Abstract
Initial gene discovery efforts through analysis of genome sequences and identification and characterization of expressed RNAs have revealed that only a relatively small portion of the genome is transcribed into protein coding mRNAs in vertebrates. However, in contrast with this paucity of protein coding ‘genes’, there is an enormous complexity in transcription and the protein coding mRNAs contribute to a very small fraction of transcripts in comparison with the different varieties of non-coding RNAs (ncRNAs). This transcriptome complexity may be hypothesized to have a regulatory role that is required for the development and function of organisms as complex as vertebrates. At the same time, it raises the fundamental question of the unequivocal definition of a gene. It is intriguing to postulate that many ncRNAs might finely modulate gene activity by acting as regulatory elements. The emerging hypotheses suggest that the gene regulatory machinery may be deeply interconnected with the world of short RNAs. These RNAs may generally act for fine-tuning of the protein-coding transcriptome.
Keywords: transcriptome, non-coding RNAs, sense–antisense transcription, cDNA annotation, RNA processing
1. Introduction
At the onset of the new millennium, the realization that the genome encoded for far fewer genes than previously anticipated was particularly surprising.1,2 In parallel with the sequencing of the genome, sequencing of DNA copied from cellular mRNA (cDNAs) made key contributions to hunting for genes encoded in the genome. Early investigations centred on the identification of protein coding mRNAs, following the traditional idea that the final products of mammalian genes are mainly proteins. This practice of limiting the gene search to mRNAs encoding proteins had its roots in classic biological studies. Built on the central dogma of molecular biology and learned from annotating the sequences of bacterial genomes, we sought out a protein-centric view of biology. Accordingly, projects such as the Mammalian Gene Collection (MGC)3 targeted only the long (protein coding) mRNAs. The MGC focused on sequencing only those full-length cDNAs which showed coding potential after an initial screening of clones with 5′-end sequence analysis. Similarly, another project that focused on expanding the collection of large protein coding cDNAs, considered coding potential as a pre-requisite for further sequencing.4 This made perfect sense, because full-length cDNA sequencing has been a very intense and expensive operation. Also, there was no evidence that other forms of RNA could be major functional components of the transcriptome. However, in the FANTOM project, the sequencing of full-length cDNAs based primarily on the novelty of end sequences regardless of coding potential brought forth a surprising new world of non-coding RNAs (ncRNAs). Indeed, the number of long ncRNAs was revealed to be greater than the protein coding RNAs, exceeding 23 000 transcriptional units in mouse.5 Analysis of public human cDNA libraries also identified ncRNAs,6 but to a much lower extent, due to its focus on protein coding RNAs.3 Subsequently, studies of expressed sequences in human cells using tiling arrays7 revealed a similar complexity of ncRNAs in human cells.8,9 The fact that the genome is pervasively transcribed into long ncRNAs has only recently been accepted. It is now known that a large fraction of the transcriptome is constituted of long and short ncRNAs (up to 93% of the genome is transcribed), with disparate functions, many still unknown. These include transcriptional regulation by sense–antisense,5 activation or inactivation of transcription through regulatory regions, and numerous other functions that have been reviewed elsewhere.10–12
In parallel, the discovery of RNA interference (RNAi) and micro-RNAs has provided the notion that at least some of the longer RNAs are cleaved into smaller, functional short RNAs (sRNAs are defined as RNAs shorter than 200 nt and therefore non-coding).13 A summary of the families of RNAs discussed in this review is provided in Table 1. For almost a decade, the research community has been focused on miRNAs, siRNAs, and few other sRNAs, allowing the field to flourish, with particular emphasis on miRNA as translational regulators.14 However, further integrated understanding of the relationship between long ncRNAs and sRNAs worlds is still in its infancy and only recently links between these two classes are finally becoming evident. In fact, novel discoveries are pointing at a much larger complexity of cleavage patterns, suggesting that the same region of the genome can encode for long mRNAs and a plethora of other long ncRNAs and sRNAs (Table 1). These RNAs are overlapping and they may partially arise from the same primary transcripts.
Table 1.
