Abstract
The development of modern neuroscience tools is critical for deciphering brain circuit organization and function. An important aspect for technical development is to enhance each technique's advantages and compensate for limitations. We developed a high-precision and fast functional mapping technique in brain slices that incorporates the spatial precision of activation that can be achieved by laser-scanning photostimulation with rapid and high-temporal resolution assessment of evoked network activity that can be achieved by voltage-sensitive dye imaging. Unlike combination of whole cell recordings with photostimulation for mapping local circuit inputs to individually recorded neurons, this innovation is a new photostimulation-based technique to map cortical circuit output and functional connections at the level of neuronal populations. Here we report on this novel technique in detail and show its effective applications in mapping functional connections and circuit dynamics in mouse primary visual cortex and hippocampus. Given that this innovation enables rapid mapping and precise evaluation of cortical organization and function, it can have broad impacts in the field of cortical circuitry.
INTRODUCTION
A key to revealing brain mechanisms is to understand the principles of circuit organization and function, thereby decoding how the circuit processes information and guides behavior. This requires use of modern tools to systematically characterize properties of neuronal components and their connections and to enable precise measurements of circuit activity. We developed a new technique based on the integration of laser scanning photostimulation and voltage-sensitive dye (VSD) imaging. This novel approach is an all-optical approach for brain circuit mapping, providing important applications in cortical circuitry studies.
Our new technique uses VSD imaging to allow for functional connectivity and spatiotemporal dynamics of neuronal population activity in large cortical regions to be examined. Fast VSD imaging has been widely used to study population neuronal activity in cortical tissue both in vivo and in vitro (Grinvald and Hildesheim 2004). Particularly for in vitro brain slices, fast VSD imaging allows mapping of circuit organization and circuit dynamics, and more recently, has been used to probe functional abnormalities in models of neurological and psychiatric disorder (Airan et al. 2007; Ang et al. 2006). One major limitation of most in vitro VSD imaging studies, however, is that sources of activation are not precisely controlled. For example, neuronal responses have been imaged either after pharmacologically induced seizure activity or electrical stimulation (Ang et al. 2006; Huang et al. 2004; Petersen and Sakmann 2001). Significant disadvantages of electric stimulation include indiscriminate activation of axons of passage, slow and inefficient placement of multiple stimulation locations, and tissue damage. To compensate this limitation, we use laser scanning photostimulation to provide spatially precise activation of neuronal circuits. The optical stimulation via glutamate uncaging enables rapid and noninvasive photoactivation of neurons with great convenience and superior spatial resolution in practical experiments (Callaway and Katz 1993; Shepherd et al. 2003; Xu and Callaway 2009). Combining whole cell recordings from single neurons with photostimulation of clusters of presynaptic neurons has been developed for extensive mapping of local functional inputs to a single recorded neuron (Schubert et al. 2003; Shepherd and Svoboda 2005; Xu and Callaway 2009). However, successful application of this whole cell recording approach requires repeated experiments from numerous recorded neurons and analysis of hundreds of synaptic currents that are recorded in each recorded cell. Results therefore tend to be limited to a small number of targeted cell types, making this method less useful for fast, unbiased surveys of overall changes in network connectivity that might occur during development or in responses to experimental manipulations.
The purpose of our technical development was to develop a new photostimulation-based technique to map cortical circuit output and functional connections at the neuronal population level by combining the advantages of VSD imaging and optical stimulation. In this report, we first describe our system and provide details to make this technique available to other investigators. Then we describe effective applications of the technique in mapping functional connections, circuit organization and dynamics in mouse primary visual cortex (V1) and hippocampus.
METHODS
Slice preparation
Wild-type C57/B6 mice were used in the experiments. Fifteen mice were used for V1 circuit studies, and five mice used for hippocampal circuit studies. Only the best cut slices were used for this study. On average, one animal yielded two successful slices. Some data obtained from these experiments will be reported separately. All animals were handled and experiments were conducted in accordance with procedures approved by the Institutional Animal Care and Use Committee at the University of California, Irvine. To prepare living brain slices, animals (postnatal day 17–23) were deeply anesthetized with pentobarbital sodium (>100 mg/kg, ip) and rapidly decapitated, and their brains removed. Visual cortical or hippocampal sections were cut 400 μm thick with a vibratome (VT1200S, Leica Systems) in sucrose-containing artificial cerebrospinal fluid (ACSF) (in mM: 85 NaCl, 75 sucrose, 2.5 KCl, 25 glucose, 1.25 NaH2PO4, 4 MgCl2, 0.5 CaCl2, and 24 NaHCO3). Slices were first incubated in sucrose-containing ACSF for 30 min to 1 h at 32°C, and after the initial incubation period, transferred to recording ACSF (in mM: 126 NaCl, 2.5 KCl, 26 NaHCO3, 2 CaCl2, 2 MgCl2, 1.25 NaH2PO4, and 10 glucose) for the dye staining at room temperature. The slices were stained for 2–3 h in a staining chamber containing ACSF with 0.02 mg/ml of an oxonol dye, NK3630 (first synthesized by R. Hildesheim and A. Grinvald as RH482; available from Nippon Kankoh-Shikiso Kenkyusho) and maintained in regular ACSF before use. The NK3630 dye has been characterized in previous studies and has its peak signal-to-noise ratio centered around 705 nm (Jin et al. 2002). Throughout the incubation, staining, and recording, the slices were continuously bubbled with 95% O2-5% CO2.
Electrophysiology and photostimulation
Slices were visualized with an upright microscope (BW51X, Olympus) with infrared differential interference contrast optics. Electrophysiological recordings, photostimulation, and imaging of the slice preparations were done in a slice perfusion chamber mounted on a motorized stage of the microscope. At low magnification (4× objective lens, 0.16 NA; UplanApo, Olympus), laminar and cytoarchitectonic features of brain slices were visualized under infrared bright-field transillumination, and the slice images were acquired by a high-resolution digital CCD camera (Retiga 2000, Q-imaging, Austin, TX). Digitized images from the camera were used for guiding and registering photostimulation sites in cortical slices.
The absorption dye, NK3630, had low toxicity for neurons in the slice, and even after long sessions of VSD recordings over several hours, the slice still remained healthy (Supplemental Fig. S1, A and B).1 The cells in the VSD-stained slices had normal spiking patterns and resting membrane potentials that did not differ from those cells recorded in control slices without the dye staining. The resting membrane potentials for the identified pyramidal cells in the VSD-stained slices (n = 11 cells) and the cells in the control slices (n = 10 cells) were −66.6 ± 2.2 and −64.9 ± 1.8 (SE) mV, respectively (P > 0.05, Mann-Whitney U test).
During experiments, simultaneous electrophysiological recordings (whole cell recordings, loose-seal patchings, or local field potential recordings) were conducted to monitor laser stimulation and correlate VSD signals with the electrical activity. To perform patch recording, cells were visualized at high magnification (60× objective, 0.9 NA; LUMPlanFl/IR, Olympus). Neurons were patched with borosilicate electrodes and recorded at room temperature in the whole cell or loose-seal mode. The patch pipettes (4–6 MΩ resistance) were filled with an internal solution containing (in mM) 126 K-gluconate, 4 KCl, 10 HEPES, 4 ATP-Mg, 0.3 GTP-Na, and 10 phosphocreatine (pH 7.2, 300 mOsm). The electrodes also contained 0.1% biocytin for cell labeling and morphological identification. Resting membrane potentials were measured immediately after electrodes broke into the cells following formation of a gigaohm seal, and current pulses were injected to examine each cell's basic electrophysiological properties. Data were acquired with a Multiclamp 700B amplifier (Molecular Devices, Sunnyvale, CA), data acquisition boards (models PCI MIO 16E-4 and 6713, National Instruments, Austin, TX), and custom modified version of Ephus software (Ephus, available at https://openwiki.janelia.org/). Data were filtered at 2 kHz using a Bessel filter, digitized at 10 kHz, and stored on a computer. Once stable whole cell recordings were achieved with good access resistance (usually <20 MΩ), the microscope objective was switched from 60× to 4× for laser scanning photostimulation. The same low-power objective lens was used for delivering the UV flash stimuli.
