Abstract
Autosomal dominant polycystic kidney disease (ADPKD) is the most common inherited kidney disease and affects 1 in 1,000 individuals. Ultrasound is most often used to diagnose ADPKD; such a modality is only useful late in the disease after macroscopic cysts are present. There is accumulating evidence suggesting that there are common cellular and molecular mechanisms responsible for cystogenesis in human and murine PKD regardless of the genes mutated, and, in the case of complex metabolomic analysis, the use of a mouse model has distinct advantages for proof of principle over a human study. Therefore, in this study we utilized a urinary metabolomics-based investigation using gas chromatography-time of flight mass spectrometry to demonstrate that the cystic mouse can be discriminated from its wild-type counterpart by urine analysis alone. At day 26 of life, before there is serological evidence of kidney dysfunction, affected mice are distinguishable by urine metabolomic analysis; this finding persists through 45 days until 64 days, at which time body weight differences confound the results. Using functional score analysis and the KEGG pathway database, we identify several biologically relevant metabolic pathways which are altered very early in this disease, the most highly represented being the purine and galactose metabolism pathways. In addition, we identify several specific candidate biomarkers, including allantoic acid and adenosine, which are augmented in the urine of young cystic mice. These markers and pathway components, once extended to human disease, may prove useful as a noninvasive means of diagnosing cystic kidney diseases and to suggest novel therapeutic approaches. Thus, urine metabolomics has great diagnostic potential for cystic renal disorders and deserves further study.
autosomal dominant polycystic kidney disease (ADPKD) is the most prevalent inherited disease of the kidney, accounting for 4.5% of the end-stage renal disease (ESRD) population (8). Renal ultrasonography is the current diagnostic method of choice for affected individuals with ADPKD. Ultrasound is preferentially used due to cost, level of invasiveness, and relative accuracy for at-risk PKD1 individuals. In those at risk, a negative ultrasound confers a >95% likelihood of not inheriting a PKD1 mutation by age 30 (39). However, in at-risk PKD2 individuals, only a 67% likelihood can be attributed to a negative ultrasound at age 30 due to the milder and later age of onset of renal cystic disease (2). Computed tomography (CT)- or magnetic resonance (MR)-based imaging provides significant but marginal improvement in disease detection and is not warranted for routine clinical screening. However, in certain situations, such as identifying potential living relative kidney donors, these imaging modalities provide increased detection of small cysts (1). Genetic testing using direct sequencing for PKD1 and PKD2 mutations is available, but the highest mutation detection rates using multiple approaches only reach 85% (40). Thus at present a reliable, specific, and accurate diagnostic test for ADPKD is not available for patients in the earliest presymptomatic, precystic stages. Given that new molecularly targeted therapies are quickly becoming available that may change the clinical course of ADPKD (1, 9), new biofluid diagnostic tests are urgently needed.
Genomics and proteomics (the first of the “omics” techniques to be described) have been studied extensively in human diseases, and rodent models thereof, including those of the kidney, and have resulted in significant advances in the understanding of the pathophysiology of a variety of maladies. However, translating these techniques into the clinical arena of biofluid diagnosis has been hampered by the fact that, while tissue samples are in many cases adequate, many disease-specific nucleic acids and proteins are not secreted into biofluids in sufficient quantities to allow for the construction of an effective diagnostic test. Furthermore, the use of genomic and transcriptomic analysis of diseased tissue is now considered controversial by some, due to the lack of substantial “payoff” of these techniques in translational medicine (41). Thus, of the three major “omics” fields, metabolomics, the study of all metabolites produced in an organism, provides the most promise for both diagnosis of and elucidating physiological responses to disease.
It is becoming increasingly clear, from work in our (18, 21, 37) and others’ (20, 24, 27, 28, 46) laboratories, that metabolomics possesses significant as yet untapped potential for biomarker discovery, since it represents the signature of the actual processes that are occurring within the body rather than compounds (such as untranscribed DNA or pre- or posttranslationally modified proteins) that may be superfluous to these processes. In addition, there are a relatively small number of metabolites to examine compared with genes, transcripts, and proteins in their respective “omics” fields. Proponents of metabolomics provide convincing justification that this technique will offer more immediate translational benefit than the other “omics” fields (10, 41), and our initial forays into this technology for kidney cancer have confirmed this likelihood for diseases of the urinary system (21, 37).
In this study, we have applied metabolomics to cystic diseases of the kidney for the first time. Using an established model of renal cystic disease, the jck mouse, a double point mutation in Nek8 and mouse model for recessive human juvenile nephronopthisis type 9 (52), we show that there is significant separation of the urine metabolome between genotypes as early as 26 days after birth when kidney function is normal and there are few cysts in the kidneys. From these data, based on pathways from the KEGG database (17), we identify biochemical pathways which are altered in the disease state and uncover several specific metabolites which contribute most to the discrimination between genotypes. When validated in other mouse models and in human disease, these data may lead to novel biomarkers as well as new pathways for therapeutic development.
MATERIALS AND METHODS
Sample Procurement
Since using urine samples would be the most expedient method of diagnosing PKD in the clinic, we obtained urine samples from female jck and wild-type animals of the same genetic background (C57BL/6J) at three different times after birth, representing a wide range of disease progression (45). Only female mice were used given the sexual dimorphism of jck disease in the mouse. Because female jck mice have milder disease compared with male jck mice (45), identified biomarkers in females would be expected to be more robust for detecting the disease than if only males or both sexes were used. Mice were handled under the appropriate institutionally approved Animal Care Committee protocols. We analyzed urine from 10, 9, and 10 female jck and 9, 10, and 9 female wild-type mice from each of the three time points: 26, 45, and 64 days of age, respectively. Mice at 26 days of age show essentially normal renal function, although the kidneys are significantly enlarged (45). Renal function had worsened by 45 days, and by 64 days, little normal kidney tissue remained (45). All urine samples were collected for 24 h as previously described (32).
Gas Chromatography-Time of Flight Mass Spectrometry Analysis
Urine samples were aliquoted into 1-ml volumes and stored at −80°C. Samples were prepared for gas chromatography-time of flight (GC-TOF) mass spectrometry analysis by first precipitating out large proteins. Briefly, for each sample, 10 μl of urine was thawed, vortexed for 10 s, and added to tubes containing 1 ml of a 3:3:2 (vol/vol/vol) acetonitrile:isopropanol:water extraction solvent mixture at −20°C. Samples were then vortexed for 10 s, shaken for 5 min at 4°C, and centrifuged for 2 min at 14,000 relative centrifugal force. For each sample, a whole supernatant was aliquoted into two tubes each containing 500 μl and dried down overnight using a speed vacuum concentration system (Labconco Centrivap cold trap). A mixture of internal retention index (RI) markers was prepared using fatty acid methyl esters of C8, C9, C10, C12, C14, C16, C18, C20, C22, C24, C26, C28, and C30 linear chain length, dissolved in chloroform at a concentration of 0.8 (C8-C16) and 0.4 mg/ml (C18-C30). Two microliters of this RI mixture were added to the dried extracts. Ten microliters of a solution of 20 mg/ml of 98% pure methoxyamine hydrochloride (Sigma, St. Louis, MO) in pyridine (silylation grade, Pierce, Rockford IL) was added and shaken at 30°C for 90 min to protect aldehyde and ketone groups. Ninety microliters of N-methyl-N-(trimethylsilyl)trifluoroacetamide (1-ml bottles, Sigma-Aldrich) was added for trimethylsilylation of acidic groups and shaken at 37°C for 30 min. The reaction mixture was transferred to a 2-ml clear glass autosampler vial with microinsert (Agilent, Santa Clara, CA) and closed by a 11-mm T/S/T crimp cap (MicroLiter, Suwanee, GA). A Gerstel automatic liner exhange system was used in conjunction with a Gerstel CIS cold injection system (Gerstel, Muehlheim, Germany). For every 10 samples, a fresh multibaffled liner was inserted (Gerstel no. 011711–010-00) using Maestro1 Gerstel software version. 1.1.4.18. Before and after each injection, the 10-μl injection syringe was washed three times with 10 μl ethyl acetate. A 1-μl sample was filled using 39-mm vial penetration at 1 μl/s fill speed, injecting 0.5 μl at 10 μl/s injection speed at initial 50°C, which was ramped by 12°C/s to a final 250°C and held for 3 min. The injector was operated in split mode at a 1:5 ratio. An Agilent 6890 gas chromatograph was controlled by the Leco ChromaTOF software version 2.32 (St. Joseph, MI). A 30-m long, 0.25-mm ID Rtx5Sil-MS column with 0.25-μm 5% diphenyl/95% dimethyl polysiloxane film and additional 10-m integrated guard column were used (Restek, Bellefonte PA). Helium (99.9999% pure) with built-in purifier (Airgas, Radnor PA) was set at a constant flow of 1 ml/min. The oven temperature was held constant at 50°C for 1 min and then ramped at 20°C/min to 330°C, at which it was held constant for 5 min. A Leco Pegasus IV time of flight mass spectrometer was controlled by the Leco ChromaTOF software version 2.32. The transfer line temperature between the gas chromatograph and mass spectrometer was set to 280°C. Electron impact ionization at −70 V was employed with an ion source temperature of 250°C. After 330-s solvent delay, filament 1 was turned on and mass spectra were acquired at a mass resolution R = 600 from m/z 85–500 at 20 spectra/s and 1,750-V detector voltage without turning on of the mass defect option. Recording ended after 1,200 s. The instrument performed autotuning for mass calibration using FC43 (perfluorotributylamine) before the start of analysis sequences.
Daily quality controls were used. These comprised two method blanks (involving all the reagents and equipment used to control for laboratory contamination) and four calibration curve samples spanning one order of dynamic range and consisting of 31 pure reference compounds.
