Abstract
The genetic code is implemented by aminoacyl-tRNA synthetases (aaRS). These twenty enzymes are divided into two classes that, despite performing same functions, have nothing common in structure. The mystery of this striking partition of aaRSs might have been concealed in their sterically complementary modes of tRNA recognition that, as we have found recently, protect the tRNAs with complementary anticodons from confusion in translation. This finding implies that, in the beginning, life increased its coding repertoire by the pairs of complementary codons (rather than one-by-one) and used both complementary strands of genes as templates for translation. The class I and class II aaRSs may represent one of the most important examples of such primordial sence-antisence (SAS) coding (Rodin and Ohno, 1995). In this report, we address the issue of SAS coding in a wider scope. We suggest a variety of advantages that such coding would have had in exploring a wider sequence space before translation became highly specific. In particular, we confirm that in Achylia klebsiana a single gene might have originally coded for an HSP70 chaperonin (class II aaRS homolog) and an NAD-specific GDH-like enzyme (class I aaRS homolog) via its sense and antisense strands. Thus, in contrast to the conclusions in (Williams et al., 2009), this could indeed be a “Rosetta stone” (eroded somewhat, though) gene for the SAS origin of the two aaRS classes (Carter and Duax, 2002).
Keywords: RNA world, genetic code, sense-antisense coding, Aminoacyl-tRNA synthetases, NAD-Gdh, HSP70, Rosetta stone
1 Introduction
Watson-Crick pairing of complementary nucleotides is the most essential feature of life. Obviously, the replication process is based on it, and so is transcription. On a more subtle level, the very paradigm of pre-protein RNA life is also founded on this pairing. Indeed, the G-C and A-U complementarities determine folding of each transcript in a 2D structure that shapes specific catalytic and other functional centers; therefore, in the “RNA world” this folding would actually unfold the genetic information contained in genes --- thus, it appears that the replication/transcription and coding were the two sides of the same coin for riboorganisms.
The emergence of the genetic code (Table 1) led to the separation of the information carriers (nucleic acids) from the functional entities (proteins), thus radically changing the dynamics of molecular evolution. Nevertheless, complementary base pairing remained fundamentally important. The code is not implemented directly, but via its adaptors, transfer RNAs (tRNA), by means of specific enzymes, the aminoacyl-tRNA synthetases (aaRS). It is the aaRSs that de facto activate amino acids for protein synthesis and implement the genetic code by recognizing and attaching a proper amino acid to the CCA3’ terminus of the acceptor stem of tRNAs with corresponding anticodon(s). Specificity of the attachment is determined mostly by the “determinator base” and first three base pairs of the acceptor, adding up to what have been defined as “RNA operational code” (Schimmel et al., 1993). Intriguingly, the aaRSs are divided into two classes with sterically complementary modes of tRNA recognition – from the minor (class I) and major (class II) groove sides of the tRNA acceptor stem (Fig. 1).
Table 1.
The conventional representation of the genetic code table with yellow and blue colors marking two modes of tRNA recognition by aaRSs – from the minor and major groove sides of the acceptor stem, respectively.
| 1 | 2 | 3 | |||
|---|---|---|---|---|---|
| U | C | A | G | ||
| U | UUU Phe | UCU Ser | UAU Tyr | UGU Cys | U |
| U | UUC Phe | UCC Ser | UAC Tyr | UGC Cys | C |
| U | UUA Leu | UCA Ser | UAA stop | UGA stop | A |
| U | UUG Leu | UCG Ser | UAG stop | UGG Trp | G |
| C | CUU Leu | CCU Pro | CAU His | CGU Arg | U |
| C | CUC Leu | CCC Pro | CAC His | CGC Arg | C |
| C | CUA Leu | CCA Pro | CAA Gln | CGA Arg | A |
| C | CUG Leu | CCG Pro | CAG Gln | CGG Arg | G |
| A | AUU Ile | ACU Thr | AAU Asn | AGU Ser | U |
| A | AUC Ile | ACC Thr | AAC Asn | AGC Ser | C |
| A | AUA Ile | ACA Thr | AAA Lys | AGA Ser/Gly | A |
| A | AUG Met | ACG Thr | AAG Lys | AGG Ser/Gly | G |
| G | GUU Val | GCU Ala | GAU Asp | GGU Gly | U |
| G | GUC Val | GCC Ala | GAC Asp | GGC Gly | C |
| G | GUA Val | GCA Ala | GAA Glu | GGA Gly | A |
| G | GUG Val | GCG Ala | GAG Glu | GGG Gly | G |
Lys is colored in lighter shade of blue in order to indicate the fact that some archaebacteria use class I synthetases for this amino acid (Ibba et al., 1997). Stop codons are colored in yellow because the known cases of their “capture” by amino acids are mostly from class I (Knight et al., 2001). Codons AGG and AGA are assigned to blue Ser or Gly, as they are in mitochondria (ibid.) Three aromatic amino acids, Phe, Tyr and Trp, with their mode of tRNA aminoacylation contradicting the class aaRS membership, are italicized (from Rodin et al., 2009).
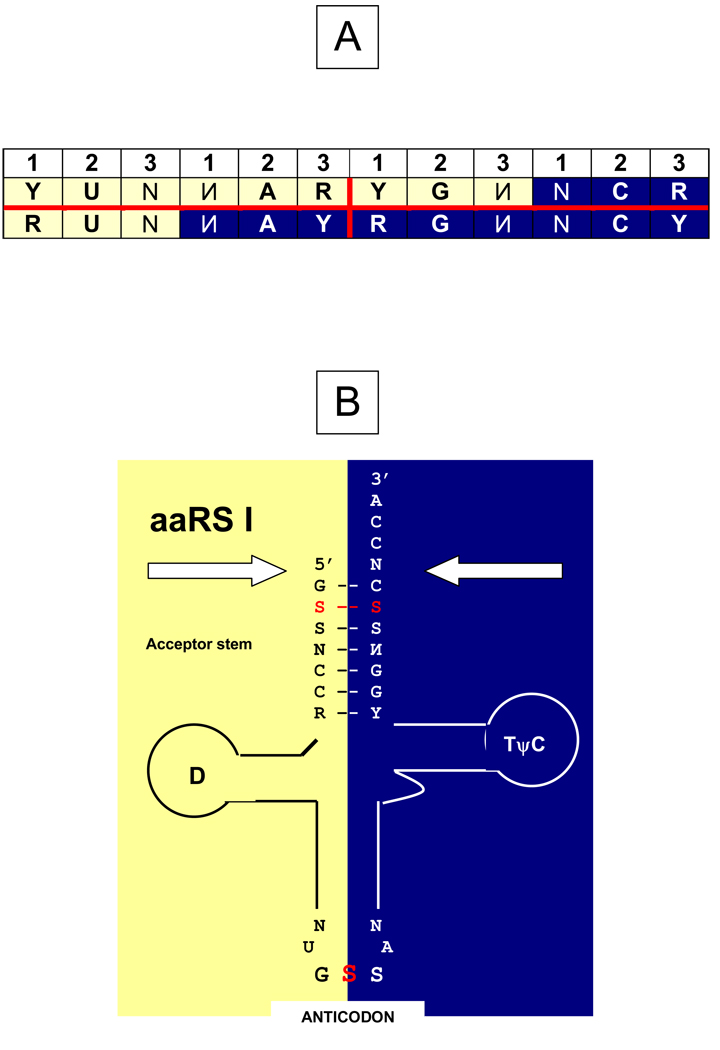
Figure 1.
The subcode for two sterically mirror modes of tRNA recognition by aminoacyl-tRNA synthetases.
A: The condensed rearranged representation of the genetic code (Table 1), in which complementary codons are put vis-à-vis each other. The yin-yang-like pattern of the representation reveals the latent sub-code for the two modes of tRNA aminoacylation: (1) If the complementary codons contain YY vs. RR at the second and adjacent (either first or third) positions, their aaRSs recognize the tRNA acceptor from the same side of the groove, namely: minor (yellow) for 5’NAR3’ – 5’YUN3’ pairs, or major (blue) for 5’RGN3’ – 5’NCY3’ pairs; (2) If these positions are occupied by RY and YR, the modes of tRNA recognition are different, namely: minor (yellow) 5’YGN3’ vs. major (blue) 5’NCR3’ and major (blue) 5’NAY3’ vs. minor (yellow) 5’RUN3’. Precisely same rules are applicable to pairs of complementary anticodons. Taking into account the anticodon flanking 5’U and R3’ nucleotides allows us to show that in fact this sub-code minimizes a risk of confusion of tRNAs with complementary anticodons by aaRS, no matter are the latter real proteins or their putative ribozymic precursors.