Definition of RNAs classes discussed in this review
| Short name of RNA classes | Full name of RNA classes | Notes | References |
|---|---|---|---|
| PALRs | Promoter-associated long RNAs | Hundreds nt long RNAs spanning regions on proximal promoters to the first exon | 29,30 |
| PASRs | Promoter-associated short RNAs | 20–70 nt long RNAs spanning regions around core promoters | 29,30 |
| TASRs | Termini-associated short RNAs | 20–70 nt long RNAs spanning regions around transcription termination sites | 29,30 |
| PROMPTs | Promoter upstream transcripts | Unstable transcripts mapping 0.5–2 kb upstream the transcription starting sites | 35 |
| TSSa-RNAs | Transcription start sites antisense RNAs | RNAs, generally short and non-coding, generated from bidirectional activity of mammalian RNA Polymerase II | 31 |
| NRO-RNAs | Nuclear run-on assay derived RNAs | Short RNA detected by nuclear run-on assays, mapping 20 to 50 downstream to transcriptions starting sites of mRNAs | 32 |
| RE RNAs | Retrotransposon-derived RNAs | A heterogeneous class of RNAs which starting sites overlap retrotransposon elements | 42,43 |
| tiRNAs | Tiny transcription initiation RNAs | RNAs about 18 nt long, positioned about 20 bp after the transcription starting sites of highly expressed mRNAs | 33 |
| snoRNAs | Small nucleolar RNAs | Small ncRNAs that guide chemical modifications of other non-coding RNAs | 22 |
| siRNAs | Small interfering RNAs | Double-stranded RNA molecules, 20–25 nucleotides in length, that act in various silencing pathways | 13,14,51,52 |
| miRNAs | microRNAs | Single-stranded RNA molecules of 21–24 nucleotides in length, which regulate gene expression | 13,51,52 |
| LincRNAs | Large intervening non-coding RNAs | Large non-coding RNAs that map in intergenic locations | 55 |
| ncRNAs | Non-coding RNAs | Generic definition for non-protein coding RNAs | 10,15,24,25,29–35,50–55 |
| sRNA | Short RNAs | Generic definition for short RNAs | 29–35,50–55 |
| snRNA | Small nuclear RNAs | Nuclear small non-coding RNAs involved in various functions including splicing | 48 |
| piRNA | Piwi interacting RNAs | 26–31 nt long RNAs involved in transcriptional gene silencing, including retrotransposons | 51–53 |
Here, I will discuss emerging aspects of transcription complexity, starting from the complexity of primary transcript production. Next, I will discuss the recent identification of a variety of novel shortened yet natural, ncRNA isoforms, their relationship with the genes they overlap, and their biogenesis. Finally, I will discuss their potential function and relate it to the classic concept of a ‘gene’, which is challenged by the identification of these novel RNAs.
2. ncRNAs and isoform complexity
There is growing evidence that genes for both coding and ncRNAs produce various RNA isoforms that are shorter than the full-length transcripts, which can be primarily produced either as sRNA or alternatively by the cleavage of longer RNAs. Main pathways have been described for the production of miRNA, as well as other RNAs.13 As miRNAs are thought to be produced from introns or from independent ncRNAs, the degradation processes that result in their biogenesis (including Drosha and Dicer cleavages) have long been recognized as those that are physiologically involved in the production of functional molecules.
To some extent, less is known about other sRNAs. Although there is also evidence that the degradation of other RNA occurs, it has been difficult to distinguish between (i) experimental artefacts, (ii) natural RNAs that are degradation products, and (iii) natural sRNAs that have a functional role. The scepticism of many investigators derives from the notorious instability of RNAs, which are susceptible to artificial degradation during experimental manipulations, resulting in experimental artefacts. The use of second generation sequencers is expected to resolve this issue. Since the number of RNAs that can be analysed per sample is very large (routinely obtaining 10–100 millions sequences/sample),15 there is a good chance to distinguish reproducible natural cleavage patterns from experimental artefacts. Together with experimental validation, this will ultimately help convince the sceptics, moving the focus from verification of existence to the search for biological function.
3. Truncated versions of ncRNAs with well established functions
There is growing evidence of functional RNAs that are truncated forms of tRNAs and snoRNAs. Fragments of specific length (30–40 nt) derived from tRNAs have been found to constitute a consistent part of the sRNA transcriptome.16,17 Several lines of evidence indicate that these are not random degradation products generated during RNA preparation, but truly, physiologically present in the cell. First, these short fragments, detectable as discrete bands by Northern blotting, have a non-random distribution, wherein only few tRNA genes contribute to a majority of the observed small RNAs. Secondly, onconase, an RNase produced by frog oocytes, was found to cleave tRNAs into smaller RNAs resulting in slowing the proliferation of cancerous cells.18 It has been shown that cleavage of tRNAs by the RNase Rny1p in yeast promotes Cell Death,19 and is involved in the stress response.6,20 Human tRNAse ZL was found to bind half-tRNA fragments in the cytosol, and it is now known that such complexes are capable of modifying mRNA expression levels, including those of genes involved in p53 and apoptosis.21 In addition, truncated tRNAs have been found to be essential for cell proliferation.16 Altogether, these data show that these RNAs can potentially act as regulators; most likely as messengers, signalling the arrest of cell proliferation. Given the number of mammalian RNAses A homologous to onconase, we can envisage novel findings related to the reutilization of processed tRNA fragments.