Files were preprocessed directly after data acquisition and stored as ChromaTOF-specific *.peg files, as generic *.txt result files, and additionally as generic ANDI MS *.cdf files. ChromaTOF version 2.32 was used for data preprocessing without smoothing, 3-s peak width, baseline subtraction just above the noise level, and automatic mass spectral deconvolution and peak detection at signal/noise levels of 10:1 throughout the chromatogram. Apex masses were used for quantification. Resulting *.txt files were exported to a data server with absolute spectra intensities and further processed by a filtering algorithm implemented in the metabolomics BinBase database (11). The following algorithm was used the settings: validity of chromatogram (<10 peaks with intensity >10^7 counts/s), unbiased retention index marker detection (MS similarity >800, validity of intensity range for high m/z marker ions), with a retention index calculation by fifth-order polynomial regression. Spectra were cut to 5% base peak abundance and matched to database entries from most to least abundant spectra using the following matching filters: retention index window ± 2,000 units (equivalent to about ±2-s retention time), validation of unique ions and apex masses (unique ion must be included in apexing masses and present at >3% of base peak abundance), mass spectrum similarity must fit criteria dependent on peak purity and signal/noise ratios (12), and a final isomer filter. Failed spectra were automatically entered as new database entries if s/n >25, purity <1.0, and presence in the biological study design class was >80%. All thresholds reflect settings for ChromaTOF version 2.32. Quantification was reported as peak height using the unique ion as a default, unless a different quantification ion was manually set in the BinBase administration software Bellerophon. A quantification report table was produced for all database entries that were positively detected in >80% of the samples of a study design class (as defined in the SetupX database) (42) for unidentified metabolites, and with a 50% threshold for structurally identified compounds. A subsequent postprocessing module was employed to automatically replace missing values from the *.cdf files using open access mzmine software. Replaced values were labeled as “low confidence” by color coding.
Raw Data Processing
GC-TOF result files were transformed by calculating the sum intensities of all structurally identified compounds for each sample and subsequently dividing all data associated with a sample by the corresponding metabolite sum. This normalization method improves the robustness of normalization-between-individuals compared with a single marker (e.g., creatinine), as any single specific metabolite may be subject to metabolic control that differs between individuals, whereas the complement of all metabolites should reflect the overall status of metabolic phenotypes. In addition, at least for the 26-day samples, both groups of mice had normal kidney function. The resulting data were multiplied by a constant factor for convenience of obtaining integer values. Intensities of identified metabolites with more than one peak (e.g., methoximated-reducing sugars) were summed to only one value in the transformed data set. The original nontransformed data set was retained.
Metabolites were unambiguously assigned by the BinBase identifier numbers, using the retention index and mass spectrum as the two most important identification criteria. Additional confidence criteria were given by mass spectral metadata, using the combination of unique ions, apex ions, peak purity, and signal/noise ratios as given in data preprocessing. All database entries in BinBase were matched against the Fiehn mass spectral library of 1,200 authentic metabolite spectra using retention index and mass spectrum information, or the NIST05 commercial library, annotating metabolites based on mass spectrum similarity alone and adding the name suffix “NIST” to indicate a lower level of confidence. BinBase entries were named manually by both matching mass spectra and retention index. PubChem numbers and KEGG identifiers were also added. In addition, all reported compounds (identified and unknown metabolites) are reported by the quantification ion and the full mass spectrum encoded as a string. All raw and processed data are downloadable at http://fiehnlab.ucdavis.edu:8080/m1/main_public.jsp.
Statistical Analysis
Processing of the raw data for all three time points at the first analysis yielded 143 peaks, corresponding to known metabolites for analysis. To evaluate the technical reproducibility of GC-TOF when applied to urine, we conducted a second GC-TOF run of the same urine samples collected from mice at 45 days. This technical replicate was processed in the second analysis and yielded 116 peaks. Correlation analysis was performed to assess the degree of the consistency of metabolite intensities between the two technical replicates. All statistical analyses were conducted in R 2.7.1 language and environment (The R Foundation for Statistical Computing, Auckland University, Auckland, New Zealand; http://www.r-project.org/).
Before statistical analyses, we applied a log (base 2) transformation to meet underlying assumptions of normality with a constant variance and to reduce the dominant effect of extreme values. The primary objective of the statistical analysis was to identity metabolites whose concentration differentiates between “genotypes” that eventually can be used as diagnostic tools for PKD. We accounted for potential confounding effects of differences in body weight between genotypes. To eliminate the variation in data resulted from the differences in body weight between the two genotype groups, we adjusted intensity measurements for body weight by regressing each measurement of body weight and using the residuals in subsequent analyses. We also conducted the analyses without this adjustment and compared the results with those adjusted for body weight to assess the effect of the body weight adjustment on biomarker discovery. In both cases, intensity measurements were centered to have a mean of 0 and scaled to a variance of 1 at each time point for use in the following statistical analyses.
Functional score analysis.
We conducted a functional score analysis (35) to identify pathways that were significantly altered in the disease state. Each metabolite was associated with KEGG pathways through their compound identifiers (e.g., “C00001” for water). Every pathway listed in the KEGG Compound data set with at least one metabolite identified by the MS/GC associated with that pathway was noted and the MS/GC-identified metabolites cross listed with the pathway. Each metabolite could belong to multiple pathways. We conducted a t-test for a difference in mean intensities between genotypes at day 26 for each metabolite using log (base 2) transformed values. Because body weight did not differ significantly between genotypes at day 26, we did not adjust intensities for body weight in this analysis. For each pathway, we calculated the pathway's functional score as the median of the squared t-statistic values of metabolites in the pathway. We used squared t-statistic values to focus on pathways with metabolites that differed between genotypes irrespective of the direction of the difference. All metabolites associated with a pathway were included regardless of whether the metabolite differed significantly between genotypes. We used a permutation null distribution to identify pathways that were significantly altered in jck mice relative to wild-type mice. To generate the null distribution, we permuted class labels 10,000 times and recalculated the functional score for each pathway. This process yielded a null distribution of the functional scores for each pathway from which we calculated P values. Pathways with a P value <0.05 were considered significantly different between genotypes.
Partial least squares and linear discriminant analysis.
We used partial least squares (PLS) regression and linear discriminant analysis (LDA) to determine whether the urine metabolome (as represented by the identified metabolites) could distinguish between jck and wild-type mice and thereby predict the genotype. First, PLS regression was used to reduce the intensity measurements of 143 peaks to a small number of latent components that explained most of the variation (33). Latent components identified through PLS were then used in a LDA to classify samples as wild-type or jck (30). To identify metabolites that were most influential in separating wild-type mice from jck mice, we examined loading scores of the first few latent components. Because the objective of this study was to identify metabolites that differentiate between genotypes (jck vs. wild-type mice) rather than between changes in metabolic profiles as a function of time, data from each time period were evaluated separately and independently from other time points.
Two methods were used to determine the appropriate number of latent components. First, leave-one-out cross validation was conducted. In this approach, intensity measurements of one mouse at a time were left out in conducting the PLS. The genotype of the excluded mouse was predicted with LDA using k latent components, k = 1,…, 8, and the misclassification rate was calculated for each k. This process was repeated for each sample. Second, Boulesteix's bootstrap cross-validation approach to choosing the number of components (4) was used. In this approach, α% of the samples, the training set, are used to identify k latent components, k = 1,…, 8, which are then used to predict the class membership of the remaining (1 − α)% observations that comprise the test set. Observations for the training set are randomly selected with replacement. The process of selecting the training set, conducting PLS, and predicting class membership of the test set is repeated B times. The misclassification rate (number of samples incorrectly classified) is calculated for each training set-test set combination and averaged over the B resamples. We used α = 66.7 and B = 50. PLS analyses were conducted with the R package pls-genomics (5).
Differential analysis.
Metabolites whose expression was associated with the genotype were identified through differential analysis. We tested for mean differences in intensity level between the genotypes with permutation t-tests to preserve the complex correlation structure of the metabolite intensity data. We permuted the class labels 10,000 times and recomputed the t-statistics of the metabolites for the permuted data to yield a null distribution. The permutation null distribution was used to determine P values to assess significance. To account for multiple hypothesis testing, a step-down maxT procedure was used to identify metabolites that differed significantly in expression between jck and wild-type mice while maintaining a family-wise type I error rate of 0.05 (54). The differential analysis was conducted using the R package multtest (Pollard KS, Ge Y, Taylor S, and Dudoit S: multtest: resampling-based multiple hypothesis testing, 2008; version 1.21.1).
RESULTS
Metabolomic Data are Reproducible When Analyzed at Different Times
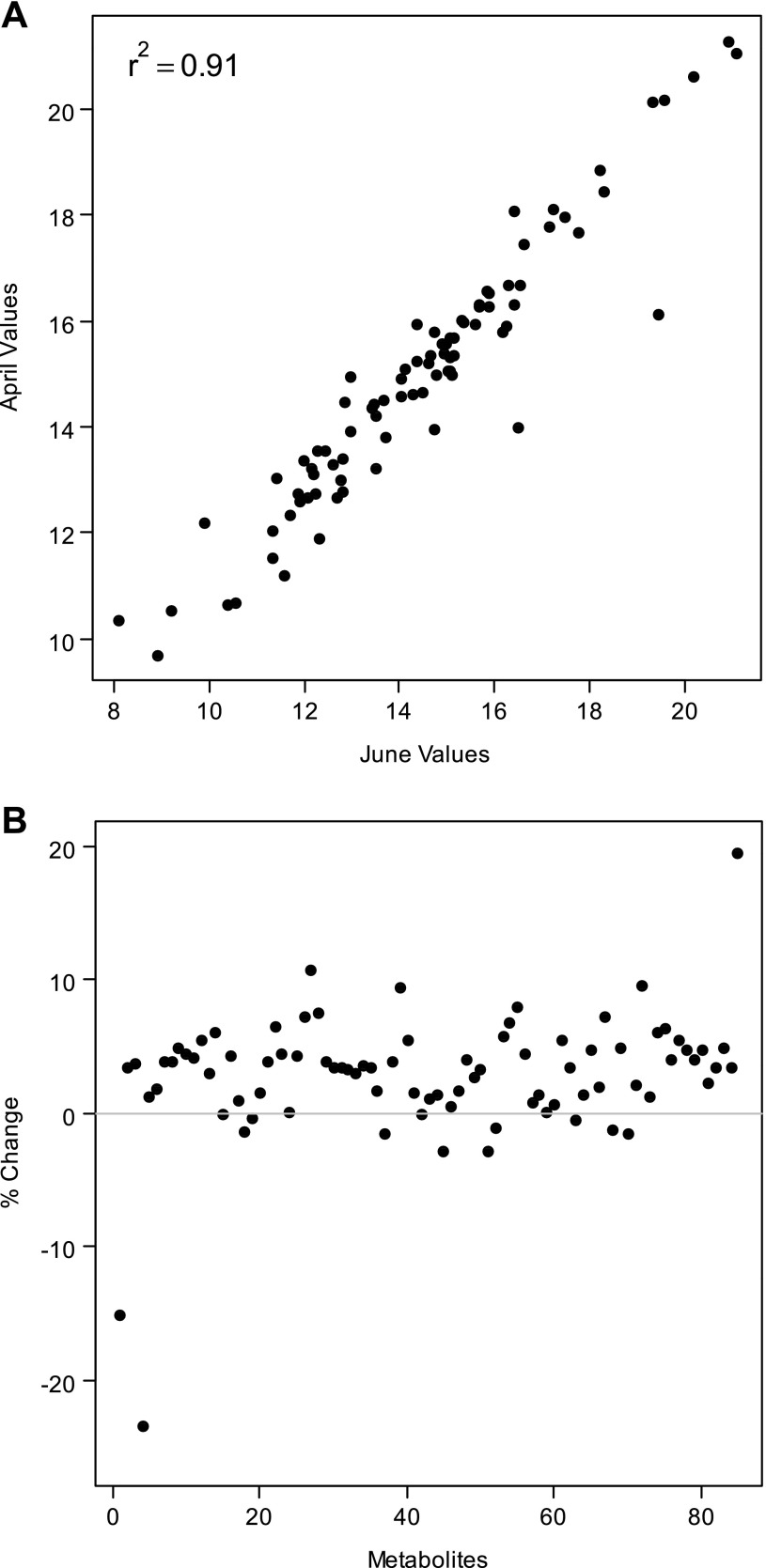
Of the 143 peaks corresponding to known compounds identified in the first analysis of both jck and wild-type 45-day samples, and 116 peaks for the second technical replicate of this analysis, 85 compounds were identified in both replicates. We found that intensities of the 85 metabolites were commonly identified in both replicates were highly correlated. Correlation coefficients between intensity measurements for each of the 19 mice ranged from 0.84 to 0.96 (Fig. 1A). Mean intensities also were consistent between runs, with mean values varying by <10% between GC-TOF analyses for nearly all metabolites (Fig. 1B). Thus the analytic equipment used to generate metabolomic data in this study yielded consistent results when operated at different and distinct times.
