, certain mixtures would form aggregates. Below, the design methodology is critically evaluated, identifying modifications likely to improve the properties of future designs.
Appropriate Pairwise Interactions
Sequences for peptides A, B and C were selected with favorable computed values: EABC = −29 kcals/mole, PABC = 1.0 and Egap = 22 kcals/mole. However, experimental characterization showed ABC was not formed while competing species were formed in A + B and B + C mixtures. A post hoc analysis of the computed stabilities using a different model (Eq. 2 and Figure 1B) that only took into account charge-pairs adjacent in structure, indicated a structure-based model was more consistent with the experimental outcome: EABC = −4 kcals/mole, PABC = 0.043 and Egap = −3 kcals/mole (Table 2). In this model, the ABC species would only account for ~4% off the total population. AAB, on the other hand, would account for 87% of the population, due to its improved stability over the target (EAAB = −7 kcals/mole). A, B or C alone and A + C mixtures that did not form triple-helices also had unfavorable, positive energies under the structure based model.
In retrospect, it is clear the structure-based model is more appropriate to achieve the stated design goals. Most host-guest peptide studies do not support a role for long range electrostatic interactions in triple-helix stability. Rather, only the pairwise interactions included in Eq.2 are considered key (17, 18, 43). By including additional interactions in the sequence model, it was assumed that non-native interactions and long range electrostatic interactions might contribute to triple-helix folding. A similar sequence based model was originally used to identify stabilizing interactions between triple-helices that could contribute to D-periodic packing observed in natural collagen (44). However, based on the results herein, a sequence based model is not applicable for triple-helix design.
In a follow-up study, we will use the structure-based model in Eq. 2 to optimize a set of sequences for A, B and C for stability and specificity of ABC. To estimate the contributions of long-range electrostatic interactions, it may be necessary to construct atomistic models of these designs and extend the discrete energy function used here to more accurate methods for calculating surface electrostatics (45).
Compositional Constraints and the Unfolded State
Although favorable Y-X and Y-X’ interactions promote triple-helical structure, they are not sufficient for optimally stable designs. A previous study demonstrated that under reducing conditions, a homotrimer of (GER)15GPCCG had a Tm of only ~22°C, despite eighty-three favorable charge pairs (46). With an Etarget = −83 kcals/mole and the additional backbone hydrogen bonding accommodated by fifteen triplets, it was surprising that the triple-helix stability was only marginally better than our designs. In contrast, the Tm of [(POG)10]3 was 68°C, and the Tm of the model heterotrimer (POG)10:(EOG)10:(PRG)10 was 54°C (23). Using the structure model, the computed stability of [(POG)10]3 is −45 kcals/mol (−1.5 kcals/mol per POG triplet). For the heterotrimer system, nineteen favorable Y-X or Y-X’ charge pairs and ten POG triplets combine add up to −34 kcals/mole. This is consistent with a critical role for imino acids at the X and Y in stabilizing triple-helices.
Compositional constraints were imposed on our designs through the sole availability of imino acid containing triplets, resulting in sequences where one third of amino acids were either X=Pro or Y=Hyp. Even at this high frequency, triple-helices were not formed without additional attractive inter-strand interactions. AAA, AAB and ACB were computed to lack attractive or repulsive interactions (E=0 kcals/mole) under low pH conditions (Table S2), yet no combination of A, B and C peptides showed experimental evidence of triple-helix formation. Gauba and Hartgerink found that neither (EOG)10 nor (PRG)10 formed homotrimer triple-helices, despite the lack of any unfavorable interactions (based on Eq.2, Etarget = 0 kcals/mole for both species). Together, these observations suggest that either net-favorable pairwise interactions or an imino acid content greater than one third are rough requirements for triple-helix formation in model collagen peptides.
