Abstract
The structural features responsible for the activities of hepatic lipase (HL) can be clarified by in vivo comparisons of naturally occurring variants. The coding sequence of HL from C57BL/6J (B6) and SPRET/EiJ (SPRET) mice differs by four amino acids (S106N, A156V, L416V, S480T); however, these changes are not predicted to influence HL function. To test for allelic effects, we generated SPRET-HL transgenics with physiological levels of HL mRNA and HL activity that was parallel in female transgenics and about 70% higher in male transgenics, toward tri-[3H]oleate, compared with B6 controls. We found no correlation between activity levels and plasma lipids. However, significant allelic effects on plasma lipids were observed. Compared with B6-HL, SPRET-HL mediated reductions in total cholesterol (TC) and VLDL-, LDL- and HDL-cholesterol and HDL-triglyceride (TG) in fed males, and SPRET-HL decreased total TG and VLDL- and HDL-TG levels in fasted males. Fasted female transgenics had reduced TC compared with controls. We also found allele and sex effects on lipoprotein particle size. Male transgenic mice had increased VLDL and decreased LDL size, and female transgenic mice had decreased HDL size compared with control animals. These findings demonstrate highly divergent effects of naturally occurring HL coding sequence variants on lipid and lipoprotein metabolism.
Keywords: 129 donor allele, allele, hepatic lipase, lipoprotein, natural variant, obesity, SPRET/Ei, transgenic
Hepatic lipase (HL) affects plasma lipid levels and remodels lipoproteins in both humans and mice (1–4). As a multifunctional protein, it demonstrates ligand binding, which reduces plasma apoB-containing lipoproteins in the absence of lipolytic activity (5–7). Additionally, HL activity is associated with promoter polymorphisms, intra-abdominal fat mass (IAF), body mass index (BMI), physical activity, and dietary fat-intake (8–11). Gender influences HL activity in humans, with males having higher activity than females (12–14).
While much is known about the function of HL, its role in cardiovascular disease is not as clear. In humans, decreased HL activity is associated with increased risk of coronary artery disease (15), but it is also associated with the generation of a beneficial lipid profile, i.e., increased high density lipoprotein-2 (HDL2) and decreased small dense-LDL concentrations (16, 17). Moreover, increased HL activity leads to pro-atherogenic lipid profiles with reductions of HDL2 and increases in small dense LDL, yet is inversely associated with extent of coronary calcification in familial hypercholesterolemia (18–20). Studies in mice also show disparate effects on cardiovascular disease, and these effects appear to be model dependent. Studies in mice overexpressing human HL on a LDL-receptor knockout background show reduced aortic cholesterol, but mice with HL deficiency on an apolipoprotein E knockout background had attenuated aortic lesion thickness (21, 22).
To identify structural features of HL that regulate function, engineered or naturally occurring variants can be studied. For example, humans with low or no HL activity (23, 24), and mice with spontaneous mutations causing combined lipase deficiency (25) and fatty liver dystrophy (26) have been successfully used to study lipid metabolism. In vitro techniques, such as site-directed mutagenesis and domain exchange methods have highlighted HL's functional characteristics, including the N-terminal domain, carboxyl-terminal domain, catalytic site, heparin-binding sites, and ligand binding specificities (27–30).
The mouse strains MOLF/EiJ, CAST/EiJ, and PWD/PhJ encode naturally occurring variants of HL, each with a single amino acid change either in exon 3 (MOLF/EiJ and CAST/EiJ; rs49265789), or exon 4 (PWD/PhJ; rs46038991). We previously sequenced the nine exons of SPRET Lipc (NCBI: AY225159) from cDNA and identified 17 variants, 4 which produced unique amino acid substitutions (S106N, A156V, L416V, and S480T) (31). Thus SPRET-HL is an ideal variant to study because of both the number and location of the substitutions. The first two changes, S106N and A156V, are in the highly conserved N-terminal domain containing the catalytic triad. The second set of amino acid differences (L416V and S480T) between B6- and SPRET-HL is located in the carboxyl-terminal domain, a region shown to be required for variety of lipase functions, including lipid binding, control of heparin binding, substrate specificity, receptor binding, and catalysis of long-chain substrates (29, 30, 32, 33). Although the amino acid substitutions are in close proximity to important structural features of HL they were not predicted to alter function by the computational protein prediction programs, PolyPhen (http://genetics.bwh.harvard.edu/pph) and SIFT (http://sift.jcvi.org). Nevertheless our initial studies using a reciprocal hemizygosity approach to examine SPRET-HL suggested variable allelic effects on measures of adiposity (31).
To investigate the in vivo effects of SPRET-HL on plasma lipids and obesity, we generated a bacterial artificial chromosome (BAC) transgenic mouse strain and backcrossed positive transgenics to two strains of mice. The first strain, 129-Lipctm1Unc/J (HL−/−), are HL null on a B6 background and demonstrate no intrinsic HL activity (2). Due to the nature of knockout construction, a large 129P2 (129) donor region could exist surrounding the structural locus of Lipc on mouse chromosome 9. Previous studies of other knockout models have demonstrated that phenotypes originally ascribed to the knocked-out gene were actually due to the linked 129 strain alleles surrounding the targeted locus (31). To rule out 129 allele effects, we conducted a second backcross with SPRET-HL transgenics and B6 animals. This backcrossed removed 129 alleles and, subsequently, any differences between mice positive for SPRET-HL and B6 control animals may be attributed to the transgene.
For the detection of allelic effects on lipoprotein composition and/or size, we conducted lipoprotein compositional analysis by computer assisted HPLC to determine cholesterol and triglyceride levels in the four main lipoproteins (CM, VLDL, LDL, and HDL) and corresponding subfractions (32, 33). Here we show similar physiologic levels of HL mRNA and HL activity in SPRET-HL and B6-HL expressing mice, and demonstrate that SPRET-HL has markedly different effects on lipoprotein composition and size when compared with endogenous B6-HL despite the absence of functional effects as forecasted by protein prediction programs. We also report the effect of homozygous 129 alleles on chromosomes 6, 9, and 13 on measures of adiposity. We identify a unique resource to study the structure-function relationship of HL in vivo.
MATERIALS AND METHODS
Generation of transgenic
The Children's Hospital Oakland Research Institute SPRET BAC genomic DNA library was screened for HL using probes designed on the basis of HL Mus musculus cDNA sequence (Accession AY228765) (http://bacpac.chori.org/home.htm). The screening probes were: HL-1: 5′- GGAGAACCACACTCTGCAGAGAAAGATAACTGTGGCCCT-3′ and HL-6: 5′-CAAAGTGCCCTCAAAGTCTCACAGGAATAGCCAAGGGTGT-3′. To determine insert size, pulse field gel electrophoresis was performed after Not1 digestion of the BAC and paired BAC-end sequencing from glycerol stock was performed using traditional DNA sequencing methods on an ABI Prism 373x1 DNA sequencer with universal primer, T7: 5′-TAATACGACTCACTATAGGG-3′, for 5′ downstream sequence determination and custom primer, BAC End-1: 5′-GGATCCTCCCGAATTGACTA-3′, for 5′ upstream sequencing (SeqWright, Huston, TX). BAC DNA was purified for microinjection using a combination of the Large Construct Kit (QIAGEN, Valencia, CA) and Endo Free Maxi Kit (QIAGEN, Valencia, CA). The purified BAC was linearized prior to injection by PI-SceI digestion and microinjected into B6CBAF1/J cells by the Murine Targeted Genomics Laboratory at the University of California, Davis.
Mice and crosses
Pups were screened with PCR primers polymorphic between SPRET, CBA/J (CBA) and B6 strains. Fig. 1 provides primer locations, and Table 1 lists primer sequences and base pair positions. Three pups were positive for the full length BAC clone genotyping assay and were backcrossed for two generations to B6.129- Lipctm1Unc/J (HL−/−) mice (Bar Harbor, ME) (2) which lack intrinsic HL activity and are on a B6 background. Progeny from a single founder demonstrated HL activity on the HL−/− background; therefore we present data from a single founder. Supplementary Fig. I illustrates the breeding schemes of the five HL genotype combinations presented here. The first group of mice studied were generated from an N4, N5 or ≥N10 backcross between HL−/− mice and mice positive for the SPRET-HL transgene on an HL−/− background (HL−/−,+S) producing a phenotyped cohort of N5, N6 or N10 littermate mice. To generate mice positive for the transgene on an HL+/+ background, N8 HL−/−,+S mice were backcrossed to HL+/+ mice for two generations. Therefore, the second group was generated by a backcross between HL+/+ and N10 SPRET-HL positive HL+/+ mice (HL+/+,+S), producing a phenotyped cohort of littermate HL+/+,+S or HL+/+ mice. The third cross was an F1 cross between HL−/− and HL+/+ mice. The fourth cross was a parental cross between HL+/+ and HL+/+ mice. A total of five different HL genotypes were generated, HL−/−, HL−/−,+S, HL+/+, HL+/+,+S, and HL+/−. All animals were on a B6 background with the HL−/− and HL−/−,+S animals homozygous and HL+/− animals heterozygous for 129 alleles on chromosome 6, 9, and 13. All animals were housed and cared for under conditions meeting the NIH standards as stated in the Guide for the Care and Use of Laboratory Animals, AAALAC accreditation standards, and the Animal Welfare Act (PL85-544).
Fig. 1.
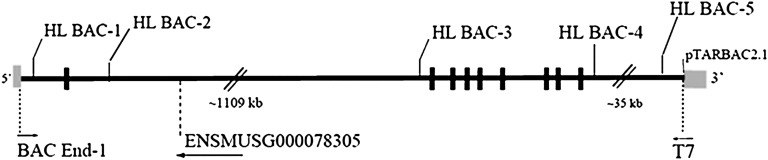
Diagram of BAC with BAC-end and genotyping primers mapped using Ensembl genome browser v53. See Table 1 for sequences and bp positions of genotyping primers. Black rectangles = exons 1–9; gray rectangle = vector (pTARBAC2.1). Sequence from T7 mapped to Chr 9: 70,612,052-70,612,192 bp and BAC End-1 to Chr 9:70,801,301-70,801,753 bp. Ensembl gene ENSMUSG00000078305 maps to Chr:9 70,700,357-70,702,269 bp. BAC, bacterial artificial chromosome.
TABLE 1.
Custom designed primers for positive founder identification
| Primer | 5′-3′ Sequence | Position, Ensembl v53, bp |
|---|---|---|
| HL BAC 1-F | TTTGTGAGATTTCCACACACATC | 70,789,525 – 70,789,547 |
| HL BAC 1-R | TGGTCTCCTGTGCCATTACA | 70,789,725 – 70,789,744 |
| HL BAC 2-F | TGCTCCTCTCACTCTCCTGTT | 70,768,934 – 70,768,954 |
| HL BAC 2-R | TGAATTAGCATATTCCCTGCAGA | 70,769,065 – 70,769,087 |
| HL BAC 3-F | TTGCAAAACATCTCCACGAA | 70,674,578 – 70,674,597 |
| HL BAC 3-R | TATGGTTTCCAGGGGGACT | 70,768,934 – 70,768,954 |
| HL BAC 4-F | AGCTCAGTTTTAAGACGGTTTTT | 70,645,771 – 70,645,793 |
| HL BAC 4-R | TAAAGGAGGGTGTGCTTTGC | 70,645,960 – 70,645,979 |
| HL BAC 5-F | CAGCATGTACCCTGTCCAGT | 70,633,017 – 70,633,036 |
| HL BAC 5-R | GGAGTGTTGGGAAATGCTGT | 70,633,296 – 70,633,315 |
Custom-designed simple sequence length polymorphic (SSLP) markers designed as described in “Materials and Methods.” Primer positions were mapped on the Ensembl v53 mouse genome assembly (www.ensembl.org).
