Abstract
As a cancer chemotherapeutic drug, arsenic acts on numerous intracellular signal transduction pathways in cancer cells. However, its mechanism of actions is still not fully understood. Previous studies suggest that arsenic reacts with closely spaced cysteine (Cys) residues of proteins with high Cys content and accessible sulfhydryl (SH) groups. In this study, human breast cancer cell line MCF-7 was examined as a cellular model to explore arsenic-binding proteins and the mechanism of binding. An arsenic-biotin conjugate was synthesized by coupling the pentafluorophenol ester of biotin with p-aminophenylarsenoxide. Arsenic-binding proteins were eluted with streptavidin resin from arsenic-biotin treated MCF-7 cells, separated by polyacrylamide gel electrophoresis, and identified by matrix assisted laser desorption ionization mass spectrometry (MALDI-MS). Arsenic-binding properties of two of these proteins, β-tubulin and pyruvate kinase M2 (PKM2), were studied further in vitro and the biological consequences of this binding was evaluated. Binding assay with Western blotting confirmed binding of β-tubulin and PKM2 by arsenic in a concentration-dependent manner. Arsenic binding inhibited tubulin polymerization, but surprisingly had no effect on PKM2 activity. Molecular modeling showed that binding of Cys12 alone or vicinal Cys residues (Cys12 and Cys213) of β-tubulin by arsenic blocked the active site for access of GTP, which is necessary for tubulin polymerization. On the contrary, all Cys residues of PKM2 were far away from the active site of the enzyme. In summary, this study confirmed β-tubulin and PKM2 as arsenic-binding proteins in MCF-7 cells. Functional consequence of such binding may depend on whether arsenic binding causes conformational changes or blocks active sites of target proteins.
Keywords: Arsenic, Cysteine, Breast cancer
Introduction
As a cancer chemotherapeutic drug, arsenic trioxide was originally developed for promyelocytic leukemia (1). It is believed that similar mechanisms mediate both the therapeutic and toxic activities of arsenic compounds (2). Many actions of arsenic compounds are mediated by oxidative stress through multiple signaling pathways (3). Direct interaction with cellular targets, such as tubulin, has also been suggested as a mechanism of action in myeloid leukemia cells (4).
Specifically, arsenic induces apoptosis in many types of cancer cells through caspase activation, changes in mitochondrial membrane potential and Bcl-2 expression. Arsenic inhibits STAT3 activity and induces growth arrest at the G1 phase to promote differentiation of leukemia cells. Arsenic inhibits angiogenesis by down-regulating vascular endothelial growth factor (2, 5, 6). Arsenic may also induce DNA hypomethylation, gene mutation, and aberrant gene expression (7, 8). In breast cancer, many studies have shown that arsenic inhibits cell growth, induces apoptosis and differentiation, antagonizes estrogen receptor signaling, and modulates immune response of MCF-7 human breast cancer cells (9-13).
Arsenic may affect cellular functions directly by interacting with cellular structures, or indirectly by altering signal transduction pathways and the expression of numerous genes. Direct binding to arsenic is believed to be important in at least two aspects: 1) binding and resulting inhibition of enzymes and other proteins may cause the chemotherapeutic effects; 2) binding to certain proteins may be a detoxification process (14). It is known that trivalent arsenic is chemically more reactive than pentavalent species, and binds many proteins, for example, tubulin and actin (15), IKKα/β (16), E. coli arsR protein (17), E. coli RI methyltransferase (18), hemoglobin (19, 20), nicotinic receptor (21), metallothionein (22), galectin 1 and thioredoxin peroxidase II (23, 24), thioredoxin and protein disulfide isomerase (25), glucocorticoid receptor (26), and estrogen receptor α (27). Some other proteins were also shown to bind arsenic, yet their identities are still unknown (25, 28).
Biological responses to arsenic have been suggested to be partially mediated through vicinal SH groups of Cys residues. In some cases, even more than two SH groups may be bound by arsenic, although two such groups would be required to produce conformational change with functional consequences (17). Binding of arsenic to proteins through Cys residues may inhibit functions of the target protein. For example, arsenic binds to Cys179 in the activation loop of IKKα/β, and mutation of Cys179 abolished arsenite sensitivity (16). Arsenic blocks the interaction of ligand and receptor by binding to critical Cys residues of glucocorticoid receptor and estrogen receptor (26, 27).
In this study, we used biotin-conjugated trivalent arsenic to identify arsenic-binding proteins in MCF-7 cells. Potential mechanism of action was explored with molecular modeling to gain insight into the functional consequences of arsenic-binding to proteins.
Materials and Methods
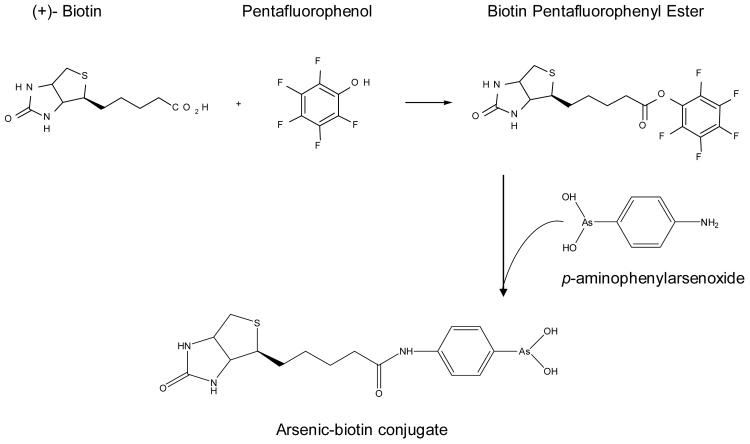
Synthesis of arsenic-biotin conjugate (29)
The biotin conjugate was synthesized in a manner similar to the published procedure (29), by coupling of p-aminophenylarsenoxide with the activated pentafluoro ester of biotin (29, 30). A mixture of 100 mg p-aminophenylarsenoxide, prepared according to the literature with minor modifications (30), in 1 ml of anhydrous dimethylformamide and 100 mg biotin pentafluorophenyl ester (31) in 1 ml of anhydrous dimethylformamide was stirred overnight at 50°C under nitrogen. Another 200 mg ester in 1 ml dimethylformamide was added and the mixture was stirred for an additional 2 days. Dimethylformamide was evaporated and the crude product was subjected to silica gel column chromatography, eluting with 10% methanol in chloroform. In total, 107 mg of the arsenic-biotin conjugate was obtained as a white solid (Figure 1). The structure was supported by TOF mass spectrometry: Calculated for C16H22AsN304S (M + 1- H2O = 410.05); found (M + 1 = 410.02).
Figure 1.
Schematic representation of synthesis of arsenic-biotin conjugate.