Other symbols: N and complementary и denote all four nucleotides; R, purine (G or A); Y, pyrimidine (C or U). For details, see (Rodin and Rodin, 2006b, 2008).
B: The tRNA cloverleaf with complementary halves that are colored yellow (5’ half) and blue (3’ half), in accordance with the sub-code (A). Arrows show the two sides from which the putative ribozymic precursors of class I and class II p-aaRSs approached the proto-tRNAs. The 2nd bases of triplets in the acceptor stem (marked red) and the anticodons show the concerted dual complementarity that may point to a common ancestor of the two codes they represent, the operational and classic ones, respectively (Rodin et al., 1996, 2009; Rodin and Rodin, 2006a). The 3’ strand of the acceptor arm represents the presumable ancestral palindrome self-templating and duplication of which readily form the extant tRNA cloverleaf (Rodin et al., 2009; see Rodin and Rodin, 2009 for details).
Avoidance of the proverbial chicken-or-egg paradox has led to the prevailing assumption that the coding system as a whole (with all tRNAs and aaRSs) could have originated solely in a complex, metabolically and catalytically rich, RNA world (Crick, 1968; Orgel, 1968; Szathmary, 1999). On the other hand, a principle of evolutionary continuity suggests that the emerging code must have inherited fundamental properties of complementary base-pairing from any preceding, protein-free RNA life (Rodin and Rodin, 2006a,b).
In particular, one would hope to find imprints of the primordial complementarity in the codon-to-aa assignment (Table 1) itself, as well as in the structure of tRNAs and aaRSs. Such imprints have indeed been observed:
In the code itself – in its complementary symmetry (Table 1) and the latent internal sub-code (Fig. 1) consistent with the existence of the two mutually complementary modes of tRNA aminoacylation (Rodin and Rodin, 2006b, 2008, Rodin et al., 2009).
In pairs of tRNAs with complementary anticodons – in the concerted complementarity of 2nd bases in their acceptor stems. This dual complementarity suggests that the operational code of tRNA aminoacylation (the one associated mostly with the acceptor stem) (Schimmel et al., 1993) and the classic genetic code per se (associated with anticodons) diverged from a common ancestor (Rodin et al., 1996). Moreover, since translation without coding is impossible, as is “foresight” evolution in general (reviewed in (Maynard Smith and Szathmary, 1995) and closely related to the notion of “retrospective coronation (Dennett 1995 )), we arrive at the hypothesis that these aspects of the contemporary code likely preceded the origin of translation (Szathmary, 1993, 1999). For example, the RNA world could use some amino acids as cofactors of ribozymes (ibid.) A closer analysis of the sub-code for two aminoacylation modes of tRNAs and possible palindromic structure of their minimal precursor strongly supports the pre-translation code-ancestor origin hypothesis (Rodin et al., 2009).
In the possible origin of two aaRS classes –their in-frame coding by complementary strands of the same primordial gene (Rodin and Ohno, 1995). This is, in fact, the central focus of the present study. First, we will consider the general issue of ancient sense-antisense (SAS) coding and how it may have contributed to the evolution of the earliest proteins. Second, we will address a more singular issue: does the HSP70 gene in Achlya klebsiana, with its presumptive NAD-specific DGH2-like complement, indeed serve, as claimed by Carter and Duax, 2002 (but recently challenged by Williams et al, 2009), as a “Rosetta stone” for sense-antisense origin of two aaRS classes? Our answer to this instructive polemic is an unequivocal Yes.
2. The hypothesis of ancient sense-antisense coding: the (dominating) pros and cons
2.1. Basic premises
There are many reasons to believe that SAS coding may have had significant selective advantages, and hence have been common at the beginning of the”ribonucleoprotein world”.
First, the codon-to-aa assignment (Table 1) is apparently non-random: similar triplets encode similar amino acids (with the third base being the least specific), thus minimizing negative effects of base substitutions. However, does the real code outrank all other (possible) codes in this respect? Since the code’s deciphering in the 60s, there was no shortage of attempts to answer that question largely by testing the codes for robustness (sensitivity) to translation errors. Remarkably, although the real code is certainly more robust than the vast majority of random ones (Freeland and Hurst, 1998), even among the codes of the same block structure and degree of degeneracy, the real code turned out to be certainly not the best (Novozhilov et al., 2007). One can propose a number of explanations for this sub-optimality (see reviewers’ comments in (Novozhilov et al., 2007); most of these apologetics invoke the event of an original frozen accident and subsequent selection for robustness to translation errors.
In our opinion, there is one further, perhaps game-changing, explanation that has not been called upon yet. The very criteria used in such comparisons are associated with how the fidelity of translation impacted protein stability. In particular, customarily the only codes chosen to perform the robustness analysis upon are those possessing the same degree of degeneracy. This choice is dictated by the fact that the third base of the codon makes the smallest contribution to the specificity of its recognition by anticodon. However, the irrationality of “foresight evolution” argues for a pre-translational origin of the code (Szathmary, 1993, 1999). Therefore, when we consider the very first (and the most crucial, actually) steps of the code’s formation, references to proteins and translation simply do not make sense. Accordingly, if the RNA life did manage to start up the code development process before translation emerged, and if we define the code “fitness” with respect to its compliance to the translation machinery, then why would one be surprised by the seeming inferiority of the real code to the possible ones more adapted to translation?
Indeed, probably long before the origin of translation, the genetic code had already fixed such basic (and, one would think, translation-ineradicable) features as the triplet codon size and the two complementary modes of tRNA aminoacylation that take into account the invariant U and A/G nucleotides flanking the anticodon from 5’ and 3’ sides, respectively (Rodin et al., 2009). If so, the translation machinery evolving to “fit” the earliest code makes no less sense than the code evolving to fit the translation. Further analyses are needed to set apart (in a more concrete fashion) these pre- and co-translational stages in the origin of the genetic code. However, there is no doubt that it was the latter, code-to-translation, co-adaptation when the 1st and 3rd codon bases have become eventually nonequivalent. Until that change, considering any constraints imposed on the SAS coding makes little sense. It thus appears that the hypothesis of a pre-translation shaping of the genetic code strengthens the hypothesis of primordial SAS coding.
Second, in-frame coding on both strands doubles the potential number of proteins in the genome – an advantage that was especially crucial for the error-prone RNA world, because frequent errors during RNA replication (transcription) imposed strong constraints on the genome growth, and translation errors likely limited proteins to molten globules with rudimentary functionality. And even though the SAS coding for the two proteins in the same frame hobbles their co-evolution, at that time, “the need to make maximum use of sequence space” (Kuhns and Joyce, 2003) might have significantly outweighed the lack of evolutionary freedom.
Although they seem substantial from a contemporary perspective, disadvantages of SAS-coding for evolution of the first proteins should not be overrated. Compared to randomized codes, by being less sensitive to mutations in flanking, 1st vs. 3rd,codon positions, the real code actually favors the SAS coding until one of the complementarily encoded proteins gains a decisive function(s) making further parallel improvements very difficult (if at all possible) to accomplish (Konechny et al., 1993: Rodin and Rodin, 2006a,b).
Moreover, the best contemporary evidence suggests that simple binary patterning of hydrophilic vs hydrophobic amino acids produces molten globules at high frequency with a rich variety of promiscuous catalytic activities (Kamtekar et al. 1993; Patel et al. 2009). Thus, at a time when selection was predominantly based on an ability to form protein secondary structures, the importance of such binary patterns, determining α-helices or β-strands, would have dominated. Further, the code in Table 1 possesses an elaborate inversion symmetry ((Zull and Smith 1990); Fig. 2) that assures the preservation of binary patterns, with inversion, on the opposite strand. Thus, gene products from opposite strands likely doubled the frequency of rudimentary activities, and served as a ‘“ feedstock” for evolution’ (Patel et al. 2009). Thus, although initially counterintuitive, sense/antisense coding appears to offer very efficient exploration of sequence space in the context of a translation system with limited fidelity, as expected for the earliest biological genes.