Fig. 1.
Analysis of technical reproducibility of gas chromatography-time of flight (GC-TOF) runs. Two GC-TOF analyses were performed at separate times (first and second analyses) on identical 45-day urine samples from both jck and wild-type mice and processed in a similar manner (see materials and methods). The intensity measurements from the first and second analyses for each mouse were strongly positively correlated. In A, a strong correlation (r2 = 0.91) between intensity measurements of each metabolite for 1 representative mouse is shown. The percent change in mean intensity values of each metabolite between replicate GC-TOF analyses of 45-day urine was within 10% for most metabolites (B).
Urinary Metabolome Distinguishes Cystic from Control Animals
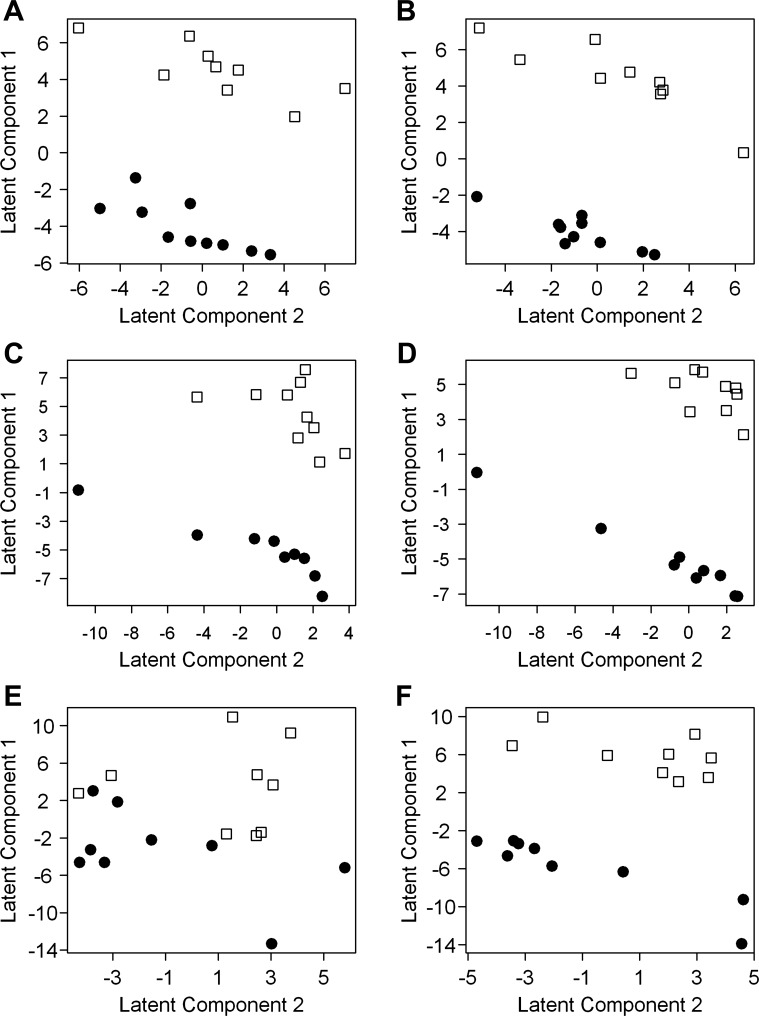
To determine whether the urine metabolome as represented by the metabolites identified in this study could discriminate the cystic (jck) mice from their wild-type counterparts, we performed PLS analysis. Because body weight (in g) differed significantly between genotypes at 64 days (means ± SD: wild-type = 21.6 ± 1.3, jck = 19.5 ± 1.15, P = 0.002), but not at 26 days (wild-type = 13.0 ± 1.9, jck = 13.3.40 ± 1.6, P = 0.77) or 45 days (wild-type = 19.0 ± 1.6, jck = 13.3 ± 1.6, P = 0.35), we conducted the PLS analysis both with and without adjusting for body weight. We used PLS to reduce the 143 spectral peaks, each representing a metabolite, to a smaller number of latent components that distinguished jck and wild-type mice for urine collected at 26, 45, and 64 days, separately. We then determined which peaks were most influential in separating jck and wild-type mice as possible biomarkers.
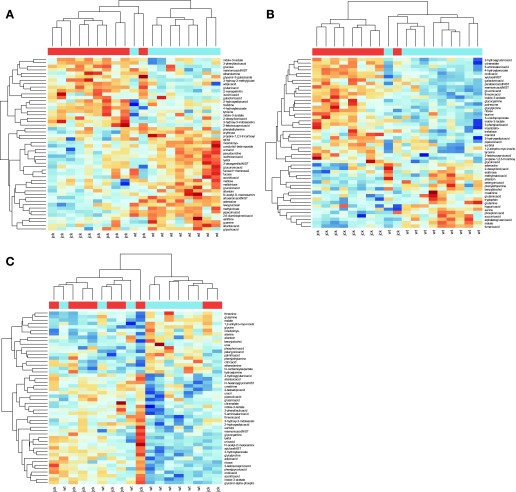
Using body weight-adjusted intensities, the metabolome from the 26-day-old mice showed significant discriminating power between the two genotypes which persisted until 45 days. This result was surprising because both 26- and 45-day-old mice exhibit normal renal function as historically measured by blood urea nitrogen (BUN) (45) and as defined by normal reference laboratory values from Research Animal Resources at the University of Minnesota (http://www.ahc.umn.edu/rar/refvalues.html). At 26 and 45 days, metabolite intensities tended to cluster according to genotype, suggesting metabolic differences between genotypes and similarities of metabolic profiles within a genotype (Fig. 2, A and B). At 64 days (Fig. 2C), clustering according to genotype was less evident. In the PLS analysis, the jck and wild-type mice were well separated by the first and second latent components at 26 and 45 days when metabolites were adjusted for body weight (Fig. 3, A and C), and these two latent components explained 96.5 and 93.2% of the variation at 26 and 45 days, respectively (Table 1). Few mice were misclassified when two latent components were used to predict genotype (Table 1). Based on leave-one-out cross-validation, the genotype of only one mouse was misclassified at 26 days and none were misclassified at 45 days (Table 1). Similarly, with the bootstrap cross-validation, the number of mice misclassified averaged less than one when first two latent components were used to discriminate between genotypes (Table 1). These results show that the urine metabolome can be reduced to a small number of components that can accurately distinguish between jck and wild-type mice at 26 and 45 days old, before the onset of significant renal failure.
Fig. 2.
Metabolite intensities of top 50 most influential metabolites for discriminating between genotypes as identified through partial least squares (PLS) regression and linear discriminant analysis (LDA). The bar at the top of the figure indicates the genotype of each individual [blue: wild-type (wt); red: jck]. Blue hues indicate higher intensities, and red hues indicate lower intensities. At 26 (A) and 45 days (B), metabolite intensities clustered according to genotype, suggesting metabolic differences between genotypes and similarities within genotypes. At 64 days (C), clustering according to genotype was less evident. Clusters were identified through hierarchical clustering (30) using Euclidean distances between body weight-adjusted intensity measurements from each mouse.
Fig. 3.
PLS graphs show separation between jck and wild-type mice. Scores of each mouse on first and second latent components are identified through PLS regression using body weight-adjusted intensity measurements at 26 (A), 45 (C), and 64 days (E) and intensity measurements that were not body weight adjusted (B: 26 days; D: 45 days; F: 64 days). Each data point represents 1 animal. Cystic (jck; ●) and wild-type (□) phenotypes were separable by urinary metabolomic analysis at 26 and 45 days regardless of adjustment for body weight. At 64 days, cystic and wild-type phenotypes were not separable with PLS using body weight-adjusted intensities but could be separated when intensities were not body weight adjusted.
Table 1.