Determination of arsenic-binding proteins by matrix assisted laser desorption ionization mass spectrometry (MALDI-MS)
MCF-7 cells (ATCC, Manassas, MA) were grown to 80% confluence, and then treated with 10 μM p-aminophenylarsenoxide or arsenic-biotin conjugate overnight. The cells were washed with PBS and lysed with RIPA buffer. The cell lysate was diluted at least 1:10 with urea buffer, mixed with 50 μl of streptavidin resin, and shaken gently at 4°C for 1 hour. The resin was briefly spun down, washed twice with urea buffer, and resuspended in 50 μl 1×SDS loading buffer for polyacrylamide gel electrophoresis. Both the flow-through fraction (non-arsenic-binding fraction) and elution faction (arsenic-binding fraction) were loaded on gel for separation.
The gel was stained with Coomassie blue. Proteins pulled down by streptavidin were identified by comparing the elution fraction of arsenic-treated cells with that of arsenic-biotin treated cells. These protein bands were cut into slices, and were subjected to in-gel tryptic digestion and MALDI-MS analysis as described previously by us (32).
Proteins were identified with an online search engine, MASCOT Peptide Mass Fingerprint (www.matrixscience.com). For each protein, a probability based Mowse score and the number of peptides matched were provided. Although a score higher than 45 indicates identity or extensive homology, the higher score and the more peptides matched, the more likely a real binding took place.
Binding assays of arsenic with β-tubulin and PKM2
Purified MCF-7 tubulin (Cytoskeleton Inc., Denver, CO) or rabbit muscle pyruvate kinase (Sigma-Aldrich, St. Louis, MO) was incubated with various concentrations of arsenic-biotin (0.1 μM, 1.0 μM, 10 μM, 100 μM) at 30°C for 15 min. p-aminophenylarsenoxide and biotin were used as controls. 50 μl streptavidin resins was added, the mixture was incubated at 4°C for 1 h, and then spun down and washed twice with urea buffer. The resin was resuspended in 100 μl 1×SDS loading buffer, boiled at 100°C for 5 min, and then 50 μl supernatant was loaded on 10% SDS-PAGE gel for electrophoresis. The proteins on the gel were transferred onto a nitrocellulose membrane. After being incubated in a blocking buffer, membranes were incubated overnight at 4°C with the following primary antibodies: mouse monoclonal anti-β-tubulin (Sigma-Aldrich), or rabbit polyclonal anti-pyruvate kinase (RDI, Parsippany, NJ). After washing, the membranes were incubated with a secondary antibody and developed.
Binding assay was also performed by treating MCF-7 cells with 10 μM arsenic-biotin, and arsenic:arsenic-biotin (10:1) and biotin. After being pulled down with streptavidin, β-tubulin and pyruvate kinase were detected by western blotting with the same procedure described above.
Tubulin polymerization assay
Tubulin polymerization kit was obtained from Cytoskeleton Inc.. G-PEM buffer was pipetted into a 96-well plate, and incubated at 37°C for 10 min. Tubulin was suspended with tubulin polymerization buffer and incubated with arsenic-biotin or p-aminophenylarsenoxide in various concentrations for 30 min at 4°C, and then pipetted into each designated well of the pre-warmed 96-well plate at 37°C. The plate was put into microreader to measure absorbance at 340 nm for 60 min at 37°C. Taxol solution (10 μm) was used as control.
Pyruvate kinase activity assay and ATP assay
Pyruvate kinase activity was measured according to the reaction: phosphoenolpyruvate + ADP → pyruvate+ATP. Pyruvate formed from phosphoenolpyruvate by pyruvate kinase was measured by the formation of NAD+ in the presence of lactate dehydrogenase. The final reaction mixture contained the following in a total volume of 1 ml: 100 mM Tris-HCl buffer pH 7.4, 5 mM MgCl2, 100 mM KCl, 5 mM phosphoenolpyruvate, 5 mM ADP, 2.5 μg/ml lactate dehydrogenase (Roche Applied Science, Indianapolis, IN), and 0.15 mM NADH. After allowing the reaction mixture to equilibrate at room temperature for 30 min, the reactions were initiated by adding purified pyruvate kinase (Sigma-Aldrich), which was pre-incubated with arsenic-biotin conjugate or p-aminophenylarsenoxide at 30°C for 1 h. The plate was read with spectrophotometer at 340 nm at 37°C.
MCF-7 cells were seeded into an opaque-walled 96-well plate at 2×104 per well, and incubated at 37 °C for 24 h. The cells were treated with 10 μM arsenic-biotin or p-aminophenylarsenoxide for 0.5 h, 1 h, 2 h, and 4 h, respectively. Cell Titer-Glo Luminescent ATP assay (Promega, Madison, WI) was performed at the indicated time according to manufacturer's instructions.
Molecular modeling
Energy minimization and molecular dynamics (MD) simulations were performed on a Silicon Graphics Origin 350 Workstation using the extensible systematic force field implemented in Insight II program (Version 2005, Accelrys Inc., San Diego, CA). The extensible systematic force field, a rule-based force field, is suitable for systems containing metals (33). For energy minimization, the steepest descent method was employed first to a 100 kcal/molÅ energy gradient and followed by the Polak and Ribiere conjugate gradients method (34), until the final convergence criterion reached the 0.01-kcal/molÅ energy gradient. The atom-based method with a cut-off value of 12.0 Å and the distance-dependent dielectric constant (ε=ε0̃r, with ε0=4.0) were used for the summation of the non-bonding interactions.
Pairwise sequence alignment of bovine β-tubulin and human β2 tubulin (accession no.: CAA26203) was performed by CLUSTALW with the BLOSUM matrix (http://align.genome.jp) (35). Based upon high homology between human β2-tubulin and bovine β-tubulin, a three-dimensional model of human β2-tubulin was constructed using the X-ray structure of bovine β-tubulin (PDB code: 1JFF) (36).
To determine the most favorable Cys residues that may be bound by arsenic, p-aminophenylarsenoxide (p-NH2-ϕ-arsenic (OH)2) was used. Four residues which are close to the GTP binding site of the β2 subunit, Cys12, Cys203, Cys213 and Cys305, were modified to form arsenic conjugates, respectively. Four other Cys residues (Cys129, Cys131, Cys241, and Cys356) were excluded due to their distant positions from the GTP binding site (see Discussion). Individual conjugates were subjected to a simulated annealing (SA) procedure, including a 50 ps MD simulation at 2000K, a series of MD simulations cooling from 2000K to 300K by reducing 200K within 2 ps, and 100 ps MD at 300K. Only side chain residues within 8 Å of the interested Cys residues were included for the simulation. The final structure was subjected to energy minimization.
