Abstract
Niemann-Pick C1-like 1 protein (NPC1L1) plays a critical role in intestinal cholesterol absorption. Our objective was to examine whether five variants (-133A>G, -18A>C, L272L, V1296V, and U3_28650A>G) at the NPC1L1 gene have effects on lipid levels, prevalence, and incidence of coronary heart disease (CHD) and lipid-lowering response to pravastatin. We studied 5,804 elderly participants from the PROSPER study, who were randomized to pravastatin 40 mg/day or placebo and were followed on average for 3.2 years. In the adjusted gender-pooled analyses, homozygous carriers of the minor alleles at four NPC1L1 sites (-18A>C, L272L, V1296V, and U3_28650A>G, minor allele frequencies 0.15–0.33) had 2–8% higher LDL-cholesterol (LDL-C) levels at baseline than homozygous carriers of the common alleles (P < 0.05). Homozygotes for the rare alleles also had a significant increase in the risk of CHD events on trial (range of hazard ratios 1.50–1.67; P < 0.02), regardless of the treatment regimen. The -133 A>G polymorphism and not other variants was associated with 6 month LDL-C lowering (P = 0.02). Our data indicate that variation in the NPC1L1 gene is associated with plasma total and LDL-C levels and CHD risk.
Keywords: statins, single nucleotide polymorphism, low density lipoprotein, coronary heart disease, cholesterol absorption
Elevated plasma LDL-cholesterol (LDL-C) and reduced HDL-cholesterol levels independently predict risk of developing coronary heart disease (CHD) (1, 2). Dietary cholesterol consumption and intestinal absorption contribute to plasma cholesterol levels. Markers of intestinal cholesterol absorption, such as β-sitosterol and campesterol, have been related to variation in levels of plasma LDL-C in human population studies (3–6). Niemann-Pick C1-like 1 (NPC1L1) is an essential protein for intestinal cholesterol absorption (7). NPC1L1 was identified based on its homology to human Niemann-Pick C1 protein, which is defective in an autosomal recessive lipid storage disorder (8, 9). NPC1L1-deficient mice exhibit a substantial reduction in cholesterol absorption, which is unaffected by dietary supplementation of bile acids (10). Recent data indicate that hepatic NPC1L1 also facilitates the retention of biliary cholesterol by hepatocytes, thus protecting the body from excessive biliary loss of cholesterol (11).
Cohen et al. (12) sequenced NPC1L1 and showed that individuals who carry rare NPC1L1 variants associated with decreased cholesterol absorption have mean LDL-C levels that are significantly lower than values in noncarriers. Moreover, NPC1L1 is the target for the drug ezetimibe, which inhibits cholesterol absorption (13). Genetic variation at NPC1L1 in humans has been shown to affect LDL-C lowering response to ezetimibe (14–16). Simon et al. (15) assessed a number of single nucleotide polymorphisms (SNPs) at NPC1L1 and reported that carriers of the rare alleles (most significantly the promoter variant at -18A>C) had a 15% greater LDL-C lowering response to ezetimibe compared with noncarriers. Furthermore, Hegele et al. (16) have shown that individuals with dyslipidemia who carry a rare NPC1L1 haplotype, including the minor alleles at L272L, V1296V, and 25342A>C, were more responsive to ezetimibe (36% LDL-C reduction) than subjects not carrying the haplotype (18% LDL-C reduction).
We and others have documented that alterations in specific plasma markers of cholesterol absorption, namely, campesterol and β-sitosterol, can affect LDL-C lowering response to statins (17–20). Moreover, it has been reported that a subset of patients placed on simvastatin in the Scandinavian Simvastatin Survival Study showed no benefit from therapy compared with placebo if they had elevated baseline levels of markers of cholesterol absorption, including cholestanol (21). Given the significant role of the NPC1L1 gene in cholesterol absorption and its reported involvement in the pharmacogenetics of ezetimibe and the ezetimibe/statin combination, our goal was to examine potential associations of the SNPs selected on the basis of previously published associations (14–16), functional relevance, and minor allele frequencies of >10%, namely, -133A>G, -18A>C, L272L, V1296V, and U_28650A>G, with baseline lipid levels, history, and incidence of CHD and lipid-lowering response to pravastatin in Prospective Study of Pravastatin in the Elderly at Risk (PROSPER).