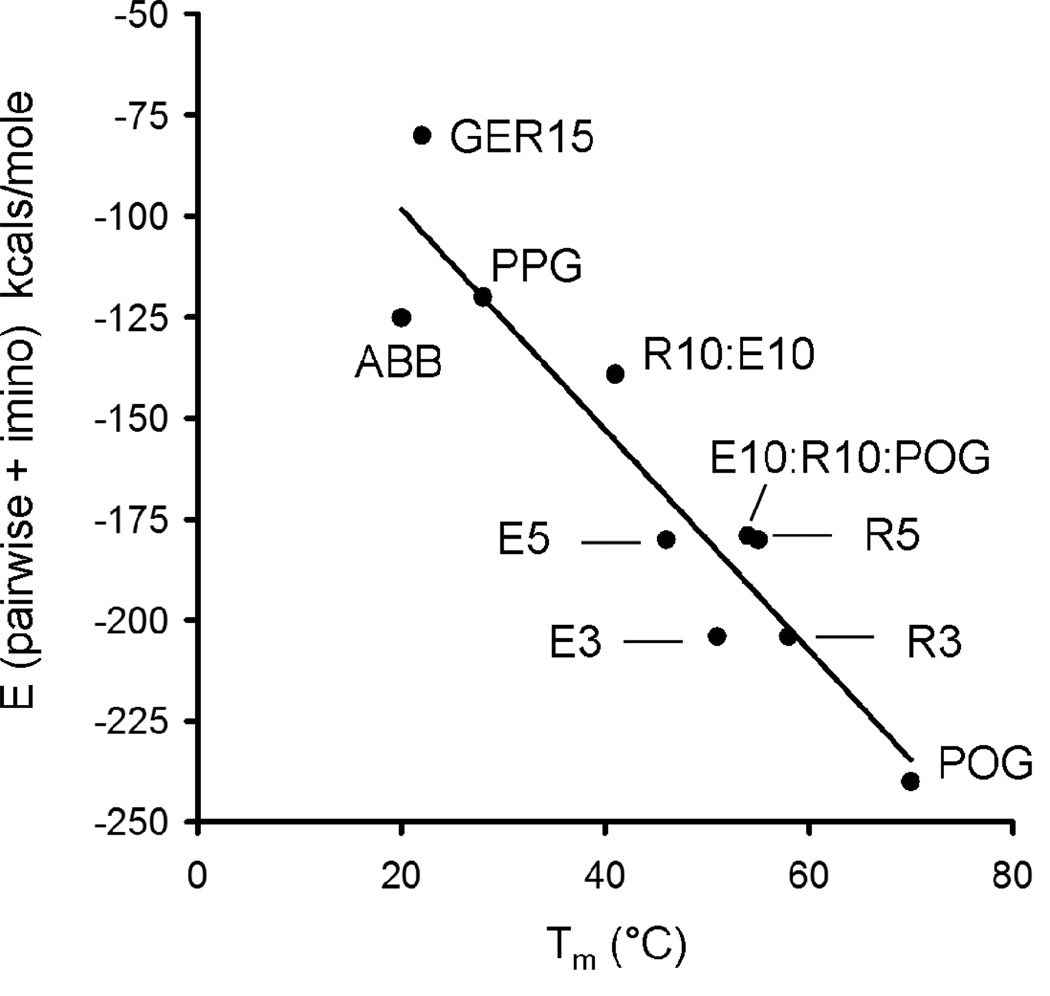
In early simulations where REG, RRG, ERG and EEG were included in addition to the allowed five triplet variations, optimal sequences for A, B and C contained no imino acids. Once triplets were limited to imino acid containing EOG, PEG, ROG, PRG and POG, no POG triplets were in the final solution. In the top five ranked solutions, only one peptide contained a single POG (Table S1). −1.5 kcals/mole per POG was not sufficient to introduce this triplet into the designs. One potential issue may be an uneven weighting of attractive charge pairs (EE/R = −1 kcal/mole) relative to POG. To explore this, a set of model collagen peptides from this and other studies were evaluated for the relative contributions of imino acid content and pairwise charge pair interactions to experimentally reported stability (Figure 10). Included were ABB from this study, (GER)15GPCCG (46), and several highly stable heterotrimer variants developed by Gauba and Hartgerink (23). The best correlation between computed energy and the observed Tm was at a ratio of pairwise energy to imino acid content of 1 to 3.8, indicating that if attractive charge pairs are −1 kcal/mole, each X=P or Y=O should contribute roughly −3.8 kcals/mole. An updated scoring function that included both pairwise terms (Eq. 2) and imino acid content would look like:
| Eq. 11 |
Figure 10.
A comparison of calculated energies using Eq. 11 versus published or experimentally determined stabilities. GER15 = (GER)15GPCCG, ref (46); ABB energy from this study, Tm from A:2B mixture; POG = (POG)10 homotrimer; PPG = (PPG)10 homotrimer. The remaining species annotated as described in ref (23): R10:E10 = 1:1 mixture of (PRG)10 and (EOG)10, association stoichiometry not specified; E10:R10:POG = 1:1:1 mixture of (EOG)10, (PRG)10 and (POG)10; E5 = (EOGPOG)5 homotrimer, R5 = (PRGPOG)5 homotrimer; E3 = (POG)2(EOGPOG)3(POG)2 homotrimer, R3 = (POG)2(EOGPOG)3(POG)3 homotrimer. Formula for the bestfit line (R2 = 0.88) in Eq. 12.