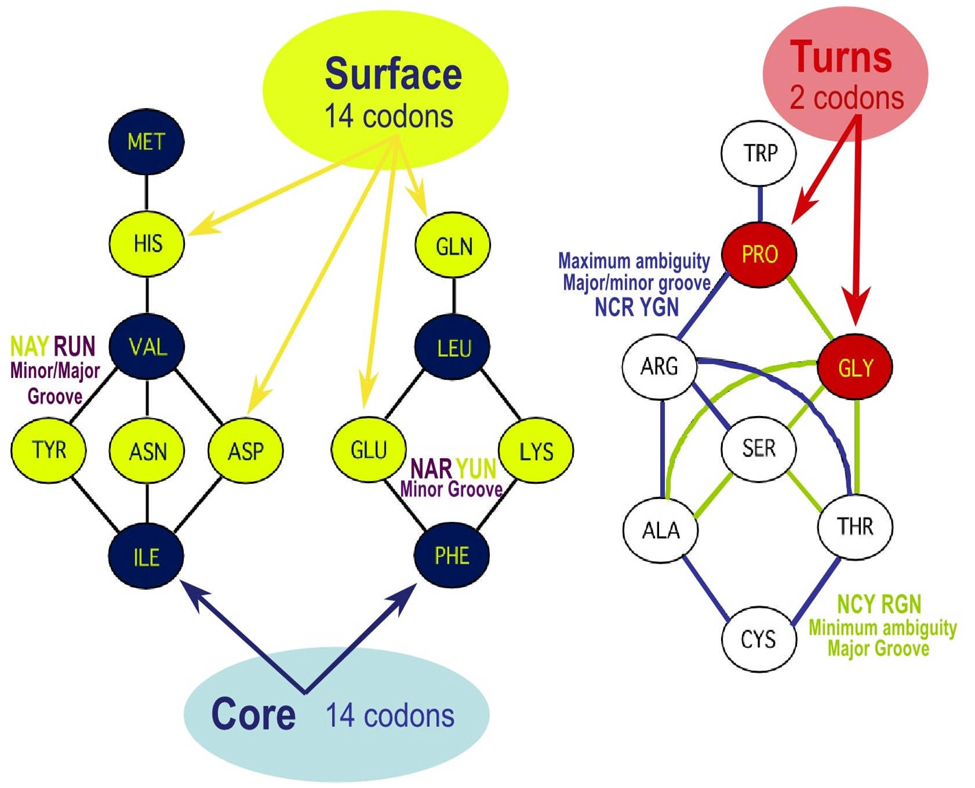
Figure 2.
Inversion symmetry in the coding properties of the genetic code (after (Zull and Smith 1990). Codons for hydrophobic “core” amino acids are invariably antisense to polar amino acids found almost exclusively on protein surfaces. This arrangement is especially suited to producing molten globular gene products from both strands of a SAS gene. Note also that Proline and Glycine share one SAS codon-anticodon pair, such that a turn specified by a Pro-Gly sequence on one strand will be preserved on the opposite strand. The three groups represent the four patterns of pairs of complementary codons (and, symmetrically, anticodons) described by Rodin and Rodin (2006b) in their discussion of their relative ambiguity in a strand-symmetric RNA world.
Third, as noted previously (Pham et al. 2007), sense/antisense coding offers the decisive advantage of linking the gene products, assuring that they are expressed together in a cell-free environment. This advantage would have been especially important for the ancestral class I and class II synthetases, whose amino acid specificities are for large nonpolar and small hydrophilic side chains, respectively, and would both have been necessary to produce molten globules.
Fourth, it is only after the 1st and 3rd codon bases have become nonequivalent that any constraints at all are imposed on SAS coding. When the first proteins appeared as relatively nonspecific, binary-patterned molten globules, it is likely that only the central codon position was crucial while the flanking 1st and 3rd ones might have been almost equal in their irrelevance(see Fig. 2). This happened long before the final shaping of the code itself, and certainly before the punctuation signs for initiation and termination of translation were established. We would like to note in this regard that perhaps it is hardly a coincidence that the starting AUG (sometimes GUG) triplet and the terminal UAR ones are complementary to each other at the central position.
Fifth, since we are talking about the very dawn of RNA + protein life, when encoded proteins and more specifically catalytic functions were only just about to emerge --- in this uninhabited protein space there was hardly any “competition” --- every RNA sequence, as well as its complement, were equally capable of serving as first templates for protein synthesis. In other words, in the beginning, it served little purpose to differentiate “+” and “ −“ RNAs into a sense (true mRNA) and antisense sequences; potentially, both could “make sense”. Above all, it is the innovative potential of this primordial SAS coding that is worthy of emphasis. Indeed, in the extant code (Table 1), mutations of the 2nd base of codons are apparently less conservative than mutations of flanking (1st and 3rd) ones, and it is the pairs of complementary codons that show the most pronounced difference between original and new amino acids (Rodin and Ohno, 1997). Furthermore, the SAS-encoded proteins would co-evolve smoothly if mutations at the flanking (1st and 3rd) codon positions were selectively equal, whereas in reality the 1st codon position is more functionally important (and, therefore, more evolutionary conservative) than the 3rd one. However, when making such judgments, we tacitly assume the pressure of the negative (purifying) selection. That only makes sense if life has already achieved something valuable to protect. We have already mentioned though that at the beginning, evolution of proteins and the coding and translation system per se was much more creative, driven by the natural selection acting not so much against as rather in favor of novelties-prone mutations (Zhu and Freeland, 2006). The remarkable feature of the SAS coding of first proteins in general, and two p-aaRSs in particular, extended the opportunity for life to experiment (by mutating the codons’ 1st positions) with one protein, and at the same time to “hold still” (due to synonymous 3rd bases) its mirror complement (Rodin and Rodin, 2006b). Furthermore, the code itself might have expanded via this route from the original complementary core (Table 1B) in the duration of a short, yet “decisive”, period of time (see also Rodin and Rodin, 2008; Rodin et al., 2009). It should be noted here that the scenario of the code genesis starting from the complementarily encoded amino acids (such as Gly, Ala, Asp and Val) is consistent with the extended co-evolution scenario (Di Giulio, 2008).
At any rate, from a purely chemical point of view, the complementary strands are undistinguishable. What makes the strands asymmetric is emerging information content for proteins. Indeed, among the other conceivable causes (reviewed in (Maynard Smith and Szathmary, 1995)), it was the fully-developed genetic code and translation machinery that developed sufficiently, to have broken the original symmetry of the two strands with regard to the ability to code for proteins (Rodin and Rodin, 2006a,b; Rodin et al., 2009).
It is exactly for these reasons that, as soon as the code’s well-known near-universal structure (Table 1) had been mostly established, translation of both strands in the same frame had become an obstacle to their evolution. This made the differentiation into the sense and anti-sense strands inevitable, and it would make it literally a miracle to encounter, in any extant genome, a pair of necessarily) very old proteins still encoded by the complementary strands in the same frame.
While not necessarily defying such a development (see below), if a gene shows any indirect sign of SAS coding (a long AS-ORF, for example) one might, at first, look for a more mundane interpretation. Of special interest in this regard is the very significant number of short chain dehydrogenase genes that have extended, in frame ORFs on their complementary strands (Duax et al. 2005).
2.2. Origins of complementary motifs in different proteins: palindromes vs. SAS coding
Two primary sources of sequence complementarity displayed by proteins are conceivable: one is direct SAS coding (Fig. 3A), and another one is indirect, due to palindromes (Fig. 3C). The difference is that palindromic sequences are self-complementary and therefore necessarily encode identical oligopeptides on both strands (Yomo & Ohno, 1989; Ohno & Yomo, 1991; Ohno, 1991: Rodin & Ohno, 1995), whereas it is not required in case of SAS. Importantly, such palindromic oligopeptides are widespread in contemporary genomes, track back to very ancient times (genetic code shaping, or even earlier, Ohno, 1991) and were most likely alternating arrays of hydrophilic and hydrophobic amino acids (Fitch and Upper, 1987; Ohno, 1987; Rodin et al., 1993a,b). SAS coding therefore provides an enhanced mechanism for the modular expansion of quite simple peptides into more elaborate and functional peptides and proteins (see, for example, (Trifonov, 2005)).