MATERIALS AND METHODS
Study subjects
The protocol of the PROSPER study has previously been published (22), as have the PROSPER results (22). Briefly, 2,804 men and 3,000 women, age range 70–82 years, with preexisting vascular disease (n = 2404) or at least one of three major vascular risk factors (diabetes n = 575, smoking n = 1433, or hypertension n = 3360) were randomized to pravastatin 40 mg/day (n = 2891) or placebo (n = 2913) and followed up on average for 3.2 years. Over this period, the mean LDL-C reduction in the active treatment group was 32%, and the risk of developing CHD was decreased by 19% (18). No significant lipid changes were noted in the placebo group. Lipid levels were similar at onset of the study in subjects randomized to pravastatin or placebo. Analysis of response to pravastatin was based on subjects reporting good compliance (65% of the subjects in the pravastatin group were in this category, with good compliance being defined as taking 90% or more of the medication).
Biochemical and DNA analysis
Total cholesterol (TC), HDL-cholesterol, and triglycerides were assessed after an overnight fast, at baseline, and at 6 months, and LDL-C was calculated by the Friedewald formula, as previously described (22, 23). Apolipoprotein B (apoB) was measured only at baseline as described (23). DNA was available from 5,783 subjects and isolated from peripheral blood. DNA and subject characteristics were available on 2,621 males and 2,796 females. ApoE phenotype was determined on plasma samples by Western blotting, using the method of Havekes et al. (24) in the central laboratory of the Royal Infirmary in Glasgow, Scotland. Subjects were classified according to the presence of apoE2, apoE3, or apoE4 bands on gel blotting.
Five SNPs in the NPC1L1 locus were genotyped in this study: two in the promoter region [-18A>C (dbSNP reference number is not available) and -133A>G (rs17655652)], two in the protein coding region [1679C>T, or L272L (rs2072183), and 27621T>C, or V1296V (rs217434)], and one in the 3′ untranslated region [U3_28650A>G (rs3187907)]. The SNPs were selected based on previously published associations (15, 16), functional relevance, and minor allele frequencies of >10%. We used Taq Man® SNP genotyping assays (Applied Biosystems, Foster City, CA). The Genbank/EMBL accession number for NPC1L1 is NC_000007.12, mim 608010. The end point read was performed using an Applied Biosystems 7900 HT sequence detection system, subsequent to PCR amplification. Genotypes with quality scores below 95% were repeated, and 5% blinded replicates for genotype determinations were performed. In addition, subjects with the apoE4/2 phenotype (n = 119; 2.2%) were excluded from further analyses, as were subjects whose apoE phenotypes had not been ascertained (n = 246). These exclusions were implemented because apoE phenotype or genotype has been demonstrated to affect statin-induced LDL lowering response, as well as CHD risk (25–29), with apoE2 and apoE4 phenotypes having opposite effects.
Statistical analysis
Observed genotype frequencies were compared with those expected under Hardy-Weinberg equilibrium using a χ2 test. For data analysis, multivariable ANCOVA was performed to detect associations of the genotypes with lipoprotein levels at baseline and with the response to treatment at 6 months, adjusted for gender, body mass index, age, alcohol, smoking, diabetes, apoE phenotype, and country of origin (Scotland, Ireland, or The Netherlands). Prevalence at baseline of myocardial infarction (MI) and all types of vascular disease (history of angina, claudication, MI, stroke, transient ischemic attack, peripheral arterial disease surgery, or amputation for vascular disease more than 6 months before study entry) as well as incidence of CHD death or nonfatal MI were compared between carriers of different NPC1L1 SNP genotypes using multivariable logistic regression analysis in the pooled sample and stratified by gender and treatment. Pooled analyses were adjusted for gender and randomized treatment, when appropriate, and age, country, history of vascular disease, body mass index, history of diabetes, and history of hypertension, alcohol use, current smoking, and apoE phenotype. To evaluate the modifying effects of genotypes and gender on the response to treatment, multiplicative gene-treatment and gene-gender interaction terms were added to the regression models. Lewontin's D value was calculated to assess the linkage disequilibrium (LD) between the SNPs of interest (30). Haplotypes were inferred using the expectation-maximization algorithm as implemented in SAS/Genetics proc haplotype. To account for allelic interaction, haplotypes were used as predictors in the regression models instead of SNPs along with the nongenetic covariates. P values were adjusted for multiple comparisons across genotypes within each SNP using the permutation test using 10,000 permutations. All analyses were performed using SAS/STAT (SAS version 9.1; SAS Institute, Cary, NC).