Misclassification rates of leave-one-out and bootstrap cross-validation for determining the number of latent components for discriminating between jck and wild-type mice
| 26 Days |
45 Days |
64 Days |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of Latent Components | Leave One Out | Bootstrap | %Variation Explained | Leave One Out | Bootstrap | %Variation Explained | Leave One Out | Bootstrap | %Variation Explained |
| 1 | 1 | 0.66 | 90.9 | 1 | 0.64 | 84.9 | 7 | 2.62 | 38.7 |
| 2 | 1 | 0.54 | 96.5 | 0 | 0.54 | 93.2 | 11 | 3.52 | 50.4 |
| 3 | 1 | 0.56 | 98.4 | 0 | 0.64 | 94.4 | 11 | 3.78 | 53.3 |
| 4 | 1 | 0.60 | 99.3 | 4 | 1.08 | 94.8 | 11 | 3.82 | 54.0 |
| 5 | 1 | 0.58 | 99.5 | 5 | 1.30 | 94.9 | 11 | 3.84 | 54.2 |
| 6 | 1 | 0.60 | 99.5 | 7 | 1.50 | 95.0 | 11 | 3.90 | 54.2 |
| 7 | 1 | 0.62 | 99.5 | 7 | 1.52 | 95.0 | 11 | 3.90 | 54.3 |
| 8 | − | − | − | 9 | 1.50 | 95.0 | 13 | 3.92 | 54.3 |
A total of 19 mice were used at 26 and 45 days; 18 mice were used at 64 days. Partial least squares (PLS) was used to identify latent components for 143 body weight-adjusted metabolite intensity measurements. Leave-one-out cross-validation values (Leave One Out) are the number of mice whose genotype was misclassified by linear discriminant analysis (LDA) using k latent components for metabolite intensity measurements at 26, 45, and 64 days. Bootstrap cross-validation values (Bootstrap) are the mean number of mice misclassified using k latent components of metabolite intensity measurements at 26, 45, and 64 days. Percent (%) variation explained is the percentage of total variation in the data that explains the portion of between-genotype variation by the latent components. Collinear variables prevented fitting (−) 8 latent components at 26 days.
At 64 days, however, when intensities were adjusted for body weight, the jck and wild-type mice were not as well separated by the first and second latent components (Fig. 3E), and the clustering of metabolite intensities according to genotype was less evident than at 26 and 45 days (Fig. 2C). Regardless of the number of latent components considered, the misclassification rates determined through either leave-one-out cross-validation or bootstrap cross-validation were high and about half of the variation between genotypes was not accounted for by the latent components (Table 1). The high misclassification rates indicate that the metabolome at 64 days was not effective at discriminating between the genotypes, when variation due to differences in body weight between the genotypes was removed.
Differences in body weight between the two genotypes at 64 days appear to have been a confounding factor in the PLS analysis. Because wild-type mice were significantly heavier than jck mice at 64 days, adjusting for body weight could have obscured variation in metabolite intensity resulting from genotype differences. To assess the effect of adjusting for body weight on the results, we recalculated the PLS on intensities that were not body weight adjusted. In contrast to the body weight-adjusted results, the PLS analysis without body weight adjustment yielded good separation of the genotypes for all three time periods, including 64 days (Fig. 3, B, D, and F). At 26 and 45 days, body weight did not differ between genotypes and the results of the PLS analyses were nearly identical (Fig. 3, A–D). However, at 64 days, body weight did differ significantly between the genotypes and the analytic results differed between the body weight-adjusted and unadjusted analyses (Fig. 3, E and F), indicating that the differences in body weight resulted in different metabolomic profiles and caused, in part, separation between genotypes. As shown in Table 1, only about half of the variation was explained by the latent components at 64 days when metabolites were adjusted for body weight, suggesting that there are other differential factors between the genotypes, most likely body weight but also perhaps kidney function, which explain the remaining variation in the data.
Urine Metabolome Identifies Biochemical Pathways Which are Significantly Altered in Cystic Mice
The strength of metabolomics lies in the ability of this technique to give a real-time assessment of metabolomic processes which are occurring in the organism, such that metabolic derangement, as that which occurs in cystic disease, can be evident by pathway analysis. To a first approximation, we hypothesize that measurement of urine metabolites mirrors tissue processes; thus it would be expected that the urine metabolome will show evidence, at least at a basic level, of alteration in those metabolomic pathways associated with cell growth (such as energy utilization) and apoptosis. For this analysis, we focused on 26-day samples where there was minimal variation in body weight and the animals had normal serum creatinines when measured in two separate experiments (Ref. 45 and data not shown).
The KEGG biochemical pathways that were significantly altered between jck and wild-type mice were identified with functional score analysis. By using the median of the squared t-statistics as the functional score, the analysis focused on pathways consisting of metabolites with intensities that differed between genotypes regardless of the direction of the difference. This is important, since defect(s) in a particular pathway can be identified by the direction of change of the metabolites surrounding the defective enzyme; for example, a decrease in metabolites or enzymes downstream of pyruvate and an increase upstream can point to a defect in glycolysis (36). However, when individual metabolites are examined for suitability as biomarkers, those which are increased are of more interest (see next section).
Seven pathways containing at least three metabolites differed significantly between genotypes (Table 2 and supplemental data; all supplementary material for this article is available on the journal web site). In four pathways (purine metabolism; amino, sugar, and nucleotide sugar metabolism; pentose and gluconuronate interconversions; and pentose phosphate pathway), most of the metabolites had higher intensities in 26-day jck mice. In two pathways (trypthophan metabolism and glucosinolate metabolism), more metabolites had lower intensities in the jck mice. Interestingly, equal numbers of metabolites were up- and downregulated in galactose metabolism. The identification of purine metabolism and galactose metabolism as differing significantly in the jck mice, despite normal renal function, is biologically highly relevant as these pathways are associated with rapid cell turnover and energy production, respectively, and could be used to mine for additional biomarkers (see below) and drug targets (see discussion).
Table 2.
Pathways that significantly (P value <0.05) differed at 26 days between jck and wild-type genotypes based on a functional score analysis
| Pathway | #Metabolites | #Sig. | #Up | #Down | #Sig. Up | #Sig. Down | P Value |
|---|---|---|---|---|---|---|---|
| Pentose phosphate pathway | 4 | 2 | 3 | 1 | 1 | 1 | 0.001 |
| Glucosinolate biosynthesis | 4 | 2 | 0 | 4 | 0 | 2 | 0.002 |
| Amino, sugar, and nucleotide sugar metabolism | 7 | 3 | 6 | 1 | 2 | 1 | 0.008 |
| Purine metabolism | 9 | 2 | 7 | 2 | 2 | 0 | 0.010 |
| Pentose and glucuronate interconversions | 7 | 2 | 6 | 1 | 2 | 0 | 0.011 |
| Galactose metabolism | 8 | 3 | 4 | 4 | 1 | 2 | 0.016 |
| Tryptophan metabolism | 5 | 2 | 0 | 5 | 0 | 2 | 0.032 |
Only pathways containing at least 3 metabolites are shown. #Metabolites is the number of metabolites identified through gas chromatography/mass spectometry in each pathway; #Sig is the number of metabolites that differed significantly (permutation P value <0.05) based on a permutation t-test; #Up and #Down are the number of metabolites up- and downregulated, respectively, in the jck genotype; #Sig. Up and #Sig. Down are the number of metabolites significantly up- and downregulated, respectively, based on P values from permutation t-tests. P values were calculated from a permutation null distribution based on 10,000 permutations.
Identification of Potential Biomarkers
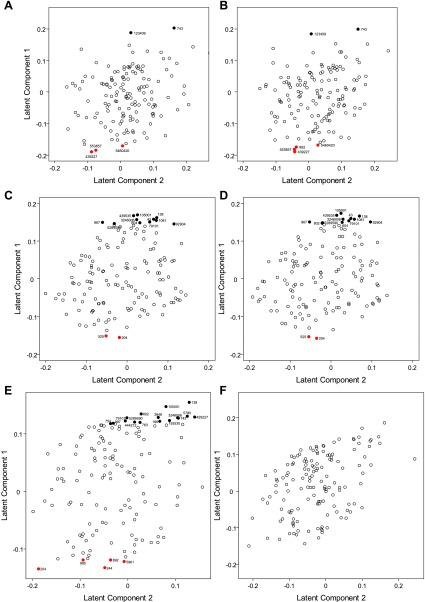
In addition to pathway analysis, we examined individual metabolites for significant differences as potential biomarkers of disease. The PLS analysis showed that the urine metabolome was effective at discriminating the two genotypes at 26 and 45 days, regardless of the adjustment for body weight. We then located the most influential metabolites for distinguishing jck and wild-type mice by looking at the loading scores of the first latent component (Fig. 4, A–D) and results of the differential analysis to determine which metabolites had significantly different intensities between the two genotypes. Using P values adjusted to account for multiple hypothesis testing, a number of metabolites were identified as significant at the adjusted P value <0.05. Six (five) metabolites differed significantly between the jck and wild-type mice at 26 days and 13 (14) at 45 days when (not) adjusted for body weight, respectively. These significantly differentiated metabolites apparently had heavy loadings in the PLS, a finding that supports these compounds as being the most promising biomarkers for PKD. With the exception of one metabolite each at 26 days [myoinositol (CID no. 892)] and 45 days [indole-3-acetate (CID no. 802)], the same metabolites were found to differ significantly between the genotypes with and without adjusting for body weight at these two time points (Fig. 4, A–D). Metabolites that differed significantly showed almost no overlap between genotypes in intensities, indicating considerable metabolic differences between cystic and wild-type animals (Fig. 5). In contrast, at 64 days, 21 metabolites differed significantly (adjusted P value <0.05) between the genotypes when the intensities were not body weight adjusted (Fig. 4F) compared with no metabolites with the body-adjusted intensities (Fig. 4E). In these 64-day samples, while some of these 21 metabolites could reflect disease-specific changes, because of the apparent confounding effect of body weight, it is unclear which result from disease-specific changes and which derive from secondary metabolic changes that are not specific to PKD, e.g., renal failure in general.
Fig. 4.