Stock solution of MNI-caged-l-glutamate (4-methoxy-7-nitroindolinyl-caged l-glutamate, Tocris Bioscience, Ellisville, MO) was prepared by dissolving MNI-glutamate in distilled water and stored in 50 μl aliquots at −20°C for up to several weeks. An aliquot was added to 20–25 ml of circulating ACSF for a concentration of 0.4 mM caged glutamate. After 5–6 h of experimentation, the bath solution and MNI-glutamate were refreshed. Care was taken to ensure a constant fluid level in the chamber of ∼2.0–2.5 mm above the slice to avoid small fluctuations in UV attenuation. For some experiments (see Fig. 2, B3 and B4), additions of 10 μM CNQX (Tocris Bioscience) and 10 μM 3-(2-carboxypiperazin-4-yl)-propyl-1-phosphonic acid (CPP; Tocris Bioscience) in solutions were used to block ionotropic glutamate receptors. In other experiments (see Supplemental Fig. S3), 2 μM TTX (Tocris Bioscience) in the bath solution was used to block voltage-gated sodium channels, preventing neuronal spiking activity. In addition, modified ACSF containing 0.2 mM Ca2+ and 4 mM Mg2+ was used to block synaptic transmission.
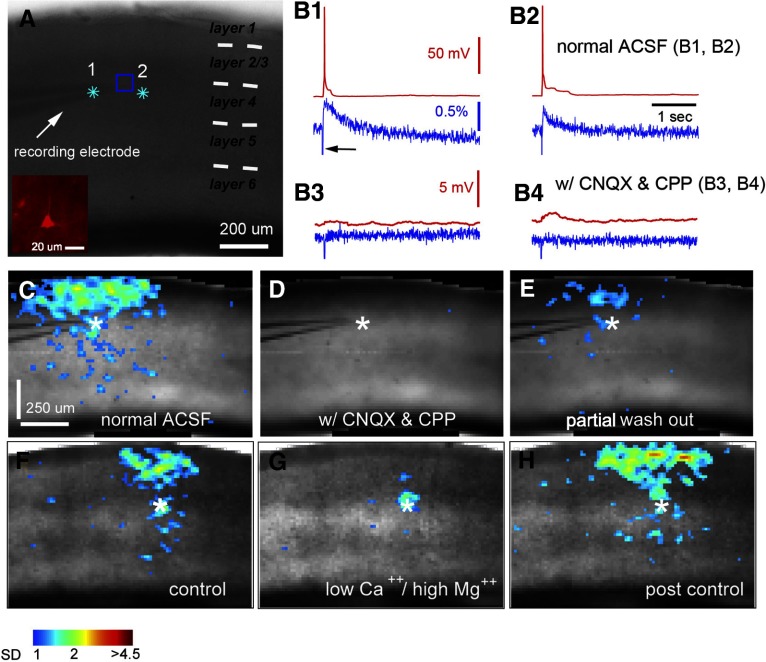
Fig. 2.
Characterization of VSD responses evoked by glutamate uncaging. A: grayscale image of V1 slice stained with NK3630, photostimulated (laser: 1 ms, 35 mW) at 2 sites (indicated by cyan stars) close to the recording electrode placed at the boundary of layers 3 and 4. As shown in the Inset, the recorded neuron was identified as an excitatory pyramidal neuron. B1 and B2: data traces of simultaneous whole cell recording and VSD imaging in response to photostimulation at sites 1 and 2 in normal artificial cerebrospinal fluid (ACSF), respectively. Red traces represent membrane potentials of the recorded neuron, and blue traces represent VSD signals that were measured from the region of interest (ROI) marked by a blue square around the electrode tip shown in A. The black arrow in B1 points to the artifact signal of laser excitation in the VSD signal trace. B3 and B4 show data traces of simultaneous whole cell recording and VSD imaging in response to photostimulation at the same sites as B1 and B2 but in ACSF with 10 μM CNQX and 10 μM 3-(2-carboxypiperazin-4-yl)-propyl-1-phosphonic acid (CPP). C–E: VSD image frames of peak activation after glutamate uncaging at site 1 (indicated by the white star) before and after bath application and after washout of CNQX and CPP, respectively. VSD signal amplitudes expressed as SD above the mean baseline signal are color coded. F–H: peak activation frames of a different V1 slice in response to laser photostimulation in a layer 4 site (indicated by the white star; laser: 1 ms, 35 mW) with perfusion of normal ACSF (control), low Ca2+ and high Mg2+ ACSF (containing 0.2 mM Ca2+, 4 mM Mg2+), and postcontrol normal ACSF, respectively. The data show that most VSD responses outside the photostimulation site are predominantly postsynaptic responses that are blocked in the low Ca2+ and high Mg2+ solution.
We adopted the design of the laser scanning photostimulation system described previously (Shepherd and Svoboda 2005; Shepherd et al. 2003). A laser unit (model 3501, DPSS Lasers, Santa Clara, CA) was used to generate a 355 nm UV laser for glutamate uncaging. The laser beam was 1.5 mm in diameter and directed through the optical path of our system. Short durations of laser flashes (e.g., 1–3 ms) were controlled by using an electro-optical modulator (i.e., pockels cell) (Conoptics, Danbury, CT) and a mechanical shutter (Uniblitz, Vincent Associates, Rochester, NY). Laser beam power was modulated by a neutral density gradient wheel and monitored by diverting a small fraction of the laser beam with a glass coverslip to a photodiode. The laser scanning system included an X-Y pair of scan mirrors, the scan lens, the tube lens, and the objective lens (Fig. 1A). The mirrors delivered the laser beam through a scan lens; the beam entered the microscope (BX51WI, Olympus) via a dichroic mirror (351DRLP, Chroma Technology, Brattleboro, VT) and was focused by a custom-made UV-transmitting tube lens. The beam underfilled the back aperture of the microscope objective to provide a more columnar (as opposed to conical) illuminating beam, keeping the mapping as two dimensional as possible by reducing the axial resolution. Various laser stimulation positions could be achieved through galvanometer-driven X-Y scanning mirrors (Cambridge Technology, Cambridge, MA), as the mirrors and the back aperture of the objective were in conjugate planes, translating mirror positions into different scanning locations at the objective lens focal plane. During uncaging, a variable number of patterned sites that covers the whole field are stimulated sequentially in a nonraster, nonrandom sequence, following a “shifting-X” pattern designed to avoid revisiting the vicinity of recently stimulated sites (Shepherd and Svoboda 2005; Shepherd et al. 2003). Ephus software was used to control photostimulation protocols and acquire photostimulation data. In most experiments, whole cell recordings or loose-seal patchings were simultaneously performed with optical recordings.
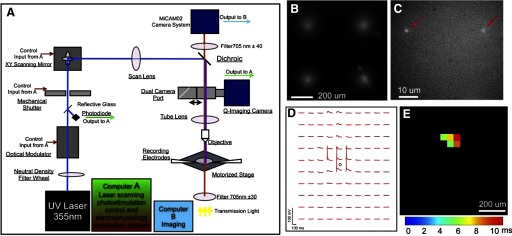
Fig. 1.