RESULTS
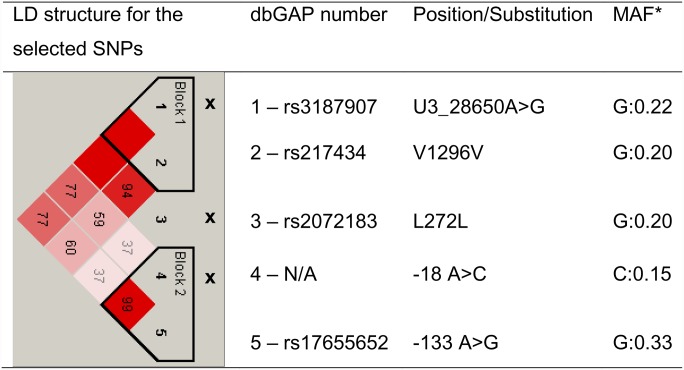
As summarized in Table 1, the participating subjects had a mean age of 75 ± 3 years at baseline. Mean LDL-C levels were in the moderate-risk category (130–160 mg/dl), as defined by the US National Cholesterol Education Program (31). Also, 1,371 men and 1,033 women reported a history of all types of vascular disease, including 508 men and 222 women with a history of MI. Data on apoE phenotype distribution in this population are also shown in Table 1. Allele frequencies for the NPC1L1 SNPs are given in Fig. 1, along with the haplotype block structure. Genotype frequencies conformed to Hardy-Weinberg equilibrium (P > 0.05; data not shown).
TABLE 1.
Subject characteristics (n = 5418)
| Study Characteristics Mean (SD)a | Men (n = 2,621) | Women (n = 2,797) |
|---|---|---|
| Age (years) | 75.0 (3.3) | 75.6 (3.4)b |
| BMI (kg/m2) | 26.6 (3.6) | 27.1 (4.6)b |
| History diabetes mellitus, n (%) | 324 (12.4) | 251 (9.0)b |
| History hypertension, n (%) | 1333 (50.9) | 2027 (72.5)b |
| History vascular disease, n (%) | 1371 (52.3) | 1033 (36.9)b |
| History of MI, n (%) | 508 (19.4) | 222 (7.9)b |
| Current smoking, n (%) | 847 (32.3) | 586 (21.0)b |
| Alcohol consumption, n (%) | 1851 (70.6) | 1165 (41.7)b |
| TC (mg/dl) | 207.0 (30.7) | 231.9 (34.5)b |
| LDL-C (mg/dl) | 138.5 (27.8) | 154.9 (35.3)b |
| HDL-cholesterol (mg/dl) | 45.6 (12.2) | 53.0 (13.4)b |
| Triglyceride (mg/dl) | 132.4 (64.3) | 140.6 (59.4)b |
| apoA-I (mg/dl) | 124.4 (22.2) | 139.9 (24.1)b |
| apoB (mg/dl) | 110.6 (21.3) | 119.1 (22.6)b |
| apoE 2/2 + 2/3 (%) | 13.05 | 11.30 |
| apoE 3/3 (%) | 63.1 | 65.7 |
| apoE 3/4 + 4/4 (%) | 23.8 | 23.0 |
BMI, body mass index.
Means (SD) unless otherwise specified; differences between men and women were assessed using a t-test for continuous traits and χ2 test for binary traits.