Truncated isoforms of snoRNAs have also been identified and there is now evidence of interplay between snoRNA and siRNA pathways particularly in cases of truncated snoRNA fragments derived from H/ACA snoRNAs (20–24 nt) and C/D snoRNAs (17–19 and longer than 27 nt).22
4. ncRNA variants of protein-coding mRNAs
Known variants of mRNAs are largely constituted of splicing variants, comprehensively reviewed elsewhere.23 Here, I will mainly focus on novel types of ncRNA variants derived from genome sequences that typically generate protein coding mRNAs. Additionally, there are also very large RNAs, likely to be ncRNAs, which span multiple genes and produce transcripts across multiple loci. For instance, a broad set of enormous ncRNA transcripts have been identified, which are often transcribed across genes, some of which are known imprinted ncRNAs,24 with multiple potential mechanisms of action.10 Previous genome-wide analyses were conservatively limited to transcripts spanning up to 2 Mb of genome length,25 identifying at least 181 000 transcripts, including >1500 transcripts that seem to contain sequences of two or more adjacent known genes. However, there is now evidence of transcripts spanning chromosomal regions larger than 2 Mb. Indeed, RACE has identified transcripts that originate and terminate in chromosomal regions that are as long as several megabases.26 It is not known whether these RNAs arise from transcription through very large regions, or from as yet not well characterized formation of RNA chimeras.27
Additionally, there are newly discovered ncRNAs that overlap known protein coding genes, with various possible biogenesis and role. These ncRNAs, discussed below in details (Fig. 1), overlap typical protein coding genes in various parts spanning from upstream regulatory and promoter regions down to the 3′ ends. A large majority of novel ncRNAs are localized around the ends and in particular, at 5′ ends, on the core promoters and to some extent also in the 3′ termination regions.
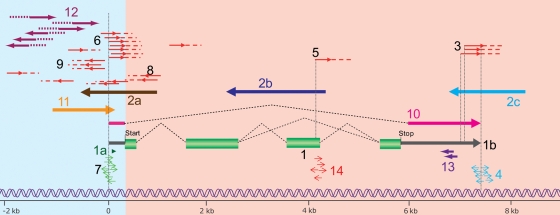
Figure 1.
Complexity of transcriptome around a hypothetical gene. Green boxes represent exons of typical protein coding gene. Not all the sRNAs and genomic elements are in scale. CAGE tags (red lines; dots beyond the arrowheads indicate than the 3′ ends are unknown) identify TSSs or other capped molecules; the dashed lines on CAGE tags indicate that the 3′ end is not determined. Various types of transcripts are indicated by numbers: (1) protein coding mRNA transcript gene (green boxes: coding exons, gray lines, 5′ and 3′ untranslated regions); (2, a–c), antisense RNAs in various relation with the transcripts (3′–3′ overlap, full overlap, 5′–5′ overlap); (3) CAGE tags identify transcript in the 3′-UTRs, likely polyadenylated; (4) termination-associated sRNAs (TASRs); (5) exonic long-capped transcripts; (6) CAGE tags identifying TSS (exact location can vary) and may overlap PALRs; (7) PASRs (green) and tiny 18 nt long RNAs (tiRNAs, arrowhead only); (8) antisense transcription events detected by CAGE, often (but not limited to) the first exons and introns; (9) bidirectionally transcribed RNAs from core promoters; (10) ncRNA splicing isoforms only partially overlapping to coding mRNA sequences; (11) PALRs; (12) PROMPTs, unstable transcripts on upstream regulatory regions; (13) miRNAs and endogenous siRNAs (deriving mostly from other loci, not perfectly matching in most cases in animal cells); (14) other sRNAs associated to exonic-capped long transcripts. The list of different types of RNAs is continuously growing and subject to revisions and further classifications.