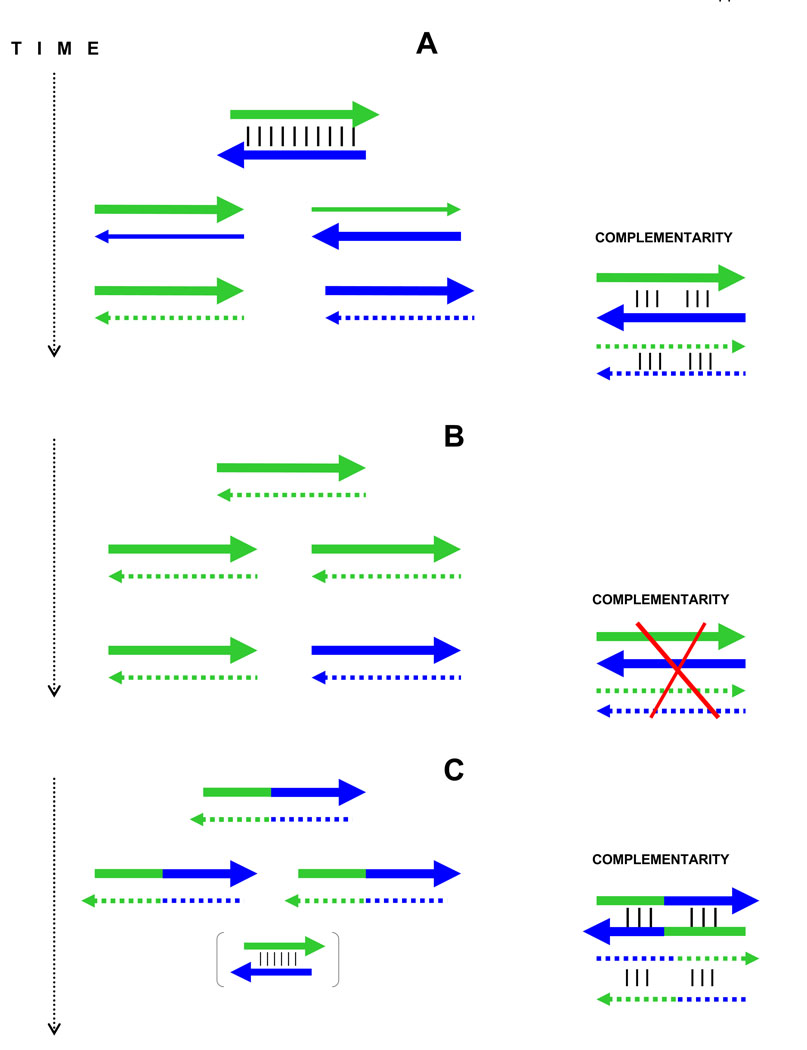
Figure 3.
The primordial SAS coding, gene duplications and complementarity of the diverged extant duplicates.
A: The ancient in-frame SAS coding imposed strong constrains on evolution of complementary strand-genes (thick green and blue arrows). A duplication releases the daughter copies from these constraints. Their subsequent divergence with gradual silencing of the opposite strands (thin arrows) results eventually in two different genes (with sense strands, shown by thick arrows and antisense strand, shown by punctuated arrows) that may retain fingerprints of the original complementarity (on the right).
B: The classic scheme of gene duplication without the primordial SAS coding: the diverged extant genes show no complementarity in anti-parallel “head-to-tail” alignment.
C: The same as B but the original gene was a self-complementary palindrome. In this case, the extant offspring genes may also still display some complementarity derived from the original palindrome even though the latter had no SAS coding. However, the palindrome per se suggests possible descent from preceding SAS-encoded pairs of segments (shown in brackets by thick mini-arrows) that merged to constitute the gene duplicated later as a whole, i.e. the variant C is in fact a combination of the primordial SAS (A) and classic scheme of gene duplication (B).
Obviously, consecutive duplications of an initial palindrome might extend the region of self-complementarity, even significantly so. If, at some step, the duplicates begin to evolve as independent protein-encoding genes, and if their independent divergence ends up eroding (but not yet unrecognizably) the originally identical oligopeptides, it eventually would become very difficult to distinguish the A and C pathways (Fig. 3). One should keep in mind, though, that the palindromic and SAS models are not fully mutually exclusive, because it is the SAS coding that provides the very origin of palindromic oligopeptide genes (Fig. 3C). After all, any hairpin in mRNA might originate by self-templating, which if it coincides with the translation frame is in essence equivalent to SAS coding (Fig. 3C).
Remarkably, the most fundamental molecule of life – tRNA – might also have originated in the same way, with the primordial short palindrome (Fig. 1B) serving as a primary building brick. The detailed models of its gradual expansion into the final cloverleaf are reviewed in (Rodin and Rodin, 2009).
2.3. SAS origin of two aaRSs
All tRNAs have the same 2D cloverleaf- and 3D L-like shape. In contrast, the two classes of aaRSs have nothing common in their 1D, 2D and 3D structures. Not surprisingly, it has been a popular opinion that two archaic independent translation systems corresponding to classes I and II (for 10 amino acids, each) operated independently and fortuitously merged later, producing a complete repertoire of aaRSs for the canonical set of 20 amino acids. However, when one looks more closely at this seemingly appealing scenario (“earliest molecular symbiosis”!), it immediately presents many serious inconsistencies (reviewed by Rodin and Ohno, 1995; see also Carter and Duax, 2002). We mention just one: the most evolutionary conserved (and, likely, oldest) catalytic signature motifs in the two aaRSs classes are made predominantly of amino acids activated by the opposite class (Rodin and Ohno, 1995). For example, the class I motifs are constructed from P,H(2),G,K(2),D, and S(2) (all activated by class II aaRS) with only a single I and M. The class II catalytic residues include E(2) and R(2–3), which are activated by class I enzymes. As acyl group activation is, by many orders of magnitude, the most significant kinetic barrier to protein synthesis in the absence of catalysts, that function seemingly must have evolved simultaneously for the two classes (Pham, et al., 2007).
Fifteen years ago a hypothesis, aiming to reconcile these contradictions, was put forward: what if the ancestors of both aaRS classes were encoded (in the same frame) by complementary strands of one primordial gene (Rodin and Ohno, 1995)? Indeed, when aligned head-to-tail, the regions with conserved signature motifs from the opposite aaRS classes do appear as complementary images of each other (ibid). Remarkably, our subsequent findings for tRNAs – the concerted complementarity of the acceptor’s 2nd bases with complementarity of anticodons (Rodin et al., 1996, 2009; Rodin and Rodin, 2006a) – independently pointed to the primary growth of codon repertoire by means of complementary pairs, thus making it perfectly consistent with the SAS origin of the two classes of synthetases.
Subsequently, after the complementary transformation of the conventional code table, we have identified the internal latent sub-code (Table 1B) that rationalizes the two sterically complementary modes of tRNA recognition by aaRSs (Rodin and Rodin, 2006b, 2008;see also Carter, 2008; Delarue, 2007): the sub-code minimizes the risk of confusing primordial adaptors with complementary anticodons. These relationships are highlighted in Fig. 2, which relates the biophysical properties associated with protein folding of the corresponding codon-anticodon pairs.
All this necessarily implies great antiquity of both modes of tRNA aminoacylation – the ribozymic precursors of both class I and class II aaRSs (r-aaRS) having already recognized the complementary halves of tRNAs (most likely via W-C pairing). Accordingly, it would make sense to assume that class I and class II r-aaRSs have been complementary to each other as themselves, at least in their tRNA-binding segments. Later, when the code’s complementary core had already been established, the iso-functional proteins (p-aaRS) replaced the r-aaRSs. The principle of evolutionary continuity dictates, and the analyses do indicate, that the p-aaRSs inherited precisely the same two modes of tRNA aminoacylation and, accordingly, had to be of two complementary types as well (Rodin and Rodin, 2008).
In our opinion, the two putative complementarily symmetric r-aaRSs coevolved concertedly, with the gradual elongation of the initial short palindromes into the eventual pair of tRNAs with complementary anticodons (Rodin et al., 2009; see Rodin and Rodin for details). A contemporary relic related to such r-aaRS may persist in contemporary biology as the “t-box” riboswitch (Henkin 2009). This riboswitch apparently recognizes both the 3’ accepter stem and the anticodon of the tRNA whose expression it regulates. It is an existence proof that ribozymes can possess these functions, and is hence substantive evidence that r-aaRS may have existed.