To determine the most plausible conjugate between arsenic and two Cys residues, a similar procedure except 250 ps MD simulation at 300K was performed. Among three candidate Cys residues for the formation of a double conjugate with the Cys12-arsenic conjugate, Cys203 and Cys305 were not considered, because their displacement toward Cys12 appeared to be energetically highly unfavorable. Cys203 was located deep in the core bundle formed by β-sheets, and Cys305 was in the region facing opposite to the GTP binding site. Thus, Cys213 was the only Cys residue to form a double conjugate with Cys12-arsenic conjugate. We moved Cys213 toward Cys12 because Cys213, part of an α-helix exposed on the surface of tubulin, appeared more flexible than Cys12, at the edge of an α-helix and a β-sheet forming a helical bundle and a parallel β-sheet segment. During the SA procedure, only the Cys213-containing motif (i.e., D205-Y224) and the side chains within 15Å of the Cα of Cys213 were freely moved while others were constrained. The final structure was subjected to energy minimization for selecting the best conformation according to energy criteria, shape, and the number of associated hydrogen bonds.
The structure of human PKM2 (PDB code: 1T5A) was obtained from the protein database. The relative positioning of the active site and the Cys residues of PKM2 was determined on the structure to estimate the potential effect of arsenic binding on enzyme activity.
Results
Identification of tentative arsenic-binding proteins in MCF-7 cells
MCF-7 cells were treated with arsenic-biotin conjugate or p-aminophenylarsenoxide as control. The arsenic-binding proteins were pulled down by streptavidin and separated by denaturing gel electrophoresis. Figure 2 showed a typical gel picture of the fractions. Multiple protein bands showed up in the elution faction of arsenic-biotin treated cells as compared with that of arsenic-treated cells.
Figure 2.
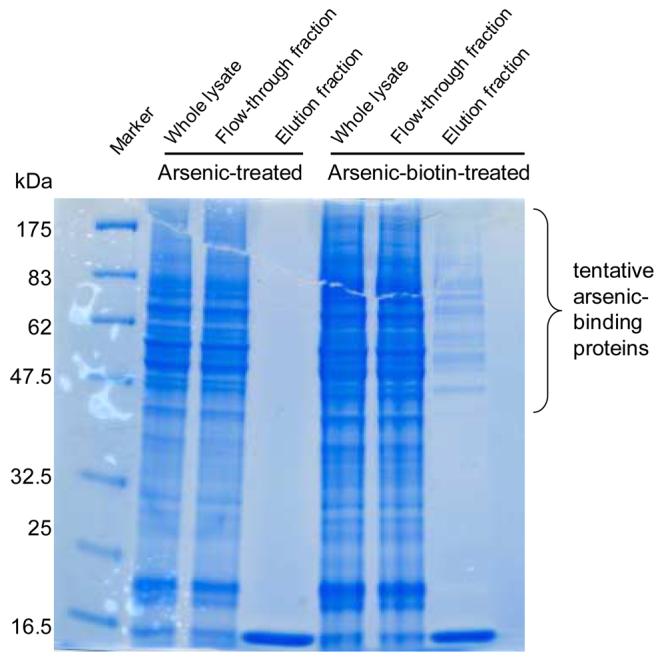
SDS-PAGE and Coomassie blue staining of the proteins from MCF-7 cell lysate. Three fractions were loaded, whole lysate, flow-through fraction which was unbound by streptavidin, and elution faction which was pulled down by streptavidin.
After digestion and MALDI-MS analysis, tentative arsenic-binding proteins were identified on the basis of their mass fingerprinting. Table 1 lists proteins whose probability based Mowse score was higher than 45. These 50 proteins mainly fell into three categories according to their functions:
Metabolic enzymes, such as glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerated kinase, PKM2, enolase, pyruvate carboxylase, and L-lactate dehydrogenase B chain, are involved in glycolysis. Transketolase and 6-phosphogluconate dehydrogenase participate in the pentose phosphate cycle, and malate dehydrogenase in the tricarboxylic cycle. Several other enzymes were also isolated, such as D-3-phosphoglycerate dehydrogenase, ubiquitin-activating enzyme E1, ATP synthase, aminopeptidase, protein disulfide isomerase, ATP-AMP transphosphorylase, triosephosphate isomerase, methylcrotonyl-CoA carboxylase, palmitoyl-protein thioesterase, and peroxiredoxin.
Structural proteins, such as tubulins, β-actin, filamin A, and cytokeratin, among which tubulins and β-actin are known to be bound by arsenic (15).
Stress response proteins, such as Hsp90β, Grp94, Hsp27, Hsp70, protein disulfide isomerase, T-complex protein 1, most of which were known to be induced by arsenic treatment (5, 37).
Table 1.
Arsenic-binding proteins in MCF-7 cells identified by MALDI-MS
| Name | MW (Da) | Total score1 |
No. of Peptides matched |
Function |
|---|---|---|---|---|
| Tubulin β2 2 | 49,799 | 990 | 33 | Structural subunit of microtubules |
| Tubulin β5 2 | 49,639 | 974 | 40 | Structural subunit of microtubules |
| Hsp 90β 3 | 83,081 | 961 | 25 | Stress response |
| Pyruvate kinase M2 (PKM2) | 57,746 | 723 | 19 | Glycolysis |
| HnRNP K | 50,944 | 644 | 27 | Mitochondria-related protein |
| Phosphoglycerate kinase 1 | 44,568 | 599 | 13 | Glycolysis |
| Pyruvate carboxylase | 129,551 | 552 | 12 | Glyconeogenesis and lipogenesis |
| Neural enolase | 47,008 | 516 | 12 | Glycolysis |
| β-tubulin 2 | 50,004 | 474 | 19 | Structural subunit of microtubules |
| β-tubulin-1 2 | 41,465 | 465 | 15 | Structural subunit of microtubules |
| Transketolase | 67,835 | 425 | 16 | Pentose phosphate cycle |
| D-3-phosphoglycerate dehydrogenase | 56,614 | 419 | 7 | Amino acid biosynthesis |
| β-actin 2 | 41,648 | 397 | 14 | Cytoskeleton |
| Malate dehydrogenase | 35,509 | 360 | 9 | Tricarboxylic acid cycle |
| Ubiquitin-activating enzyme E1 | 117,774 | 321 | 8 | Proteolysis |
| 6-phosphogluconate dehydrogenase | 52,975 | 319 | 9 | Pentose phosphate cycle |
| Filamin A | 280,586 | 311 | 8 | Cytoskeleton |
| T-complex protein 1, theta subunit | 59,582 | 301 | 7 | Stress response |