A variety of long RNAs have been identified mapping around the 5′ ends of genes, with as yet undefined function and biogenesis (Fig. 1). Of particular note is the antisense transcription activity at core promoters. There is evidence that both long and short, most likely ncRNAs, are transcribed starting from the core promoter in the direction opposite to the promoter and overlapping the regions upstream of the promoters. Such long RNAs have been identified by cap-analysis gene-expression (CAGE),28 which does not allow the identification of their 3′ ends; however, these signals overlap tiling arrays signals, which have been instrumental to the discovery of the promoter-associated long RNAs (PALRs; longer than 100–200 nt) and promoter-associated sRNAs (PASRs; 20–100 nt long).29 It is likely that at least some of these RNAs are produced by the RNA polymerase II (Pol II). It is known that during transcription of the typical mRNAs and long ncRNAs, Pol II produces antisense RNAs for at least 50% of the promoters, and other defective short-sense RNAs in the direction of the main mRNA transcription, which might be the result of Pol II stalling on the promoters.30 At least a fraction of these RNAs are capped,30 but this capping is not limited to bidirectionally transcribed RNAs.
It is likely that PASRs and PALRs include heterogeneous classes of ncRNAs of various differing lengths and mapping positions. Three other groups have identified other types of short ncRNAs overlapping the promoters: TSSa-RNAs, NRO-RNAs, and the tiRNAs (Table 1). The TSSa-RNAs are sRNAs transcribed in the opposite direction to the transcripts of known genes.31 NRO-RNAs are transcribed bidirectionally and are sRNA elongating from the transcription starting sites (TSSs). They overlap with PASRs and PALRs, but they may be shorter and less stable.32 Additionally, a class of shorter RNAs associated with promoters (tiny RNAs or tiRNAs) has been recently discovered. tiRNAs have a narrow size distribution, on average 18 nt, and are specifically located about 20 nt downstream of the transcriptions start site.33 Their lack of overlap with the TSS suggests that tiRNAs are not simply abortive transcripts but are produced by some alternative mechanisms. In general, caution is advised in interpreting these new classes based on size, as the length may correspond to preferential sizes targeted in the experiments.
Not all PASRs necessarily overlap the TSSs, suggesting that at least some of these RNAs are not abortive Pol II products, but rather processed RNAs, some of which may have been cleaved and perhaps recapped30 to serve as yet unknown functions.
Further experiments are necessary to clarify and distinguish their function. Besides the possibility that they are simply by-products of RNA transcription and processing, there are exciting possibilities that they may be involved, together with various proteins, in the local and global regulation of the epigenomic machinery at the promoter level. It is likely that they are involved in triple helix regulation,34 or bound to various yet unknown proteins to regulate transcription (potential mechanisms discussed elsewhere10). Identification of their interactors (protein, RNA, promoters, 3D structures, other structures of the chromatin) will allow a more functional classification and a better understanding of whether these RNAs can be considered genes, because of their potential role as regulatory nuclear RNAs.
5. Transcripts diverging from the core promoters
Genomic regions that are transcribed on either or both of the two genomic strands, producing overlapping sets of transcripts, can be defined as transcriptional forests. Transcriptional forests can extend up to 1–2 kb upstream of the TSS of known mRNAs and PASRs. Transcripts upstream of core promoters, known as PROMPTs, are inherently unstable and could only be unambiguously identified by stabilizing the exosome.35 PROMPTs are thought to be longer than 500 nt, although their exact length has not been fully investigated. Because of their location and instability, they resemble a class of yeast RNA called cryptic unstable transcripts.36,37 PROMPTs are intriguing in their biogenesis. They are found to be transcribed in both orientations; pointing away from the mRNA coding gene and pointing towards the gene from 1 to 2 kb upstream regions. It seems probable that PROMPTs transcribed diverging away from the gene (or the main mRNA promoter) could be promoted from the bidirectional activity inherent of promoters. However, PROMPTs are also identified as being transcribed in the same orientation of the downstream sense mRNA (transcribed from the distal region towards the TSS of mRNAs). Intriguingly, chromatin immunoprecipitation (ChIP) analyses reveal that such PROMPTs do not have an upstream, distal promoter that could possibly transcribe RNAs towards the gene. This phenomenon suggests the existence of alternative mechanisms to produce PROMPTs, which do not depend on known types of promoters. One possibility is that these RNAs are produced by a mechanism which depends on RNA-dependent RNA polymerase (RDRP). Indeed, the human TERT, generally involved in the elongation of the telomeres, has recently been discovered in human cells to have RDRP38 activity. RNA could be transcribed outwards using the inherent bidirectional transcriptional activity of Pol II promoters. Next, RDRP activity could use any RNA secondary structure to prime transcription towards the original promoters. Such RNAs, or a part of them, would be degraded by the exosome, while some unknown fractions might be reprocessed to generate shorter RNAs. Indeed long PROMPTs were identified only upon stabilization of the exosome, but it seems likely that these transcripts may otherwise be processed into shorter RNA, such as the PASRs or the TSSa-RNAs, and act locally. In this regard, it is intriguing that CAGE signals identified capped RNAs overlapping PROMPTs and PASRs. Since cap structures functionally stabilize RNAs, it is likely that at least some of the PASRs may be produced by cleavage of long RNA followed by re-capping. It may well be that all these RNAs are a part of a complex machinery to produce regulatory RNAs around core promoters and upstream regions. Alternatively, PROMPTs might be transcribed without a promoter. The structure of the chromatin, genome-wide organized in loops,39,40 might be such that the core promoter elements are in close proximity not only with the upstream regulatory elements but also to upstream regions, which could then generate RNAs due to the high concentration of transcription machinery elements.