Mouse phenotyping
At weaning, mice were group housed with ad libitum access to water and standard rodent chow (Purina Rodent Chow 5001, Dean's Animal Feed, Redwood City, CA). At 133-147 days of age, mice were fasted overnight for fasted studies or maintained free access to chow for fed studies, weighed, and anesthetized with inhaled isofluorane. A retro-orbital sinus bleed was conducted using heparinized capillary tubes and blood was collected into serum separator tubes (Microtainer, Becton Dickinson, New Jersey) containing 2 µl of EDTA. A subset of mice was fasted overnight 14 days prior to sacrifice and tail bled to obtain preheparin plasma. For postheparin plasma collection the same mice were fasted overnight and administered tissue culture grade heparin (Elkin-Sinns, Cherry Hill, NJ) via intra-peritoneal injection at 100 U/kg body weight. Five minutes post injection mice were anesthetized and a retro-orbital bleed conducted. All samples were centrifuged for 10 min at 8,000 RPM, placed on ice, aliquoted, and stored at -80°C. Anal-nasal (AN) length was measured and mice were sacrificed by cervical dislocation. Liver, kidney, skeletal muscle, and gonadal white adipose tissue (GWAT) were dissected and flash frozen in liquid nitrogen. The weights of liver and the femoral (FWAT), retroperitoneal (RWAT), and mesenteric (MWAT) white adipose tissue depots were recorded.
Genotyping.
Toe clips from 8-12 day old mice were collected and DNA was extracted by an overnight incubation at 55°C in digestion buffer (0.5% NP40, 0.5% Tween-20, 40 mM Tricine-KOH, 150 mM KOAc, 3.5 mM(oAc)2) and 100 ug/ml proteinase K. Custom simple sequence length polymorphic (SSLP) markers were used to detect differences between the B6, CBA, 129, and SPRET-HL by PCR, see Table 1 for primer sequences and positions. For genotyping the 129 donor region surrounding the structural locus of Lipc in HL−/− and HL−/−,+S mice polymorphic MIT markers between B6 and 129 were used. All PCR products were run on a 2% agarose gel and visualized with ethidium bromide staining.
To assess genome wide CBA and 129 strain passenger alleles we isolated genomic DNA (gDNA) from the liver of four unrelated mice positive for the transgene (>N10) using a QIAGEN DNeasy kit (QIAGEN Inc., Valencia, CA). Genomic DNA was provided to the Murine Genetic Analysis Laboratory at UC Davis and the samples were genotyped using an Illumina medium density mouse 1449 SNP chip and Illumina BeadStation 500G Genotyping system (Illumina, Inc., San Diego, CA). The SNP panel has known genotypes for all domestic strains including B6, 129, and CBA.
Sequencing.
To partially sequence ENSMUSG00000078305, PCR was performed with primers designed using Primer3 (http://frodo.wi.mit.edu/primer3/) that amplified 400-600 bp fragments of SPRET gDNA. PCR products were gel extracted and purified using the QIAquick Gel Extraction Kit (QIAGEN Inc., Valencia, CA). DNA sequencing was performed on an ABI 3730 XL capillary sequencer using ABI BigDye Terminator v3.1 Cycle Sequencing chemistry (Applied Biosystems, Foster City, CA) and Geneious software was used for sequence assembly (Biomatters Ltd, New Zealand).
Quantitative real-time PCR.
Quantitative PCR (qPCR) was performed in liver and liver, kidney, skeletal muscle, and adipose tissue in triplicate to test for differential ENSMUSG00000078305 and HL expression, respectively, using the SYBR® Green 1 dye detection system (Applied Biosystems, Foster City, CA) and run on a 7900HT (Applied Biosystems, Foster City, CA). Primer Express software (Applied Biosystems, Foster City, CA) was used for primer design. The primers sequences for ENSMUSG00000078305 were PG Lipc-1F: 5′ - AGTCTTGCAGACCAAGAGAGGCA - 3′ and PG Lipc-1R 5′ – TAGCATCACCAAACACCCCAGG – 3′ and were located in the 3′ UTR. The HL primers spanned exons 4 and 5 over a nonpolymorphic region between B6- and SPRET-HL. The primer sequences were, HL SYBR-F: 5′- GACGGG AAGAACAAGATTGGAA- 3′and HL SYBR-R: 5′- TTGGCATCAGGAGAAAGG- 3′. Two control genes, Gapdh and Gusb, corrected for input RNA. Relative expression was determined by calculating 2−ΔCt for all HL genotypes and defined as fold-change compared with 2−ΔCt of the HL+/+ mice. Relative expression for Gusb and Gapdh was found to be similar; however we only report data normalized to Gusb here.
Hepatic lipase activity
Pre- and postheparin plasma was collected as previously described and glycerol tri-[3H]oleate in 2 M NaCl was added as a substrate and liberated free fatty acid radioactivity was extracted and counted. Under these conditions, other lipases do not show activity (34). For postheparinized samples, the assay was assessed for linearity. As discussed in detail by Nilsson-Ehle et. al. we used the equation K(HL) = (cpm × 9.67)/[time of assay (min) X specific activity of tri-[3H]oleate (cpm/nmol) X sample volume (ml)] to calculate HL activity in mU/ul toward tri-[3H]oleate (35).
Lipoprotein determination.
Lipoprotein cholesterol and TG levels for chylomicron (CM), VLDL, LDL, HDL, and 20 subclasses were determined using published methods (32, 33) by Skylight Biotech (Skylight Biotech, Inc., Japan). Briefly, plasma from individual mice was subjected to gel filtration HPLC on two tandem connected TSK-Gel Lipopropak XL columns. Triglycerides and cholesterol were measured simultaneously by enzymatic treatment following continuous size fractionation on HPLC columns. Particle size was estimated by the elution time on the column and comparison to appropriate sized markers.
Statistics
Data are presented as means ± SEM and transformed for statistical analyses when necessary to meet requirements for normality and homogeneity of variance. A paired Student's t-test was used to determine significance between pre and postheparin samples. ANCOVA was used to assess genotype effects for HL activity with HL genotype as the main predictor and age at sacrifice and body weight as covariates. For pairwise comparisons, sexes were analyzed separately and Sidak t-test used for confidence interval adjustment. To assess genotype effects on lipoprotein composition and particle size ANOVA was used with HL genotype and sex as main factors. Body weight was initially included as a covariate for the lipoprotein analyses and found to be nonsignificant and therefore excluded. Scheffe's posthoc testing was used for multiple comparisons with sexes analyzed separately. To test for associations of HL activity with mRNA levels, lipoprotein composition, and lipoprotein size Pearson's correlation was used and all HL genotypes, with the exception of HL−/− mice, were included. A P < 0.05 was considered significant. All analyses were conducted using the SPSS 16.0 program (SPSS, inc., Chicago, IL).
RESULTS
Characterization of the SPRET- HL BAC clone
The BAC (pTARBAC2.1) containing SPRET-HL was obtained and subjected to paired BAC-end sequencing (see “Materials and Methods” for details). End-sequencing results indicated that pTARBAC2.1 contained a 190 kb insert which was consistent with data from pulsed-field gel electrophoresis of the BAC after Not1 digestion (185 kb, data not shown) or ∼19 kb of upstream promoter and ∼35 kb downstream DNA, plus HL itself. Fig. 1 maps the sequences obtained from primers T7 (Chr 9: 70,612,052-70,612,192 bp) and BAC End-1 (Chr 9:70,801,301-70,801,753 bp). We determined the BAC also contained the Ensembl protein coding gene ENSMUSG00000078305, which is transcribed antisense to HL, see Fig. 1. The single exon coding sequence of ENSMUSG00000078305 is 98% identical to the seven exon rRNA processing 7 homolog A gene (Rrp7a) located on mouse chromosome 15.
Production of transgenic mice expressing SPRET-HL
To examine allelic effects of HL, we created a transgenic mouse strain expressing SPRET-HL. To ensure positive founders had integrated the full-length BAC, genotyping primers from the 5′ end, middle, and 3′ end of the BAC were used (Fig. 1). To determine SPRET-HL activity, transgenic lines were backcrossed to HL−/− mice for two generations and preheparin HL activity was assessed. HL−/− mice demonstrate no intrinsic HL activity, thus any measured HL activity was due to expression of the SPRET-HL transgene. Pups from only one founder were positive for HL activity, and animals derived from this founder were used in all subsequent studies.
To check for ectopic expression of the transgene, we used qPCR to measure relative HL mRNA levels in liver, kidney, skeletal muscle, and GWAT, in HL−/−, +S mice. Negligible amounts of HL mRNA were detected in the kidney, skeletal muscle, and adipose tissue (data not shown). This brief tissue survey revealed no evidence of ectopic HL expression in our SPRET-HL transgenic animals. Next, to assess levels of ENSMUSG0000078305, we partially sequenced and used qPCR to identify any transgene driven expression differences in the liver. Fig. 2 shows similar relative expression levels for HL−/−, HL+/+, HL−/−.+S, and HL+/+,+S mice, demonstrating no influence of the transgene on ENSMUSG00000078305 expression.
Fig. 2.
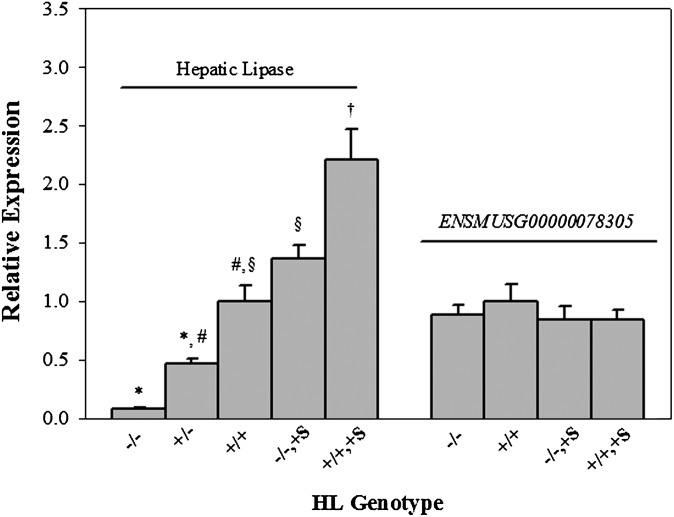
Relative expression fold change of liver HL mRNA as determined by qPCR in fasted mice sacrificed at 140 ± 14 days. For HL expression, 6 biologic replicates were used per genotype except for HL+/+ mice, which had 4 biologic replicates. For ENSMUSG00000078305 expression, a minimum of 8 and a maximum of 12 biological replicates were used. HL−/− = mice lacking endogenous HL; HL+/− = F1 (C57BL6/J X HL−/− mice); HL+/+ = C57BL6/J mice; HL−/−,+S = mice lacking endogenous HL and positive for the SPRET-HL transgene; HL+/+,+S = C57BL6/J mice positive for the SPRET-HL transgene; Relative expression = Ct(specified HL genotype)-Ct(control gene)/ Ct(HL+/+) – Ct(Gusb). Data are means ± SEM. Bars not sharing a symbol are different by P < 0.05 by Sidak's t-test for multiple comparisons.