| Glyceraldehyde-3-phosphate dehydrogenase | 35,899 | 258 | 8 | Glycolysis |
| Importin 90 | 97,108 | 248 | 7 | Nuclear import |
| Annexin II | 38,449 | 226 | 5 | Phospholipids-binding protein |
| Cytokeratin 2-1 | 65,847 | 216 | 4 | Late cytoskeletal intermediate filament |
| Lactate dehydrogenase B chain | 36,484 | 176 | 4 | Glycolysis |
| ATP synthase alpha chain | 59,714 | 174 | 3 | Mitochondrial ATP synthesis |
| Hsp27 3 | 22,768 | 158 | 3 | Stress response |
| Elongation factor 2 | 95,146 | 146 | 3 | Protein biosynthesis |
| Puromycin-sensitive aminopeptidase | 103,237 | 140 | 3 | Aminopeptidase |
| HnRNP C1/2 | 33,667 | 137 | 3 | Mitochondrial protein |
| Nucleolar protein p120 | 94,020 | 131 | 3 | Ribonucleoprotein |
| GTP-binding nuclear protein RAN | 24,408 | 122 | 3 | Nucleocytoplasmic transport |
| Ribophorin I (glycosylation) | 68,527 | 121 | 3 | Protein amino acid glycosylation |
| DNA replication licensing factor MCM6 | 92,831 | 107 | 3 | DNA replication |
| Eukaryotic translation initiation factor 4A1/2 | 46,125 | 99 | 3 | Protein translation |
| Protein disulfide isomerase precursor 2 | 57,081 | 97 | 3 | Stress response |
| ATP-AMP transphosphorylase 2 | 26,330 | 71 | 3 | Adenylate kinase |
| Proteasome subunit alpha type 6 | 27,382 | 70 | 3 | Proteolysis |
| Rho GDP-dissociation inhibitor 1 | 23,193 | 56 | 3 | GDP/GTP exchange |
| Triosephosphate isomerase | 26,522 | 140 | 2 | Isomerase |
| Clathrin heavy chain 1 | 191,493 | 112 | 2 | Coated pits and vesicles |
| Methylcrotonyl-CoA carboxylase alpha chain | 80,382 | 102 | 2 | Leucine catabolism |
| 14-3-3 protein tau | 27,747 | 105 | 2 | Adapter protein |
| Tumor rejection antigen 1 (GRP94) 3 | 92,411 | 94 | 2 | Stress response |
| HnRNP A2/B1 | 37,407 | 90 | 2 | Mitochondrial protein |
| Cytokeratin e2 | 65,825 | 84 | 2 | Cytoskeleton |
| Elongation factor 1-delta | 31,103 | 73 | 2 | Protein biosynthesis |
| Palmitoyl-protein thioesterase | 34,171 | 78 | 1 | Protein modification |
| Alpha-actinin 4 | 104,788 | 76 | 1 | F-actin cross-linking protein |
| Hsp70-4 3 | 94,240 | 49 | 1 | Stress response |
| Peroxiredoxin 4 2 | 30,540 | 48 | 1 | Redox regulation |
| T-complex protein 1, delta subunit | 57,803 | 46 | 1 | Stress response |
Probability based Mowse score is −10*Log(P), where P is the probability that the observed match is a random event. Individual ions scores >45 indicate identity or extensive homology (p<0.05)(http://www.matrixscience.com/).
Known arsenic-binding proteins (15, 25). Arsenic has been shown to bind peroxiredoxin-1 (thioredoxin peroxidase 2) (24). Its active site is the redox-active Cys52 oxidized to Cys-SOH. Cys-SOH rapidly reacts with Cys173-SH of the other subunit to form an intermolecular disulfide with a concomitant homodimer formation. Similar to peroxiredoxin-1 in structure and function, peroxiredoxin-4 (thioredoxin peroxidase AO372) has its active site, the redox-active Cys124, oxidized to Cys-SOH. Cys-SOH rapidly reacts with Cys245-SH of the other subunit to form an intermolecular disulfide with a concomitant homodimer formation.
Stress response proteins known to be induced by arsenic treatment (37).
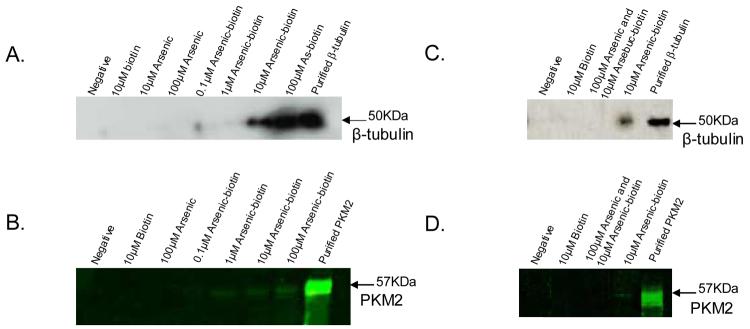
Confirmation of binding of β-tubulin and PKM2 by arsenic
Two proteins, β-tubulin and PKM2, were further confirmed as arsenic-binding proteins using in vitro binding assays. β-tubulin was chosen because it has already been known to be bound by arsenic (15), although the mechanism of binding was not clear. As an isoform of pyruvate kinase, PKM2 plays a key role in glycolysis, and is dramatically increased in a metabolic state characteristic of tumor cells (38).
Arsenic-biotin conjugate was first incubated with either purified β-tubulin (from MCF-7 cells) and purified PKM2 (from rabbit muscle) and then treated with streptavidin. Under these condtions, arsenic-biotin conjugate (1, 10, and 100 μM) pulled down detectable β-tubulin or PKM2 in a concentration-dependent manner (Fig. 3A, 3B). As expected, neither p-aminophenylarsenoxide nor biotin alone could pull down proteins. Similar to this experiment, we also treated MCF-7 cells with arsenic-biotin conjugate. After being pulled down by streptavidin, β-tubulin and PKM2 were identified by specific antibodies (Fig. 3C, 3D). Biotin itself failed to pull down these proteins. Excessive free phenylarsenoxide (10:1 ratio) competed with the arsenic-biotin conjugate in binding to both β-tubulin and PKM2. These data confirmed both β-tubulin and PKM2 as arsenic-binding proteins in MCF-7 cells.
Figure 3.
Binding assay with purified proteins and MCF-7 cell lysate. A) binding assay with purified MCF-7 tubulin preparation; B) binding assay with purified rabbit muscle pyruvate kinase; C) binding assay with MCF-7 cells for β-tubulin; and D) binding assay with MCF-7 cells for PKM2.