Transcription through a given area, like PROMPTs do, has consequences. Transcription often activates neighbouring genes, for instance, genes that respond early to external stimuli.41 There are several examples where transcription through regulatory regions can have either a repressor or an activator role, as reviewed elsewhere.10
Expanding on upstream transcripts, we have recently found that retrotransposon elements (REs) are transcribed in a tissue-specific manner, providing at least 200 000 TSS in the human genome. These RE elements show low-to-moderate expression levels.42,43 More than 117 000 human TSSs are positioned upstream of genes producing RNAs that are in the same orientation with the downstream protein coding mRNA. In more than two-third of the cases, these RNAs overlap the first exons, implying their potential as alternative promoters.43 However, further studies are necessary to comprehensively map their termination sites, it is plausible that a part of them constitute ncRNAs spanning promoters such as the PALRs or PROMPTs. The expression of these RE RNAs is mostly positively correlated with the expression of downstream genes. It is likely that the REs not working as alternative promoters might function by producing RNAs that activate the downstream chromatin regions. RE-derived RNAs may also form sense–antisense and sRNAs.43 Since the mRNAs that contain repeat elements in their 3′-UTR ends are generally less expressed than those devoid of them, these RNAs may also act in trans as negative regulators of transcription (and perhaps translation).
6. Shortened RNA recapping: is stabilization functional?
Surprisingly, at least a fraction of the PASRs and other shortened RNAs, whose 5′ ends map in the middle of mRNAs, have been found to carry a cap structure, most likely due to recapping after cleavage of larger RNAs.30 However, the cap has been considered a peculiarity of full-length mRNAs and some of the RNA polymerase III RNAs. Cap is known to stabilize RNAs, is this then suggesting that truncated RNAs are functionally relevant? There is evidence that transfection of different PASRs, mapping either upstream or downstream of the TSS of the MYC gene, negatively regulate the expression of the MYC mRNA, irrespective of the PASRs sense or antisense orientation.30
Identification of recapped RNAs is not limited to promoter regions. The CAGE technology44,45 identifies capping sites along various regions of long mRNAs. Besides known and novel promoters, CAGE signals also identify capped molecules overlapping exons. Existence of these capped RNAs was further verified by isolating them using an antibody directed against the cap, followed by sequencing.30 These capped exonic transcripts do not correlate with gene expression measured at the main promoter, suggesting that they are not randomly degraded products. Instead, truncated RNA CAGE signals correlate with a specific type of promoter. Promoters can be grouped based on their shape as broad or sharp. The TSSs are distributed over 50–100 bp and are often associated with CpG islands in broad promoters. In sharp promoters, TSSs originate mainly at a single genomic position and are often associated with TATA boxes.46 CAGE tags corresponding to truncated exonic transcripts are more frequently associated with the sharp TATA-box promoters.47 Although no mechanisms have so far been identified, it is tempting to hypothesize that the regulation of sharp promoters may require these seemingly aberrant exonic transcripts.
Interestingly, Gingeras group at CSHL have observed clusters of both long and sRNAs that overlap these exonic CAGE tag clusters.30 The sRNAs (<200 nt) are likely to be produced by cleavage and reprocessing of the longer RNAs into shorter RNAs.