Since the founding transgenic embryo was a B6CBAF1/J, there is a 50% chance that the BAC integrated into CBA alleles. If the transgene integrated into CBA alleles, then passenger DNA remaining even after 10 generations of backcrossing could influence measured phenotypes. Therefore we used a 1449 SNP based chip assay on four unrelated mice positive for the transgene that had been backcrossed a minimum of 10 times to probe for the presence of CBA alleles. We identified only one instance of CBA alleles in the entire panel. Two of the four animals were heterozygous for CBA at a single SNP located on the X chromosome. It is important to rule out an X chromosome transgene integration site and our breeding schemes allowed for this as we bred sire HL−/−,+S and HL+/+,+S mice to HL−/− and HL+/+ dams and collected males positive for the transgene (data not shown); therefore male to male transmission rules out the possibility of an X chromosome integration. Additionally, and exclusive to mice on an HL−/− background, we observed a region on chromosome 9, the structural locus of Lipc, that was homozygous for 129 strain alleles, consistent with a microsatellite marker determined donor region ranging from D9Mit297 to D9MIT269 or ∼33.8 Mb to ∼87.7 Mb. Using the SNP chip assay to fine map the region, we identified 129 strain chromosome 9 alleles ranging from rs13480122 at 30,964,211 bp to rs13480365 at 98,479,731 bp. We additionally identified two separate 129 strain passenger loci located on chromosomes 6 and 13. The maximal extent of the chromosome 6 loci mapped to mCV23042866 at 115,587,647 bp and to rs3672808 at139,805,730 bp while the chromosome 13 loci had proximal and distal boundaries of rs6244558 at 47,911,360 bp and rs3693942 at 55,054,140 bp.
SPRET hepatic lipase is expressed at physiological levels
To confirm physiologic expression of SPRET-HL, both liver HL mRNA and plasma enzymatic HL activity were assessed. Hepatic lipase mRNA was detected at similar levels for males and females in liver (data not shown). Fig. 2 shows the relationship between HL relative expression in five HL genotypes, HL−/−, HL+/−, HL+/+, HL−/−,+S, and HL+/+,+S mice. Compared with HL+/+ mice, HL+/− mice expressed roughly half (0.47-fold; P=0.042), HL−/−,+S mice showed similar levels (1.37-fold; NS), and HL+/+,+S mice had approximately 2-fold greater expression (2.13-fold; P<0.0001) of HL mRNA. These findings suggest that elements present in the BAC produced physiological or near-physiological levels of HL mRNA.
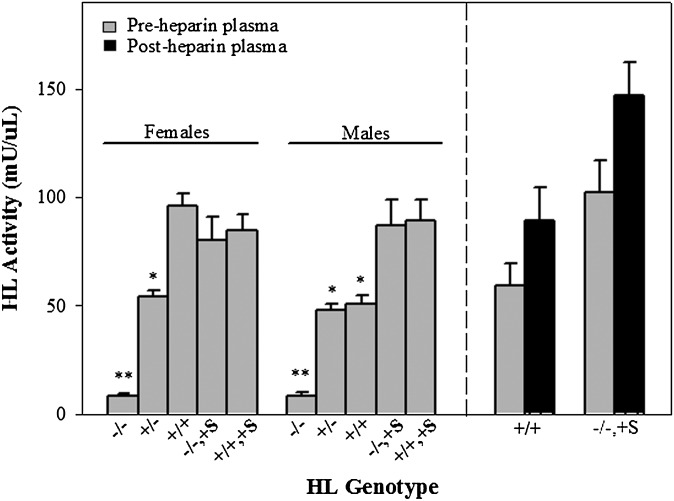
In mice, the majority of HL activity is found in preheparin plasma (36, 37). To test for allelic differences in HL activity levels or differences in the plasma versus liver pools of HL we measured HL activity in pre and postheparinized plasma (Fig. 3). In preheparin plasma HL−/− mice demonstrated little residual HL activity and mice expressing SPRET-HL had HL activity levels roughly comparable to wild-type (Fig. 3). In positive transgenic mice, regardless of the presence of endogenous HL, differences in HL activity levels under optimal conditions were minimal and there were no gender differences in mice expressing the transgene. Female HL−/−,+S and HL+/+,+S mice had activities of 88.7 ± 6.6 and 72.1 ± 1.0 mU/ml, respectively, and male HL−/−,+S and HL+/+,+S mice had HL activities of 84.0 ± 12.7 and 89.1 ± 9.97 mU/ml, respectively. Reports in humans show disparity between HL activity levels in the sexes (12–14) and here male and female HL+/+ mice from a parental cross, had significant differences in HL activity levels. Female HL+/+ mice had roughly 2-fold more activity than male HL+/+ mice, but comparable to male and female mice expressing SRPET-HL (Fig. 3). Male HL−/−,+S and HL+/+,+S mice had significantly increased HL activities (65.6% and 75.2%, respectively; P < 0.05 for both) compared with HL+/+ males. Despite these gender effects on serum HL activity, when sexes and HL genotypes were combined, liver HL mRNA expression was significantly correlated with HL activity (N = 30, r=0.607; P=0.003). To test for allelic differences in B6 and SPRET liver versus plasma pools of HL, we subjected a subset of mice to intra-peritoneal heparin injection. Our results indicated that the majority of HL activity was found in preheparin plasma independently of genotype. Fig. 3 shows a nonsignificant increase in postheparin plasma HL activity in both female HL−/−,+S (N = 4, 43.4%) and male HL+/+ mice (N = 3, 50.6%).
Fig. 3.
Hepatic lipase activity in fasted mice measured in pre and postheparin plasma. Activity units are mU/ul. HL−/− = mice lacking endogenous HL; HL+/− = F1 (C57BL6/J X HL−/− mice); HL+/+ = C57BL6/J mice; HL−/−,+S = mice lacking endogenous HL and positive for the SPRET-HL transgene; HL+/+,+S = C57BL6/J mice positive for the SPRET-HL transgene. Ns are as follows for preheparin plasma: females HL−/− 14; HL+/− 9; HL+/+ 15; HL−/−,+S 12; HL+/+,+S 10; and males HL−/− 13; HL+/− 7; HL+/+ 10; HL−/−,+S 9; HL+/+,+S 9. For postheparin plasma, N = 3 for HL+/+ and N = 4 for HL−/−,+S mice. Data shown are means ± SEM. Sexes were analyzed separately. **P < 0.0001 compared with all other HL genotypes and *P ≤ 0.008 compared with HL+/+; HL−/−,+S; and HL+/+,+S for females; and HL−/−,+S and HL+/+,+S for males by Sidak's t-test for multiple comparisons.
Obesity phenotypes
We previously used a reciprocal hemizygosity approach to explore HL allelic derived adiposity differences (38). Our initial study contained linked 129 and SPRET alleles surrounding the Lipc locus and to rule out the effect of linked alleles we phenotyped SPRET-HL transgenic mice for measures of obesity. In the present experiments we observed that 129 alleles rather than the SPRET-HL transgene influenced obesity (Table 2). The 129 alleles promoted obesity in both sexes and in four white adipose depots in females and in GWAT, FWAT and RWAT in males. We calculated adiposity index (AI) which is a useful measure of obesity because it is highly correlated with total body fat as determined by body composition analysis. Adiposity index is the sum of four white adipose depots in grams divided by body weight in grams and multiplied by 100. Adiposity index was increased by 79.9% and 95.0% and 54.9% and 68.0% for HL−/− and HL−/−,+S mice respectively compared with either HL+/+ or HL+/+,+S mice for males with even larger increases observed in females. That is, the 129 strain alleles in HL−/− mice produced obesity, but the SPRET-HL transgene had no effect.
TABLE 2.
Obesity phenotypes in fasted mice sacrificed at 140 ± 14 days
| Lipc Locus | Transgenec | Sex | N | BW (g) | AN (cm) | FWAT (g) | GWAT (g) | RWAT (g) | MWAT (g) | AId |
|---|---|---|---|---|---|---|---|---|---|---|
| B6/B6 | — | M | 12 | 23.9 ± 0.63a | 9.79 ± 0.09 | 0.165 ± 0.02a | 0.301 ± 0.03a | 0.048 ± 0.01a | 0.075 ± 0.01 | 2.44 ± 0.22a |
| B6/B6 | + | M | 13 | 24.5 ± 0.68a,b | 9.75 ± 0.08 | 0.135 ± 0.01a | 0.268 ± 0.03a | 0.040 ± 0.01a | 0.119 ± 0.05 | 2.25 ± 0.28a |
| 129/129 | — | M | 21 | 26.1 ± 0.56b | 9.80 ± 0.06 | 0.262 ± 0.03b | 0.596 ± 0.05b | 0.117 ± 0.02b | 0.191 ± 0.03 | 4.39 ± 0.33b |
| 129/129 | + | M | 12 | 24.9 ± 0.79a,b | 9.75 ± 0.09 | 0.233 ± 0.03b | 0.516 ± 0.08b | 0.108 ± 0.02b | 0.115 ± 0.02 | 3.78 ± 0.38b |
| Overall P | Lipc Locus | NS | NS | 0.001 | <0.0001 | <0.0001 | NS | <0.0001 | ||
| Transgene | NS | NS | NS | NS | NS | NS | NS | |||
| Interaction | NS | NS | NS | NS | NS | NS | NS | |||
| B6/B6 | — | F | 16 | 19.13 ± 0.59a | 9.38 ± 0.08 | 0.118 ± 0.01a | 0.151 ± 0.01a | 0.016 ± 0.00a | 0.048 ± 0.01a | 1.73 ± 0.09a |
| B6/B6 | + | F | 12 | 20.39 ± 0.80a,b | 9.51 ± 0.08 | 0.123 ± 0.01a | 0.174 ± 0.03a | 0.024 ± 0.01a | 0.048 ± 0.01a | 1.78 ± 0.15a |
| 129/129 | — | F | 23 | 20.92 ± 0.33b | 9.46 ± 0.07 | 0.256 ± 0.0b | 0.332 ± 0.03b | 0.063 ± 0.01b | 0.131 ± 0.02b | 3.63 ± 0.23b |
| 129/129 | + | F | 8 | 21.47 ± 0.48b | 9.64 ± 0.12 | 0.248 ± 0.02b | 0.406 ± 0.03b | 0.060 ± 0.01b | 0.156 ± 0.02b | 4.03 ± 0.26b |
| Overall P | Lipc Locus | 0.020 | NS | <0.0001 | <0.0001 | 0.001 | <0.0001 | <0.0001 | ||
| Transgene | NS | NS | NS | NS | NS | NS | NS | |||
| Interaction | NS | NS | NS | NS | NS | NS | NS |
AI, adipose index; AN, anal-nasal length; BW, live fasted body weight; FWAT, femoral white adipose tissue; GWAT, gonadal white adipose tissue; MWAT, mesenteric white adipose tissue; NS, not significant; RWAT, retroperitoneal white adipose tissue; SPRET, SPRET/EiJ. All animals are ≥N10. HL−/−,+S and HL−/− are littermates, and HL+/+,+S and HL+/+ are littermates. Data shown are means ± SEM. Values not sharing a superscript (a,b) are different by P < 0.05 by Sidak's t-test for multiple comparisons.