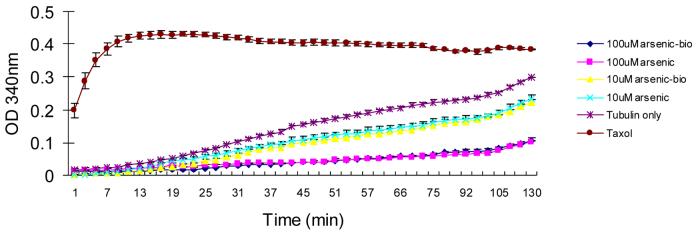
Functional consequence of arsenic binding with β-tubulin and PKM2
An in vitro microtubule assembly assay was performed to determine the functional consequence of arsenic binding on β-tubulin. Prior treatment of monomeric tubulin with arsenic-biotin conjugate or p-aminophenylarsenoxide markedly inhibited GTP-induced polymerization and microtubule formation in a dose-dependent effect (Fig. 4). Arsenic-biotin conjugate had similar effects as p-aminophenylarsenoxide suggesting that conjugation with biotin did not affect the activity of arsenic at either dose (10 or 100μM). Taxol served as a control since it is known to enhance tubulin polymerization (39).
Figure 4.
Arsenic and arsenic-biotin conjugate inhibit GTP-induced tubulin polymerization in vitro. Monomeric tubulin (5μg/μl) was incubated with different concentrations of arsenic-biotin conjugate or p-aminophenylarsenoxide. The formation of microtubules was determined by measuring OD340nm at room temperature. Tubulin only or addition of taxol (10μM) was used as controls, and taxol is known to promote tubulin polymerization.
Pyruvate kinase activity was analyzed with a classical enzyme assay using purified enzymes. It was surprising to find that neither arsenic nor arsenic-biotin conjugate inhibited the activity of purified pyruvate kinase (data not shown). Since several putative arsenic-binding proteins were enzymes related to glycolysis, ATP assay was then conducted to detect potential effects of arsenic treatment on glycolysis. Again, neither arsenic-biotin conjugate nor arsenic had any apparent effects on ATP production in MCF-7 cell (data not shown).
Molecular modeling of arsenic binding with β-tubulin and PKM2
In order to determine how arsenic might bind β-tubulin and PKM2, and explain why the binding had inhibitory effects on β-tubulin, but no effects on PKM2 activity, we conducted molecular modeling of the binding between arsenic and β-tubulin or PKM2. Since arsenic is known to bind Cys residues, we aimed to determine whether arsenic binding to certain Cys residues might interfere with the conformations or block the active sites of these proteins.
To determine the most favorable Cys residues which are close to the GTP-binding site of β2 subunit of human tubulin and may be bound by arsenic, the potential energy values of p-aminophenylarsenoxide-Cys conjugates were calculated. We found the energy of arsenic-Cys12 was −6731.62 kcal/mol, arsenic-Cys203, −6701.91 kcal/mol, arsenic-Cys213, −6711.52 kcal/mol, and arsenic-Cys305, −6732.74 kcal/mol, respectively. This order of potential energy, Cys12 ≈ Cys305 > Cys213 > Cys203, suggested that Cys12 or Cys305 of β-tubulin was likely the Cys residue of choice for arsenic binding. It appeared that arsenic binding to Cys12 alone would be able to block GTP binding, whereas arsenic binding to Cys305 would not due to its distance from the GTP-binding site (Fig. 5A). According to the model of human tubulin, two Cys residues (Cys241 and Cys356) are embedded inside the molecule, and thus are not accessible to arsenic. Cys129 and Cys131 are far away from the GTP-binding site, and therefore are not likely to be the target residues of arsenic binding.
Figure 5.

Three-dimensional structures of human β-tubulin, PKM2 and their interactions with arsenic. A) Homology model of human β2-tubulin based on the X-ray structure of bovine β-tubulin (PDB code: 1JFF), and its binding by arsenic at Cys12. For comparison, all the Cys residues of human β2-tubulin are represented in ball-and-stick depiction: Cys12, Cys203, Cys213 and Cys305 are located near the GTP binding site, while Cys129, Cys131, Cys241 and Cys356 are distantly located from the GTP binding site and near the interface with the α subunit. The α-helical motif containing Cys213 and the adjacent loop labile to structural modification are colored in blue. B) Plausible conformation of human β-tubulin with Cys12 and Cys213 bound by arsenic. The α-helical motif containing Cys213 (Glu207-Tyr224) has been modified accordingly. This motif and the adjacent loop labile to structural modification are colored in red. Hydrogen atoms are omitted to clarify the view. C) GTP-binding site of human β-tubulin indicated by dot depiction (in magenta). The closest Cys residue from the active site is Cys12 with a distance of 2.60Å between the GDP sugar ring oxygen and the side chain sulfur of Cys12. D) Active site of human PKM2 (PDB code: 1T5A), indicated by dot depiction (in magenta) for oxalate and phosphate. The closest Cys residue from the active site is Cys326 with a distance of 13.80Å between the active site Mg2+ and the side chain sulfur of Cys326. Color coding: green, C; red, O; blue, N; yellow, S; and magenta, phosphate and arsenic.
We also investigated the possibility of binding two vicinal Cys residues by arsenic. Theoretically, such a binding could be more efficient and stable in blocking the access of GTP-binding site. Three Cys residues (Cys203, Cys213 and Cys305) are relatively close to Cys12 (40). Cys203 was a less likely candidate because it is part of the β sheet bundle in the core region, which makes it least labile to form the arsenic conjugate. Based on the structure of human tubulin, the structural motif containing Cys213 (Asp205-Tyr224) is likely to be modified to form a double conjugate to arsenic, i.e., arsenic-(Cys12)(Cys213)(Fig. 5B). This conformation contained a total of 54 hydrogen bonds and showed the lowest energy when the motif was folded toward Cys12 as compared with other potential conformations.
We also outlined the active sites of β-tubulin and PKM2 (Fig. 5C, 5D). We found that the distance between GDP sugar ring oxygen and the side chain sulfur of β-tubulin Cys12 was 2.60 Å. The Cys residue of PKM2 closest to its active site was Cys326, and the distance between the active site Mg2+ and the side chain sulfur of Cys326 was 13.80 Å. It suggested that arsenic was not likely to block the active site and thus inhibit enzyme activity of PKM2, even when Cys326 was bound by arsenic.
Discussion
Since arsenic is a well-known toxin and a cancer chemotherapeutic drug, dissection of its mechanism of action has been an area of active investigation. In this study, we isolated 50 potential arsenic-binding proteins in MCF-7 cells using a p-aminophenylarsenoxide-biotin conjugate. Binding of arsenic with β-tubulin and PKM2 was further confirmed by binding assays. Molecular modeling suggested that functional consequences of such binding depended on the position of target Cys residues relative to the active sites of arsenic-binding proteins.