The system design, calibrations, and tests for combining laser scanning photostimulation with voltage-sensitive dye (VSD) imaging. A: the system diagram of a laser scanning photostimulation and VSD imaging setup. B: the physical calibration measurement of the laser scanning photostimulation system with a 2 × 2 (spacing at 500 μm) uncaging pattern generated by single 100 μs laser pulses (32 mW) on a thin layer of 0.2% caged fluorescein dextran through a 4× objective. C: the result of fluorescein uncaging by 10-μs laser pulses (32 mW) using a 60× objective. Red arrows point to fluorescent spots resulting from uncaging. We estimated the laser physical excitation size with the full width at the half height (FWHH) from the cross profile of the uncaging spot with the height referring to fluorescence intensity. D and E: the functional calibration of laser scanning photostimulation by measurement of a single neuron's excitation profile. The pyramidal neuron in visual cortex (V1) layer 4 was recorded in whole cell patch-clamp configuration and photostimulated with 1-ms laser pulses (32 mW). The array of 16 × 16 stimulation sites spanned different V1 cortical layers, and the sites were spaced 50 μm apart. D: the neuronal response traces depicting changes in the membrane potential in response to photostimulation at the 10 × 10 stimulation sites centered at the recorded cell (indicated by the small black circle). Note that suprathreshold spikes were evoked only in the perisomatic region. E: the spiking locations of the neuron recorded in D across the 16 × 16 stimulation sites. The color code indicates latencies of spiking in response to stimulation at different locations (see the color bar).
After all physiological assays had been completed, the brain slices were fixed in 4% paraformaldehyde in PBS overnight and transferred to 30% sucrose solution in PBS. The slices were stained against biocytin with 1:1,000 Cy3-conjugated streptavidin (Jackson ImmunoResearch, West Grove, PA) to show the morphology of the recorded cells. Sections were also stained for 4′-6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, St. Louis, MO) to identify laminar boundaries. Neuron reconstructions were computer-assisted and based on stacks of optical sections acquired by a confocal microscope (LSM510-META, Carl Zeiss).
VSD imaging and data analysis
As indicated in Fig. 1A, a dual camera port was used to couple the Q-imaging camera and the laser scanning photostimulation system to a MiCAM02 fast imaging system (SciMedia USA, Costa Mesa, CA) for VSD imaging. The VSD imaging computer (B) was independent from the computer (A), but its image acquisitions were triggered and synchronized with computer A through output TTL pulses from computer A. On triggering, optical recording of VSD signals was performed by the MiCAM02 system with a sampling rate of 2.2 ms/frame [frame resolution 88 (w) × 60 (h) pixels]. Under the 4× objective, the imaging field covered the area of 1.28 × 1.07 mm2 with a spatial resolution of 14.6 × 17.9 μm/pixel. With the 60× objective, the imaging field covered the area of 84.5 × 70.8 μm2 with a resolution of 0.96 × 1.18 μm/pixel. The trials were obtained every 8 s, and the recording periods were either 400 or 1,000 frames for each photostimulation trial. VSD images were smoothed by convolving images with a Gaussian spatial filter (kernel size: 3 × 3 pixels; δ size: 1 × 1 pixel) and a Gaussian temporal filter (kernel size: 3 frames; δ size: 1 frame). VSD signals were originally measured by the percent change in pixel light intensity [ΔI/I%; the % change in the intensity (ΔI) at each pixel relative to the initial intensity (I)]. In addition, signal amplitudes were expressed as SD above the mean baseline signal for display and quantification. In this study, single-trial VSD signals were of sufficiently high amplitudes and could be discerned from background noise; hence no averaging over multiple trials was necessary. Images were displayed and initially analyzed using an acquisition and analysis software (BV-Analyzer, BrainVision). Further quantification and measurements were performed with custom-made Matlab Programs.
As for quantitative analysis of evoked activation in image frames, the mean and SD of the baseline activity of each pixel across the 50 frames preceding photostimulation was first calculated, and then activated pixels were measured. The activated pixel was empirically defined as the pixel with the amplitude ≥1 SD above the mean of the corresponding pixel's amplitude preceding the stimulation (equivalent to the detectable signal level in the original VSD maps of ΔI/I%). The overall activation size in image frames was defined as the fraction of activated pixels, expressed as a percentage of the image frame size.
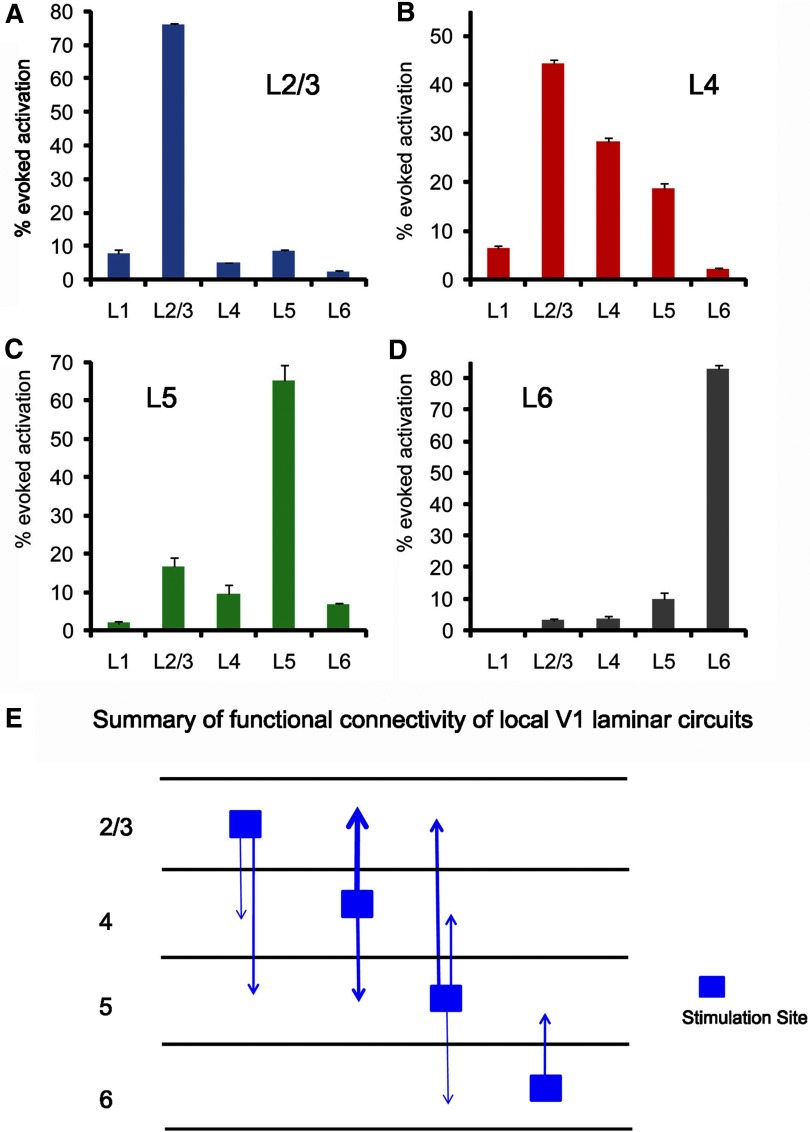
To quantify the layer-specific evoked activation in V1, the average numbers of activated pixels in different cortical layers across the 10 frames around the peak VSD activation frame (e.g., at 50 ms after photostimulation) were measured, and the mean VSD activation strength of individual layers was calculated in percent of the total number of activated pixels of all the cortical layers (i.e., % total evoked activation; Fig. 5, A–D).
Fig. 5.