SPRET/EiJ hepatic ipase
(Sum of four white adipose depots/BW) × 100
Lipoproteins composition by HPLC determination
Hepatic lipase affects lipoprotein composition; therefore, we tested for allelic variations in cholesterol and TG concentrations in CM, VLDL, LDL, and HDL as determined by enzymatic treatment following continuous size fractionation on HPLC columns (32, 33, 39). In this report male mice were sacrificed fed or fasted and female mice were sacrificed fasted only. Overall, in fed males SPRET-HL mediated significant reductions in total cholesterol (TC) (-30.9%) and VLDL (-51.9%), LDL (-55.9%) and HDL (-25.3%) cholesterol and HDL (-34.6%) triglyceride levels. In fasted males SPRET-HL produced significant reductions in total TG (-56.0%) and VLDL (-63.1%) and HDL (-50.0%) triglycerides. In fasted females SPRET-HL produced significant reductions in TC (-27.1%) compared with B6-HL. Tables 3 and 4 show cholesterol concentrations and Tables 5 and 6 show TG concentrations of the major lipoprotein classes in fasted and fed mice respectively. Fig. 4 shows fasted lipoprotein subfractions. Results demonstrate allele and sex effects specific to lipoprotein class.
TABLE 3.
Plasma cholesterol levels in fasted mice sacrificed at 140 ± 14 days
| TC |
CM-C |
VLDL-C |
LDL-C |
HDL-C |
|||
|---|---|---|---|---|---|---|---|
| Genotype | Sex | N | (mg/dl) | (mg/dl) | (mg/dl) | (mg/dl) | (mg/dl) |
| HL−/− | M | 6 | 89.5 ± 9.6a | 0.13 ± 0.04 | 2.0 ± 0.4 | 5.1 ± 0.7a,b | 82.9 ± 9.2a |
| HL+/+ | M | 8 | 70.1 ± 4.8a,b | 0.12 ± 0.02 | 3.5 ± 0.3 | 6.5 ± 0.5a | 59.9 ± 4.4a,b |
| HL−/−,+S | M | 7 | 59.1 ± 2.4b | 0.13 ± 0.03 | 1.5 ± 0.2 | 3.7 ± 0.2b | 53.7 ± 2.1b |
| HL+/+,+S | M | 5 | 57.7 ± 4.4b | 0.19 ± 0.05 | 1.9 ± 0.2 | 6.4 ± 0.3a | 49.2 ± 4.2b |
| HL−/− | F | 6 | 93.3 ± 6.4a | 0.12 ± 0.04 | 1.8 ± 0.4 | 5.5 ± 0.8a | 85.9 ± 5.5a |
| HL+/+ | F | 5 | 60.8 ± 2.0b | 0.16 ± 0.06 | 2.3 ± 0.3 | 8.4 ± 0.4b | 50.0 ± 2.3b |
| HL−/−,+S | F | 4 | 44.3 ± 2.7c | 0.09 ± 0.04 | 1.5 ± 0.3 | 4.4 ± 0.7a | 38.2 ± 2.4b,c |
| HL+/+,+S | F | 5 | 47.6 ± 3.2b,c | 0.07 ± 0.01 | 1.7 ± 0.2 | 8.2 ± 0.3b | 37.7 ± 3.3c |
| Overall P | |||||||
| HL | <0.0001 | NS | NS | <0.0001 | <0.0001 | ||
| Sex | 0.017 | NS | NS | 0.005 | 0.004 | ||
| HL x Sex | NS | NS | NS | NS | NS |
CM-C, chylomicron-cholesterol; HDL-C, HDL-cholesterol; LDL-C, LDL-cholesterol; TC, total cholesterol; VLDL-C, VLDL-cholesterol. HL−/− and HL−/−,+S are N5 and N6 littermates. HL+/+ mice are from a parental cross. HL+/+,+S mice are from a backcross between N10 HL+/+,+S and HL+/+ mice. Data presented are means ± SEM. Analyses were run on natural log transformed data. For posthoc analysis, sexes were analyzed separately. Values not sharing superscript (a,b,c) are different by Scheffe's posthoc test (P < 0.05).
TABLE 4.
Plasma cholesterol levels in nonfasted mice sacrificed at 140 ± 14 days
| Genotype | Sex | N | TC (mg/dl) | CM-C (mg/dl) | VLDL-C (mg/dl) | LDL-C (mg/dl) | HDL-C (mg/dl) |
|---|---|---|---|---|---|---|---|
| HL−/− | M | 5 | 109.90 ± 2.5a | 0.454 ± 0.1 | 3.71 ± 0.4a | 7.91 ± 0.3a | 97.82 ± 2.4a |
| HL+/+ | M | 4 | 79.87 ± 0.8b | 0.658 ± 0.2 | 4.95 ± 0.6a | 9.51 ± 0.4b | 64.74 ± 0.8b |
| HL−/−,+S | M | 4 | 55.21 ± 2.4c | 0.250 ± 0.0 | 2.38 ± 0.2b | 4.19 ± 0.2c | 48.39 ± 0.2c |
| Overall P | <0.0001 | NS | 0.008 | <0.0001 | <0.0001 |
CM-C, chylomicron-cholesterol; HDL-C, HDL-cholesterol; LDL-C, LDL-cholesterol; TC, total cholesterol; VLDL-C, VLDL-cholesterol. All animals are ≥N10. HL−/− and HL−/−,+S mice are littermates. HL+/+ mice are from a backcross between N10 HL+/+,+S and HL+/+ mice. Data presented are means ± SEM. Analyses were run on natural log transformed data. For posthoc analysis, sexes were analyzed separately. Values not sharing superscript (a,b,c) are different by Scheffe's posthoc test (P < 0.05).
TABLE 5.
Plasma triglyceride levels in fasted mice sacrificed at 140 ± 14 days
| Genotype | Sex | N | Total TG (mg/dl) | CM-TG (mg/dl) | VLDL-TG (mg/dl) | LDL-TG (mg/dl) | HDL-TG (mg/dl) |
|---|---|---|---|---|---|---|---|
| HL−/− | M | 6 | 27.4 ± 4.0a,b | 1.2 ± 0.2 | 12.4 ± 2.4b | 10.1 ± 1.5a | 3.7 ± 0.4a |
| HL+/+ | M | 8 | 50.5 ± 6.3a | 1.7 ± 0.3 | 30.9 ± 4.3a | 15.3 ± 1.6b | 2.6 ± 0.4a |
| HL−/−,+S | M | 7 | 22.2 ± 4.0b | 1.1 ± 0.2 | 11.4 ± 3.1b | 8.4 ± 0.5a | 1.3 ± 0.2b |
| HL+/+,+S | M | 5 | 30.8 ± 2.8a,b | 1.2 ± 0.3 | 9.6 ± 1.3b | 18.1 ± 1.1b | 1.8 ± 0.3a,b |
| HL−/− | F | 6 | 24.6 ± 4.0 | 1.0 ± 0.3a | 9.8 ± 2.1a | 9.6 ± 1.2 | 4.2 ± 0.5a |
| HL+/+ | F | 5 | 19.1 ± 1.1 | 0.42 ± 0.3a,b | 5.1 ± 0.8a,b | 12.9 ± 0.8 | 0.73 ± 0.1b |
| HL−/−,+S | F | 4 | 18.8 ± 1.6 | 0.68 ± 0.3a,b | 8.3 ± 0.9a | 8.5 ± 0.5 | 1.4 ± 0.2b |
| HL+/+,+S | F | 5 | 16.1 ± 1.6 | 0.10 ± 0.0b | 3.3 ± 0.7b | 12.1 ± 1.1 | 0.59 ± 0.1c |
| Overall P | |||||||
| HL | 0.018 | 0.022 | 0.003 | <0.0001 | <0.0001 | ||
| Sex | <0.0001 | <0.0001 | <0.0001 | NS | <0.0001 | ||
| HL x Sex | 0.007 | 0.004 | <0.0001 | NS | <0.0001 |
CM-TG, chylomicron-triglyceride; HDL-TG, HDL-triglyceride; LDL-TG, LDL-triglyceride; TG = triglyceride; VLDL-TG = VLDL-triglyceride. HL−/− and HL−/−,+S are N5 and N6 littermates. HL+/+ mice are from a parental cross. HL+/+,+S mice are from a backcross between N10 HL+/+,+S and HL+/+ mice. Data presented are means ± SEM. Analyses were run on natural log transformed data. For posthoc analysis, sexes were analyzed separately. Values not sharing superscript (a,b,c) are different by Scheffe's posthoc test (P < 0.05).
TABLE 6.
Plasma triglyceride levels in nonfasted mice sacrificed at 140 ± 14 days
| Genotype | Sex | N | Total TG (mg/dl) | CM-TG (mg/dl) | VLDL-TG (mg/dl) | LDL-TG (mg/dl) | HDL-TG (mg/dl) |
|---|---|---|---|---|---|---|---|
| HL−/− | M | 5 | 69.47 ± 10.1 | 8.40 ± 2.7 | 37.59 ± 6.5 | 14.95 ± 0.9a,b | 8.54 ± 0.4a |
| HL+/+ | M | 4 | 71.09 ± 10.8 | 10.34 ± 3.5 | 38.36 ± 6.4 | 17.57 ± 1.2a | 4.82 ± 0.4b |
| HL−/−,+S | M | 4 | 42.38 ± 2.3 | 4.95 ± 2.2 | 22.91 ± 0.8 | 11.38 ± 0.8b | 3.15 ± 0.1c |
| Overall P | NS | NS | NS | 0.006 | <0.0001 |
CM-TG, chylomicron-triglyceride; HDL-TG, HDL-triglyceride; LDL-TG, LDL-triglyceride; TG = triglyceride; VLDL-TG = VLDL-triglyceride. All animals are ≥N10. HL−/− and HL−/−,+S mice are littermates. HL+/+ mice are from a backcross between N10 HL+/+,+S and HL+/+ mice. Data presented are means ± SEM. Analyses were run on natural log transformed data. For posthoc analysis, sexes were analyzed separately. Values not sharing superscript (a,b,c) are different by Scheffe's posthoc test (P < 0.05).
Fig. 4.