Additionally, an empirical correlation between Etot and Tm is determined as:
| Eq. 12 |
Assuming no attractive pairwise interactions, designed peptides with Tm ≈ 40°C would require 39 imino acids per triple helix, or 13 per peptide. This can be accommodated by including three POG triplets and one imino acid in the remaining seven triplets of each peptide.
Based on the above analysis, the contribution of POG to stability was significantly underestimated in the current scoring function. Another obstacle to increasing imino acid frequency was the contribution to specificity. In the Monte Carlo optimization scheme employed, adding POG to one of the peptides would lower the specificity of ABC for that iteration. For example, if an ROG to POG mutation improved ABC stability by −3.8 kcals/mole without disrupting pairwise interactions in any species, AAA would be stabilized by −11.4 kcals/mole and 2A:B and 2A:C species would be stabilized by −7.6 kcals/mole. If one assumed an Egap of −4 kcals/mole, on the same magnitude as the stability change, then addition of one POG would PABC drop from 0.65 to 0.02, resulting in a large unfavorable score change using Eq. 4. As such, single POG substitutions are inherently unfavorable during an MCSA optimization.
In order to maintain imino acid content at a level sufficient for target thermal stability, it is necessary to appropriately weight pairwise and sequence contributions to the computed energy. This may be accomplished by adding POG to or removing POG from all three sequences simultaneously in one MCSA cycle, which would not affect PABC unless pairwise interactions were disrupted. Alternatively, a compositional constraint that requires each peptide to contain a fixed number of POG triplets, as estimated by Eq. 12, would promote stability. However, overuse of POG to achieve stability could drive the stability of competing states above the threshold of folding, unless compensated by unfavorable pairwise interactions.
By including compositional constraints on imino acid content, competition between the target and unfolded, monomeric states is implicitly addressed. In the related study by Havranek and Harbury (41), competition with the unfolded state was addressed using α-helix secondary structure propensity term. A similar sequence-based triple-helix stability calculator has been developed that includes the position specific propensities of all the amino acids (47). For example, X=Glu is more favorable than Y=Glu. The inverse is true for Arg. This stability calculator can be modified to develop a reference energy that more accurately reflects energetic contributions of the unfolded state.
Misfolding and Aggregation
An unanticipated outcome of this design was the aggregation prone nature of peptide C. Multiple features of C may contribute to this. First, the net charge on this peptide is zero at neutral pH, and many proteins aggregate near their pI. However, these peptides lack any significant hydrophobicity, which is often the origin of aggregation in globular proteins.
A second feature is the distribution of charge. C consists of five basic triplets followed by five acidic triplets, suggesting it may form staggered interactions of five overlapping triplets allowing attractive interactions between acidic and basic regions. Consider a circular permutation of C called P, where the two regions are swapped: -EOGPEGPEGEOGEOG|PRGROGROGROGPRG- . While the energy of CCC using the structure-based model is 20 kcals/mole, the energies of PPC, CCP and PCC are −5 kcals/mole, i.e. the net pairwise interactions are favorable for multiple staggered configurations. The addition of peptide B in low amounts was found to significantly accelerate aggregation, potentially through stabilization of a staggered intermediate species.
Periodic sequences are used to promote staggered associations of natural collagens through repetitive electrostatic and hydrophobic features (44, 48). Model collagen peptide systems have also employed periodic electrostatics to drive ordered fibril formation (49). Similar staggered electrostatics interactions have been utilized extensively in designing self-assembling α-helical fibrils (50).
Therefore, two strategies present themselves in to avoid aggregation. One is to reject solutions where designed sequences are neutral at the operative pH, although this may not be useful if hydrophobic amino acids are not significantly represented in the design. The other is to compute the energies of staggered, competing states either during sequence optimization or at the end of an MCSA trajectory, and reject solutions where offset pairings are stable. This has the disadvantage of increasing the computational burden for each cycle of optimization.