Functional connectivity of V1 laminar circuits shown by quantitative analysis of VSD activation evoked by laser scanning photostimulation (1 ms, 30–35 mW). A–D: laminar distributions of VSD activation in response to photostimulation in V1 layers 2/3, 4, 5, or 6, respectively. In the histogram, the x-axis indicates different cortical layers, and the y-axis shows VSD activation strength (mean ± SE, in % of the total evoked activation). Data were based on the representative cases shown in Figs. 3 and 4. E: summary of functional connectivity of local V1 laminar circuits based on quantitative analysis of VSD activation patterns. Given deeper layer neurons (particularly layer 2/3 neurons) have extensive dendritic branches in layer 1 (e.g., Fig. 4C) and some layer 1 VSD activation might result from direct neuronal activation in other layers, we excluded layer 1 connectivity in our diagram. In addition, only the laminar projection with an average VSD activation strength ≥5% is included in the summary. The thicker projection lines correspond to stronger interlaminar projections.
For statistical comparisons across more than two groups, we used the Kruskal-Wallis test (nonparametric 1-way ANOVA) and the Mann-Whitney U test for group comparisons. α levels of P ≤ 0.05 were considered significant.
RESULTS
General system design, calibration, and VSD response characterization
Our overall system is shown in Fig. 1A, consisting of laser scanning photostimulation, VSD imaging, and electrophysiological recording systems (see methods for detailed descriptions of the system). The absorption dye, NK3630 (RH 482), was used for our VSD imaging experiments. The dye was chosen because of its good sensitivity, low bleaching, and phototoxicity, as well as its preferential staining of neurons with a low affinity for glial cells (Jin et al. 2002; Konnerth et al. 1987). During experiments, a 705 nm light trans-illuminated brain slices and voltage-dependent changes in the light absorbance of the dye were captured by the MiCAM02 camera (Fig. 1A). The photostimulation and imaging systems were aligned and calibrated, and uncaging tests were visualized by exciting caged fluorescein dextran on a glass slide (Fig. 1, B and C). Under the 4× objective, the laser beam formed uncaging spots, each approximating a Gaussian profile with the estimated full width at the half height (FWHH) of 153 μm laterally at the focal plane (Fig. 1B), and the laser beam caused uncaging of fluorescent dextran at ∼100 μm in depth. It is important to note that the physical laser excitation size in the glass slide does not directly translate into the effective spatial resolution of physiological uncaging in brain slices. In comparison, under the 60× objective, the UV laser induced uncaging was focused to a smaller spot with FWHH of 2.3 μm. Although imaging through the 60× objective is useful for exciting smaller numbers of neurons or possibly even single cell stimulation, this study focused on imaging and excitation both through the 4× objective to map propagation of activity reflecting interlaminar connectivity within local cortical circuits.
In our experiments, VSD imaging was routinely performed with simultaneous whole cell recordings or loose-seal patching of single neurons to monitor the effectiveness and spatial precision of laser photostimulation in cortical slices. For example, in the slice of mouse primary visual cortex (V1), through the 4× objective, laser stimulation (1 ms, 35 mW) had a spatial resolution of 50–100 μm in evoking suprathreshold spikes from the recorded neuron (Fig. 1D) and spike latencies were 5.4–10.7 ms (Fig. 1E). Under the photostimulation conditions (power level: 30–35 mW; pulse duration: 1 ms; caged glutamate concentration: 0.4 mM) used for our data set (10 excitatory pyramidal cells), spikes evoked by photostimulation occurred with an average latency of 11.5 ± 0.7 (SE) ms with somata located within 100 μm from the stimulation site. These excitation profile data were consistent with previous studies (Dantzker and Callaway 2000; Shepherd and Svoboda 2005; Xu and Callaway 2009). Therefore with proper experimental conditions, photostimulation-evoked action potentials only from neurons located at or near the stimulation site and thus can provide spatially restricted activation. However, under stronger photostimulation conditions (power level: 30–35 mW; pulse duration: 2 ms or above), the spatial resolution of laser photostimulation was lowered as spikes could be evoked from cells located 100–300 μm away from the stimulation sites with an average spike latency of 23.9 ± 3.4 ms (6 cells). The stronger laser stimulation evoked neuronal spikes from the cells located >100 μm away from the photostimulation sites possibly either through direct activation of distant dendrites or through strong synaptic drive (Dantzker and Callaway 2000; Weiler et al. 2008). The example data shown in Supplemental Fig. S2 strongly supports that under the experimental condition with a longer laser duration, stimulating the presynaptic neurons in the strong excitatory pathways likely evoked spikes in the recorded postsynaptic neuron through synaptic drive (e.g., from layer 4 to layer 2/3). Hence in some imaging experiments conducted for this study, strong laser stimulation was used to further assess trans-synaptic spread of activity.
In our VSD imaging experiments, the observed properties of photostimulation evoked VSD signals were closely related to membrane potential depolarization of individual neurons (Fig. 2, B1–B4). Spikes responded faster to photostimulation than VSD signals, because the average spike peak time in response to photostimulation was 11.8 ± 3 ms, and the average VSD signal peak time was 29.9 ± 6.3 ms (P < 0.001; data from 3 similar experiments as shown in Fig. 2A). In these experiments, the average VSD response latency across different V1 layers was 15.4 ± 0.7 ms.
Because photostimulation and VSD imaging are combined, separation of the laser excitation and the VSD recording light is important. Given the UV laser and the VSD absorption light have differing wavelengths (355 vs. ∼705 nm), the laser and its excitation artifact had been significantly reduced using a band-pass filter centered at 705 nm right before the imaging camera (Fig. 1A). Furthermore, when using short laser pulses (i.e., 1–3 ms), the laser artifact signal only existed in the initial two image frames (2.2 ms/frame). Compared with the VSD signal reflecting neural activity (0.1–0.5%), the artifact signal was large, up to 2% change from baseline (Fig. 2, B1–B4), however, did not interfere with true VSD signals, considering detectable VSD responses occur approximately five to six frames after the laser onset.
As exemplified in Fig. 2, A–E, the photostimulation-evoked VSD responses were mediated by glutamate and its receptors, as neuronal spiking and VSD responses to glutamate uncaging were essentially abolished by the ionotropic glutamate receptor antagonists (CPP and CNQX; n = 5 slices). There was no VSD signal without glutamate uncaging; laser flashes alone did not activate neurons or induce VSD responses (Supplemental Fig. S1, C and D). Furthermore, reducing neurotransmitter release and synaptic spread from stimulated neurons using low Ca2+ and high Mg2+ ACSF solution restricted VSD changes to the region near the stimulation site. This indicates that most activity far from the stimulation site reflected synaptic spread of activity to postsynaptic neurons rather than activity in the axons and distant dendrites of directly stimulated cells (Fig. 2, F–H). This is similar to the result observed in TTX, which blocks both synaptic spread and conduction of activity with the axons of stimulated cells (Supplemental Fig. S3).
Unlike electrical stimulation, a significant advantage of photostimulation is such that uncaged glutamate does not activate passing axon fibers (Supplemental Fig. S1, E and F), thus ensuring spatial specificity of stimulation and avoiding antidromic presynaptic activation. In addition, VSD imaging of the photostimulation evoked network activity showed good repeatability and low variations, because maps were consistent across multiple repetitions.
Through VSD imaging of large areas of cortical activity probed with laser scanning photostimulation in various locations, our new technique is a powerful tool for functional circuit analysis.
Mouse V1 cortical circuit mapping
We first show high-precision mapping of V1 circuits with our new technique. Compared with previous studies of rat V1 circuits that used VSD imaging with electrical stimulation or puffed glutamate at different cortical locations (Yuste et al. 1997), our new technique allowed us to evaluate laminar propagation of evoked excitation and probe functional connectivity of mouse V1 laminar circuits (n = 10 slices), with a much improved precision and speed.