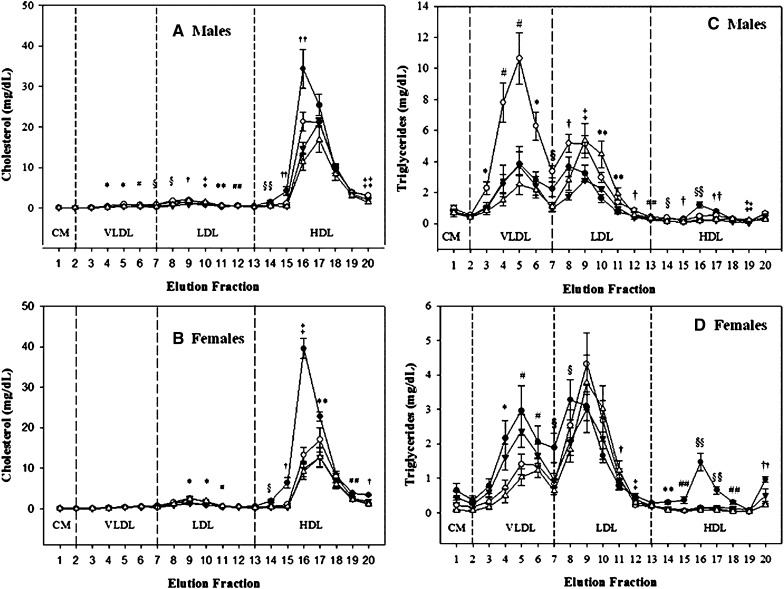
Lipoprotein composition determined in 20 continuously eluted fractions of fasted male and female plasma determined by dual detection HPLC. Filled circles = HL−/−; Filled triangles = HL+/+; white circles = HL−.-,+S; white triangles = HL+/+,+S; HL−/− = mice lacking endogenous HL; HL−/−,+S = mice lacking endogenous HL and positive for the SPRET-HL transgene; HL+/+ = C57BL6/J mice; HL+/+,+S = C57BL6/J mice positive for the SPRET-HL transgene. A: Male cholesterol. B: Female cholesterol. C: Male triglyceride. D: Female triglyceride. Elution fractions correspond to the following lipoprotein subfractions: VLDL, large, 3–5; medium, 6; and small, 7. LDL, large, 8; medium, 9; small, 10; and very small, 11–13. HDL, very large, 14–15; large, 16; medium, 17; small 18; very small, 19–20. Sample numbers are as follows: HL−/− 6, HL+/+ 8, HL−/−,+S 7, HL+/+,+S 5. Data shown are means ± SEM and significance determined by Scheffe's posthoc test. A: * P < 0.005 HL+/+ compared to all other genotypes; # P < 0.005 HL+/+ compared to HL−/− and HL−/−,+S; § P < 0.05 HL+/+ compared to HL−/−,+S and HL+/+,+S; † P < 0.0001 HL−/−,+S compared to HL+/+ and HL+/+,+S; ‡ P < 0.01 HL+/+,+S compared to all other genotypes; ** P < 0.05 HL−/− compared to HL+/+,+S; ## P < 0.05 HL−/− compared to HL−/−,+S; §§ P < 0.05 HL−/− compared to HL−/−,+S and HL+/+,+S; †† P < 0.05 HL−/− compared to all other genotypes; ‡‡ P < 0.01 HL+/+,+S compared to HL−/− and HL+/+. B: * P < 0.0001 HL+/+ compared to HL−/− and HL−/−,+S; # P < 0.05 HL−/− compared to HL+/+ and HL+/+,+S; § P < 0.05 HL−/− compared to all other genotypes; † P < 0.005 HL−/− compared to all other genotypes; ‡ P < 0.0001 HL−/− compared to all other genotypes; ** P < 0.001 HL−/− compared to HL+/+ and HL+/+,+S; #,# P < 0.01 HL−/− compared to HL+/+,+S. C: * P < 0.05 HL+/+ compared to all other genotypes; # P < 0.005 HL+/+ compared to all other genotypes; § P < 0.05 HL+/+ compared to HL−/−,+S and HL+/+,+S; † P < 0.05 HL+/+ compared to HL−/−,+S; ‡ P < 0.005 HL+/+,+S compared to HL−/− and HL−/−,+S; ** P < 0.0001 HL+/+,+S compared to all genotypes; ## P < 0.05 HL−/−,+S compared to HL+/+ and HL+/+.+S; §§ P < 0.001 HL−/− compared to all other genotypes; †† P < 0.02 HL−/− compared to HL−/−,+S and HL+/+,+S; ‡‡ P < 0.05 HL+/+,+S compared to HL+/+ and HL−/−,+S. D: * P < 0.05 HL+/+,+S compared to HL−/− and HL−/−,+S; # P < 0.05 HL+/+,+S compared to HL−/−; § P < 0.05 HL+/+ compared to HL−/− and HL−/−,+S; † P < 0.05 HL+/+,+S compared to HL−/− and HL−/−,+S; ‡ P < 0.05 HL+/+ compared to HL−/−; ** P < 0.05 HL−/− compared to all other genotypes; ## P < 0.01 HL−/− compared to all other genotypes; §§ P < 0.0001 HL−/− compared to all other genotypes; †† P < 0.0001 HL−/− compared to HL+/+ and HL+/+,+S.
Hepatic lipase participates in remnant particle reuptake (40, 41); however, we found no differences in CM-cholesterol (CM-C) for either fed or fasted males or fasted females (Tables 3 and 4). The TG component of CMs however, was reduced by B6- and SPRET-HL coexpression in females where HL+/+,+S mice had a 90% reduction in CM-TG (1.0 ± 0.3 vs. 0.1 ± 0.0 mg/dl; P=0.03) compared with HL−/− mice. Feeding increased CM-TG in all HL genotypes, but with no allelic effect (Table 6). Hepatic lipase activity was not correlated with CM-TG, although it tended toward significance in females (N = 14, r = -0.505; P=0.066) and was not correlated in males (N = 16, r = 0.179; NS). As such the greater activity in HL+/+,+S females may have produced the decreases in CM-TG. Since activity and mRNA levels were positively correlated, this reduction could potentially be due to an increase in HL mRNA.
As reviewed by Zambon et al., HL participates in the remodeling of VLDL, the primary TG carrying lipoprotein during the fasted state (42). We report significant allelic effects for VLDL- cholesterol (VLDL-C) and allelic and sex effects for VLDL-TG levels. In fasted males the expression of SPRET-HL was sufficient to reduce cholesterol in the small VLDL subfraction (Fig. 4A) and total VLDL-C in fed males (HL−/−,+S, 2.38 ± 0.2 vs. HL+/+, 3.71 ± 0.4 mg/dl). Unlike males, females showed no differences in VLDL-C in any subfraction (Fig. 4B). Here alleles of HL significantly influenced VLDL-TG in fasted mice. Our results are in agreement with a recently published paper that reported wild-type B6 mice had increased VLDL-TG compared with HL knockout mice (43). In males, B6-HL mediated dramatic increases in VLDL-TG by 2.5-, 2.7-, and 3.2-fold compared with HL−/−, HL−/−,+S and HL+/+,+S mice, respectively (Table 5). These increases were not specific to a particular subfraction, (Fig. 4C), as HL+/+ mice had significant TG increases in large, medium and small VLDL. The effect of feeding only mildly increased VLDL-TG in HL+/+ mice, but substantially increased VLDL-TG in HL−/− and HL−/−,+S mice (Table 6). In fed mice SPRET-HL reduced VLDL-TG by -39.1% and -40.3% compared with HL−/− and HL+/+ mice; however, these reductions were nonsignificant.
Hepatic lipase activity differences in males and premenopausal females have been reported in humans, which may partly explain gender differences in lipoprotein profiles (14). Our results demonstrate significant sexual dimorphism in regards to HL activity and VLDL-TG. Female mice expressing B6-HL had decreased total VLDL-TG (Table 5) resulting in a highly significant two-way interaction between HL and sex (P < 0.0001). The greatest reduction of VLDL-TG was confined to the small VLDL subfraction. Fig. 4D shows a significant decrease (P < 0.05) in TG concentrations in small VLDL in HL+/+ mice compared with HL−/− or HL−/−,+S mice.
Unlike humans, where the majority of circulating cholesterol is found in LDL, in mice HDL is the major cholesterol carrying lipoprotein. Tables 3 and 5 show a clear pattern in both sexes where HL+/+ and HL+/+,+S mice had increased LDL-C and LDL-TG compared with either HL−/− or HL−/−,+S mice. The effect after feeding was similar where B6-HL expressing mice had increased LDL-TG compared with HL−/−,+S mice (Table 6). These results may indicate structural differences in B6- and SPRET-HL that alter specificity toward LDL. Another possibility is that linked 129 alleles in HL−/− and HL−/−,+S mice are reducing LDL-C and LDL-TG in this model. However, in fed mice, SPRET-HL produced a significant decrease in LDL-C compared with the absence of HL (4.19 ± 0.2 vs. 7.91 ± 0.3 mg/dl; P < 0.0001), which supports an HL allelic effect (Table 4).
In this study, SPRET-HL promoted greater reductions in HDL-cholesterol (HDL-C) and HDL-TG than B6-HL. HDL-C in HL−/−,+S and HL+/+,+S mice were decreased by −10.3% (NS) and −17.9% (NS) for fasted males, respectively; −23.6% (NS) and −24.6% (P < 0.05) for females, respectively; and −16.7% (P < 0.05) for fed males (HL−/−,+S only) compared with HL+/+ mice (Tables 3 and 4). HDL-C and HL activity were not significantly correlated in this study (males, N = 16, r = -0.234; NS and females, N = 14, r = −0.183; NS). SPRET-HL was even more potent at reducing HDL-TG. Male HL−/−,+S fasted mice had 50.0% less (P < 0.05) and fed mice had 34.6% less (P < 0.05) HDL-TG compared with HL+/+ mice. Interestingly, male HL+/+,+S mice, which express both B6- and SPRET-HL, had an intermediate phenotype with nonsignificant differences in HDL-TG compared with either HL+/+ mice or HL−/−,+S. In contrast, females expressing B6-HL showed a large decrease in HDL-TG (Table 3) and HL+/+,+S females had the greatest reduction in HDL-TG (−86.0%; P < 0.05) compared with HL−/− mice, demonstrating a potential additive effect of both HL alleles,. This resulted in a highly significant two-way interaction between sex and HL (P < 0.0001) for HDL-TG.
Allele specific effects on lipoprotein particle size
To determine CM, VLDL, LDL, and HDL sizes, effluents from both cholesterol and triglyceride HPLC plots were analyzed separately and particle size was determined based on appropriate sized controls (32, 33). Particle size may be an important predictor of cardiovascular disease risk (44, 45) and there is evidence that different sized lipoproteins of the same class reach alternative metabolic fates (42, 46). Smaller VLDL undergo plasma remodeling by lipid transfer proteins as well as HL and are more likely converted to LDL and thus less likely to participate in particle reuptake at the level of the liver (46, 47). We present particle size data from the cholesterol elution plot for the three major lipoproteins present during the fasted state as summarized in Table 7.
TABLE 7.
Lipoprotein particle size in fasted mice as determined by dual detection HPLC
|
Cholesterol Plot |
|||||
|---|---|---|---|---|---|
| Genotype | Sex | N | VLDL (30–80nm) | LDL (16–30nm) | HDL (8–16nm) |
| HL−/− | M | 6 | 40.2 ± 0.7a | 23.9 ± 0.3a,b | 11.2 ± 0.1a |
| HL+/+ | M | 8 | 41.3 ± 0.3a,b | 24.3 ± 0.1a | 10.9 ± 0.1b |
| HL−/−,+S | M | 7 | 43.4 ± 0.4c | 23.2 ± 0.2b | 10.9 ± 0.0b |
| HL+/+,+S | M | 5 | 43.2 ± 0.4b,c | 23.8 ± 0.1a,b | 10.9 ± 0.1b |
| HL−/− | F | 6 | 39.1 ± 0.6a | 24.0 ± 0.3 | 11.4 ± 0.1a |
| HL+/+ | F | 5 | 42.1 ± 0.5b | 24.4 ± 0.1 | 11.0 ± 0.1b |
| HL−/−,+S | F | 4 | 40.4 ± 0.4a,b | 24.2 ± 0.1 | 10.7 ± 0.1c |
| Overall P | |||||
| HL | <0.0001 | 0.014 | <0.0001 | ||
| Sex | <0.0001 | 0.004 | NS | ||
| HL x Sex | 0.002 | 0.05 | NS | ||
Average particle size in nm as determined by dual detection HPLC using plots generated by the cholesterol eluted fractions. Data presented are means ± SEM. Analyses were run on natural log transformed data. For posthoc analysis, sexes were analyzed separately. Values not sharing superscript (a,b,c) are different by Scheffe's posthoc test (P < 0.05).