At some point, well before the code gained its complete codon repertoire, the first complementarily encoded oligopeptides appeared, including precursors of the two p-aaRS classes, which apparently accelerated the rate of amino acid activation by a substantial amount (Pham et al. 2007). Most likely, being better catalysts, these two minimalist p-aaRSs accelerated the evolution of the code itself (Rodin and Rodin, 2006a, 2008; Schimmel and Beebe, 2006; Pham et al., 2007). Furthermore, it does not seem too much of a speculation to propose that the gene for the very first p-aaRSs was just a r-aaRS gene duplicate. Importantly, in the RNA world, because of W-C pairing, any RNA sequence always carries a complementary message. Therefore, elongation (by self-templating and/or duplicating) of one r-aaRS necessarily entails elongation of its complement, and the same is true for the proteins they are coding for. This is also consistent with (1) the r-aaRS → p-aaRS transitions maintaining the same mode of tRNA aminoacylation (Rodin and Rodin, 2006b, 2008), and (2) the stereochemical theory of direct aa-anticodon affinity (Yarus, 1998; Yarus et al., 2005).
To conclude this overview of the primordial SAS coding, recall that, as we have already emphasized, an extant gene showing an indirect sign of SAS coding (such as a long AS-ORF) does not necessarily imply its SAS origins. At first glance, the recent analysis of the HSP70 gene in Achlya klebsiana (Williams et al., 2009) gives such a lesson. However, at a closer inspection, the reality appears much more intriguing.
3. Is the HSP70 gene in Achlya klebsiana a Rosetta stone for the sense-antisense origin of class I and class II aaRS?
In our opinion, the arguments presented below provide a positive answer.
This gene (that codes for the heat shock protein) is of particular interest because: 1) its antisense strand has a long reading frame, supposedly coding for a stress-inducible, NAD-specific glutamate dehydrogenase (NAD-GDH) (LeJohn et al., 1994) and 2) the NAD-GDH and HSP70 are homologous to the class I and class II aaRSs, respectively (Carter and Duax, 2002). However, recently Williams et al (2009) have claimed this antisense reading frame to be a spurious consequence of the high conservation of the HSP70 gene, casting doubt on the homology of its (supposed) protein product to the active members of the NAD-GDH family. Thus, unraveling a mystery of fundamental importance is at stake.
3.1. Long AS–NRF
Following (Rother et al., 1997), we shall distinguish truly functional open reading frames (ORF) from non-stop reading frames (NRF) that have only a potential to be translated.
There is a strong selection for maintenance of a very long AS-NRF in the HSP70 gene in Achyla klebsiana. Quite telling in this regard are the excesses of serine TCG (16 per 652 positions) and leucine TTGcodons. Due to high mutability of CpGs, one would expect frequent transitions of this TCG into synonymous TCA, which is complementary to the TGA stop codon. It may seem reasonable to ascribe this excess of silent CpGs in coding regions to the methylation-mediated epigenetic control of gene expression (Rauch et al., 2009; Branchiamore et al., 2009). However, the question arises: Why is the preference of such CpGs not observed beyond the gene proper, at least in the 5’UTR? Immediately upstream of the NRF region we do see, in the same reading frame, 18 TCAs vs. 8 TCGs (out of 520 positions total). Furthermore, the CpG methylation hypothesis obviously does not work for the leucine TTG/TTA pair (TTA is complementary to the TAA stop-codon). Even though the leucine TTG is apparently not as mutable as the serine TCG, the 12:0 TTGs vs. TTA ratio within the 652 codon-long gene, as contrasted to the 7:9 ratio within the aforementioned 520 positions of the 5’UTR, speaks for itself. Interestingly, neither the leucine CTG nor its likely mutational derivative CTA (complement of the TAG nonsense-codon) are found within the gene proper, whereas we see them in both upstream and downstream vicinities: in total 6 CTAs vs. 4 CTGs per 858 positions.
All of the above unambiguously point to selection against these Leu and Ser codons in the sense strand and, complementarily, against stop-codons in the antisense strand. Could not the selection work through biases in nucleotide composition (towards G and C), and thus in codon usage, just as certain other commonly cited general factors of the sort? It certainly could, but it would be very unlikely to completely account for the phenomenon of long AS-NRFs. There is something else at work here, more HSP70-specific. Indeed, let us take into consideration the fact that long NRFs are also found in other paralogs and orthologs of the A. klebsiana AS-HSP70. Williams et al (2009) mention this as evidence of the “simple” conservation of HSP70 genes on the opposite strand and refer the readers to (Rother et al., 1997; Silke, 1997; reviewed in Culbertson, 1999) for explanation(s) alternative to the SAS coding. Furthermore, it is telling that Duax has shown that the dehydrogenase family of genes also is replete with long, in-frame AS-NRFs (Duax, et al. 2005), despite the fact that the dehydrogenase family has diverged considerably more than has the HSP70 family.
The primary (and most popular) alternative hypothesis to account for the prevalence of long AS-NRFs suggests that they do not function (and, therefore, are not preserved by selection) at the protein level – they exist because the antisense RNA might directly regulate the level of sense RNA via sense-antisense duplex formation (ibid). Note in this regard that antisense transcription was indeed detected for the HSP70 gene in A. klebsiana (LeJohn et al., 1994) and this important fact is also mentioned in (Williams et al., 2009). Further, according to this hypothesis, the presence of in-frame stop codons on the antisense RNA would result in its premature degradation, hence inability to regulate activity of the sense mRNA. However, the nonsense-mediated mRNA decay (NMD) hypothesis in fact tacitly implies not only the existence of the long AS-NRF but also its reading by the ribosome machinery during translation. Thus, the NMD hypothesis is not actually alternative to the SAS coding; on the contrary, it appears to logically suggest the latter!
Importantly, if even we assume that the NMD of antisense transcripts does occur and, more, that the ribosome reads (and recognizes stop-codons) but does not translate the anti-sense RNA, one still wonders: Why should this strange “translation-innocent” reading of AS transcripts occur in the same frame? Indeed, it seems self-evident that a sense-antisense duplex does not presume any (same or not) reading frame for its formation. However, shifting the reading frame by one or two base(s) in the sense HSP70 sequence of A. klebsiana (as well as in all other HSP70 (and short-chain dehydrogenase) genes with long AS-NRF) reveals numerous CTA, TTA and TCA triplets that are complemented by stop-codons on the corresponding anti-sense sequences.
We see only one reasonable explanation for all of the above: the NMD and SAS hypotheses do not exclude each other. Moreover, the NMD-based explanation of long AS-NRF in the HSP70 gene of Achlya klebsiana makes sense if and only if this explanation actually implies that the AS-HSP70 was recently (or even still is) not just an AS-NRF, but a true (not spurious) AS-ORF --- and, importantly, in the same reading frame. If it is a true, translatable antisense gene, then the next question is: What protein could it code for?
3. 2. Homology of AS-HSP70 to NAD-GDH
The essence of the argument advanced by Williams et al (2009) is their skepticism that the AS-HSP70 is a functional gene for the canonical NAD-specific GDH in A. klebsiana (shown aligned with its closest counterpart from the oomycete Aphanomyces euteiches in Fig. 4). True or not, this is largely immaterial. What does matter is the evidence presented below that this AS-HSP70 gene belongs to the NAD-GDH family and is still either functionally active (for example as a stress protein) or just starting on the road of degradation into a pseudogene. This evidence unifies the diverse threads on the conservation of AS-NRFs opposite both HSP70 and short chain dehydrogenase genes by reinforcing the proposal that these two protein families descended from a single SAS ancestral gene.