Under the stimulation conditions used, laser scanning photostimulation (1 ms, 30–35 mW) offered spatially restricted neuronal activation in a specific cortical layer so that we were able to map direct projections from the stimulated layer to its targeted layer(s) by VSD imaging of evoked activation. Photostimulation-evoked action potentials were restricted to neurons with cell bodies at or close to the stimulation site; activity propagated through the axons of the stimulated neurons and generated postsynaptic responses in the neurons that were connected to the stimulated cells. The measured VSD signals reflected the combined contributions of these sources, but responses distant from the stimulation site were dominated by postsynaptic changes, as evidenced by control experiments in which synaptic transmission was blocked by using low Ca2+ and high Mg2+ ACSF.
Figure 3, A–C, show VSD image frames in response to laser photostimulation at cortical layers 2/3, 4, or 5 in a V1 coronal slice, respectively, whereas Fig. 3D shows VSD image frames in response to electrical stimulation at V1 layer 4. In comparison, layer 4 photostimulation was effective as electrical stimulation in evoking population neuronal responses, but its activation was more restricted and specific (cf. Fig. 3, B6 and D6).
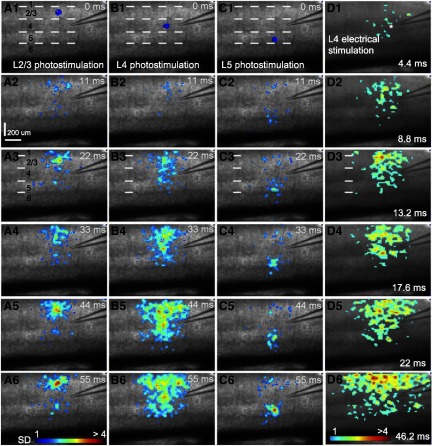
Fig. 3.
Spatially restricted neuronal activation via laser photostimulation enables high-resolution mapping of interlaminar connections in mouse V1 local circuits. A–C: sequences of VSD image frames in response to photostimulation (laser duration: 1 ms; power: 32 mW) at cortical layers 2/3, 4, or 5 in a V1 coronal slice, respectively, whereas D shows VSD image frames in response to electrical stimulation (1 ms, 50 μA current injection) through a microelectrode placed at V1 layer 4. VSD images were acquired at the rate of 2.2 ms/frame during the experiment and are displayed at specific intervals. Time progresses from top to bottom in the column, and color code is used to indicate VSD signal amplitudes expressed as SD above the mean baseline. The map pixels with amplitudes ≥1 SD are plotted and included for further quantification. Warmer colors indicate greater excitation. The displayed maps are from 1 trial without trial averaging, because single-trial VSD signals were of sufficiently high amplitudes in our study. The site of photostimulation can be identified by the laser excitation artifact (the blue spot) in the initial frames of the sequences. The short dashed white lines in the first image of A, B, and C denote the laminar boundaries of V1 layers 1, 2/3, 4, and 5 and 6. VSD images in D1–D6 are color coded differently from A–C. In D1, the white star indicates the tip of the glass pipette for current injections. Note that the CCD camera images have a slightly different aspect ratio. Under the 4× objective, the camera covers an area of 1.28 (w) × 1.07 (h) mm2, with a spatial resolution of 14.6 (w) × 17.9 (h) μm/pixel.
During photostimulation and VSD imaging experiments, stimulation in V1 cortical layers initiated excitation that resulted in VSD signals first localized to the stimulation site at around 10–20 ms after laser exposure; excitation propagated to functionally connected cortical regions. For layer 2/3 stimulation, the activation in layer 2/3 was mostly localized in layer 2/3; however, relatively small but clear activation propagated from layer 2/3 to layer 5, bypassing most of layer 4 (Fig. 3, A1–A6). There was essentially no excitation from layer 2/3 propagating to layer 6. Stimulation in layer 4 caused excitatory activity to spread vertically to layers 2/3 and 5, but with little excitation in layer 6 (Fig. 3, B1–B6). Strong activation in layers 2/3 and 5 caused by layer 4 stimulation indicates these layers receive strong direct projections from layer 4. Layer 5 stimulation resulted in distinct foci of activation in layer 2/3 (Fig. 3, C1–C6), and some activation spread into layers 6 and 4. When increased photostimulation was applied, evoked activation tended to be more robust (Fig. 4, B–D), and the projection of layer 2/3 to layer 5 or layer 5 to layer 2/3 had consistent patterns. Within mouse V1 intracortical circuits, the relatively weak functional activation in layer 5 from L2/3 photostimualtion or in layer 2/3 from layer 5 photostimulation fits with previous anatomical and physiological work (Burkhalter 1989; Callaway 1998; Gilbert 1983; Yuste et al. 1997), reflecting the property of the interlaminar connectivity. Layer 6 stimulation resulted in mostly localized responses, but some activation could spread into upper layers with stronger phostimulation (Fig. 4E).
Fig. 4.
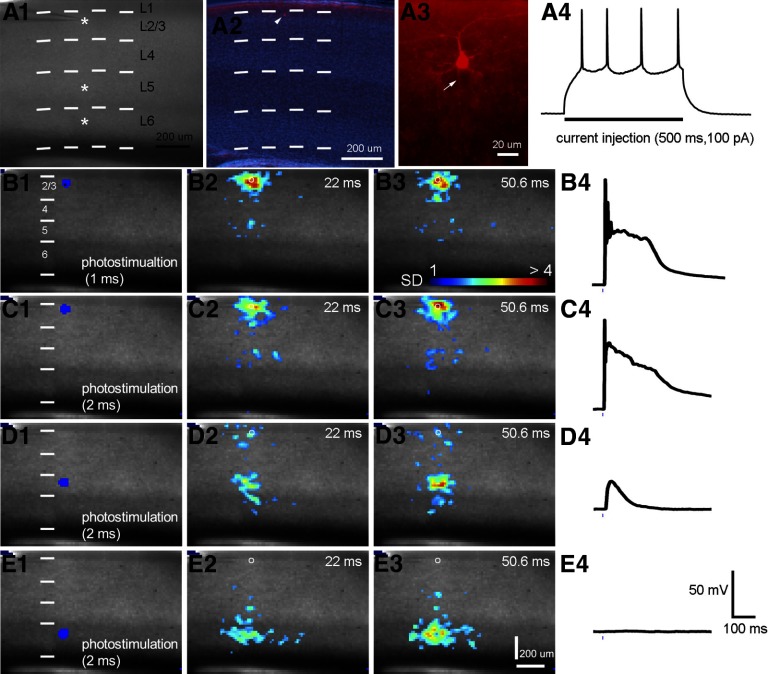
Simultaneous VSD imaging and electrophysiological recordings in response to photostimultaion in layers 2/3, 5, and 6. A1: the slice image with white stars indicating photostimulation sites. The patch pipette was placed in the upper layer 2/3 (close to the layer 2/3 photostimulation site) to record from a single pyramidal neuron. A2: the 4′-6-diamidino-2-phenylindole (DAPI)-stained image of the same V1 slice in A1, with an overlay image of the cell stained against biocytin (pointed by the arrowhead). Combination of DAPI staining patterns and DIC slice images helps to accurately determine V1 laminar boundaries. The short dashed white lines in A1, A2, B1, C1, D1, and E1 denote the laminar boundaries of V1 layers 1, 2/3, 4, 5, and 6. A3: the morphology of the recorded pyramidal cell under high magnification. The white arrow points to the cell's axon. A4: the regular spiking pattern of the cell in response to a current injection (500 ms, 100 pA). B1–B3: VSD image frames in response to photostimulation (1 ms, 32 mW) in layer 2/3 at 0, 22, and 50.6 ms, respectively. The small white circles in B2 and B3 indicate the recorded cell location in the data maps. B4: the suprathreshold spikes from the recorded cell in response to the photostimualtion at layer 2/3. The short blue line below the response trace indicates the laser stimulation. C1–C4: data in response to stronger photostimualtion (2 ms, 32 mW) in layer 2/3. D1–D3: VSD image frames in response to photostimulation (2 ms, 32 mW) in layer 5 at 0, 22, and 50.6 ms, respectively, with D4 showing the subthreshold response of the cell to photostimulation in layer 5. E1–E3: VSD image frames in response to photostimulation (2 ms, 32 mW) in layer 6 at 0, 22, and 50.6 ms, respectively, with E4 showing no response to photostimualtion at layer 6.