What is the connection between associations of exonic-capped RNAs with TATA box, sharp promoters, and cleavage-recapping events? The association of exonic transcripts with genes having sharp promoters would suggest that these transcripts are involved in regulating specific type of transcription. However, it is not known if these transcripts are localized in the nucleus or in the cytoplasm. Intriguingly, the CAGE exonic tags can be aligned to spliced mRNAs implying that the secondary processing and recapping takes place after splicing. Do these secondary-capped RNAs have the same 7-methyl-guanosine cap of mRNAs? The determination of the type of cap will be of primary importance, as the tri-methylated cap is used to import some snRNAs into the nucleus.48 Interestingly, a cytoplasmic capping enzyme has recently been identified,49 which has been shown to recap processed longer mRNA fragments transcribed from the globin locus. It is likely that this enzyme is involved in recapping other cleaved RNAs as well, some of which may be imported into the nucleus for further processing or signalling. Alternatively, processed recapped cytoplasmic RNAs may interact with the translation machinery, thus regulating translation. Processed capped RNAs could be produced in association with the splicing machinery in the nucleus, acting locally or being later exported to the cytoplasm.
The ENCODE project is providing additional transcript maps, which will help address these genome-wide questions by mapping the specific cell compartments, including cytoplasm, polysomes, and other nuclear compartments in which these capped RNAs are specifically located. A further issue concerns the potential new roles of known RNA-processing and binding proteins, such as the RNAi and piRNA pathways and their specific role in nuclear compartments together with these novel short ncRNAs.
7. Capped transcripts near the 3′ ends
The regions near the 3′ ends of genes also show a notable transcriptome complexity. Here, longer antisense RNAs have been identified.5,50 This region is known to generate the termination-associated RNAs (TASRs)29 which are a class of heterogeneous size RNAs that span the regions upstream and downstream to the termination site, identified by tiling arrays using sRNA fractions.29 In addition, CAGE has identified capped transcripts that originate close to the transcription termination site. These were verified by RACE to be at least 100–200 nt long, thus being longer than TASRs. These transcripts originating from the 3′-UTR appear to differ from TASRs, as they do not extend beyond the known polyadenylation site. A lack of extensive overlap between CAGE-identified transcripts and TASRs, downstream of the termination site, suggests that a proportion of the TASRs may be non-capped transcripts mapping outside the conventional borders of the genes. The fact that 3′-UTR transcripts are preferentially identified in oligo-dT primed libraries (and poorly in random primed ones) indicates that they terminate at known polyadenylation sites with a poly-A tail. Notably, these are RNAs overlapping to 3′-UTRs, which are main targets of a plethora of regulatory mechanisms including RNA localization, control of stabilization, miRNA targets, and may be the target of RE-derived RNAs.43 There have been relatively fewer studies regarding these ncRNAs at the 3′ ends compared with the ncRNAs at the 5′ ends. However, the genes in the nucleus are not located linearly but rather the chromatin is organized in loops and interacting zones. Thus, it is likely that the analysis of global chromatin interactions39,40 integrated with ncRNAs and other DNA binding proteins may provide evidence of larger complexes that regulate gene expression, which may include at least some of these ncRNAs.
8. Are these RNA genes?
It is important to note that not all the known sRNAs, such as miRNAs or piRNAs, have been described here, as others have extensively reviewed the field.51–53 The role of miRNAs as a gene is not disputed any longer. However, the novel types of short or shortened RNAs identified in large scale projects are challenging current gene models, because of their abundance, likely partial redundancy of action, and assessment of phenotype. Due to the particularly large number of these ncRNAs, new high-throughput strategies are needed to test their function, since current mutagenesis approaches are unlikely to resolve their role.
It has been relatively straightforward to define a ‘gene’ as the genomic region, which produces a given mRNA, which in turn produces one or more protein isoforms. Similarly, the identification of a genomic region that produces a RNA that is further processed to produce a miRNA could also be unambiguously defined as a ‘miRNA gene’. Similarly, large ncRNAs, called LincRNAs54,55 that were identified in intergenic regions and therefore far from complicated genic regions with sense–antisense transcription, can also be easily called ‘genes’, mainly because a function could be identified for several of them. However, a large part of the job ahead requires understanding and classifying the most challenging cases, the ncRNAs of various sizes overlapping intermingled known genes discussed in this review.
Overlapping transcripts probably account for a majority of the genomic output making phenotype and functional assessment far more complicated. For most, if not all genes, we are identifying sets of long to short transcripts overlapping the regulatory or coding regions. Are we genomicists simply dissecting the background generated around the transcription machinery, composed of a large fraction of sRNA by-products? Or are we dissecting a whole microcosm of functional RNA components around regulatory regions, which are acting with an unforeseen complexity? It is unlikely that these ncRNAs are just experimental noise. Also, it is incorrect to label them as ‘biological noise’ for the reason that we have not yet understood their functions.