Overall, HL deficiency produced smaller VLDL and larger HDL in both male and female mice, SPRET-HL increased VLDL size and B6-HL decreased LDL size in males. However, in females, SPRET-HL had the opposite effect and reduced VLDL particle size and B6-HL had no effect on LDL size. Therefore, we report a significant interaction of sex and HL for VLDL size (P = 0.002) and a marginally significant interaction of sex and HL for LDL size (P = 0.05). Here HL+/+,+S males show an intermediate phenotype with VLDL and LDL particle sizes not significantly different from either HL+/+ or HL−/−,+S mice (Table 7). However, female mice expressing both HL alleles mirrored HL−/−,+S mice and produced smaller VLDL than mice expressing B6-HL alone, although the size differences did not reach statistical significance. HL activity was positively correlated with VLDL size (N = 16, r = 0.634; P = 0.008) and inversely correlated with LDL size (N = 16, r = −0.656; P = 0.007) in males. For females, the correlation of VLDL size and HL activity only trended toward significance (N = 14, r = −0.498; P = 0.070), but HL activity was inversely correlated with LDL size (N = 14, r = -0.716; P = 0.004). In a previous report, mice lacking HL had elevated levels of HDL1 when compared with wild-type littermates (2). Here the absence of HL promoted earlier elution peaks (Fig. 4), which correspond to large HDL particles in both male and female mice, yet HDL size was not significantly correlated with HL activity in either sex (data not shown).
DISCUSSION
We generated a BAC transgenic mouse model expressing SPRET-HL at physiologic levels and show that naturally occurring variants of HL affect lipoprotein composition and size with no direct effects on obesity with the present breeding scheme. The current model system was used to test for an intrinsic difference between B6- and SPRET-HL function attributable to the 17 variants in the coding region of HL, four of which produce either conservative or semi-conservative amino acid substitutions (38). We provide a unique resource to study the structure-function relationship of HL in vivo.
Plasma lipid results are consistent with published reports that demonstrated reductions in cholesterol and TG levels when HL knockout animals were compared with wild-type (1, 2, 21, 43, 48, 49). However, we present novel information regarding allelic derived differences in lipoprotein composition. Overall, compared with B6-HL, SPRET-HL mediated significant reductions in TC (−30.9%) and VLDL (−51.9%), LDL (−55.9%), and HDL (−25.3%) cholesterol and HDL (−34.6%) triglyceride levels in fed males. In fasted males, SPRET-HL produced significant reductions in total TG (−56.0%) and VLDL (−63.1%) and HDL (−50.0%) triglycerides. In fasted females, SPRET-HL produced significant reductions in TC (−27.1%) compared with B6-HL. The largest difference between the two alleles was for VLDL-TG in fasted mice whereby HL+/+ mice had large increases compared with either positive transgenic or HL deficient mice. This is in agreement with a recently published study that compared HL knockout mice to a parental B6 cross (43). Our fed data, however, was not in concordance with our fasted data, but these differences may be explained by the variability of food intake of each mouse prior to sacrifice.
In humans, sex influences HL activity, which could potentially affect lipoprotein profiles (14).We report here an allele-specific sex effect, such that male HL+/+ mice had increased total TG, VLDL-, and HDL-TG compared with positive transgenics, while female HL+/+ mice had decreased total TG, VLDL-TG, and HDL-TG compared with mice with the SPRET-HL transgene. This resulted in a highly significant two-way interaction between HL and sex. Since female HL+/+ mice had ∼2-fold increase in activity compared with male HL+/+ mice, this may explain the difference in TG levels in the various lipoprotein fractions. SPRET-HL did not produce a similar sex-specific effect for TG levels or HL activity levels.
In our study we report both sex and allelic effects on particle size. Hepatic lipase activity in males was positively correlated with VLDL particle size but was negatively correlated in females. In males, LDL particle size was negatively correlated with HL activity and not correlated in females. This resulted in significant two-way interactions for both VLDL and LDL particle size between sex and HL activity. To our knowledge, this is the first report of a sex by HL allele interaction in mice. Previous reports indicate increased HL activity in transgenic mice positive for human HL resulted in reduced HDL particle size (1). However, that finding may have been due to supra-physiologic expression of human HL that does not mimic endogenous mouse HL.
Because BAC technology was used, SPRET-HL in the transgenic was controlled by the endogenous promoter and all coding and regulatory SPRET polymorphisms were faithfully reproduced. Although we did not sequence the regulatory region from the BAC clone we determined by BAC-end sequencing and full length BAC genotyping assay that the clone was large enough (∼190 kb) to include HL regulatory elements. We found no support for SPRET promoter polymorphisms influencing the amount or regulation of HL mRNA. Liver mRNA levels showed no difference between B6- and SPRET-HL. Temporal or spatial expression differences may exist that we did not detect, but minimally there were no differences at time of sacrifice.
While HL protein levels were not directly measured, female HL+/+, HL−/−,+S and HL+/+,+S mice demonstrated parallel HL activity levels and male SPRET- and B6-HL expressing mice had a 60–70% difference in HL activity. This difference is comparable to variation detected in a human population with naturally occurring alleles (50) rather than the dramatic 1000-fold differences in activity that have been reported in models that overexpress human HL (1). We also measured postheparin plasma HL activity to test for allelic differences in liver versus plasma pools of B6- and SPRET-HL expressing mice. Our results indicate that the majority of HL activity was found in preheparin plasma, independently of genotype. Fig. 3 showed a parallel increase in postheparin plasma HL activity in both female HL−/−,+S (43.4%) and male HL+/+ (50.6%) mice. This is similar to published reports showing that in mice, the majority of HL is found unbound in circulating plasma (36, 51). Moreover, mice in this study demonstrated parallel HL activity levels and no correlations between activity and VLDL-C, LDL-C, and HDL-C levels. This lack of correlation supports the hypothesis that allele differences rather than enzyme amount is driving the plasma lipid differences.
The amino acid substitutions (S106N, A156V, L416V, and S480T) in SPRET-HL were not predicted to alter function by the computational protein prediction programs, PolyPhen (http://genetics.bwh.harvard.edu/pph) and SIFT (http://sift.jcvi.org); however, their proximity to critical regions could affect HL's function. Supplementary Fig. II provides a sequence alignment of B6-, SPRET-, and human-HL and highlights the location of the SPRET amino acid changes. The first two mutations are in the highly conserved N-terminal domain, which contains the catalytic triad. The domain is found in mammalian, bacterial, and fungal lipases suggesting a long and preserved evolutionary lineage. Because of preserved structure and function, lipase-specific characteristics are unlikely to be conferred by the N-terminal domain, but rather by the second smaller carboxyl-terminal domain, which only the mammalian lipases possess. Lower-order organisms lack a domain that corresponds to the HL carboxyl-terminal domain, suggesting that the domain was added later in evolution for accessory functions. The second set of amino acid differences between B6- and SPRET-HL is located in the carboxyl-terminal domain, a region shown to be required for variety of lipase functions, including lipid binding, control of heparin binding, substrate specificity, receptor binding and catalysis of long-chain substrates (29, 30, 51, 52). Mutations in this domain are of particular interest, since there is greater sequence variability within this domain in the lipase gene family as a whole. It is likely that variations located in the carboxyl-terminal domain bestow the unique character of each lipase family member. Therefore, although the amino acid substitutions in SPRET-HL compared with that of B6 are conservative they could be in critical regions, which could substantially alter enzyme function.
BAC clones reproduce polymorphisms and are under the control of the endogenous promoter; however they integrate randomly in the genome with (potential) multiple copies and may display ectopic expression or knock out a gene (53). Our evidence is consistent with the hypothesis that SPRET-HL is expressed with correct tissue specificity and is integrated at a single site. We conducted a tissue survey in GWAT, kidney, and skeletal muscle and report no detectable HL mRNA (data not shown). In this study, mice were backcrossed for either N5, N6, and ≥N10 times and it is unlikely that more than one integration site remains after ten generations of backcross. Mice backcrossed for ≥N10 generations had similar cholesterol and triglyceride phenotypes as N5 or N6 mice and provide direct functional evidence that there has been limited colony drift over time. We also used a 1449 SNP panel to identify passenger CBA alleles from the B6CBAF1/J founders and two or more transgene integration sites would have increased the likelihood of finding CBA alleles. We found no possible CBA integration site passenger alleles so there is likely a single integration site. While it is not possible to rule out integration site effects because we obtained a single founder, evidence indicates that the transgene rather than a knocked-out gene is responsible for the observed lipoprotein alterations. Overall, positive transgenic mice were not obese or thin, healthy, bred well, and were otherwise normal except for lipoprotein alterations and other HL related phenotypes. We show mRNA levels comparable in HL+/+ and HL−/−,+S mice, similar HL activity levels in females, and roughly comparable HL activity in males. Additionally, N5 mice have similar HL activity levels of N10 mice, which indicated stable functional HL over time (data not shown). Thus, even if a gene is disrupted it is not having any measurable effects on phenotypes discussed in this study.
In addition to SPRET-HL, the BAC insert included a single exon protein, ENSMUSG00000078305. This gene is 98% identical to the seven exon gene, Rrp7a, located on mouse chromosome 15. Rrp7a is a hypothetical RNA-binding protein because it contains an RNA binding domain consensus sequence. No publications for Rrp7a were identified; however, the Gene Ontology database identified 4,104 protein products classified as RNA binding across multiple species and 148 specific to mouse. A two-way BLASTN revealed that Rrp7A is not transcribed antisense to HL and therefore is unlikely to convey regulatory properties (54). We conducted qPCR with custom primers designed to amplify the 3′ UTR region of ENSMUSG00000078305. It was possible that the primers amplified both endogenous Rrp7a and the intronic gene; however, our results show similar expression levels in four genotypes of mice and no effect of transgene mediated expression differences.
The problems concerning passenger alleles in transgenic mice have been previously discussed (55). To rule out CBA allelic effects, we used transgenic and nontransgenic littermate mice and thus effects of unlinked CBA strain genes are randomized in our data. We also include data from ≥N10 backcrossed mice, which showed similar changes in lipoprotein composition and size as those backcrossed for N5 or N6 generations. Finally, with a 1449 SNP chip assay, we identified a single CBA SNP located on the X chromosome in two of the four mice, one on the HL−/− background and one on the HL+/+ background. Since we report significant interactions between sex and HL for TG levels in VLDL and HDL it was important to rule an X chromosome transgene integration site. Our breeding schemes allowed for this as we bred sire HL−/−,+S and HL+/+,+S mice to HL−/− and HL+/+ dams and collected males positive for the transgene (data not shown); therefore male to male transmission rules out the possibility of an X chromosome integration. Our data are consistent with the hypothesis that the original BAC clone integrated into a B6 chromosome in the founding embryo, which means it is unlikely there are linked CBA alleles in our transgenic line.
The HL−/− strain used in these studies provided an added complication to our analyses since the knockout contained known 129 alleles proximal and distal to the structural locus of Lipc. In theory, the 129 strain congenic donor region surrounding the Lipc locus in the HL−/− mice may have influenced lipoprotein composition and size and/or obesity in our mouse model. Previous studies of other models have demonstrated that phenotypes originally ascribed to a targeted gene were actually due to the linked 129 strain alleles surrounding the knocked-out locus (31). Therefore, we genotyped the surrounding region using microsatellite makers and identified linked 129 alleles and determined the 129 strain congenic donor region ranged from D9Mit297 to D9MIT269 or ∼53.3 Mb. Subsequently we further fine mapped the 129 alleles using a 1449 SNP chip. We determined that the maximal size of the 129 donor region ranged from rs13480122 at 30,964,211 bp to rs13480365 at 98,479,731 bp on chromosome 9. We also identified 129 passenger alleles on chromosomes 6 and 13. These alleles are absent in HL+/+ and HL+/+,+S mice.