Loading diagrams of metabolites. Loadings of each metabolite on first and second latent components determined through PLS with body weight-adjusted intensities for 26 (A), 45 (C), and 64 days (E) and not body weight-adjusted intensities for 26, (B), 45 (D), and 64 days (F). Red-labeled points show metabolites that were significantly higher in jck mice than wild-type; black-labeled points indicate metabolites that were significantly lower in jck mice. The metabolites are the following: pipecolic acid (CID no. 439227), alloxonic acid NIST (CID no. 553857), methylcitrate (CID no. 5460420), myoinositol (CID no. 892), glutaric acid (CID no. 743), 2-monopalmitin (CID no. 123409), allantoic acid (CID no. 204); malate (CID no. 525), 4-hydroxybenzoate (CID no. 105001), threonic acid (CID no. 439535), 5-aminovaleric acid (CID no. 138), 2-hydroxyglutaric acid (CID no. 43), mannonic acid NIST (CID no. 3246006), citramalate (CID no. 1081); gluconic acid (CID no.604), orotic acid (CID no. 967), indole-3-lactate (CID no. 92904), glycylproline (CID no. 79101), xyulose NIST (CID no. 5289590), indole-3-acetate (CID no. 802), benzylalcohol (CID no. 244); palmitic acid (CID no. 985), sorbitol (CID no. 5780), 3-phenyllactic acid (CID no. 3848), xylulose NIST (CID no. 5289590), 2-hydroxyadipic acid (CID no. 193530), glutamine (CID no. 5961), adipic acid (CID no. 196); and aconitic acid (CID no. 444212), glycocyamine (CID no. 763), phenylpyruvic acid (CID no. 997), and glycerol-α-phosphate (CID no. 754).
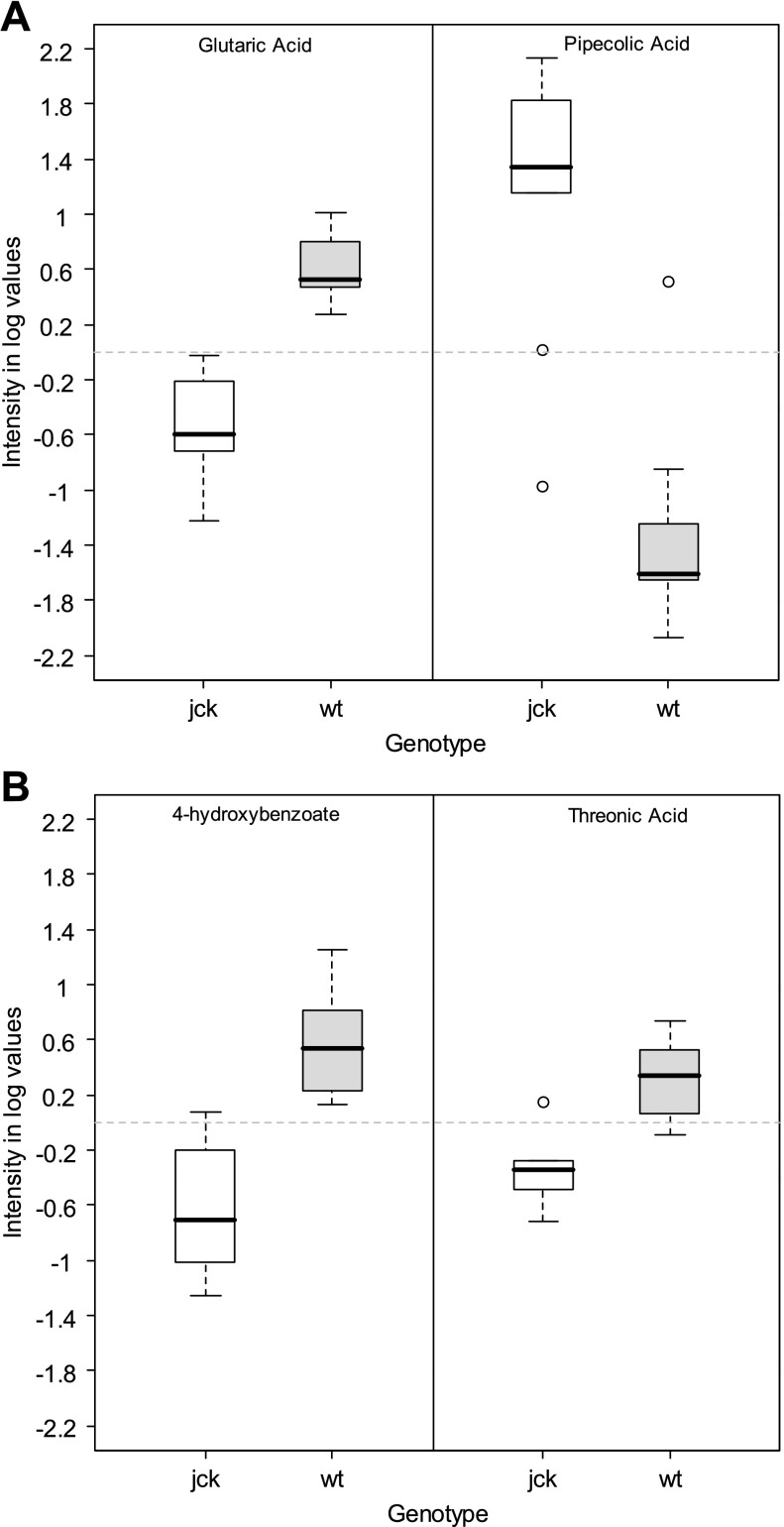
Fig. 5.
Distribution of intensity measurements. Distribution of intensity measurements [body weight adjusted and log (base 2) transformed] for glutaric acid (CID no. 743) and pipecolic acid (CID no. 439227) at 26 days and 4-hydroxybenzoate (CID no. 105001) and threonic acid (CID no. 439535) at 45 days for cystic (jck) and wild-type (wt) genotypes. Intensity measurements for these metabolites [body weight adjusted and log (base 2) transformed] had the largest significant (adjusted P < 0.05) differences between genotypes for their respective time points.
Because, similar to human ADPKD, the cystic disease that occurs in jck mice is a progressive disease that consistently reduces renal filtration function over time [as evidenced by increasing BUN reported for affected mice in a previous (45) and subsequent (data not shown) study], intensity measurements of specific metabolites from jck mice in general may be expected to be lower than in wild-type mice as a result of decreasing kidney function. Thus metabolites with higher intensity measurements in cystic mice compared with wild-type were considered to be the best choice as first-iteration (but not exclusive) candidates for biomarkers of the disease itself rather than for kidney function in general. Of the six metabolites found to differ significantly between jck and wild-type mice at 26 days, four [pipecolic acid (CID no. 439227), alloxanoic acid NIST (CID no. 553857), myoinositol (CID no. 892), methylcitrate (CID no. 5460420)] had higher mean intensities for jck mice than wild-type (adjusted P value <0.05). At 45 days, two [allantoic acid (CID no. 525) and malate (CID no. 525)] of the 13 metabolites that differed significantly between genotypes were higher in jck mice (adjusted P value <0.05). Of these metabolites, allantoic acid was also present in the pathway analysis discussed in the previous section and appeared in the highly significant pathway of purine metabolism (see supplemental data), thus emphasizing its importance in cystic disease (see below and discussion).
Of those metabolites with higher intensities in jck mice, we focused on those that are changed consistently over time as those would be the most plausible candidates for early and robust biomarkers for ultimate patient applicability. While metabolites that are decreased at later times are possibly due to changes in glomerular filtration, those that are increased are more likely to be biomarkers specific to the disease. After correcting for multiple hypothesis testing, no metabolites met the nominal threshold for statistical significance (i.e., adjusted P value <0.05) over all three time points. This result could be because the relevant biological differences between the genotypes are modest relative to the noise inherent to the analytic technology. Hence, to increase the likelihood of identifying potential biologically meaningful biomarkers, we also considered metabolites with raw P values <0.05 at increasing risk that some of the metabolites in this group are false positives. However, it is unlikely that an individual metabolite found to be significantly higher at multiple time points would be a false positive. Thus we considered the metabolites found to be higher at all the three time points for downstream biological validation experiments. We prioritized these metabolites in order of their PLS loading scores and adjusted P values. Of primary interest was allantoic acid, which was significantly greater in jck mice than in wild-type mice at all time points (Fig. 6A). We further found that intensity measurements of allantoic acid [body weight adjusted and log (base 2) transformed] could distinguish jck mice from wild-type mice with high sensitivity and specificity (Fig. 6B), particularly at 45 days. As these data strongly implicate the purine metabolic pathway as a basis for further study of urinary biomarkers in cystic disease, both murine and human, we also evaluated adenosine for sensitivity and specificity given its role in cAMP production and in the purine metabolic pathway (see supplementary data), although unlike allantoic acid it was not significantly altered at all time points (Fig. 7 and see discussion).
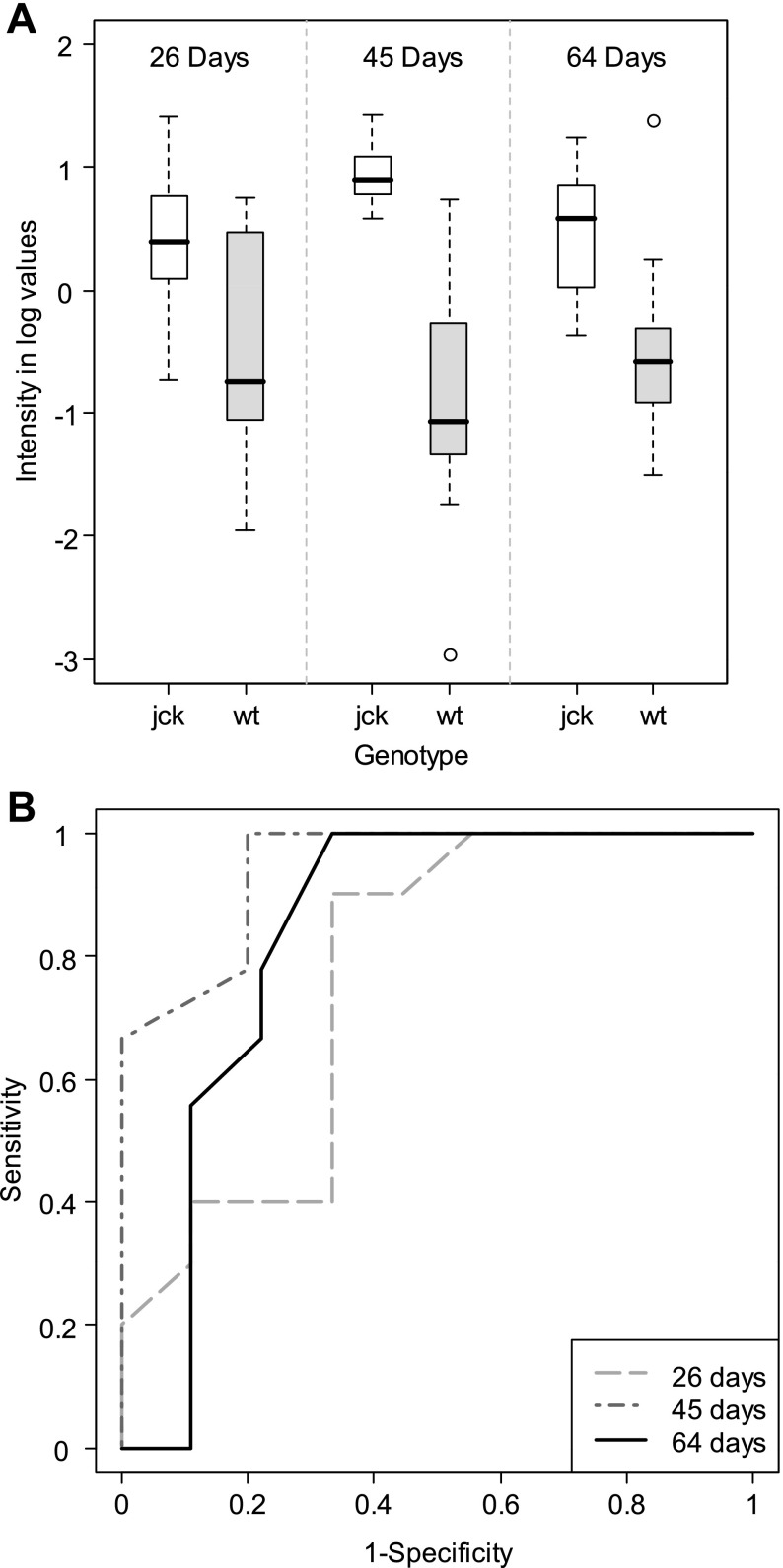
Fig. 6.