Dissection of the function of individual sRNAs is particularly challenging, because individual sRNAs may only moderately regulate gene activity; below the statistically significant range of our experimental standards. However, these ncRNAs may cooperatively contribute to gene expression and thus cellular fates and phenotypes. Indeed biology is not composed of single molecules, but networks of molecules, thus creative assays will be required to test multiple ncRNAs in cooperation. Biological networks are usually robust,56 therefore functional perturbation studies might require to target multiple RNAs at once.
Perhaps sRNAs and other ncRNAs, including overlapping ncRNAs, might not be after all independent genes but a part of a ‘RNA regulome’ of gene expression. Gene regulation in eukaryotes is much more complex than the model of a bacterial operon controlled by a single promoter, and these ncRNAs could be the emerging components of this regulatory machine. If we think of multiple transcription factor binding sites (TFBS) regulating a eukaryotic promoter, a single TFBS might be dispensed with under certain conditions to produce an mRNA or a phenotype. sRNAs or ncRNAs overlapping specific genes may have subtle effects at an individual level; however, several such ncRNAs could push the system beyond a certain threshold causing differential gene regulation and potentially altered phenotype, acting for instance on the epigenomic regulation of promoters. Altogether, these sets of ncRNAs could be minor components of the larger ‘regulome’ of specific genes, acting in cis or in trans and interacting with the epigenomic machinery.
Should we move towards a global recognition of these RNA regulons as counterparts of protein coding genes, for all of those overlapping transcripts? Or is a gene composed of multiple elements including the mRNA and isoforms, splicing factors, promoters, enhancers, and ‘RNA regulons’? After all, what defines a gene and its borders?
Funding
The author is supported by a grant of the 7th Framework of the EU commission to the Dopaminet consortium, a Grant-in-Aids for Scientific Research (A) No. 20241047, a Research Grant for RIKEN Omics Science Center from MEXT and by an U54 ENCODE grant from NIH-NHGRI.
Acknowledgements
The authors would like to thank Miki Nishikawa for support in the preparation of the manuscript and Alistair Forrest and Alka Saxena for the critical reading of the manuscript.
Footnotes
Edited by Osamu Ohara
References
- 1.Waterston R.H., Lindblad-Toh K., Birney E., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 2.Lander E.S., Linton L.M., Birren B., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Strausberg R.L., Feingold E.A., Grouse L.H., et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl Acad. Sci. USA. 2002;99:16899–903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohara O., Nagase T., Ishikawa K., et al. Construction and characterization of human brain cDNA libraries suitable for analysis of cDNA clones encoding relatively large proteins. DNA Res. 1997;4:53–9. doi: 10.1093/dnares/4.1.53. [DOI] [PubMed] [Google Scholar]
- 5.Katayama S., Tomaru Y., Kasukawa T., et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–6. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 6.Imanishi T., Itoh T., Suzuki Y., et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:e162. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng J., Kapranov P., Drenkow J., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–54. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 8.Carninci P. Tagging mammalian transcription complexity. Trends Genet. 2006;22:501–10. doi: 10.1016/j.tig.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Carninci P. Is sequencing enlightenment ending the dark age of the transcriptome? Nat Methods. 2009;6:711–3. doi: 10.1038/nmeth1009-711. [DOI] [PubMed] [Google Scholar]
- 10.Carninci P., Yasuda J., Hayashizaki Y. Multifaceted mammalian transcriptome. Curr. Opin. Cell Biol. 2008;20:274–80. doi: 10.1016/j.ceb.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Wilusz J.E., Sunwoo H., Spector D.L. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 13.Kim V.N. Small RNAs: classification, biogenesis, and function. Mol. Cells. 2005;19:1–15. [PubMed] [Google Scholar]
- 14.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 15.Forrest A.R., Carninci P. Whole genome transcriptome analysis. RNA Biol. 2009;6:107–12. doi: 10.4161/rna.6.2.7931. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y.S., Shibata Y., Malhotra A., Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–49. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawaji H., Nakamura M., Takahashi Y., et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ardelt B., Ardelt W., Darzynkiewicz Z. Cytotoxic ribonucleases and RNA interference (RNAi) Cell Cycle. 2003;2:22–4. doi: 10.4161/cc.2.1.232. [DOI] [PubMed] [Google Scholar]
- 19.Williams R. Blood, stress, and tiRNAs. J. Cell Biol. 2009 [Google Scholar]