Among the potential arsenic-binding proteins in MCF-7 cells (Table 1), some are known to be bound by arsenic (e.g., tubulin, actin, protein disulfide isomerase, peroxiredoxin), and some induced by arsenic (e.g., stress proteins). It was interesting that several glycolytic enzymes (e.g., glyceraldehydes-3-phosphate dehydrogenase, phosphoglycerate kinase, enolase, PKM2, lactate dehydrogenase, glucose 6-phosphate dehydrogenase) were isolated in this study, and they were known to form glycolytic enzyme complex in MCF-7 cells (41, 42). Tubulin has been reported to interact with glycolytic enzymes with its C-terminal region (43), and such interaction destabilizes the microtubule (44). Furthermore, PKM2 has been associated with cancer in human patients and animal models (38, 45, 46). Arsenic inhibited the activity of pyruvate dehydrogenase complex, which catalyzes the oxidative decarboxylation of the end product of glycolysis, pyruvate, to form acetylCoA. Enzyme inhibition may take place mainly as a result of oxidative damage to the enzyme (47). Based on these evidence, we hypothesized that the biological effects of arsenic may be mediated by its binding to Cys residues of β-tubulin and PKM2, since arsenic is known to bind proteins containing vicinal SH groups (48, 49).
Previous studies with synthetic peptides conjugated with phenylarsenoxide have shown that no amino acids other than Cys bind arsenic. Binding depends on the number, accessibility and relative positioning of Cys residues (50). For example, rat hemoglobin has three Cys residues in each α chain and two in each β chain, while human hemoglobin contains only one Cys residue in each α chain and two in each β chain. As a result, rat hemoglobin had a much higher binding affinity to arsenic than the human protein (20). Human β-tubulin has a total of eight Cys residues. Based upon our human tubulin model (Figure 5A), Cys12, Cys213 and Cys305, which are close to the GTP binding site, appeared to be accessible to arsenic. In particular, Cys12 is right at the GTP binding site, and the potential energy at Cys12 was one of the lowest, suggesting that Cys12 would be a critical residue for inhibition of tubulin polymerization by arsenic. This is consistent with a previous study with S. cerevisiae, which showed that Cys12 played important roles in the control of tubulin function and structure. C12S mutation was lethal probably by destabilizing the interaction of tubulin with GTP (51). From the three-dimensional model of β-tubulin-Cys12-arsenic complex (Figure 5A), it appeared that binding of arsenic to Cys12 alone would be sufficient to block GTP binding.
An alternative model is that arsenic binds to Cys12 and then another Cys residue, Cys213 or Cys305, which is relatively close to Cys12, to block the GTP binding site, as previously proposed by Li and Broome (4). It is generally believed that cyclic dithioarsenites are more stable than the noncyclic products formed from trivalent arsenicals and monothiols. The possibility of forming the binding complex with both Cys12 and Cys213 was also supported by the experimental observation that a cross-linkage could be formed between these two residues, which was estimated to be ∼9 Å (40). Based on our tubulin model, it is likely that the α-helical structural motif containing Cys213, which exists on the solvent-accessible surface of the proteins, would be able to modify its structure and be accessible to Cys12 through the formation of arsenic binding complex (Figure 5B). In this model, the altered conformation of the motif that contains Cys213 would efficiently block the GTP binding site. This is consistent with previous observation that arsenic binding can modulate α-helical secondary structure (52). However, hydrophobicity may contribute to the affinity of arsenic for proteins. In the case of hemoglobin, binding to one SH group was stronger than binding to two SH groups (20). Further studies are needed to confirm the exact mode of arsenic binding to β-tubulin.
We further speculate that formation of the arsenic-Cys binding conjugate may inhibit the functions of the target proteins, if the binding causes conformational changes or blocks active sites (52, 53). However, if critical Cys residues are not in the active site, Cys305 of β-tubulin for example, of an arsenic-binding protein, or binding does not change the conformation of a target protein, such binding may have no functional consequences. The present study showed that arsenic binding failed to inhibit enzyme activity of PKM2. It could be explained by the PKM2 structure (54): Cys326, the closest Cys residue to the active site, is still far away from the active site of PKM2, and Cys326 appeared to be buried inside the protein and thus inaccessible to arsenic. This is in agreement with our data that arsenic binding had no effect on pyruvate kinase activity of either purified enzyme or MCF-7 cell lysate. Furthermore, arsenic treatment had no effect on the level of ATP in MCF-7 cells. These observations are reminiscent of several arsenic-binding enzymes (e.g., DNA polymerase β, DNA ligase I and DNA ligase III), whose activies were insensitive to arsenic even though they had critical SH groups for arsenic binding (55).
The approach using compound-biotin conjugate and proteomic techniques was successful in identifying methionine aminopeptedase as a molecular target of an angiogenesis inhibitor, fumagillin (56, 57). A similar approach was also used to identify protein disulfide isomerase as an arsenic-binding protein (25). Since most proteins identified in this study were relatively abundant in MCF-7 cells, we believe further improvement of this technique is needed to identify low-abundance arsenic-binding proteins.
Acknowledgement
This work is supported by a research grant from NIH (U56 CA092077) and by the intramural research program of NIDDK and NCI.
The abbreviations used are
- Cys
cysteine
- MALDI-MS
matrix assisted laser desorption ionization mass spectrometry
- MD
molecular dynamics
- PKM2
pyruvate kinase M2
- SA
simulated annealing
- SH
sulfhydryl
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Hayakawa F, Privalsky ML. Phosphorylation of PML by mitogen-activated protein kinases plays a key role in arsenic trioxide-mediated apoptosis. Cancer Cell. 2004;5:389–401. doi: 10.1016/s1535-6108(04)00082-0. [DOI] [PubMed] [Google Scholar]