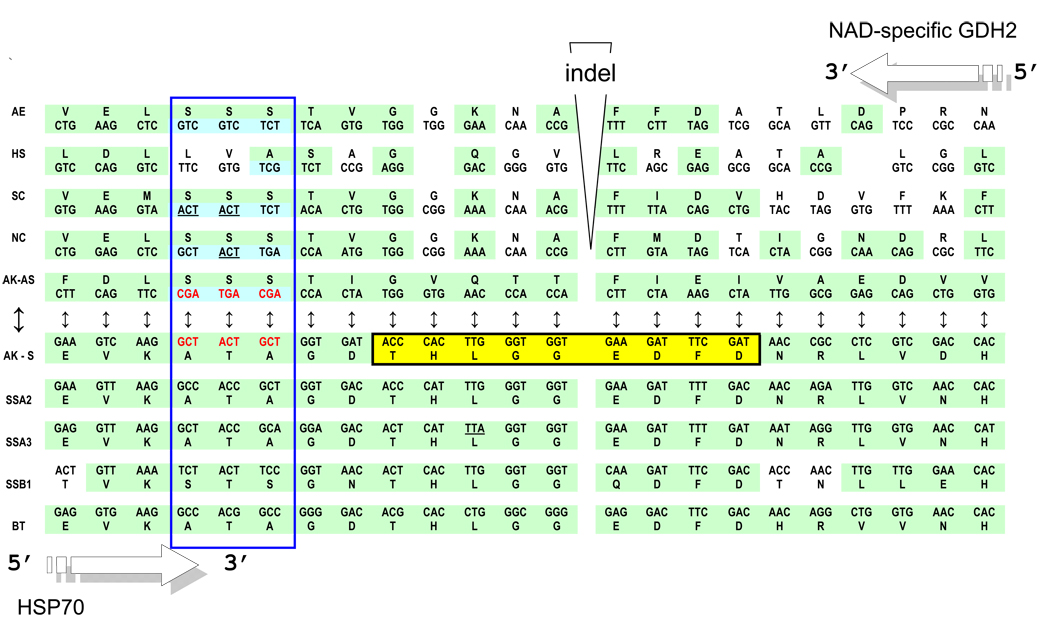
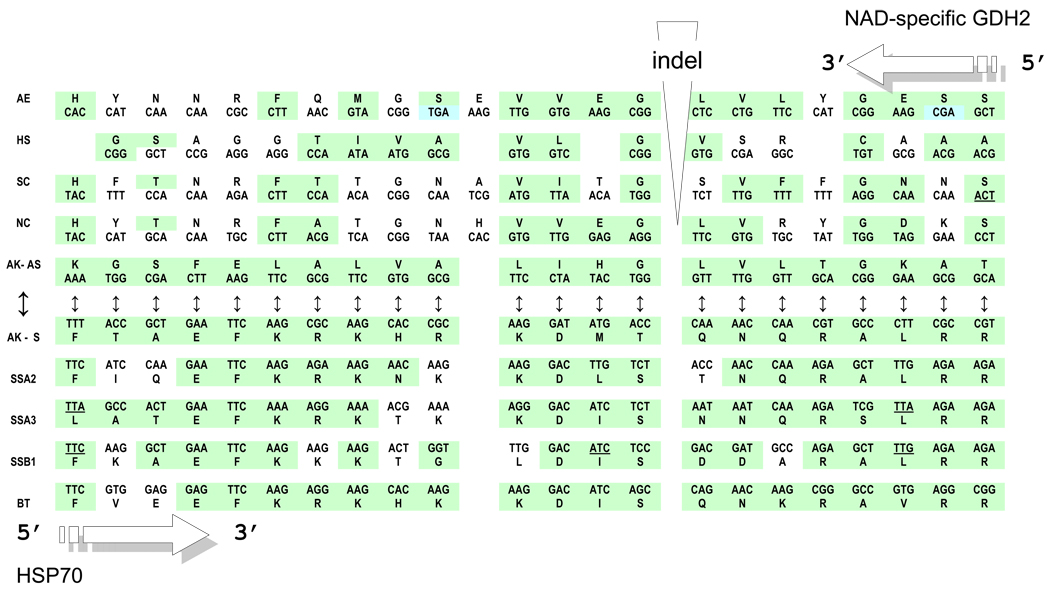
Figure 4.
Hsp70 vs. NAD-Gdh complementarity. Shown in the center (opposite directions) are two complementary sequences: the fragment of the HSP70 gene of the Achlya klebsiana and its (head-to-tail aligned) antisense complement, denoted AK-s and AK-as, respectively. Under the AK-s, the homologous sequence fragments of yeast (SSA2, SSA3 and SSB1) and bovine (Bos taurus, BT) HSP70 genes are aligned from left to right. Above the AK-as, from right to left, we mapped the homologous pieces of NAD-specific GDH genes of N. crassa (NC) (following the alignment in Fig. 2 from (Williams et al., 2009)) and S. cerevisiae (SC), as well as the 3-hydroxyacyl-CoA dehydrogenase (Type II HADH) of H. sapiens (HS). The putative (according to Williams et al., 2009) NAD-GDH homolog from the A.klebsiana’s close relative, the oomycete Aphanomyces euteiches (AE), is also shown, at the very top. Codons that share the same, complementary central nucleotide in the SAS alignment, according to the hypothesis of ancient SAS complementary coding, are colored green. Similarly marked are the cognate amino acids (some of them are identical). For the sense and anti-sense homologies of this A.klebsiana HSP70 region with two classes of aaRS, see (Carter and Duax, 2002).
Paralogous pairs of genes descended from a common ancestor by a trivial duplication and pairs of genes that originated from a common SAS ancestor (usually in the very remote past, with subsequent duplication when the simultaneous SAS co-evolution became too constraining) differ in a fundamental respect. SAS-originated gene pairs would be expected to show a significantly greater complementarity at the central codon positions. In other words, we have to check the genes of interest not just on the aa identity index but also the complementarity of their codons’ second positions in head-to-tail alignments. This approach results in more than 50% of the 2nd bases showing complementarity for the 404-position region (see Fig. 2 in Williams et al., 2009), where all three sequences in question (Achlya klebsiana AS-NRF, Neurospora crassa NAD-GDH, and Aphanomyces euteiches contig) are aligned. This is significantly higher than ~ 25–30% expected by chance alone (Pham et al. 2007).
The alignment in Figure 4 represents perhaps the most convincing evidence in support of the origin of genes encoding NAD-specific GDH and HSP70 (homologs of the class I and class II aaRS, respectively) from complementary, sense and anti-sense, strands of one ancestral gene.
Common ancestry of all HSP70 (only four are presented in Figure 4, just for the visualization purposes) is patently obvious and does not require significance testing. Figure 4 shows a key 46-residue segment of the putative SAS alignment derived from the region of HSP70 homologous to Motif 2 of the class II aaRS. The alignment suggests that the same can be said about their possible antisense homologs – NAD-specific GDHs shown above the AS-HSP70 of A. klebsiana.
Adding to that, of all proteins known to date, the heat shock protein is one of the oldest and most evolutionarily conserved (Gupta and Golding, 1993; Gupta, 1998). Consistent with this antiquity is its main function – protection from heat- and other stress-induced damages that very likely threatened young life in the primitive harsh conditions of early Earth (high temperature and deficient oxygen). Therefore, it would be unwise to discount any reliably established SAS similarity (even fragmental) between HSP70 and other evolutionarily old proteins.
The HSP70 gene fragment and its anti-sense complementary replica from A. klebsiana (two sequences in the center of Fig. 4) are important and worthy of a closer analysis. Indeed:
- Within this region, the AS-ORF of the HSP70 gene from A. klebsiana shows the maximum, obviously nonrandom, similarity with NAD-specific GDH and other related proteins of the Rossmann fold (including those from the very remote species) (Fig. 4).
- One small “patch” of SAS complementarity (enclosed in the blue vertical rectangle in Fig. 4) is particularly fascinating: the rather conservative sense Ala-Thr-Ala tripeptide from HSP70 is converted into the anti-sense Ser-Ser-Ser, which actually represents a signature tripeptide of fungal NAD-specific GDH enzymes. The uniqueness of this SAS case is that serine is the only amino acid encoded by triplets with complementary central bases – four triplets TCN (the code table, 2nd column) and two triplets AGY (the code table, 4th column). Only the latter two have complementary partners from the 2nd columns of the genetic code table – GCT and ACT coding for Ala and Thr, respectively. Obviously, one conservative motif, Ala-Thr-Ala, is complementarily transformable into another, Ser-Ser-Ser, if and only if it uses GCT (Ala) and ACT (Thr), and this is exactly what is observed in reality!
- Of all HSP70 genes available at present, only two – the one from A. klebsiana shown in Fig. 4 and the SSA4 from S. cerevisiae --- demonstrate this remarkable complementary transformation of one signature motif, Ala-Thr-Ala on the sense strand, into another one, Ser-Ser-Ser, on the opposite, anti-sense, strand. However, in contrast to the A. klebsiana gene, its yeast’s ortholog, SSA4, contains numerous TTA (Leu), CTA (Leu) and TCA (Ser) codons, i.e. has no sufficiently long AS-ORFs. Nonetheless, the GDH and HSP70 genes in S. cerevisiae are coded by sequences that are >95% SAS complementary.
- Consistently with this remarkable Ala-Thr-Ala vs. Ser-Ser-Ser complementarity, in the classic scenario of the code origin (Crick et al, 1976: Eigen and Schuster, 1979; and many others, see also Rodin and Rodin, 2008, 2009) the two “extra” codons for serine, AGC and AGT, actually represent the primordial RNY code, thus being (presumptively) older than its main TCN coding tetrade. Worthy of mention is the fact that the genes made of solely RNY codons are SAS coding-prone since they are free of stop triplets and their complements.