- 20.Thompson D.M., Lu C., Green P.J., Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elbarbary R.A., Takaku H., Uchiumi N., et al. Modulation of gene expression by human cytosolic tRNase Z(L) through 5′-half-tRNA. PLoS One. 2009;4:e5908. doi: 10.1371/journal.pone.0005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taft R.J., Glazov E.A., Lassmann T., Hayashizaki Y., Carninci P., Mattick J.S. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–40. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartmann B., Valcarcel J. Decrypting the genome's alternative messages. Curr. Opin. Cell Biol. 2009;21:377–86. doi: 10.1016/j.ceb.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Furuno M., Pang K.C., Ninomiya N., et al. Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet. 2006;2:e37. doi: 10.1371/journal.pgen.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carninci P., Kasukawa T., Katayama S., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 26.Djebali S., Kapranov P., Foissac S., et al. Efficient targeted transcript discovery via array-based normalization of RACE libraries. Nat. Methods. 2008;5:629–35. doi: 10.1038/nmeth.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gingeras T.R. Implications of chimaeric non-co-linear transcripts. Nature. 2009;461:206–11. doi: 10.1038/nature08452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carninci P. Cap-Analysis Gene Expression (CAGE) The Science of Decoding Gene Transcription. Singapore: Pan Stanford Publishing; 2009. [Google Scholar]
- 29.Kapranov P., Cheng J., Dike S., et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 30.Affymetrix ENCODE Transcriptome Project; Cold Spring Harbor Laboratory ENCODE Transcriptome Project. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–32. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seila A.C., Calabrese J.M., Levine S.S., et al. Divergent transcription from active promoters. Science. 2008;322:1849–51. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Core L.J., Waterfall J.J., Lis J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–8. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taft R.J., Glazov E.A., Cloonan N., et al. Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 2009;41:572–8. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 34.Martianov I., Ramadass A., Serra Barros A., Chow N., Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–70. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 35.Preker P., Nielsen J., Kammler S., et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–4. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 36.Neil H., Malabat C., d'Aubenton-Carafa Y., Xu Z., Steinmetz L.M., Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–42. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 37.Xu Z., Wei W., Gagneur J., et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–7. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maida Y., Yasukawa M., Furuuchi M., et al. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–5. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lieberman-Aiden E., van Berkum N.L., Williams L., et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fullwood M.J., Liu M.H., Pan Y.F., et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ebisuya M., Yamamoto T., Nakajima M., Nishida E. Ripples from neighbouring transcription. Nat. Cell Biol. 2008;10:1106–13. doi: 10.1038/ncb1771. [DOI] [PubMed] [Google Scholar]
- 42.Faulkner G.J., Carninci P. Altruistic functions for selfish DNA. Cell Cycle. 2009;8:2895–900. doi: 10.4161/cc.8.18.9536. [DOI] [PubMed] [Google Scholar]
- 43.Faulkner G.J., Kimura Y., Daub C.O., et al. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009;41:563–71. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 44.Valen E., Pascarella G., Chalk A., et al. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res. 2009;19:255–65. doi: 10.1101/gr.084541.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kodzius R., Kojima M., Nishiyori H., et al. CAGE: cap analysis of gene expression. Nat. Methods. 2006;3:211–22. doi: 10.1038/nmeth0306-211. [DOI] [PubMed] [Google Scholar]
- 46.Sandelin A., Carninci P., Lenhard B., Ponjavic J., Hayashizaki Y., Hume D.A. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat. Rev. Genet. 2007;8:424–36. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- 47.Carninci P., Sandelin A., Lenhard B., et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–35. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 48.Huber J., Cronshagen U., Kadokura M., et al. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 1998;17:4114–26. doi: 10.1093/emboj/17.14.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otsuka Y., Kedersha N.L., Schoenberg D.R. Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol. Cell Biol. 2009;29:2155–67. doi: 10.1128/MCB.01325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He Y., Vogelstein B., Velculescu V.E., Papadopoulos N., Kinzler K.W. The antisense transcriptomes of human cells. Science. 2008;322:1855–7. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 52.Siomi H., Siomi M.C. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 53.Kim V.N. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev. 2006;20:1993–7. doi: 10.1101/gad.1456106. [DOI] [PubMed] [Google Scholar]
- 54.Ponjavic J., Oliver P.L., Lunter G., Ponting C.P. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5:e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guttman M., Amit I., Garber M., et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masel J., Siegal M.L. Robustness: mechanisms and consequences. Trends Genet. 2009;25:395–403. doi: 10.1016/j.tig.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]