Simultaneous electrophysiological recordings of an excitatory pyramidal neuron were performed with VSD imaging in response to photostimultaion in layers 2/3, 5, and 6 (Fig. 4). Stronger laser photostimualtion in layer 2/3 evoked stronger VSD responses and larger membrane potential depolarization of the recorded layer 2/3 neuron (Fig. 4, B1–B4 and C1–C4). Laser photostimualtion in layer 5 evoked subthreshold input responses in the recorded neuron of the layer 2/3, matching VSD activation in layer 2/3 (Fig. 4, D1–D4). Layer 6 photostimulation did not evoke any detectable input to the layer 2/3 neuron, supporting lack of VSD activation in layer 2/3 in response to layer 6 photostimulation (Fig. 4, E1–E4). The use of single cell recordings further complemented imaging results by correlating single-cell activity with the VSD imaged population response.
We quantified patterns of evoked activation in individual cortical layers to analyze interlaminar functional connectivity of local V1 circuits. Figure 5, A–D, summarizes laminar distributions of evoked activation in response to photostimulation (1 ms, 30–35 mW) in V1 layers 2/3, 4, 5, or 6, respectively. For layer 2/3 photostimulation, the mean VSD activation strength (in % of the total evoked activation) for layers 1, 2/3, 4, 5, and 6 were 7.9 ± 1.1, 76.2 ± 0.3, 5.0 ± 0.3, 8.7 ± 0.4, and 2.3 ± 0.6% (SE), respectively (Fig. 5A). The measurement indicated that within V1 intracortical circuits, layer 2/3 neurons were predominantly connected to their counterparts in layer 2/3; compared with other cortical layers, there was a relatively strong projection from layer 2/3 to layer 5. For layer 4 stimulation, the mean VSD activation strength were 6.5 ± 0.5, 44.3 ± 0.7, 28.4 ± 0.6, 18.8 ± 1.0, and 2.1 ± 0.2%, respectively, for layers 1, 2/3, 4, 5, and 6 (Fig. 5B). The quantification indicated a particularly strong functional projection from layer 4 to layer 2/3, which is expected from the known dense axonal fiber projections from layer 4 to layer 2/3 in rodent visual cortex (Burkhalter 1989). In addition, there was a strong connection from layer 4 to layer 5. For layer 5 stimulation, the mean VSD activation strength was 1.9 ± 0.4, 16.6 ± 2.3, 9.6 ± 2.3, 65.1 ± 4.2, and 6.7 ± 0.4%, respectively, for layers 1, 2/3, 4, 5, and 6 (Fig. 5C). The data indicated that layer 5 had a relatively strong projection to upper layers, particularly to layer 2/3. For layer 6 stimulation, the mean VSD activation strength was 0.5 ± 0.2, 3.2 ± 0.8, 3.7 ± 0.8, 9.9 ± 2.0, and 82.8 ± 1.2%, respectively, for layers 1, 2/3, 4, 5, and 6 (Fig. 5D). Thus the layer 6 stimulation responses were localized, and layer 6 only had a moderate connection with layer 5. Provided that VSD activation outside the stimulation site mostly reflects synaptic spread of activity to postsynaptic neurons rather than activity in the axons and distant dendrites of directly stimulated cells in the photostimulated layer, we constructed a functional connectivity diagram for mouse V1 local circuits based on quantitative analysis of propagation activity from multiple laminar locations (Fig. 5E).
Next we show rapid mapping of neuronal circuitry with our technique by patterned photostimulation and VSD imaging of multiple V1 locations (n = 5 slices). As shown in Fig. 6, an array of 4 × 4 photostimulation sites (cyan stars, spacing 200 μm, in Fig. 6A1) covered the V1 cortical area at different laminar locations. Laser scanning photostimulation allowed rapid stimulation for assessment of activity propagation at multiple locations because it was possible to activate many different sites with very short time intervals. This approach is much more efficient than mapping with electrical stimulation at different locations, considering the time required for proper electrode placement on the order of minutes.
Fig. 6.
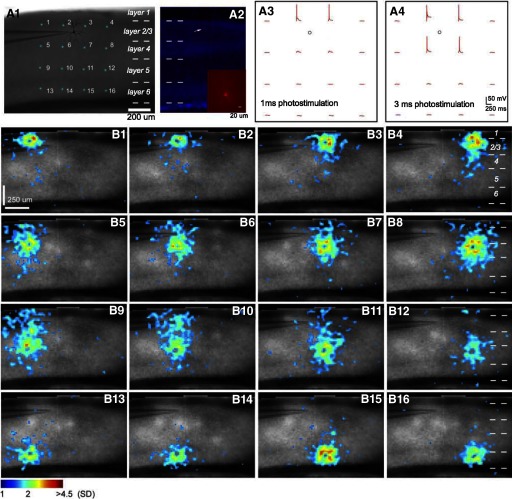
Rapid mapping of V1 local circuits through combination of VSD imaging and laser scanning photostimulation at multiple sites. A1: the slice image with cyan stars indicating a 4 × 4 stimulus pattern covering V1 from cortical layer 2/3 to layer 6. The patch pipette was placed in the middle of layer 2/3 to record from a single pyramidal neuron duringphotostimulation and imaging. A2: the DAPI-stained image of the same V1 slice recorded in A1, with an overlay image of the cell stained against biocytin (pointed by the white arrow). Inset in A2: the morphology of the recorded pyramidal cell under a higher magnification. The cell's morphological reconstruction with major dendrites is shown in A1. The short dashed white lines in A1, A2, B4, B8, B12, and B16 denote the laminar boundaries of V1 layers 1, 2/3, 4, 5, and 6. A3 and A4: the excitation profiles of the recorded neuron in response to 1 and 3 ms photostimulation (laser power 35 mW) of the 4 × 4 sites shown in A1, respectively. The small circle indicates the cell body location. The red traces displayed at the stimulation sites depict membrane potential changes in response to photostimulation in the whole cell recording mode. B1–B16: single frames corresponding to the times of peak activation from the VSD map sequences at each of the 16 stimulation sites indicated in A1. VSD signal amplitudes expressed as SD above the mean baseline signal are color coded.