Our data demonstrate that most of the lipoprotein phenotypes cannot be explained by the 129 strain donor region alleles. This is most obvious in the fed animal data where there are significant posthoc differences between all three genotypes: HL+/+, HL−/− and HL−/−,+S for TC, VLDL-C, LDL-C, HDL-C, and HDL-TG. Fasted animal data also shows significant posthoc tests differentiating all three genotypes in a related series: HL−/−, HL+/+ and HL+/+,+S for HDL-C in females. There is also a highly significant posthoc difference between HL+/+ and HL+/+,+S in males for VLDL-TG. These transgene effects cannot be due to 129 strain alleles and therefore it is more likely that all allelic effects are due to HL.
Several quantitative trait loci for adiposity have been reported near the Lipc structural locus (56–59). We observed here that 129 alleles rather than the SPRET-HL transgene influenced obesity. The 129 alleles promoted increases in all of the four white adipose depots in females and in GWAT, FWAT and RWAT in males. AI, a surrogate for total body fat determined by body composition analysis, was increased by as much as 95.0% in HL−/− and HL−/−,+S mice compared with either HL+/+ or HL+/+,+S mice in males with even larger increases observed in females. The corollary conclusion is that none of the lipoprotein profile phenotypes observed in this study can be due to effects of SPRET-HL on weight and obesity. While these data are not consistent with our earlier report of SPRET-HL influencing obesity there are several important differences in the experimental protocol that might cause divergent results. The mice reported in this manuscript are hemizygous for the SPRET-HL transgene and homozygous for 129 alleles on chromosomes 6, 9, and 13 all on a B6 background. The earlier work included animals heterozygous for SPRET alleles and 129 alleles on chromosomes 6, 9, and 13. As such if the 129 allele effect on obesity is recessive in nature the earlier test cross would not have shown the effect. Additionally, the cohort of mice used in the previous study included an average of 25% SPRET genes in the backcross, which allowed for epistatic interactions between SPRET-HL and unlinked SPRET chromosome 9 genes that are absent in this model. This difference in breeding schemas may explain the disparity in obesity results.
Here we report a unique resource to identify structural features responsible for the multiple activities of HL. The transgenic mouse model expressed SPRET-HL at physiological levels on backgrounds with or without endogenous B6-HL and differentially influenced lipoprotein composition and size. Compared with B6-HL, SPRET-HL mediated significant reductions in TC and VLDL, LDL, and HDL cholesterol and HDL triglyceride levels in fed males and in fasted males SPRET-HL produced significant reductions in total TG and VLDL and HDL triglycerides. In females, SPRET-HL produced significant reductions in TC compared with B6-HL. B6-HL expression produced decreases in females and increases in males for VLDL-TG and HDL-TG resulting in a significant sex and HL genotype interaction. Finally, we show allelic differences that influenced particle size such that SPRET-HL increased VLDL size and produced smaller LDL in males; while in females SPRET-HL produced smaller HDL. Currently, we cannot conclude whether the effects of SPRET-HL were mediated by different affinities or specificities for lipoprotein composition, size, or apolipoprotein associations. Nonetheless, our results demonstrate that there are functional allelic differences between B6- and SPRET-HL. Moreover, they demonstrate there is no substitute for in vivo investigation of variant proteins regardless of nonsignificant predications as determined by in silico prediction programs.
Supplementary Material
Acknowledgments
The authors thank Pieter de Jong for probing the BAC clone, Peter Cung for his genotyping expertise, and Kari Haus for her critical reading of the manuscript.
Footnotes
Abbreviations:
- AI
- adiposity index
- AN
- anal-nasal
- apoB
- apolipoprotein B
- B6
- C57BL6/J
- BAC
- bacterial artificial chromosome
- CM
- chylomicron
- CM-C
- chylomicron-cholesterol
- FWAT
- femoral white adipose tissue
- gDNA
- genomic DNA
- GWAT
- gonadal white adipose tissue
- HDL-C
- HDL-cholesterol
- LDL-C
- LDL-cholesterol
- MWAT
- mesenteric white adipose tissue
- qPCR
- quantitative PCR
- RWAT
- retroperitoneal white adipose tissue
- SPRET
- SPRET/EiJ
- SSLP
- simple sequence length polymorphism
- TC
- total cholesterol
- TG
- triglyceride
- VLDL-C
- VLDL-cholesterol
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases Grants DK-5281 and DK-69978. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures.
REFERENCES
- 1.Busch S. J., Barnhart R. L., Martin G. A., Fitzgerald M. C., Yates M. T., Mao S. J., Thomas C. E., Jackson R. L. 1994. Human hepatic triglyceride lipase expression reduces high density lipoprotein and aortic cholesterol in cholesterol-fed transgenic mice. J. Biol. Chem. 269: 16376–16382. [PubMed] [Google Scholar]
- 2.Homanics G. E., de Silva H. V., Osada J., Zhang S. H., Wong H., Borensztajn J., Maeda N. 1995. Mild dyslipidemia in mice following targeted inactivation of the hepatic lipase gene. J. Biol. Chem. 270: 2974–2980. [DOI] [PubMed] [Google Scholar]
- 3.Jensen G. L., Daggy B., Bensadoun A. 1982. Triacylglycerol lipase, monoacylglycerol lipase and phospholipase activities of highly purified rat hepatic lipase. Biochim. Biophys. Acta. 710: 464–470. [DOI] [PubMed] [Google Scholar]
- 4.Little J. A., Connelly P. W. 1986. Familial hepatic lipase deficiency. Adv. Exp. Med. Biol. 201: 253–260. [DOI] [PubMed] [Google Scholar]
- 5.Dichek H. L., Qian K., Agrawal N. 2004. The bridging function of hepatic lipase clears plasma cholesterol in LDL receptor-deficient “apoB-48-only” and “apoB-100-only” mice. J. Lipid Res. 45: 551–560. [DOI] [PubMed] [Google Scholar]
- 6.Krapp A., Ahle S., Kersting S., Hua Y., Kneser K., Nielsen M., Gliemann J., Beisiegel U. 1996. Hepatic lipase mediates the uptake of chylomicrons and beta-VLDL into cells via the LDL receptor-related protein (LRP). J. Lipid Res. 37: 926–936. [PubMed] [Google Scholar]
- 7.de Faria E., Fong L. G., Komaromy M., Cooper A. D. 1996. Relative roles of the LDL receptor, the LDL receptor-like protein, and hepatic lipase in chylomicron remnant removal by the liver. J. Lipid Res. 37: 197–209. [PubMed] [Google Scholar]
- 8.Mendoza S. G., Carrasco H., Zerpa A., Briceno Y., Rodriguez F., Speirs J., Glueck C. J. 1991. Effect of physical training on lipids, lipoproteins, apolipoproteins, lipases, and endogenous sex hormones in men with premature myocardial infarction. Metabolism. 40: 368–377. [DOI] [PubMed] [Google Scholar]
- 9.Despres J. P., Ferland M., Moorjani S., Nadeau A., Tremblay A., Lupien P. J., Theriault G., Bouchard C. 1989. Role of hepatic-triglyceride lipase activity in the association between intra-abdominal fat and plasma HDL cholesterol in obese women. Arteriosclerosis. 9: 485–492. [DOI] [PubMed] [Google Scholar]
- 10.Nie L., Wang J., Clark L. T., Tang A., Vega G. L., Grundy S. M., Cohen J. C. 1998. Body mass index and hepatic lipase gene (LIPC) polymorphism jointly influence postheparin plasma hepatic lipase activity. J. Lipid Res. 39: 1127–1130. [PubMed] [Google Scholar]
- 11.Ordovas J. M., Corella D., Demissie S., Cupples L. A., Couture P., Coltell O., Wilson P. W., Schaefer E. J., Tucker K. L. 2002. Dietary fat intake determines the effect of a common polymorphism in the hepatic lipase gene promoter on high-density lipoprotein metabolism: evidence of a strong dose effect in this gene-nutrient interaction in the Framingham Study. Circulation. 106: 2315–2321. [DOI] [PubMed] [Google Scholar]
- 12.Applebaum D. M., Goldberg A. P., Pykalisto O. J., Brunzell J. D., Hazzard W. R. 1977. Effect of estrogen on post-heparin lipolytic activity. Selective decline in hepatic triglyceride lipase. J. Clin. Invest. 59: 601–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brinton E. A. 1996. Oral estrogen replacement therapy in postmenopausal women selectively raises levels and production rates of lipoprotein A-I and lowers hepatic lipase activity without lowering the fractional catabolic rate. Arterioscler. Thromb. Vasc. Biol. 16: 431–440. [DOI] [PubMed] [Google Scholar]
- 14.Tan C. E., Foster L., Caslake M. J., Bedford D., Watson T. D., McConnell M., Packard C. J., Shepherd J. 1995. Relations between plasma lipids and postheparin plasma lipases and VLDL and LDL subfraction patterns in normolipemic men and women. Arterioscler. Thromb. Vasc. Biol. 15: 1839–1848. [DOI] [PubMed] [Google Scholar]
- 15.Dugi K. A., Brandauer K., Schmidt N., Nau B., Schneider J. G., Mentz S., Keiper T., Schaefer J. R., Meissner C., Kather H., et al. 2001. Low hepatic lipase activity is a novel risk factor for coronary artery disease. Circulation. 104: 3057–3062. [DOI] [PubMed] [Google Scholar]
- 16.Baynes C., Henderson A. D., Anyaoku V., Richmond W., Hughes C. L., Johnston D. G., Elkeles R. S. 1991. The role of insulin insensitivity and hepatic lipase in the dyslipidaemia of type 2 diabetes. Diabet. Med. 8: 560–566. [DOI] [PubMed] [Google Scholar]
- 17.Watson T. D., Caslake M. J., Freeman D. J., Griffin B. A., Hinnie J., Packard C. J., Shepherd J. 1994. Determinants of LDL subfraction distribution and concentrations in young normolipidemic subjects. Arterioscler. Thromb. 14: 902–910. [DOI] [PubMed] [Google Scholar]
- 18.Dugi K. A., Feuerstein I. M., Hill S., Shih J., Santamarina-Fojo S., Brewer H. B., Jr., Hoeg J. M. 1997. Lipoprotein lipase correlates positively and hepatic lipase inversely with calcific atherosclerosis in homozygous familial hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 17: 354–364. [DOI] [PubMed] [Google Scholar]
- 19.Moriguchi E. H., Tamachi H., Goto Y. 1990. Hepatic lipase activity and high density lipoproteins in familial hypercholesterolemia: adaptational mechanisms for LDL-receptor deficient state. Tokai J. Exp. Clin. Med. 15: 401–406. [PubMed] [Google Scholar]