Distribution of intensity measurements and receiver-operating characteristic (ROC) curves for allantoic acid (CID no. 204), a possible biomarker for PKD in mice. Intensity measurements for allantoic acid [body weight adjusted and log (base 2) transformed] were significantly greater in cystic (jck) mice than wild-type (wt) mice at 26, 45, and 64 days (A). Intensity measurements of allantoic acid can distinguish jck mice from wild-type mice with high sensitivity and specificity (B), particularly at 45 days.
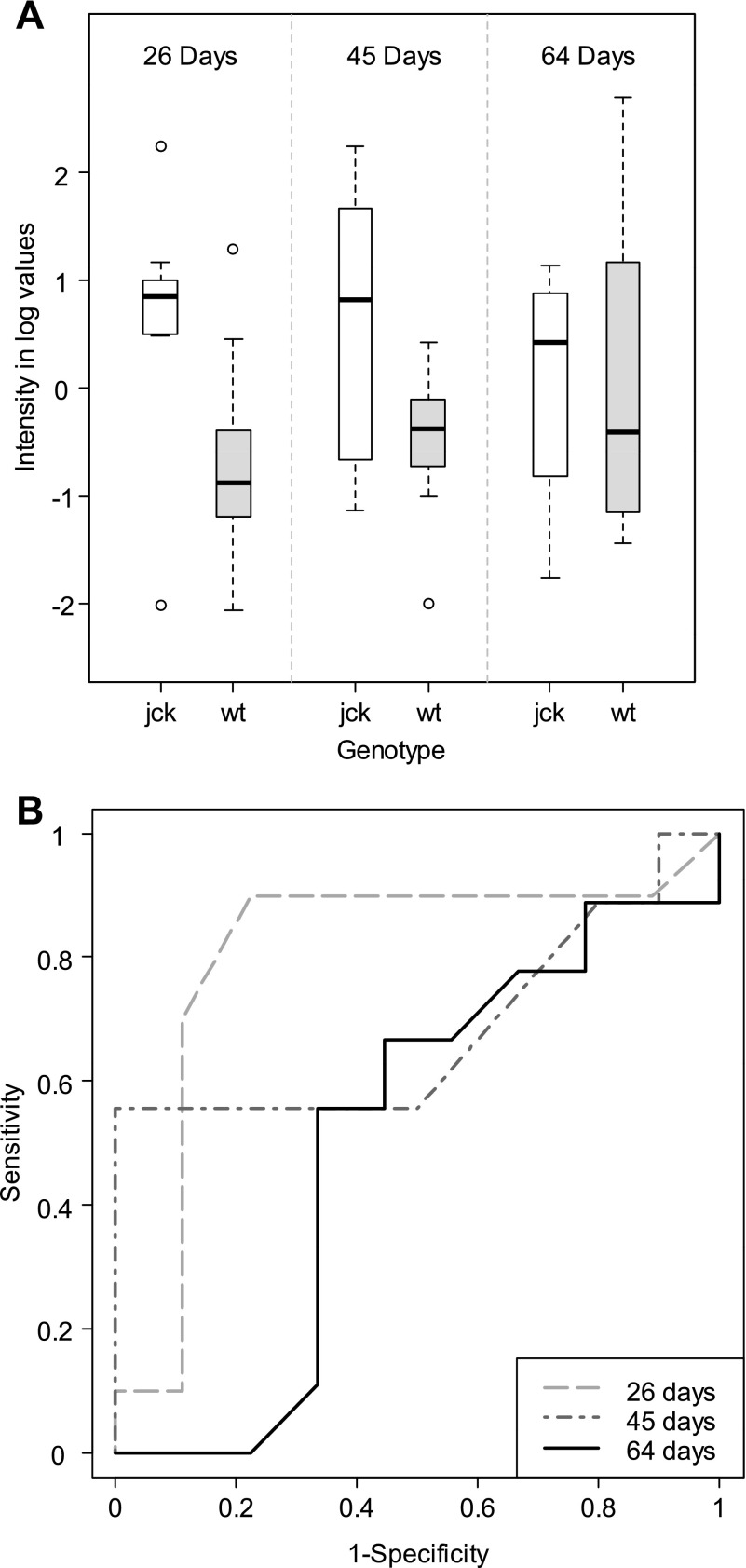
Fig. 7.
Distribution of intensity measurements and receiver-operating characteristic (ROC) curves for adenosine (CID no. 6022). Intensity measurements [body weight adjusted and log (base 2) transformed] in cystic (jck) and wild-type (wt) mice at 26, 45, and 64 days (A) and ROC curves (B) for adenosine are shown.
DISCUSSION
ADPKD is an inherited disorder with a long presymptomatic stage and an almost 100% disease penetrance by the age of 30. However, multiple ADPKD registries show significant disease progression, as measured by magnetic resonance (MR) determinations of total kidney volume, before diagnosis (8, 43). The relative frequency of screening of symptomatic family members is ∼40% and has not changed over the last four decades (49). Part of the reluctance for early diagnostic procedures has been due to lack of efficacious therapies for this disorder. However, with recently improved methods utilizing MR-based measurements of total kidney volume that demonstrate significant disease progression in a relatively short period of time (6–12 mo) (8, 43), interest in pharmacological intervention has soared. Given these technological advances, multiple molecular-targeted therapies that may improve outcomes in high-risk ADPKD individuals are being evaluated. Recently, mathematical modeling of rate of cyst growth and development in ADPKD indicates that the most accelerated phase of cystic disease progression occurs very early, before adult life (13). This suggests that time for intervention in ADPKD is before radiological evidence of renal cystic disease. Therefore, in a very short period of time, accurate and reliable diagnostic tests that do not require the presence of macrocysts will be needed in ADPKD.
The application of metabolomics is in theory ideally suited for biomarker development for early diagnosis of a variety of human diseases, since it seeks to discover and capitalizes on the metabolic derangements that occur in the body as a result of the mutated genotype which can occur well before gross phenotypic changes. The study of the metabolic profile of urine is most likely to lead to clues for pathogenesis as well as the biomarker for renal diseases, like PKD and kidney cancer, which involve derangements in tubular epithelial function (18, 21). Indeed, the study described here confirms that a mutation of a gene regulating ciliary signaling is sufficient to significantly alter the urinary metabolome and thus segregate jck from wild-type animals with an identical genetic background.
In the case of complex metabolomic analysis, the use of a mouse model has distinct advantages for proof of principle over a human study, not the least of which are convenience and ready availability of samples. Due to the similarity in background genetics between the mutant (jck) and wild-type mice used here, there are minimal confounding differences in genotype other than the mutation causing cystic kidney disease. Thus, considering minimal heterogeneity in samples, fewer subjects are required to lead to confident conclusions. Conversely, studying human ADPKD in this manner would require a much higher number of subjects due to the massive number of genetic variations present in human subjects. In addition, and more importantly, it would be considerably more difficult to guarantee that other confounding factors are evenly and randomly distributed across samples, introducing the possibility that (false) differences in metabolomic profiles would result from unknown biases present in human samples. To assess potential effects of several bias factors on metabolomic profiles and to ultimately extend the current findings to ADPKD, several studies are currently underway in our laboratories as an ancillary part of an ongoing National Institutes of Health clinical trial.
While the murine jck model of renal cystic disease possesses a mutation in a different gene (Nek8) than either PKD1 or PKD2, the mechanisms responsible for cyst formation in jck are similar to those in ADPKD, including intracellular calcium dysregulation, Wnt signaling, cAMP-activated Ras/Raf/ERK signaling, and the Akt/mammalian target of rapamycin pathway (44, 53, 55). Importantly, common to all cystic diseases, increased proliferation and apoptosis of cystic epithelia, secretory phenotype, loss of cellular polarity, and dedifferentiation exist in both jck and ADPKD (51). However, polycystin-1 and -2 expression in cilia are increased in the jck model; therefore, differences found in this study should be interpreted with caution.
Using urine from animals at day 26 of life, when there is normal kidney function and minimal renal cysts, the pathways that show significance are related to energy metabolism as well as cell proliferation. The galactose metabolism pathway (P = 0.016) is of fundamental importance for cellular energy production and proper modification of glycoproteins and glycolipids, such that deficient activity of each of the major galactose metabolic enzymes results in a human disease (reviewed in Ref. 38). While the known phenotypes of these enzyme-deficient states are not characterized by cell proliferation, galactose is a major component of glycoproteins and glycosphingolipids, which function in cell-cell communication as well as cell cycle control (38), so it is highly likely that this pathway may be important in the pathogenesis of cysts and thus should be investigated as a possible drug target.