During the experiments, a single pyramidal cell was recorded in the middle of layer 2/3; as shown in Fig. 6, A3 and A4, photostimulation (1 ms, 35 mW) triggered spikes of the recorded cell only in layer 2/3 sites (i.e., sites 2 and 3 in Fig. 6A1), but stronger photostimulation (3 ms, 35 mW) triggered spikes of the recorded cell at a larger area around the cell soma including layer 4 sites (i.e., sites 6 and 7 in Fig. 6A1). The stronger photostimulation excited more neurons at stimulation sites and evoked stronger VSD responses. When 3-ms photostimulation (laser power: 35 mW; duration: 3 ms) was used to map all 16 locations in the V 1 slice, stronger photostimulation yielded clear columnar activity seen at stimulation sites, and increased vertical propagation of activity (Fig. 6, B1–B16). Although activation patterns from stronger photostimulation in layers 2/3, 4, and 6 generally resembled those evoked by using 1-ms laser stimulation, stronger layer 5 photostimulation evoked activity that propagated across all V1 layers, ascending into layers 4, 2/3, and 1 and descending into layer 6. The average peak activation size (in % of the whole image size) for layer 5 stimulation was larger than that of layer 4 stimulation (12.3 ± 0.36 vs. 11.2 ± 0.21%), and both of them differed significantly from stimulation in layers 2/3 and 6 (7.31 ± 0.13 and 7.72 ± 0.26%; P < 0.001, Kruskal-Wallis test). The large activation derived from layer 5 stimulation is likely to result from both direct neuronal responses induced at layer 5 as well as strong indirect suprathreshold activation of layer 4 neurons via connections from layer 5 to layer 4 neurons (Schubert et al. 2003).
As shown above, our new technique enables cortical circuits to be further probed by varying the strength of stimulation, by changing either laser duration or power. This method can therefore also be used to assess the polysynaptic spread of activity when laser activation is sufficiently strong.
Mouse hippocampal circuit mapping
Considering that the hippocampus is an important cortical structure and its trisynaptic pathway is a fundamental network, we further extended our technique to study the mouse hippocampal circuitry. We are particularly interested in understanding whether or how a restricted population of dentate neurons (e.g., granule cells) can engage the entire trisynaptic circuit. We photostimulated different locations in the dentate gyrus (DG) such as the molecular layer, the granule cell layer, and the hilus, and monitored how evoked responses initiated and propagated throughout the hippocampal circuitry (n = 5 slices).
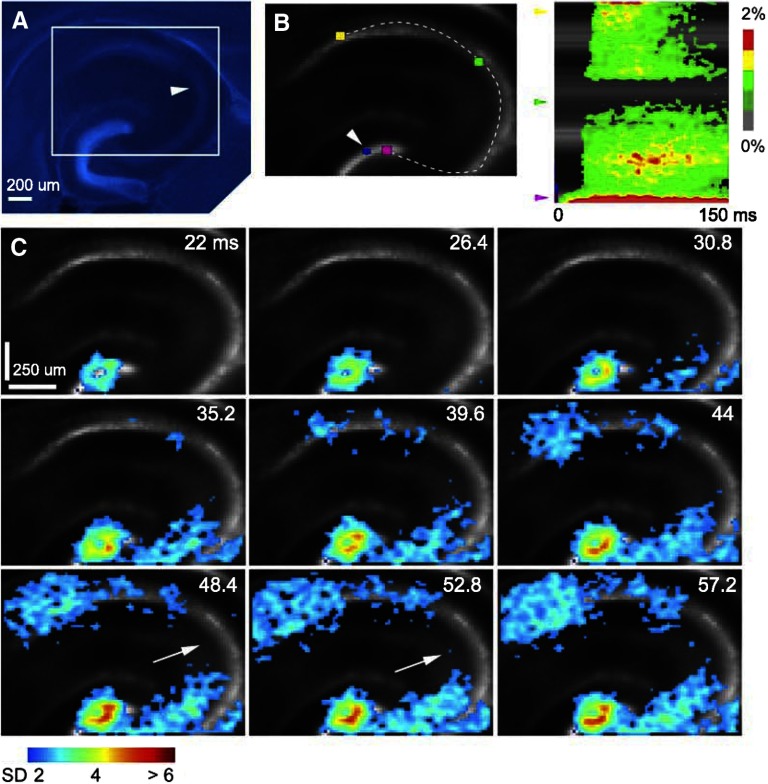
For the purpose of this study, we present example data of stimulation in the DG granule cell layer as shown in Fig. 7. During the experiment, strong laser photostimulation (laser power: 35 mW; duration: 3 ms) was used to activate the trisynaptic circuitry. With the restricted photostimulation in the upper portion DG granule cell layer (indicated by the blue spot of laser excitation artifact in Fig. 7B), the VSD response originated locally in the granule cell layer and after a short delay, traversed through the hilus, and reached CA3, followed by CA1 (Fig. 7C). The localized DG neuronal activation therefore induced serial excitatory propagation in DG, CA3, and CA1 (Fig. 7, B and C). The excitation in CA3 and CA1 was extensive, but the CA2 region had little excitation (Fig. 7, B and C), which supports evidence that CA2 lacks granule cell input (Nakagami et al. 1997; Swanson et al. 1978). The data showed a localized DG neuronal population effectively engaged in the excitatory flow of information throughout DG and the hippocampus proper collectively, thus providing a comprehensive perspective of the functional circuit organization and dynamics of the hippocampal pathway.
Fig. 7.
Hippocampal circuit mapping. A: DAPI-stained image of the hippocampus slice with the white rectangule marking the VSD imaging area shown in B and C. Overlay and alignment of DAPI staining with DIC living slice images located dentate gyrus (DG), CA3, CA2 (pointed by the arrowhead), and CA1. B: the space-time analysis indicating serial excitatory propagation in DG, CA3, and CA1. B: left: the background reference image with an analysis curve aligned with the hippocampal circuitry. The white arrowhead in the left panel points to the restricted photostimulation site (the blue spot) in the DG granule cell layer. B: right: the space-time map with the y-axis indicating the location along the analysis curve shown in the left panel (e.g., the pink, green, and yellow arrowheads correspond to the colored locations in the left panel) and with the x-axis denoting the time progression after the photostimulation. C: time frame series of the VSD response after photostimulation and a clear excitation flow through the hippocampal trisynaptic pathway. VSD signal amplitudes expressed as SD above the mean baseline signal are color coded. In this experiment, the map pixels with amplitudes ≥2 SD are plotted for analysis. The maps are identically color coded with warmer colors indicating greater excitatory activity. The white arrows in C point to the CA2 region, which had little activation.
DISCUSSION
Optical-based techniques become increasingly important for cortical circuit studies, as modern technology continues to improve spatiotemporal resolutions of imaging and provide exquisite control of the delivery of optical stimuli (Luo et al. 2008). We developed a new technique that combines laser scanning photostimulation with fast VSD imaging for high precision and rapid mapping of in vitro functional circuits. The incorporation of laser scanning photostimulation has greatly enhanced the ability of assessment of evoked network activity by fast VSD imaging. Photostimulation provides spatially restricted neuronal activation and avoids the interpretation difficulties associated with electrical stimulation of neural tissue. Furthermore, compared with microelectrode electric stimulation, laser scanning photostimulation permits rapid evaluation of multiple locations with no physical damage to the tissue under the experimental conditions. In addition, we anticipate that glutamate uncaging can be replaced with photoactivation via channelrhodopsin or other genetically encoded photosensitive molecules in specific cell types (Boyden et al. 2005; Kuhlman and Huang 2008). Thus our technique will be possible to target not only specific cortical regions but also specific subset of neurons within their participating circuits.