Conclusion
In this study, the established concepts of positive and negative computational protein design were applied for the first time to a collagen-like triple-helix. Using a simple, discrete electrostatics model, the energy landscape for an ensemble of heterotrimers was optimized to favor one target state over twenty-six alternative associations. Simultaneous optimization of energy and specificity components resulted in large energy gaps between the target and competing states. Experimental characterization of one design using this approach highlighted flaws in the pairwise electrostatics energy function, relative weighting of stability contributions of charge pairs versus secondary structure promoting imino acids, and issues with staggered interchain associations which may have promoted aggregation. A post hoc analysis of the experimental results suggests many improvements to be implemented in the next generation of collagen peptide designs.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Barbara Brodsky and Karuntekar Kar for useful discussions. VN acknowledges support from the NIH Director’s New Innovator Award Program, 1-DP2-OD006478-01. VN and FX acknowledge support from the NSF BMAT program DMR-0907273. RLK acknowledges supported by the following grants: MCB-0920448 from the NSF, MCB-5G12 RR03060 toward support for the NMR facilities at the City College of New York, P41 GM-66354 to the New York Structural Biology Center and infrastructure support from NIH 5G12 RR03060 from the National Center for Research Resources.
Footnotes
Mixtures are referred to by the ratio of peptides (e.g. A:2B or A:B:C), while species are designated explicitly (e.g. BAB or ABC).
Supporting Information Available. Additional tables and figures are provided in the supplement.
REFERENCES
- 1.Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nat Rev Cancer. 2003;3:422–433. doi: 10.1038/nrc1094. [DOI] [PubMed] [Google Scholar]
- 2.Heino J. The collagen family members as cell adhesion proteins. Bioessays. 2007;29:1001–1010. doi: 10.1002/bies.20636. [DOI] [PubMed] [Google Scholar]
- 3.Khoshnoodi J, Pedchenko V, Hudson BG. Mammalian collagen IV. Microsc Res Tech. 2008;71:357–370. doi: 10.1002/jemt.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rich A, Crick FH. The molecular structure of collagen. J Mol Biol. 1961;3:483–506. doi: 10.1016/s0022-2836(61)80016-8. [DOI] [PubMed] [Google Scholar]
- 5.Bella J, Eaton M, Brodsky B, Berman HM. Crystal and molecular structure of a collagen-like peptide at 1.9 A resolution. Science. 1994;266:75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- 6.Bella J, Brodsky B, Berman HM. Hydration structure of a collagen peptide. Structure. 1995;3:893–906. doi: 10.1016/S0969-2126(01)00224-6. [DOI] [PubMed] [Google Scholar]
- 7.Kuznetsova N, Rau DC, Parsegian VA, Leikin S. Solvent hydrogen-bond network in protein self-assembly: solvation of collagen triple helices in nonaqueous solvents. Biophys J. 1997;72:353–362. doi: 10.1016/S0006-3495(97)78674-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaramillo A, Wodak SJ. Computational protein design is a challenge for implicit solvation models. Biophys J. 2005;88:156–171. doi: 10.1529/biophysj.104.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pokala N, Handel TM. Energy functions for protein design I: efficient and accurate continuum electrostatics and solvation. Protein Sci. 2004;13:925–936. doi: 10.1110/ps.03486104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bretscher LE, Jenkins CL, Taylor KM, DeRider ML, Raines RT. Conformational stability of collagen relies on a stereoelectronic effect. J Am Chem Soc. 2001;123:777–778. doi: 10.1021/ja005542v. [DOI] [PubMed] [Google Scholar]
- 11.Holmgren SK, Taylor KM, Bretscher LE, Raines RT. Code for collagen's stability deciphered. Nature. 1998;392:666–667. doi: 10.1038/33573. [DOI] [PubMed] [Google Scholar]
- 12.Baum J, Brodsky B. Real-time NMR investigations of triple-helix folding and collagen folding diseases. Fold Des. 1997;2:R53–R60. doi: 10.1016/S1359-0278(97)00028-X. [DOI] [PubMed] [Google Scholar]