- 2.Bode AM, Dong Z. The paradox of arsenic: molecular mechanisms of cell transformation and chemotherapeutic effects. Crit Rev Oncol Hematol. 2002;42:5–24. doi: 10.1016/s1040-8428(01)00215-3. [DOI] [PubMed] [Google Scholar]
- 3.Liu SX, Davidson MM, Tang X, et al. Mitochondrial damage mediates genotoxicity of arsenic in mammalian cells. Cancer Res. 2005;65:3236–3242. doi: 10.1158/0008-5472.CAN-05-0424. [DOI] [PubMed] [Google Scholar]
- 4.Li YM, Broome JD. Arsenic targets tubulins to induce apoptosis in myeloid leukemia cells. Cancer Res. 1999;59:776–780. [PubMed] [Google Scholar]
- 5.Andrew AS, Warren AJ, Barchowsky A, et al. Genomic and proteomic profiling of responses to toxic metals in human lung cells. Environ Health Perspect. 2003;111:825–835. doi: 10.1289/ehp.111-1241504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller WH, Jr., Schipper HM, Lee JS, Singer J, Waxman S. Mechanisms of action of arsenic trioxide. Cancer Res. 2002;62:3893–3903. [PubMed] [Google Scholar]
- 7.Hei TK, Liu SX, Waldren C. Mutagenicity of arsenic in mammalian cells: role of reactive oxygen species. Proc Natl Acad Sci U S A. 1998;95:8103–8107. doi: 10.1073/pnas.95.14.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao CQ, Young MR, Diwan BA, Coogan TP, Waalkes MP. Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression. Proc Natl Acad Sci U S A. 1997;94:10907–10912. doi: 10.1073/pnas.94.20.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen GC, Guan LS, Hu WL, Wang ZY. Functional repression of estrogen receptor a by arsenic trioxide in human breast cancer cells. Anticancer Res. 2002;22:633–638. [PubMed] [Google Scholar]
- 10.Chow SK, Chan JY, Fung KP. Suppression of cell proliferation and regulation of estrogen receptor alpha signaling pathway by arsenic trioxide on human breast cancer MCF-7 cells. J Endocrinol. 2004;182:325–337. doi: 10.1677/joe.0.1820325. [DOI] [PubMed] [Google Scholar]
- 11.Chow SK, Chan JY, Fung KP. Inhibition of cell proliferation and the action mechanisms of arsenic trioxide (As2O3) on human breast cancer cells. J Cell Biochem. 2004;93:173–187. doi: 10.1002/jcb.20102. [DOI] [PubMed] [Google Scholar]
- 12.Li X, Ding X, Adrian TE. Arsenic trioxide causes redistribution of cell cycle, caspase activation, and GADD expression in human colonic, breast, and pancreatic cancer cells. Cancer Invest. 2004;22:389–400. doi: 10.1081/cnv-200029068. [DOI] [PubMed] [Google Scholar]
- 13.Baj G, Arnulfo A, Deaglio S, et al. Arsenic trioxide and breast cancer: analysis of the apoptotic, differentiative and immunomodulatory effects. Breast Cancer Res Treat. 2002;73:61–73. doi: 10.1023/a:1015272401822. [DOI] [PubMed] [Google Scholar]
- 14.Aposhian HV, Aposhian MM. Arsenic toxicology: five questions. Chem Res Toxicol. 2006;19:1–15. doi: 10.1021/tx050106d. [DOI] [PubMed] [Google Scholar]
- 15.Menzel DB, Hamadeh HK, Lee E, et al. Arsenic binding proteins from human lymphoblastoid cells. Toxicol Lett. 1999;105:89–101. doi: 10.1016/s0378-4274(98)00380-4. [DOI] [PubMed] [Google Scholar]
- 16.Kapahi P, Takahashi T, Natoli G, et al. Inhibition of NF-kappa B activation by arsenite through reaction with a critical cysteine in the activation loop of Ikappa B kinase. J Biol Chem. 2000;275:36062–36066. doi: 10.1074/jbc.M007204200. [DOI] [PubMed] [Google Scholar]
- 17.Shi W, Dong J, Scott RA, Ksenzenko MY, Rosen BP. The role of arsenic-thiol interactions in metalloregulation of the ars operon. J Biol Chem. 1996;271:9291–9297. doi: 10.1074/jbc.271.16.9291. [DOI] [PubMed] [Google Scholar]
- 18.Lam WC, Tsao DH, Maki AH, Maegley KA, Reich NO. Spectroscopic studies of arsenic(III) binding to Escherichia coli RI methyltransferase and to two mutants, C223S and W183F. Biochemistry. 1992;31:10438–10442. doi: 10.1021/bi00158a004. [DOI] [PubMed] [Google Scholar]
- 19.Winski SL, Carter DE. Interactions of rat red blood cell sulfhydryls with arsenate and arsenite. J Toxicol Environ Health. 1995;46:379–397. doi: 10.1080/15287399509532043. [DOI] [PubMed] [Google Scholar]
- 20.Lu M, Wang H, Li XF, et al. Evidence of hemoglobin binding to arsenic as a basis for the accumulation of arsenic in rat blood. Chem Res Toxicol. 2004;17:1733–1742. doi: 10.1021/tx049756s. [DOI] [PubMed] [Google Scholar]
- 21.Loring RH, Dou YM, Lane W, Jones GS, Jr., Stevenson KJ. Aromatic trivalent arsenicals: covalent yet reversible reagents for the agonist binding site of nicotinic receptors. Brain Res Mol Brain Res. 1992;15:113–120. doi: 10.1016/0169-328x(92)90158-8. [DOI] [PubMed] [Google Scholar]
- 22.Ngu TT, Stillman MJ. Arsenic binding to human metallothionein. J Am Chem Soc. 2006;128:12473–12483. doi: 10.1021/ja062914c. [DOI] [PubMed] [Google Scholar]
- 23.Lin CH, Huang CF, Chen WY, et al. Characterization of the interaction of galectin-1 with sodium arsenite. Chem Res Toxicol. 2006;19:469–474. doi: 10.1021/tx0503348. [DOI] [PubMed] [Google Scholar]
- 24.Chang KN, Lee TC, Tam MF, et al. Identification of galectin I and thioredoxin peroxidase II as two arsenic-binding proteins in Chinese hamster ovary cells. Biochem J. 2003;371:495–503. doi: 10.1042/BJ20021354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donoghue N, Yam PT, Jiang XM, Hogg PJ. Presence of closely spaced protein thiols on the surface of mammalian cells. Protein Sci. 2000;9:2436–2445. doi: 10.1110/ps.9.12.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stancato LF, Hutchison KA, Chakraborti PK, Simons SS, Jr., Pratt WB. Differential effects of the reversible thiol-reactive agents arsenite and methyl methanethiosulfonate on steroid binding by the glucocorticoid receptor. Biochemistry. 1993;32:3729–3736. doi: 10.1021/bi00065a027. [DOI] [PubMed] [Google Scholar]
- 27.Stoica A, Pentecost E, Martin MB. Effects of arsenite on estrogen receptor-alpha expression and activity in MCF-7 breast cancer cells. Endocrinology. 2000;141:3595–3602. doi: 10.1210/endo.141.10.7704. [DOI] [PubMed] [Google Scholar]
- 28.Bogdan GM, Sampayo-Reyes A, Aposhian HV. Arsenic binding proteins of mammalian systems: I. Isolation of three arsenite-binding proteins of rabbit liver. Toxicology. 1994;93:175–193. doi: 10.1016/0300-483x(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 29.Moaddel R, Sharma A, Huseni T, et al. Novel biotinylated phenylarsonous acids as bifunctional reagents for spatially close thiols: studies on reduced antibodies and the agonist binding site of reduced Torpedo nicotinic receptors. Bioconjug Chem. 1999;10:629–637. doi: 10.1021/bc9801575. [DOI] [PubMed] [Google Scholar]