Photostimulation-based mapping techniques have been widely applied for analyzing cortical circuits. Laser scanning photostimulation combined with single cell recordings or calcium imaging is an effective method for mapping local circuit inputs to single neurons, because the simultaneous recording from a postsynaptic neuron with photostimulation of clusters of presynaptic neurons at many different locations provides quantitative measures of spatial distribution of excitatory and inhibitory inputs impinging onto single recorded neurons (Callaway and Katz 1993; Nikolenko et al. 2007; Schubert et al. 2003; Shepherd and Svoboda 2005; Xu and Callaway 2009). Different from the aforementioned approach, our newly developed technique is intended to assess circuit activation and network connectivity at the neuronal population level through fast VSD imaging and photostimulation. Because our technique is often performed with simultaneous electrophysiological recordings of single neurons, the method is readily combined with whole cell patch-clamp measurements of electrical signals in brain slices so that local cortical circuits can be examined in the same brain slice at both single cell and population levels.
Our technique can have important applications in the field of cortical circuitry as shown in our studies of mouse V1 and hippocampal circuits. Because the mouse is an important model system for cortical circuit studies and mouse V1 circuits are less well understood, we imaged and probed V1 with spatially restricted photostimulation and mapped interlaminar functional connectivity and circuit dynamics. This technique enabled direct visualization of interlaminar functional connections in V1 circuits at a previously unattainable precision (Burkhalter 1989; Callaway 1998; Yuste et al. 1997). Compared with previous rat V1 findings, our functional circuit analysis showed that mouse V1 layers 2/3 and 4 had little projections to layer 6 and found that mouse V1 layers 4 and 5 had relatively strong reciprocal projections and that mouse layer 6 had weak projections to upper layers. Overall, our mouse V1 data fit with and extend previous anatomical and physiological observations of laminar patterns of axonal projections in rodent V1 local circuits (Burkhalter 1989; Yuste et al. 1997), and is generally consistent with proposed V1 laminar operational schemes (Callaway 1998; Gilbert 1983).
Alternative approaches would be far more difficult and time consuming to obtain similar connectivity data using other methods such as intracellular dye labeling and reconstruction of axonal arbors or by recording sequentially from many possible postsynaptic partners. Although our V1 results are as expected from known anatomy and connectivity in the rodent visual cortex (Burkhalter 1989; Yuste et al. 1997), this method would be valuable for assaying brain areas, species, or genetically modified mouse lines in which these data are not presently available. In addition, the application of this technique in the hippocampal mapping further validated its technical power and effectiveness. The circuit dynamics and functional connection in the hippocampal circuitry were mapped through spatially restricted activation of a subset of DG neuronal population. With strong photostimulation, we were able to examine the complete perspectives of the polysynaptic spread of activity in detail.
Future study using this technique can be extended for screening circuit alterations in transgenic animal models recapitulating specific neurological diseases. Our new technique will allow for the rapid evaluation of alterations in circuitry of mutant animals compared with normal animals and will guide more detailed studies. The technique can be further developed as a methodology for identification and monitoring of real-time responses in vitro (e.g., cell cultures and slice preparations) to drugs or therapeutic interventions.
GRANTS
This work was funded by the National Institute of Drug Abuse Grants DA-023700 and DA-023700-04S1 to X.Xu.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Gordon Shepherd at Northwestern University, Dr. Karel Svoboda at Howard Hughes Medical Institute Janelia Farm Research Campus, Dr. Hongtao Ma at Weill Medical College of Cornell University, S. Strayer at the University of California, Irvine, and A. Miller at Olympus USA for technical help and Drs. Edward Callaway, David Lyon, Ivan Soltesz, and Ralph Siegel for critical reading of our manuscript.
Footnotes
The online version of this article contains supplemental data.
REFERENCES
- Airan RD, Meltzer LA, Roy M, Gong Y, Chen H, Deisseroth K. High-speed imaging reveals neurophysiological links to behavior in an animal model of depression. Science 317: 819–823, 2007. [DOI] [PubMed] [Google Scholar]
- Ang CW, Carlson GC, Coulter DA. Massive and specific dysregulation of direct cortical input to the hippocampus in temporal lobe epilepsy. J Neurosci 26: 11850–11856, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci 8: 1263–1268, 2005. [DOI] [PubMed] [Google Scholar]
- Burkhalter A. Intrinsic connections of rat primary visual cortex: laminar organization of axonal projections. J Comp Neurol 279: 171–186, 1989. [DOI] [PubMed] [Google Scholar]
- Callaway EM. Local circuits in primary visual cortex of the macaque monkey. Annu Rev Neurosci 21: 47–74, 1998. [DOI] [PubMed] [Google Scholar]
- Callaway EM, Katz LC. Photostimulation using caged glutamate reveals functional circuitry in living brain slices. Proc Natl Acad Sci USA 90: 7661–7665, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzker JL, Callaway EM. Laminar sources of synaptic input to cortical inhibitory interneurons and pyramidal neurons. Nat Neurosci 3: 701–707, 2000. [DOI] [PubMed] [Google Scholar]
- Gilbert CD. Microcircuitry of the visual cortex. Annu Rev Neurosci 6: 217–247, 1983. [DOI] [PubMed] [Google Scholar]
- Grinvald A, Hildesheim R. VSDI: a new era in functional imaging of cortical dynamics. Nat Rev 5: 874–885, 2004. [DOI] [PubMed] [Google Scholar]
- Huang X, Troy WC, Yang Q, Ma H, Laing CR, Schiff SJ, Wu JY. Spiral waves in disinhibited mammalian neocortex. J Neurosci 24: 9897–9902, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Zhang RJ, Wu JY. Voltage-sensitive dye imaging of population neuronal activity in cortical tissue. J Neurosci Methods 115: 13–27, 2002. [DOI] [PubMed] [Google Scholar]
- Konnerth A, Obaid AL, Salzberg BM. Optical recording of electrical activity from parallel fibres and other cell types in skate cerebellar slices in vitro. J Physiol 393: 681–702, 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman SJ, Huang ZJ. High-resolution labeling and functional manipulation of specific neuron types in mouse brain by Cre-activated viral gene expression. PLoS ONE 3: e2005, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron 57: 634–660, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami Y, Saito H, Matsuki N. Optical recording of trisynaptic pathway in rat hippocampal slices with a voltage-sensitive dye. Neuroscience 81: 1–8, 1997. [DOI] [PubMed] [Google Scholar]
- Nikolenko V, Poskanzer KE, Yuste R. Two-photon photostimulation and imaging of neural circuits. Nat Methods 4: 943–950, 2007. [DOI] [PubMed] [Google Scholar]
- Petersen CC, Sakmann B. Functionally independent columns of rat somatosensory barrel cortex revealed with voltage-sensitive dye imaging. J Neurosci 21: 8435–8446, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D, Kotter R, Zilles K, Luhmann HJ, Staiger JF. Cell type-specific circuits of cortical layer IV spiny neurons. J Neurosci 23: 2961–2970, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd GM, Pologruto TA, Svoboda K. Circuit analysis of experience-dependent plasticity in the developing rat barrel cortex. Neuron 38: 277–289, 2003. [DOI] [PubMed] [Google Scholar]
- Shepherd GM, Svoboda K. Laminar and columnar organization of ascending excitatory projections to layer 2/3 pyramidal neurons in rat barrel cortex. J Neurosci 25: 5670–5679, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson LW, Wyss JM, Cowan WM. An autoradiographic study of the organization of intrahippocampal association pathways in the rat. J Comp Neurol 181: 681–715, 1978. [DOI] [PubMed] [Google Scholar]
- Weiler N, Wood L, Yu J, Solla SA, Shepherd GM. Top-down laminar organization of the excitatory network in motor cortex. Nat Neurosci 11: 360–366, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Callaway EM. Laminar specificity of functional input to distinct types of inhibitory cortical neurons. J Neurosci 29: 70–85, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R, Tank DW, Kleinfeld D. Functional study of the rat cortical microcircuitry with voltage-sensitive dye imaging of neocortical slices. Cereb Cortex 7: 546–558, 1997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.