- It should also be noted that the aforementioned possible SAS origin of the Ala-Thr-Ala vs. Ser-Ser-Ser complementarity represents only a small fraction of the nonrandom complementarity that the entire region certainly displays (Fig. 4).
- The segment shown in Figure 4 is a special one in that it appears to be highly (if not most) important evolutionary. Indeed, in the upstream proximity, all HSP70 contain a signature motif of nine amino acids, V/IDLGGGD/EFE that is extremely old and seems to have already occurred in a common ancestor to all life (Gupta and Golding, 1993). Moreover, this nine aa-long motif is in fact a multiple repeated building unit of HSP70 – its modified duplicate THLGGEDFD (enclosed in a black-bounded horizontal yellow rectangle in Fig. 4) is located just downstream of the aforementioned Ala-Thr-Ala that complements the Ser-Ser-Ser tripeptide of GDH2 genes.
- Furthermore, the putative complement of the PxxxHIGH of class I aaRSs, i.e. the motif 2 of class II aaRSs (Rodin and Ohno, 1995) is located just downstream, in close vicinity to the EVKATAGD-THLGGEDFD signature motif of HSP70 proteins,. This location reflects the still-evident complementarity of primary coding sequences (about 70% of 2nd bases) and, especially, some 2D elements (Carter and Duax, 2002). This is what the ancestral “frozen complementarity” of the two catalytic modules (and the principle of evolutionary continuity) would predict for the present-day homologs of class I and class II aaRSs, including GDH and HSP70.
4. Conclusion
Of all the HSP70 genes available today, the only one that has a very long AS-NRF and, at the same time, is complemented by the NAD-specific GDH signature motif G(I/V)TSSSLDF (with uniquely encoded three serines in the middle) on the anti-sense strand appears to be the HSP70 gene of Achlya klebsiana. The SAS coding readily explains the Leu and Ser codon usage pattern in this gene and its orthologs and paralogs, with and without CTA, TTA and TCA complements of stop codons. In fact, the NMD-based explanation of long AS-NRFs does not exclude the SAS coding, but rather implies it. However, whether this AS-NRF is still translated (being actually AS-ORF), or has recently ceased to be, is not that important. What really matters is the substantive evidence for SAS homology between HSP70 and NAD-GDH genes, and the ease with which, for instance, A.klebsiana, having a long AS-ORF, might “explore” the validity of anti-sense proteins. Moreover, such “exploration” might have been very commonplace with primordial life, while the genetic code was in the process of being molded. We believe, though, that evolution continues to be a highly opportunistic process and should not miss any novelty provided by the SAS coding. We share the well-grounded opinion of Yomo et al (1992) that AS-NRFs are still the cradle of new proteins.
Apparently, the HSP70 gene of A.klebsiana is a very good candidate to represent this “atavistic” source of novelties. After all, one should not ignore the following facts: 1) this gene in A. klebsiana is exceptional in that its lack of CTA, TTA and TCA codons does not actually follow the general correlation with C and G in synonymous positions, i.e. here we, indeed, face an abnormally long AS-NRF (Silke, 1997); and 2) although we agree with Williams et al.(2009) that the gene from oomycete Aphanomyces euteiches (Fig. 4) represents the canonical NAD-GDH gene (not sequenced in A. klebsiana), they did not take into account that of two different possible glutamate dehydrogenase genes in A. klebsiana and closely-related species, only the one expressed under nitrogen stress being relevant as a functional gene product from the HSP70 AS-NRF.
At any rate, this updated analysis confirms that in A. klebsiana a single gene might have originally coded for an HSP70-like chaperonin (class II aaRS homolog) and a dehydrogenase (class I aaRS homolog) via its sense and antisense strands. This “Rosetta stone” for the simultaneous sense-antisense origin of aaRS classes (Carter and Duax, 2002) might be partly eroded by now, but it is still unmistakably there.
References
- Branchiamore S, Riggs AD, Rodin SN. On the role of epigenetic silencing in evolution by gene duplication: Selection favors CpGs in coding regions of HOX genes – in preparation. 2009 [Google Scholar]
- Carter CW., Jr Whence the Genetic Code?: Thawing the 'Frozen Accident'. Heredity. 2008;100:339–340. doi: 10.1038/hdy.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CW, Jr, Duax WL. Did tRNA synthetase classes arise on opposite strands of the same gene? Mol. Cell. 2002;10:705–708. doi: 10.1016/s1097-2765(02)00688-3. [DOI] [PubMed] [Google Scholar]
- Crick FHC. The origin of the genetic code. J. Mol. Biol. 1968;38:367–380. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Crick FHC, Brenner S, Klug A, Pieczenik G. A speculation on the origin of protein synthesis. Orig. Life. 1976;7:389–397. doi: 10.1007/BF00927934. [DOI] [PubMed] [Google Scholar]
- Culbertson MR. RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet. 1999;15:74–80. doi: 10.1016/s0168-9525(98)01658-8. [DOI] [PubMed] [Google Scholar]
- Delarue M. An asymmetric underlying rule in the assignment of codons: Possible clue to a quick early evolution of the genetic code via successive binary choices. RNA. 2007;13:1–9. doi: 10.1261/rna.257607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennett DC. Darwin's Dangerous Idea: Evolution and the Meanings of Life. New York: Simon and Schuster; 1995. [Google Scholar]
- Di Giulio M. An extension of the coevolution theory of the origin of the genetic code. Biol. Direct. 2008 Sep 5;:3–37. doi: 10.1186/1745-6150-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duax WL, Huether R, Pletnev V, Langs D, Addlagatta A, Connare S, Habegger L, Gill J. Rational Genomes:Antisense Open Reading Frames and Codon Bias In Short Chain Oxido Reductase Enzymes and the Evolution of the Genetic Code. PROTEINS: Structure, Function, and Bioinformatics. 2005;61:900–906. doi: 10.1002/prot.20687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen M, Schuster P. Hypercycle: A principle of natural self-organization. Heidelberg: Springer-Verlag; 1979. [DOI] [PubMed] [Google Scholar]
- Fitch WM, Upper K. The phylogeny of tRNA sequences provides evidence for ambiguity reduction in the origin of the genetic code. Cold Spring Harbor Symp. Quant. Biol. 1987;52:759–-767. doi: 10.1101/sqb.1987.052.01.085. [DOI] [PubMed] [Google Scholar]
- Freeland SJ, Hurst LD. The Genetic Code is One in a Million. Journal of Molecular Evolution. 1998;47:238–248. doi: 10.1007/pl00006381. [DOI] [PubMed] [Google Scholar]
- Gupta R. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol. Mol. Biol. Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS, Golding GB. Evolution of HSP70 gene and its implications regarding relationships between Archaebacteria, Eubacteria, and Eukaryotes. J. Mol. Evol. 1993;37:573–582. doi: 10.1007/BF00182743. [DOI] [PubMed] [Google Scholar]
- Henkin TM. RNA-dependent RNA switches in bacteria. Meth. Mol. Biol. 2009;540:207–214. doi: 10.1007/978-1-59745-558-9_15. [DOI] [PubMed] [Google Scholar]
- Ibba M, Morgan S, Curnow AW, Pridmore DR, Vothknecht UC, Gardner W, Lin W, Woese CR, Soll D. Euryarchael lysyl-tRNA synthetase: Resemblance to class I synthetases. Science. 1997;278:1119–1122. doi: 10.1126/science.278.5340.1119. [DOI] [PubMed] [Google Scholar]
- Kamtekar S, Schiffer JM, Xiong H, Babik JM, Hecht MH. Protein Design by Binary Patterning of Polar and Non-polar Amino Acids. Science. 1993;262:1680–1685. doi: 10.1126/science.8259512. [DOI] [PubMed] [Google Scholar]
- Knight RD, Freeland SJ, Landweber LF. Rewriting the keyboard: evolvability of the genetic code. Nature Rev Genet 2001. 2001;2:49–58. doi: 10.1038/35047500. [DOI] [PubMed] [Google Scholar]