The purine metabolism pathway (P = 0.01) has been studied in human ADPKD. Of note, most of the metabolites in this pathway are increased, including uric acid, adenosine, allantoic acid, and xanthine, suggesting high cellular turnover as well as specific alterations in signaling which play a role in ADPKD (Refs. 16 and 34 and see below). In addition, the finding that the serum and urine uric acid levels are higher in PKD patients with normal renal function (31), and that urate nephrolithiasis is present in up to 25% of ADPKD patients (35), supports the contention that these pathway data are central and relevant to cystic disease. Importantly, clinical trials of existing drugs targeting interruption of this pathway, such as allopurinol, have not been reported.
In evaluating individual metabolites for this study, we focused our efforts on those which were increased, rather than those which were decreased, in the urine of affected mice. While it is quite likely that some of those metabolites which are decreased in jck mice may in fact represent possible biomarkers, it is more likely that metabolites which are increased are true biomarkers, due to the influence of attenuated glomerular filtration on urine composition as kidney disease progresses. However, the possibility that there is specific tubular secretion of some of these increasing metabolites with progressive disease, as is seen with creatinine, although unlikely cannot be completely discounted.
We were able to identify potential biomarkers in each of the three time points as well as those that spanned several time points. Of the leading candidates of metabolite biomarkers which are increased at 26 days (Fig. 5), pipecolic acid has physiological relevance. This metabolite is a byproduct of lysine catabolism (47), and degradation of pipecolic acid (which occurs in the peroxisome) produces hydrogen peroxide, which results in the production of reactive oxygen species. Elevated pipecolic acid levels are associated with peroxisome biogenesis disorders and may result in the production of renal cysts as seen with Zellweger's syndrome. Increased allantoic acid levels indicate peroxisome activity and are indicative of oxidative stress (14), which has been postulated to contribute to cyst progression through loss of heterozygosity in specific tubular epithelial cells.
Perhaps more relevant to clinical application of the findings in this study, we identified classes of potential metabolite biomarkers which spanned several time points. Both of the potential biomarkers identified in this study are elevated during the early times when renal dysfunction was minimal. Allantoic acid and adenosine are related to purine metabolism and appear to have immediately obvious biological relevance to PKD. Allantoic acid, which was increased in all the three observed time points (Fig. 6), is the terminal step of purine nucleotide metabolism in most mammals, except humans. However, it has been known for many years that this compound can in fact be considered a read-out of adenosine levels (see below), since addition of adenosine results in increased levels of allantoic acid, both in vitro (3) and in vivo (7). In addition, despite the absence of the uricase enzyme, which converts uric acid to allantoic acid in humans, this compound has in fact been measured in human plasma and urine in numerous studies and has been proposed as a measure of oxidative stress (Refs. 14, 19, and 50 and data not shown from kidney cancer patients in our laboratory); such a finding would be consistent with current models of PKD pathogenesis and would further suggest that urinary allantoic acid (in addition to other purine pathway metabolites) should be studied as a urinary biomarker for ADPKD.
Although not significant at a nominal level in this study, urinary adenosine (raw P value = 0.0143 at 26 days; raw P value = 0.0543 at 45 days; see Fig. 7) has been proposed as a possible marker of renal injury (15), although it has not been reported in PKD. However, cAMP has been thoroughly implicated in PKD cyst progression in human disease as well as in animal models, and there are novel therapeutic approaches based on attenuating the increased levels of cAMP, which can occur primarily as a result of low intracellular calcium due to calcium channel defects seen in ADPKD. For example, cAMP is elevated in cysts and is responsible for cyst enlargement and epithelial cell proliferation (6), since maneuvers which increase cAMP enhance cyst formation as well as cell proliferation in an in vivo model (29). Elevated levels of adenosine, which we consider a potential PKD biomarker, may be indicative of either a baseline increase in cAMP synthesis, rapid turnover of cAMP, or both. In addition, individuals with PKD show increased activity of the renin-angiotensin-aldosterone system (25). Aldosterone regulates sodium transport in part by stimulating S-adenosyl-homocysteine-hydrolase. This enzyme cleaves S-adenosyl-homocysteine to produce adenosine and cysteine. Adenosine, through SAHH, is potentially involved in sodium regulation as well as cell proliferation in ADPKD (26). Interestingly, a simple quantitative assay for measuring urinary adenosine which could be immediately clinically useful is available (48). Although adenosine can also be converted into uric acid, the latter metabolite did not reach statistical significance after correcting for multiple testing at any time point; this fact may explain why urinary uric acid has not been reported to correlate with PKD. For these reasons, urinary adenosine and other potential markers of purine metabolism in humans (possibly including uric acid) are extremely promising biomarkers for which thorough biological validation is currently underway in our laboratory.
While ours is the first study to our knowledge (based on a PubMed search as of 11 August 2009) to take a metabolomic approach to identify cystic disease biomarkers in any biofluid, a recent study has used urine proteomics to identify potential biomarkers in ADPKD patients (23). These investigators utilized capillary electrophoresis and mass spectrometry to identify urine proteins and fragments thereof in young ADPKD patients. Although many of these proteins were unidentified, a majority of them were fragments of collagen type I or type III. Unlike in our study, however, small-molecule metabolites were not examined by the above-noted investigators.
In summary, we have shown, using advanced chromatographic mass spectrometry analysis of urine to identify small-molecule metabolites, that cystic animals are separable from wild-type animals (of identical genetic background) before the onset of renal insufficiency. We have determined which biochemical pathways are altered in murine cystic disease and propose several biologically plausible biomarkers which will require further validation. Given the common molecular and cellular mechanisms present in a variety of cystic disease, both murine and human, further pursuit and ultimately application of metabolomics in cystic disease have a high likelihood of yielding a simple urine diagnostic test with general clinical applicability.
GRANTS
This work was supported by National Institutes of Health Grants R01 ES13932 (to O. Fiehn) to generate BinBase; 5UO1CA86402 (Early Detection Research Network), 1R01CA135401-01A1, and 1R01DK082690-01A1 (all to R. H. Weiss); and the Medical Service of the US Department of Veterans’ Affairs (R. H. Weiss).
Supplementary Material
ACKNOWLEDGMENTS
We appreciate help by Fiehn laboratory staff members Sevini Shahbaz with GC-TOF analysis and Gert Wohlgemuth with programming and curating BinBase, Weiss laboratory member Kristine Leenders, and Sarah Moreno at Genzyme.
REFERENCES
- 1. Bae KT, Zhu F, Chapman AB, Torres VE, Grantham JJ, Guay-Woodford LM, Baumgarten DA, King BF, Jr, Wetzel LH, Kenney PJ, Brummer ME, Bennett WM, Klahr S, Meyers CM, Zhang X, Thompson PA, Miller JP. Magnetic resonance imaging evaluation of hepatic cysts in early autosomal-dominant polycystic kidney disease: the Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease cohort. Clin J Am Soc Nephrol 1: 64–69, 2006. [DOI] [PubMed] [Google Scholar]
- 2. Belibi FA, Edelstein CL. Unified ultrasonographic diagnostic criteria for polycystic kidney disease. J Am Soc Nephrol 20: 6–8, 2009. [DOI] [PubMed] [Google Scholar]
- 3. Bontemps F, Van den Berghe G, Hers HG. Evidence for a substrate cycle between AMP and adenosine in isolated hepatocytes. Proc Natl Acad Sci USA 80: 2829–2833, 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boulesteix AL. PLS dimension reduction for classification with microarray data. Stat Appl Genet Mol Biol 3: 33, 2004. [DOI] [PubMed] [Google Scholar]
- 5. Boulesteix AL. WilcoxCV: an R package for fast variable selection in cross-validation. Bioinformatics 23: 1702–1704, 2007. [DOI] [PubMed] [Google Scholar]
- 6. Calvet JP. Strategies to inhibit cyst formation in ADPKD. Clin J Am Soc Nephrol 3: 1205–1211, 2008. [DOI] [PubMed] [Google Scholar]
- 7. Cerecedo LR, Allen FW. Studies on purine metabolism. III. The fate of guanosine and adenosine in the dog. J Biol Chem 107: 421–424, 1934. [Google Scholar]
- 8. Chapman AB. Approaches to testing new treatments in autosomal dominant polycystic kidney disease: insights from the CRISP and HALT-PKD studies. Clin J Am Soc Nephrol 3: 1197–1204, 2008. [DOI] [PubMed] [Google Scholar]
- 9. Chapman AB, Torres VE, Grantham JJ, Shoaf SE, Ouyang JJ, Czerweic FS. A phase IIB pilot study of the safety and efficacy of tolvaptan, a vasopressin V2 receptor antagonist (V2RA), in patients with ADPKD (Abstract). J Am Soc Nephrol 16: 68A, 2005.15537870 [Google Scholar]
- 10. Dettmer K, Hammock BD. Metabolomics—a new exciting field within the “omics” sciences. Environ Health Perspect 112: A396–A397, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fiehn O, Wohlgemuth G, Scholz M. Setup and annotation of metabolomic experiments by integrating biological and mass spectrometric metadata. Proc Lect Notes Bioinformatics 3615: 224–239, 2005. [Google Scholar]