- 30.Stevenson KJ, Hale G, Perham RN. Inhibition of pyruvate dehydrogenase multienzyme complex from Escherichia coli with mono- and bifunctional arsenoxides. Biochemistry. 1978;17:2189–2192. doi: 10.1021/bi00604a026. [DOI] [PubMed] [Google Scholar]
- 31.Korshun V, Pestov N, Nozhevnikova E. Reagents for multiple non-radioactive labeling of oligonucleotides. Syn Commun. 1996;26:2531–2547. [Google Scholar]
- 32.Chen X, Ding Y, Liu CG, Mikhail S, Yang CS. Overexpression of glucose-regulated protein 94 (Grp94) in esophageal adenocarcinomas of a rat surgical model and humans. Carcinogenesis. 2002;23:123–130. doi: 10.1093/carcin/23.1.123. [DOI] [PubMed] [Google Scholar]
- 33.Shi S, Yan L, Yang Y, Fisher-Shaulsky J, Thacher T. An extensible and systematic force field, ESFF, for molecular modeling of organic, inorganic, and organometallic systems. J Comput Chem. 2003;24:1059–1076. doi: 10.1002/jcc.10171. [DOI] [PubMed] [Google Scholar]
- 34.Press WHTS, Vetterling WT. Flannery BP Numerical recipes in C: the art of scientific computing. Cambridge University; New York: 1992. pp. 420–424. [Google Scholar]
- 35.Higgins DG, Thompson JD, Gibson TJ. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 36.Lowe J, Li H, Downing KH, Nogales E. Refined structure of alpha beta-tubulin at 3.5 A resolution. J Mol Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 37.Del Razo LM, Quintanilla-Vega B, Brambila-Colombres E, et al. Stress proteins induced by arsenic. Toxicol Appl Pharmacol. 2001;177:132–148. doi: 10.1006/taap.2001.9291. [DOI] [PubMed] [Google Scholar]
- 38.Zhang B, Chen JY, Chen DD, Wang GB, Shen P. Tumor type M2 pyruvate kinase expression in gastric cancer, colorectal cancer and controls. World J Gastroenterol. 2004;10:1643–1646. doi: 10.3748/wjg.v10.i11.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snyder JP, Nettles JH, Cornett B, Downing KH, Nogales E. The binding conformation of Taxol in beta-tubulin: a model based on electron crystallographic density. Proc Natl Acad Sci U S A. 2001;98:5312–5316. doi: 10.1073/pnas.051309398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Little M, Luduena RF. Location of two cysteines in brain beta 1-tubulin that can be cross-linked after removal of exchangeable GTP. Biochim Biophys Acta. 1987;912:28–33. doi: 10.1016/0167-4838(87)90243-3. [DOI] [PubMed] [Google Scholar]
- 41.Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 42.Mazurek S, Hugo F, Failing K, Eigenbrodt E. Studies on associations of glycolytic and glutaminolytic enzymes in MCF-7 cells: role of P36. J Cell Physiol. 1996;167:238–250. doi: 10.1002/(SICI)1097-4652(199605)167:2<238::AID-JCP7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 43.Volker KW, Knull H. A glycolytic enzyme binding domain on tubulin. Arch Biochem Biophys. 1997;338:237–243. doi: 10.1006/abbi.1996.9819. [DOI] [PubMed] [Google Scholar]
- 44.Kovacs J, Low P, Pacz A, et al. Phosphoenolpyruvate-dependent tubulin-pyruvate kinase interaction at different organizational levels. J Biol Chem. 2003;278:7126–7130. doi: 10.1074/jbc.M210244200. [DOI] [PubMed] [Google Scholar]
- 45.Steinberg P, Klingelhoffer A, Schafer A, et al. Expression of pyruvate kinase M2 in preneoplastic hepatic foci of N-nitrosomorpholine-treated rats. Virchows Arch. 1999;434:213–220. doi: 10.1007/s004280050330. [DOI] [PubMed] [Google Scholar]
- 46.Mazurek S, Grimm H, Boschek CB, Vaupel P, Eigenbrodt E. Pyruvate kinase type M2: a crossroad in the tumor metabolome. Br J Nutr. 2002;87(Suppl 1):S23–29. [PubMed] [Google Scholar]
- 47.Samikkannu T, Chen CH, Yih LH, et al. Reactive oxygen species are involved in arsenic trioxide inhibition of pyruvate dehydrogenase activity. Chem Res Toxicol. 2003;16:409–414. doi: 10.1021/tx025615j. [DOI] [PubMed] [Google Scholar]
- 48.Gebel T. Arsenic and antimony: comparative approach on mechanistic toxicology. Chem Biol Interact. 1997;107:131–144. doi: 10.1016/s0009-2797(97)00087-2. [DOI] [PubMed] [Google Scholar]
- 49.Snow ET. Metal carcinogenesis: mechanistic implications. Pharmacol Ther. 1992;53:31–65. doi: 10.1016/0163-7258(92)90043-y. [DOI] [PubMed] [Google Scholar]
- 50.Kitchin KT, Wallace K. Arsenite binding to synthetic peptides based on the Zn finger region and the estrogen binding region of the human estrogen receptor-alpha. Toxicol Appl Pharmacol. 2005;206:66–72. doi: 10.1016/j.taap.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 51.Gupta ML, Jr., Bode CJ, Dougherty CA, Marquez RT, Himes RH. Mutagenesis of beta-tubulin cysteine residues in Saccharomyces cerevisiae: mutation of cysteine 354 results in cold-stable microtubules. Cell Motil Cytoskeleton. 2001;49:67–77. doi: 10.1002/cm.1021. [DOI] [PubMed] [Google Scholar]
- 52.Cline DJ, Thorpe C, Schneider JP. Effects of As(III) binding on alpha-helical structure. J Am Chem Soc. 2003;125:2923–2929. doi: 10.1021/ja0282644. [DOI] [PubMed] [Google Scholar]
- 53.Kitchin KT, Wallace K. Dissociation of arsenite-peptide complexes: triphasic nature, rate constants, half-lives, and biological importance. J Biochem Mol Toxicol. 2006;20:48–56. doi: 10.1002/jbt.20108. [DOI] [PubMed] [Google Scholar]
- 54.Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44:9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- 55.Hu Y, Su L, Snow ET. Arsenic toxicity is enzyme specific and its affects on ligation are not caused by the direct inhibition of DNA repair enzymes. Mutat Res. 1998;408:203–218. doi: 10.1016/s0921-8777(98)00035-4. [DOI] [PubMed] [Google Scholar]
- 56.Griffith EC, Su Z, Niwayama S, et al. Molecular recognition of angiogenesis inhibitors fumagillin and ovalicin by methionine aminopeptidase 2. Proc Natl Acad Sci U S A. 1998;95:15183–15188. doi: 10.1073/pnas.95.26.15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sin N, Meng L, Wang MQ, et al. The anti-angiogenic agent fumagillin covalently binds and inhibits the methionine aminopeptidase, MetAP-2. Proc Natl Acad Sci U S A. 1997;94:6099–6103. doi: 10.1073/pnas.94.12.6099. [DOI] [PMC free article] [PubMed] [Google Scholar]