- 13.Persikov AV, Xu Y, Brodsky B. Equilibrium thermal transitions of collagen model peptides. Protein Sci. 2004;13:893–902. doi: 10.1110/ps.03501704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salem G, Traub W. Conformational Implications of Amino Acid Sequence Regularities in Collagen. FEBS Lett. 1975;51:94–99. doi: 10.1016/0014-5793(75)80861-1. [DOI] [PubMed] [Google Scholar]
- 15.Traub W, Fietzek PP. Contribution of the A2 Chain to the Molecular Stability of Collagen. FEBS Lett. 1976;68:245–249. doi: 10.1016/0014-5793(76)80446-2. [DOI] [PubMed] [Google Scholar]
- 16.Hulmes DJ, Miller A, Parry DA, Piez KA, Woodhead-Galloway J. Analysis of the primary structure of collagen for the origins of molecular packing. J Mol Biol. 1973;79:137–148. doi: 10.1016/0022-2836(73)90275-1. [DOI] [PubMed] [Google Scholar]
- 17.Venugopal MG, Ramshaw JA, Braswell E, Zhu D, Brodsky B. Electrostatic interactions in collagen-like triple-helical peptides. Biochemistry. 1994;33:7948–7956. doi: 10.1021/bi00191a023. [DOI] [PubMed] [Google Scholar]
- 18.Yang W, Chan VC, Kirkpatrick A, Ramshaw JA, Brodsky B. Gly-Pro-Arg confers stability similar to Gly-Pro-Hyp in the collagen triple-helix of host-guest peptides. J Biol Chem. 1997;272:28837–28840. doi: 10.1074/jbc.272.46.28837. [DOI] [PubMed] [Google Scholar]
- 19.Persikov AV, Ramshaw JA, Kirkpatrick A, Brodsky B. Electrostatic interactions involving lysine make major contributions to collagen triple-helix stability. Biochemistry. 2005;44:1414–1422. doi: 10.1021/bi048216r. [DOI] [PubMed] [Google Scholar]
- 20.Ottl J, Battistuta R, Pieper M, Tschesche H, Bode W, Kuhn K, Moroder L. Design and synthesis of heterotrimeric collagen peptides with a built-in cystine-knot. Models for collagen catabolism by matrix-metalloproteases. FEBS Lett. 1996;398:31–36. doi: 10.1016/s0014-5793(96)01212-4. [DOI] [PubMed] [Google Scholar]
- 21.Fiori S, Sacca B, Moroder L. Structural properties of a collagenous heterotrimer that mimics the collagenase cleavage site of collagen type I. J Mol Biol. 2002;319:1235–1242. doi: 10.1016/S0022-2836(02)00365-0. [DOI] [PubMed] [Google Scholar]
- 22.Gauba V, Hartgerink JD. Surprisingly high stability of collagen ABC heterotrimer: evaluation of side chain charge pairs. J Am Chem Soc. 2007;129:15034–15041. doi: 10.1021/ja075854z. [DOI] [PubMed] [Google Scholar]
- 23.Gauba V, Hartgerink JD. Self-assembled heterotrimeric collagen triple helices directed through electrostatic interactions. J Am Chem Soc. 2007;129:2683–2690. doi: 10.1021/ja0683640. [DOI] [PubMed] [Google Scholar]
- 24.Gauba V, Hartgerink JD. Synthetic collagen heterotrimers: structural mimics of wild-type and mutant collagen type I. J Am Chem Soc. 2008;130:7509–7515. doi: 10.1021/ja801670v. [DOI] [PubMed] [Google Scholar]
- 25.Nautiyal S, Woolfson DN, King DS, Alber T. A designed heterotrimeric coiled coil. Biochemistry. 1995;34:11645–11651. doi: 10.1021/bi00037a001. [DOI] [PubMed] [Google Scholar]
- 26.Kirkpatrick S, Gelatt CD, Vecchi MP. Optimization by Simulated Annealing. Science. 1983;220:671–680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- 27.Hellinga HW, Richards FM. Optimal Sequence Selection in Proteins of Known Structure by Simulated Evolution. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:5803–5807. doi: 10.1073/pnas.91.13.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E. Equations of State Calculations by Fast Computing Machines. Journal of Chemical Physics. 1951;21:1087–1092. [Google Scholar]
- 29.Huang CY. Determination of binding stoichiometry by the continuous variation method: the Job plot. Methods Enzymol. 1982;87:509–525. doi: 10.1016/s0076-6879(82)87029-8. [DOI] [PubMed] [Google Scholar]
- 30.Huang SS, Koder RL, Lewis M, Wand AJ, Dutton PL. The HP-1 maquette: From an apoprotein structure to a structured hemoprotein designed to promote redox-coupled proton exchange. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:5536–5541. doi: 10.1073/pnas.0306676101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Summa CM, Rosenblatt MM, Hong JK, Lear JD, DeGrado WF. Computational de novo design, and characterization of an A(2)B(2) diiron protein. J Mol Biol. 2002;321:923–938. doi: 10.1016/s0022-2836(02)00589-2. [DOI] [PubMed] [Google Scholar]