- 20.Marques-Vidal P., Jauhiainen M., Metso J., Ehnholm C. 1997. Transformation of high density lipoprotein 2 particles by hepatic lipase and phospholipid transfer protein. Atherosclerosis. 133: 87–95. [DOI] [PubMed] [Google Scholar]
- 21.Mezdour H., Jones R., Dengremont C., Castro G., Maeda N. 1997. Hepatic lipase deficiency increases plasma cholesterol but reduces susceptibility to atherosclerosis in apolipoprotein E-deficient mice. J. Biol. Chem. 272: 13570–13575. [DOI] [PubMed] [Google Scholar]
- 22.Dichek H. L., Johnson S. M., Akeefe H., Lo G. T., Sage E., Yap C. E., Mahley R. W. 2001. Hepatic lipase overexpression lowers remnant and LDL levels by a noncatalytic mechanism in LDL receptor-deficient mice. J. Lipid Res. 42: 201–210. [PubMed] [Google Scholar]
- 23.Knudsen P., Antikainen M., Uusi-Oukari M., Ehnholm S., Lahdenpera S., Bensadoun A., Funke H., Wiebusch H., Assmann G., Taskinen M. R., et al. 1997. Heterozygous hepatic lipase deficiency, due to two missense mutations R186H and L334F, in the HL gene. Atherosclerosis. 128: 165–174. [DOI] [PubMed] [Google Scholar]
- 24.Knudsen P., Antikainen M., Ehnholm S., Uusi-Oukari M., Tenkanen H., Lahdenpera S., Kahri J., Tilly-Kiesi M., Bensadoun A., Taskinen M. R., et al. 1996. A compound heterozygote for hepatic lipase gene mutations Leu334→Phe and Thr383→Met: correlation between hepatic lipase activity and phenotypic expression. J. Lipid Res. 37: 825–834. [PubMed] [Google Scholar]
- 25.Paterniti J. R., Jr., Brown W. V., Ginsberg H. N., Artzt K. 1983. Combined lipase deficiency (CLD): a lethal mutation on chromosome 17 of the mouse. Science. 221: 167–169. [DOI] [PubMed] [Google Scholar]
- 26.Langner C. A., Birkenmeier E. H., Ben-Zeev O., Schotz M. C., Sweet H. O., Davisson M. T., Gordon J. I. 1989. The fatty liver dystrophy (FLD) mutation. A new mutant mouse with a developmental abnormality in triglyceride metabolism and associated tissue-specific defects in lipoprotein lipase and hepatic lipase activities. J. Biol. Chem. 264: 7994–8003. [PubMed] [Google Scholar]
- 27.Wong H., Davis R. C., Nikazy J., Seebart K. E., Schotz M. C. 1991. Domain exchange: characterization of a chimeric lipase of hepatic lipase and lipoprotein lipase. Proc. Natl. Acad. Sci. USA. 88: 11290–11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dichek H. L., Parrott C., Ronan R., Brunzell J. D., Brewer H. B., Jr., Santamarina-Fojo S. 1993. Functional characterization of a chimeric lipase genetically engineered from human lipoprotein lipase and human hepatic lipase. J. Lipid Res. 34: 1393–1401. [PubMed] [Google Scholar]
- 29.Hata A., Ridinger D. N., Sutherland S., Emi M., Shuhua Z., Myers R. L., Ren K., Cheng T., Inoue I., Wilson D. E., et al. 1993. Binding of lipoprotein lipase to heparin. Identification of five critical residues in two distinct segments of the amino-terminal domain. J. Biol. Chem. 268: 8447–8457. [PubMed] [Google Scholar]
- 30.Hill J. S., Yang D., Nikazy J., Curtiss L. K., Sparrow J. T., Wong H. 1998. Subdomain chimeras of hepatic lipase and lipoprotein lipase. Localization of heparin and cofactor binding. J. Biol. Chem. 273: 30979–30984. [DOI] [PubMed] [Google Scholar]
- 31.Peirce J. 2001. Looking at old tools in new ways: using knockouts as congenics to study QTLs. Genome Res. 11: 1469–1471. [DOI] [PubMed] [Google Scholar]
- 32.Okazaki M., Usui S., Ishigami M., Sakai N., Nakamura T., Matsuzawa Y., Yamashita S. 2005. Identification of unique lipoprotein subclasses for visceral obesity by component analysis of cholesterol profile in high-performance liquid chromatography. Arterioscler. Thromb. Vasc. Biol. 25: 578–584. [DOI] [PubMed] [Google Scholar]
- 33.Usui S., Hara Y., Hosaki S., Okazaki M. 2002. A new on-line dual enzymatic method for simultaneous quantification of cholesterol and triglycerides in lipoproteins by HPLC. J. Lipid Res. 43: 805–814. [PubMed] [Google Scholar]
- 34.Twu J. S., Garfinkel A. S., Schotz M. C. 1984. Hepatic lipase. Purification and characterization. Biochim. Biophys. Acta. 792: 330–337. [DOI] [PubMed] [Google Scholar]
- 35.Nilsson-Ehle P., Schotz M. C. 1976. A stable, radioactive substrate emulsion for assay of lipoprotein lipase. J. Lipid Res. 17: 536–541. [PubMed] [Google Scholar]
- 36.Peterson J., Bengtsson-Olivecrona G., Olivecrona T. 1986. Mouse preheparin plasma contains high levels of hepatic lipase with low affinity for heparin. Biochim. Biophys. Acta. 878: 65–70. [DOI] [PubMed] [Google Scholar]
- 37.Dallinga-Thie G. M., Zonneveld-de Boer A. J., van Vark-van der Zee L. C., van Haperen R., van Gent T., Jansen H., De Crom R., van Tol A. 2007. Appraisal of hepatic lipase and lipoprotein lipase activities in mice. J. Lipid Res. 48: 2788–2791. [DOI] [PubMed] [Google Scholar]
- 38.Farahani P., Fisler J. S., Wong H., Diament A. L., Yi N., Warden C. H. 2004. Reciprocal hemizygosity analysis of mouse hepatic lipase reveals influence on obesity. Obes. Res. 12: 292–305. [DOI] [PubMed] [Google Scholar]
- 39.Okazaki M., Usui S., Fukui A., Kubota I., Tomoike H. Component analysis of HPLC profiles of unique lipoprotein subclass cholesterols for detection of coronary artery disease. Clin Chem. 52: 2049–2053. [DOI] [PubMed] [Google Scholar]
- 40.Jansen H., Verhoeven A. J., Weeks L., Kastelein J. J., Halley D. J., van den Ouweland A., Jukema J. W., Seidell J. C., Birkenhager J. C. 1997. Common C-to-T substitution at position -480 of the hepatic lipase promoter associated with a lowered lipase activity in coronary artery disease patients. Arterioscler. Thromb. Vasc. Biol. 17: 2837–2842. [DOI] [PubMed] [Google Scholar]
- 41.Jansen H., Chu G., Ehnholm C., Dallongeville J., Nicaud V., Talmud P. J. 1999. The T allele of the hepatic lipase promoter variant C-480T is associated with increased fasting lipids and HDL and increased preprandial and postprandial LpCIII:B: European Atherosclerosis Research Study (EARS) II. Arterioscler. Thromb. Vasc. Biol. 19: 303–308. [DOI] [PubMed] [Google Scholar]
- 42.Zambon A., Bertocco S., Vitturi N., Polentarutti V., Vianello D., Crepaldi G. 2003. Relevance of hepatic lipase to the metabolism of triacylglycerol-rich lipoproteins. Biochem. Soc. Trans. 31: 1070–1074. [DOI] [PubMed] [Google Scholar]
- 43.van Haperen R., Samyn H., van Gent T., Zonneveld A. J., Moerland M., Grosveld F., Jansen H., Dallinga-Thie G. M., van Tol A., de Crom R. 2009. Novel roles of hepatic lipase and phospholipid transfer protein in VLDL as well as HDL metabolism. Biochim. Biophys. Acta. 1791: 1031–1036. [DOI] [PubMed] [Google Scholar]
- 44.Williams P. T., Superko H. R., Haskell W. L., Alderman E. L., Blanche P. J., Holl L. G., Krauss R. M. 2003. Smallest LDL particles are most strongly related to coronary disease progression in men. Arterioscler. Thromb. Vasc. Biol. 23: 314–321. [DOI] [PubMed] [Google Scholar]
- 45.Scanu A. M., Edelstein C. 2008. HDL: bridging past and present with a look at the future. FASEB J. 22: 4044–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldberg I. J. 2009. Hypertriglyceridemia: impact and treatment. Endocrinol. Metab. Clin. North Am. 38: 137–149. [DOI] [PubMed] [Google Scholar]
- 47.Berneis K. K., Krauss R. M. 2002. Metabolic origins and clinical significance of LDL heterogeneity. J. Lipid Res. 43: 1363–1379. [DOI] [PubMed] [Google Scholar]
- 48.Freeman L., Amar M. J., Shamburek R., Paigen B., Brewer H. B., Jr., Santamarina-Fojo S., Gonzalez-Navarro H. 2007. Lipolytic and ligand-binding functions of hepatic lipase protect against atherosclerosis in LDL receptor-deficient mice. J. Lipid Res. 48: 104–113. [DOI] [PubMed] [Google Scholar]
- 49.Dichek H. L., Qian K., Agrawal N. 2004. Divergent effects of the catalytic and bridging functions of hepatic lipase on atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 24: 1696–1702. [DOI] [PubMed] [Google Scholar]
- 50.Scriver C. R., Sly W. S. 2001. The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill, YN. [Google Scholar]
- 51.Brown R. J., Schultz J. R., Ko K. W., Hill J. S., Ramsamy T. A., White A. L., Sparks D. L., Yao Z. 2003. The amino acid sequences of the carboxyl termini of human and mouse hepatic lipase influencecell surface association. J. Lipid Res. 44: 1306–1314. [DOI] [PubMed] [Google Scholar]
- 52.Davis R. C., Wong H., Nikazy J., Wang K., Han Q., Schotz M. C. 1992. Chimeras of hepatic lipase and lipoprotein lipase. Domain localization of enzyme-specific properties. J. Biol. Chem. 267: 21499–21504. [PubMed] [Google Scholar]
- 53.Heintz N. 2001. BAC to the future: the use of BAC transgenic mice for neuroscience research. Nat. Rev. Neurosci. 2: 861–870. [DOI] [PubMed] [Google Scholar]
- 54.Werner A., Carlile M., Swan D. 2009. What do natural antisense transcripts regulate? RNA Biol. 6: 43–48. [DOI] [PubMed] [Google Scholar]
- 55.Lusis A. J., Yu J., Wang S. S. 2007. The problem of passenger genes in transgenic mice. Arterioscler. Thromb. Vasc. Biol. 27: 2100–2103. [DOI] [PubMed] [Google Scholar]
- 56.Cheverud J. M., Vaughn T. T., Pletscher L. S., Peripato A. C., Adams E. S., Erikson C. F., King-Ellison K. J. 2001. Genetic architecture of adiposity in the cross of LG/J and SM/J inbred mice. Mamm. Genome. 12: 3–12. [DOI] [PubMed] [Google Scholar]
- 57.Reed D. R., McDaniel A. H., Li X., Tordoff M. G., Bachmanov A. A. 2006. Quantitative trait loci for individual adipose depot weights in C57BL/6ByJ x 129P3/J F2 mice. Mamm. Genome. 17: 1065–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mehrabian M., Wen P. Z., Fisler J., Davis R. C., Lusis A. J. 1998. Genetic loci controlling body fat, lipoprotein metabolism, and insulin levels in a multifactorial mouse model. J. Clin. Invest. 101: 2485–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor B. A., Tarantino L. M., Phillips S. J. 1999. Gender-influenced obesity QTLs identified in a cross involving the KK type II diabetes-prone mouse strain. Mamm. Genome. 10: 963–968. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.