- 12. Fiehn O, Wohlgemuth G, Scholz M, Kind T, Lee dY, Lu Y, Moon S, Nikolau B. Quality control for plant metabolomics: reporting MSI-compliant studies. Plant J 53: 691–704, 2008. [DOI] [PubMed] [Google Scholar]
- 13. Grantham JJ, Cook LT, Torres VE, Bost JE, Chapman AB, Harris PC, Guay-Woodford LM, Bae KT. Determinants of renal volume in autosomal-dominant polycystic kidney disease. Kidney Int 73: 108–116, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gruber J, Tang SY, Jenner AM, Mudway I, Blomberg A, Behndig A, Kasiman K, Lee CY, Seet RC, Zhang W, Chen C, Kelly F, Halliwell B. Allantoin in human plasma, serum and nasal lining fluids as a biomarker of oxidative stress; avoiding artifacts and establishing real in vivo concentrations. Antioxid Redox Signal 2009. [DOI] [PubMed] [Google Scholar]
- 15. Heyne N, Benohr P, Muhlbauer B, Delabar U, Risler T, Osswald H. Regulation of renal adenosine excretion in humans—role of sodium and fluid homeostasis. Nephrol Dial Transplant 19: 2737–2741, 2004. [DOI] [PubMed] [Google Scholar]
- 16. Ibraghimov-Beskrovnaya O, Bukanov N. Polycystic kidney diseases: from molecular discoveries to targeted therapeutic strategies. Cell Mol Life Sci 65: 605–619, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 38: D355–D360, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim K, Aronov P, Zakharkin SO, Anderson D, Perroud B, Thompson IM, Weiss RH. Urine metabolomics analysis for kidney cancer detection and biomarker discovery. Mol Cell Proteomics 8: 558–570, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim KM, Henderson GN, Frye RF, Galloway CD, Brown NJ, Segal MS, Imaram W, Angerhofer A, Johnson RJ. Simultaneous determination of uric acid metabolites allantoin, 6-aminouracil, and triuret in human urine using liquid chromatography-mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 877: 65–70, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim YS, Maruvada P, Milner JA. Metabolomics in biomarker discovery: future uses for cancer prevention. Future Oncol 4: 93–102, 2008. [DOI] [PubMed] [Google Scholar]
- 21. Kind T, Tolstikov V, Fiehn O, Weiss RH. A comprehensive urinary metabolomic approach for identifying kidney cancer. Anal Biochem 363: 185–195, 2007. [DOI] [PubMed] [Google Scholar]
- 23. Kistler AD, Mischak H, Poster D, Dakna M, Wuthrich RP, Serra AL. Identification of a unique urinary biomarker profile in patients with autosomal dominant polycystic kidney disease. Kidney Int 76: 89–96, 2009. [DOI] [PubMed] [Google Scholar]
- 24. Lanz C, Patterson AD, Slavik J, Krausz KW, Ledermann M, Gonzalez FJ, Idle JR. Radiation metabolomics. 3. Biomarker discovery in the urine of gamma-irradiated rats using a simplified metabolomics protocol of gas chromatography-mass spectrometry combined with random forests machine learning algorithm. Radiat Res 172: 198–212, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lawson CR, Doulton TW, MacGregor GA. Autosomal dominant polycystic kidney disease: role of the renin-angiotensin system in raised blood pressure in progression of renal and cardiovascular disease. J Renin Angiotensin Aldosterone Syst 7: 139–145, 2006. [DOI] [PubMed] [Google Scholar]
- 26. Leal JF, Ferrer I, Blanco-Aparicio C, Hernandez-Losa J, Ramon YC, Carnero A, Lleonart ME. S-adenosylhomocysteine hydrolase downregulation contributes to tumorigenesis. Carcinogenesis 29: 2089–2095, 2008. [DOI] [PubMed] [Google Scholar]
- 27. Lewis GD, Asnani A, Gerszten RE. Application of metabolomics to cardiovascular biomarker and pathway discovery. J Am Coll Cardiol 52: 117–123, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maletic-Savatic M, Vingara LK, Manganas LN, Li Y, Zhang S, Sierra A, Hazel R, Smith D, Wagshul ME, Henn F, Krupp L, Enikolopov G, Benveniste H, Djuric PM, Pelczer I. Metabolomics of neural progenitor cells: a novel approach to biomarker discovery. Cold Spring Harb Symp Quant Biol 73: 389–401, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mangoo-Karim R, Uchic ME, Grant M, Shumate WA, Calvet JP, Park CH, Grantham JJ. Renal epithelial fluid secretion and cyst growth: the role of cyclic AMP. FASEB J 3: 2629–2632, 1989. [DOI] [PubMed] [Google Scholar]
- 30. Mardia KV, Kent JT, Bibby JM. Multivariate Analysis. Amsterdam: Academic, 1979. [Google Scholar]
- 31. Mejias E, Navas J, Lluberes R, Martinez-Maldonado M. Hyperuricemia, gout, and autosomal dominant polycystic kidney disease. Am J Med Sci 297: 145–148, 1989. [DOI] [PubMed] [Google Scholar]
- 32. Moreno S, Ibraghimov-Beskrovnaya O, Bukanov NO. Serum and urinary biomarker signatures for rapid preclinical in vivo assessment of CDK inhibition as a therapeutic approach for PKD. Cell Cycle 7: 1856–1864, 2008. [DOI] [PubMed] [Google Scholar]
- 33. Nguyen DV, Rocke DM. Partial least squares proportional hazard regression for application to DNA microarray survival data. Bioinformatics 18: 1625–1632, 2002. [DOI] [PubMed] [Google Scholar]
- 34. Park JY, Schutzer WE, Lindsley JN, Bagby SP, Oyama TT, Anderson S, Weiss RH. p21 is decreased in polycystic kidney disease and leads to increased epithelial cell cycle progression: roscovitine augments p21 levels. BMC Nephrol 8: 12, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pavlidis P, Qin J, Arango V, Mann JJ, Sibille E. Using the gene ontology for microarray data mining: a comparison of methods and application to age effects in human prefrontal cortex. Neurochem Res 29: 1213–1222, 2004. [DOI] [PubMed] [Google Scholar]
- 36. Perroud B, Ishimaru T, Borowsky AD, Weiss RH. Grade-dependent proteomic characterization of kidney cancer. Mol Cell Proteomics 8: 971–985, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Perroud B, Lee J, Valkova N, Dhirapong A, Lin PY, Fiehn O, Kultz D, Weiss RH. Pathway analysis of kidney cancer using proteomics and metabolic profiling. Mol Cancer 5: 64, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Petry KG, Reichardt JK. The fundamental importance of human galactose metabolism: lessons from genetics and biochemistry. Trends Genet 14: 98–102, 1998. [DOI] [PubMed] [Google Scholar]
- 39. Ravine D, Gibson RN, Walker RG, Sheffield LJ, Kincaid-Smith P, Danks DM. Evaluation of ultrasonographic diagnostic criteria for autosomal dominant polycystic kidney disease 1. Lancet 343: 824–827, 1994. [DOI] [PubMed] [Google Scholar]
- 40. Rossetti S, Consugar MB, Chapman AB, Torres VE, Guay-Woodford LM, Grantham JJ, Bennett WM, Meyers CM, Walker DL, Bae K, Zhang QJ, Thompson PA, Miller JP, Harris PC. Comprehensive molecular diagnostics in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 18: 2143–2160, 2007. [DOI] [PubMed] [Google Scholar]
- 41. Schmidt C. Metabolomics takes its place as latest up-and-coming “omic” science. J Natl Cancer Inst 96: 732–734, 2004. [DOI] [PubMed] [Google Scholar]
- 42. Scholz M, Fiehn O. SetupX—a public study design database for metabolomic projects. Pacific Symp Biocomp 12: 169–180, 2007. [PubMed] [Google Scholar]
- 43. Serra AL, Kistler AD, Poster D, Struker M, Wuthrich RP, Weishaupt D, Tschirch F. Clinical proof-of-concept trial to assess the therapeutic effect of sirolimus in patients with autosomal dominant polycystic kidney disease: SUISSE ADPKD study. BMC Nephrol 8: 13, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Simons M, Gloy J, Ganner A, Bullerkotte A, Bashkurov M, Kronig C, Schermer B, Benzing T, Cabello OA, Jenny A, Mlodzik M, Polok B, Driever W, Obara T, Walz G. Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nat Genet 37: 537–543, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smith LA, Bukanov NO, Husson H, Russo RJ, Barry TC, Taylor AL, Beier DR, Ibraghimov-Beskrovnaya O. Development of polycystic kidney disease in juvenile cystic kidney mice: insights into pathogenesis, ciliary abnormalities, and common features with human disease. J Am Soc Nephrol 17: 2821–2831, 2006. [DOI] [PubMed] [Google Scholar]
- 46. Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457: 910–914, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 47. Steinberg SJ, Dodt G, Raymond GV, Braverman NE, Moser AB, Moser HW. Peroxisome biogenesis disorders. Biochim Biophys Acta 1763: 1733–1748, 2006. [DOI] [PubMed] [Google Scholar]
- 48. Taniai H, Sumi S, Ito T, Ueta A, Ohkubo Y, Togari H. A simple quantitative assay for urinary adenosine using column-switching high-performance liquid chromatography. Tohoku J Exp Med 208: 57–63, 2006. [DOI] [PubMed] [Google Scholar]
- 49. Taylor M, Johnson AM, Tison M, Fain P, Schrier RW. Earlier diagnosis of autosomal dominant polycystic kidney disease: importance of family history and implications for cardiovascular and renal complications. Am J Kidney Dis 46: 415–423, 2005. [DOI] [PubMed] [Google Scholar]
- 50. Torres VE. New insights into polycystic kidney disease and its treatment. Curr Opin Nephrol Hypertens 7: 159–169, 1998. [DOI] [PubMed] [Google Scholar]
- 51. Torres VE, Harris PC. Polycystic kidney disease: genes, proteins, animal models, disease mechanisms and therapeutic opportunities. J Intern Med 261: 17–31, 2007. [DOI] [PubMed] [Google Scholar]
- 52. Valkova N, Yunis R, Mak SK, Kang K, Kultz D. Nek8 mutation causes overexpression of galectin-1, sorcin, and vimentin and accumulation of the major urinary protein in renal cysts of jck mice. Mol Cell Proteomics 4: 1009–1018, 2005. [DOI] [PubMed] [Google Scholar]
- 53. Wahl PR, Le HM, Vogetseder A, Arcaro A, Starke A, Waeckerle-Men Y, Serra AL, Wuthrich RP. Mitotic activation of Akt signalling pathway in Han:SPRD rats with polycystic kidney disease. Nephrology (Carlton) 12: 357–363, 2007. [DOI] [PubMed] [Google Scholar]
- 54. Westfall PH, Young SS. Resampling-Based Multiple Testing: Examples and Methods for P-Value Adjustment. York: Wiley, 1993. [Google Scholar]
- 55. Yamaguchi T, Wallace DP, Magenheimer BS, Hempson SJ, Grantham JJ, Calvet JP. Calcium restriction allows cAMP activation of the B-Raf/ERK pathway, switching cells to a cAMP-dependent growth-stimulated phenotype. J Biol Chem 279: 40419–40430, 2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.