- 32.Sindelar CV, Hendsch ZS, Tidor B. Effects of salt bridges on protein structure and design. Protein Sci. 1998;7:1898–1914. doi: 10.1002/pro.5560070906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berezovsky IN, Zeldovich KB, Shakhnovich EI. Positive and negative design in stability and thermal adaptation of natural proteins. PLoS Comput Biol. 2007;3:e52. doi: 10.1371/journal.pcbi.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrissey MP, Shakhnovich EI. Design of proteins with selected thermal properties. Fold Des. 1996;1:391–405. doi: 10.1016/S1359-0278(96)00054-5. [DOI] [PubMed] [Google Scholar]
- 35.Shakhnovich EI, Gutin AM. A new approach to the design of stable proteins. Protein Eng. 1993;6:793–800. doi: 10.1093/protein/6.8.793. [DOI] [PubMed] [Google Scholar]
- 36.Shakhnovich EI, Gutin AM. Engineering of stable and fast-folding sequences of model proteins. Proc Natl Acad Sci U S A. 1993;90:7195–7199. doi: 10.1073/pnas.90.15.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seno F, Vendruscolo M, Maritan A, Banavar JR. Optimal protein design procedure. Phys. Rev. Lett. 1996;77:1901–1904. doi: 10.1103/PhysRevLett.77.1901. [DOI] [PubMed] [Google Scholar]
- 38.Persikov AV, Ramshaw JAM, Kirkpatrick A, Brodsky B. Amino Acid Propensities for the Collagen Triple-Helix. Biochemistry. 2000;39:14960–14967. doi: 10.1021/bi001560d. [DOI] [PubMed] [Google Scholar]
- 39.Koehl P, Levitt M. De novo protein design. II. Plasticity in sequence space. J Mol Biol. 1999;293:1183–1193. doi: 10.1006/jmbi.1999.3212. [DOI] [PubMed] [Google Scholar]
- 40.Bolon DN, Grant RA, Baker TA, Sauer RT. Specificity versus stability in computational protein design. Proc Natl Acad Sci U S A. 2005;102:12724–12729. doi: 10.1073/pnas.0506124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Havranek JJ, Harbury PB. Automated design of specificity in molecular recognition. Nat Struct Biol. 2003;10:45–52. doi: 10.1038/nsb877. [DOI] [PubMed] [Google Scholar]
- 42.Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458:859–864. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan VC, Ramshaw JA, Kirkpatrick A, Beck K, Brodsky B. Positional preferences of ionizable residues in Gly-X-Y triplets of the collagen triple-helix. J Biol Chem. 1997;272:31441–31446. doi: 10.1074/jbc.272.50.31441. [DOI] [PubMed] [Google Scholar]
- 44.Hulmes DJ, Miller A, Parry DA, Woodhead-Galloway J. Fundamental periodicities in the amino acid sequence of the collagen alpha1 chain. Biochem Biophys Res Commun. 1977;77:574–580. doi: 10.1016/s0006-291x(77)80017-x. [DOI] [PubMed] [Google Scholar]
- 45.Strickler SS, Gribenko AV, Keiffer TR, Tomlinson J, Reihle T, Loladze VV, Makhatadze GI. Protein stability and surface electrostatics: a charged relationship. Biochemistry. 2006;45:2761–2766. doi: 10.1021/bi0600143. [DOI] [PubMed] [Google Scholar]
- 46.Mechling DE, Bachinger HP. The collagen-like peptide (GER)15GPCCG forms pH-dependent covalently linked triple helical trimers. J Biol Chem. 2000;275:14532–14536. doi: 10.1074/jbc.275.19.14532. [DOI] [PubMed] [Google Scholar]
- 47.Persikov AV, Ramshaw JA, Brodsky B. Prediction of collagen stability from amino acid sequence. J Biol Chem. 2005;280:19343–19349. doi: 10.1074/jbc.M501657200. [DOI] [PubMed] [Google Scholar]
- 48.Knupp C, Squire JM. A new twist in the collagen story--the type VI segmented supercoil. Embo J. 2001;20:372–376. doi: 10.1093/emboj/20.3.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rele S, Song Y, Apkarian RP, Qu Z, Conticello VP, Chaikof EL. D-periodic collagen-mimetic microfibers. J Am Chem Soc. 2007;129:14780–14787. doi: 10.1021/ja0758990. [DOI] [PubMed] [Google Scholar]
- 50.Pandya MJ, Spooner GM, Sunde M, Thorpe JR, Rodger A, Woolfson DN. Sticky-end assembly of a designed peptide fiber provides insight into protein fibrillogenesis. Biochemistry. 2000;39:8728–8734. doi: 10.1021/bi000246g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.