- Konechny J, Eckert M, Schoniger M, Hofacker GL. Neutral adaptation of the genetic code to double-strand coding. J. Mol. Evol. 1993;36:407–416. doi: 10.1007/BF02406718. [DOI] [PubMed] [Google Scholar]
- Kuhns ST, Joyce GF. Perfectly complementary nucleic acid enzymes. J. Mol. Evol. 2003;56:711–717. doi: 10.1007/s00239-002-2445-7. [DOI] [PubMed] [Google Scholar]
- LeJohn HB, Cameron LE, Yang B, Rennie SL. Molecular characterization if an NAD-specific glutamate dehydrogenase gene inducuble by L-glutamine (antisense gene pair arrangement with L-glutamine-inducible heat shock 70-like protein gene) J. Biol. Chem. 1994;269:4523–4531. [PubMed] [Google Scholar]
- Maynard Smith J, Szathmary E. The Major Transitions in Evolution. Oxford: Freeman; 1995. [Google Scholar]
- Novozhilov AS, Wolf Yu, Koonin EV. Evolution of the genetic code: partial optimization of a random code for robustness to translation error in a rugged fitness landscape. Biol. Direct. 2007;2:24. doi: 10.1186/1745-6150-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution from primordial oligomeric repeats to modern coding sequences. J. Mol. Evol. 1987;25:325–329. doi: 10.1007/BF02603117. [DOI] [PubMed] [Google Scholar]
- Ohno S, Yomo T. The grammatical rule for all DNA: junk and coding sequences. Electrophoresis. 1991;12:103–108. doi: 10.1002/elps.1150120203. [DOI] [PubMed] [Google Scholar]
- Ohno S. The grammatical rule of DNA anguage: Messages in palindromic verses. In: Osawa S, Honjo T, editors. Evolution of Life: Fossils, Molecules and Culture. Tokyo: Springer; 1991. pp. 97–108. [Google Scholar]
- Orgel LE. Evolution of the genetic apparatus. J. Mol. Biol. 1968;38:381–393. doi: 10.1016/0022-2836(68)90393-8. [DOI] [PubMed] [Google Scholar]
- Patel SC, Bradley LH, Jinadasa SP, Hecht MH. Cofactor binding and enzymatic activity in an unevolved superfamily of de novo designed 4-helix bundle proteins. Protein Science. 2009;18:1388–1400. doi: 10.1002/pro.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham Y, Li L, Kim A, Erdogan O, Weinreb V, Butterfoss GL, Kuhlman B, Carter CW., Jr A minimal Trp RS catalytic domain supports sense/antisense ancestry of class I and II aminoacyl-tRNA synthetases. Mol. Cell. 2007;25:851–862. doi: 10.1016/j.molcel.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100-base pair resolution. Proc. Natl. Acad. Sci. USA. 2009;106:671–678. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin S, Ohno S, Rodin A. Transfer RNAs with complementary anticodons: Could they reflect early evolution of discriminative genetic code adapters? Proc. Natl. Acad. Sci. USA. 1993;90:4723–4727. doi: 10.1073/pnas.90.10.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin S, Ohno S, Rodin A. On concerted origin of transfer RNAs with complementary anticodons. Origins of Life and Evolution of Biosphere. 1993;23:393–418. doi: 10.1007/BF01582088. [DOI] [PubMed] [Google Scholar]
- Rodin S, Ohno S. Two types of aminoacyl-tRNA synthetases could be originally encoded by complementary strands of nucleic acids. Origins of Life and Evolution of the Biosphere. 1995;25:565–589. doi: 10.1007/BF01582025. [DOI] [PubMed] [Google Scholar]
- Rodin S, Rodin A, Ohno S. The presence of codon-anticodon pairs in the acceptor stem of tRNAs. Proc. Natl. Acad. Sci. USA. 1996;93:4537–4542. doi: 10.1073/pnas.93.10.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin S, Ohno S. Four primordial modes of tRNA-synthetase recognition, determined by the (G,C) operational code. Proc. Natl. Acad. Sci. USA. 1997;93:4537–4542. doi: 10.1073/pnas.94.10.5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin SN, Rodin AS. Origin of the genetic code: First aminoacyl-tRNA syntheatses could replace isofunctional ribozymes when only the second base of codons was established. DNA Cell Biol. 2006a;25:365–375. doi: 10.1089/dna.2006.25.365. [DOI] [PubMed] [Google Scholar]
- Rodin SN, Rodin AS. Partitioning of aminoacyl-tRNA synthetases in two classes could have been encoded in a strand-symmetric RNA world. DNA Cell Biol. 2006b;25:617–626. doi: 10.1089/dna.2006.25.617. [DOI] [PubMed] [Google Scholar]
- Rodin SN, Rodin AS. On the origin of the genetic code: Signatures of its primordial complementarity in tRNAs and aminoacyl-tRNA synthetases. Heredity. 2008;100:341–355. doi: 10.1038/sj.hdy.6801086. [DOI] [PubMed] [Google Scholar]
- Rodin AS, Szathmary E, Rodin SN. One ancestor for two codes viewed from the perspective of two complementary modes of tRNA aminoacylation. Biology Direct. 2009 Jan 27;4(4) doi: 10.1186/1745-6150-4-4. doi: 10.1186/1745-6150-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin AS, Rodin SN. Frozen complementarity of the genetic code: Relics of primordial mirror symmetry in tRNAs and aminoacyl-tRNA synthetases. – in preparation. Cell Mol Life Sci. 2009 [Google Scholar]
- Rother KI, Clay OK, Bourquin J-P, Silke J, Schaffner W. Long non-stop reading frames on the antisense strand of heat shock protein 70 genes and prion protein (PrP) genes are conserved between species. Biol. Chem. 1997;378:1521–1530. doi: 10.1515/bchm.1997.378.12.1521. [DOI] [PubMed] [Google Scholar]
- Schimmel P, Giege R, Moras D, Yokoyama S. An operational RNA code for amino acids and possible relation to genetic code. Proc Natl, Acad Sci USA. 1993;90:8763–8768. doi: 10.1073/pnas.90.19.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P, Beebe K. Aminoacyl tRNA synthetases: from the RNA world to the theater of proteins. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. Cold Spring Harbor Laboratory Press; 2006. pp. 227–255. [Google Scholar]
- Silke J. The majority of long non-stop reading frames on the antisense strand can be explained by biased codon usage. Gene. 1997;194:143–155. doi: 10.1016/s0378-1119(97)00199-6. [DOI] [PubMed] [Google Scholar]
- Szathmary E. Coding coenzyme handles: A hypothesis for the origin of the genetic code. Proc. Natl. Acad. Sci. USA. 1993;90:9916–9920. doi: 10.1073/pnas.90.21.9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szathmary E. The origin of the genetic code: amino acids as cofactors in an RNA world. Trends Genet. 1999;15:223–229. doi: 10.1016/s0168-9525(99)01730-8. [DOI] [PubMed] [Google Scholar]
- Trifonov EN. Theory of early molecular evolution: predictions and confirmations. In: Eisenhaber F, editor. Discovering Biomolecular Mechanisms with Computational Biology. Georgetown: Landes Bioscience; 2005. pp. 107–116. [Google Scholar]
- Williams TA, Wolfe KH, Fares MA. No Rosetta stone for a sense-antisense origin of aminoacyl tRNA synthetase classes. Mol.Biol.Evol. 2009;26:445–450. doi: 10.1093/molbev/msn267. [DOI] [PubMed] [Google Scholar]
- Yarus M. Amino acids as RNA ligands: A direct-RNA-template theory for the code’s origin. J. Mol. Evol. 1998;47:109–117. doi: 10.1007/pl00006357. [DOI] [PubMed] [Google Scholar]
- Yarus M, Caporaso JG, Knight R. Origins of the genetic code: The escaped triplet theory. Annu. Rev. Biochem. 2005;74:125–151. doi: 10.1146/annurev.biochem.74.082803.133119. [DOI] [PubMed] [Google Scholar]
- Yomo T, Ohno S. Concordant evolution of coding and noncoding regions of DNA made possible by the universal rule of TA/CG deficiency – TG/CT excess. Proc. Natl. Acad. Sci. USA. 1989;81:2650–2654. doi: 10.1073/pnas.86.21.8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yomo T, Urabe I, Okada H. No stop codons in the antisense strands of the genes for nylon oligomer degradation. Proc Natl Acad Sci U S A. 1992;89:3780–3784. doi: 10.1073/pnas.89.9.3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Freeland S. The standard genetic code enhances adaptive evolution of proteins. J. Theor. Biol. 2006;239:63–70. doi: 10.1016/j.jtbi.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Zull JE, Smith SK. Is genetic code redundancy related to retention of structural information in both DNA strands? Trends in Biochemical Sciences. 1990;15:257–261. doi: 10.1016/0968-0004(90)90048-g. [DOI] [PubMed] [Google Scholar]