P < 0.001, apoE 2/4 carriers were excluded (see Materials and Methods).
Fig. 1.
LD structure and allele frequencies of the NPC1L1 gene polymorphisms. Numbers on the diagram represent pairwise D’; missing numbers indicate D’ = 1. x, SNPs selected for haplotype analysis.
Association with baseline levels
Baseline lipid and apoB levels stratified by genotype and gender are shown in Table 2. In the sex-pooled adjusted model, the minor alleles of -18A>C, L272L, V1296V, and U3_28650A>G were associated with slightly higher TC, LDL-C, and apoB levels in both men and women (differences ranged between 2% and 8% when comparing homozygotes for the minor allele with homozygotes for the common allele; P < 0.05).
TABLE 2.
Adjusted baseline lipid levels (mean ± SE, mg/dl) by gender and genotype
| n |
TC |
LDL-C |
apoB |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Genotype | Men | Women | Men | Women | Pa | Men | Women | Pa | Men | Women | Pa |
| −133 A>G | AA | 1131 | 1280 | 203.6 ± 1.3 | 222.8 ± 1.5 | 0.04 | 134.6 ± 1.1 | 146.1 ± 1.3 | 0.03 | 108.8 ± 0.9 | 113.8 ± 1.0 | 0.08 |
| AG | 1185 | 1223 | 202.1 ± 1.2 | 220.7 ± 1.5 | 132.7 ± 1.1 | 143.9 ± 1.3 | 107.1 ± 0.8 | 113.3 ± 1.0 | ||||
| GG | 292 | 281 | 203.3 ± 2.0 | 217.0 ± 2.3 | 135.9 ± 1.8 | 139.7 ± 2.1 | 108.9 ± 1.3 | 110.4 ± 1.5 | ||||
| −18 A>C | AA | 1882 | 2007 | 202.4 ± 1.1 | 220.4 ± 1.4 | 0.03 | 133.3 ± 1.4 | 143.9 ± 1.2 | 0.02 | 107.7 ± 0.8 | 113.2 ± 0.9 | 0.08 |
| AC | 665 | 718 | 204.4 ± 1.5 | 223.4 ± 1.7 | 135.3 ± 1.3 | 146.0 ± 1.5 | 108.7 ± 1.0 | 113.4 ± 1.1 | ||||
| CC | 64 | 62 | 205.8 ± 3.9 | 224.9 ± 4.4 | 138.9 ± 3.4 | 146.3 ± 3.9 | 112.9 ± 2.6 | 115.6 ± 2.8 | ||||
| L272L | CC | 1611 | 1685 | 202.2 ± 1.2 | 219.9 ± 1.4 | 0.007 | 133.1 ± 1.0 | 143.8 ± 1.3 | 0.02 | 107.4 ± 0.8 | 113.0 ± 0.9 | 0.04 |
| CG | 872 | 968 | 203.8 ± 1.4 | 223.3 ± 1.6 | 134.8 ± 1.2 | 145.6 ± 1.4 | 108.8 ± 0.9 | 114.0 ± 1.0 | ||||
| GG | 129 | 130 | 207.0 ± 2.8 | 224.5 ± 3.2 | 138.1 ± 2.5 | 148.0 ± 2.8 | 111.6 ± 1.9 | 114.8 ± 2.0 | ||||
| V1296V | CC | 1686 | 1746 | 202.8 ± 1.2 | 220.8 ± 1.4 | 0.006 | 133.6 ± 1.0 | 143.9 ± 1.3 | 0.004 | 108.0 ± 0.8 | 113.1 ± 0.9 | 0.04 |
| CT | 822 | 924 | 202.5 ± 1.4 | 221.5 ± 1.6 | 133.6 ± 1.2 | 145.1 ± 1.4 | 107.6 ± 0.9 | 113.4 ± 1.0 | ||||
| TT | 103 | 113 | 211.9 ± 3.1 | 227.5 ± 3.4 | 143.6 ± 2.7 | 148.7 ± 3.0 | 114.7 ± 2.1 | 115.2 ± 2.2 | ||||
| U3_28650 | AA | 1677 | 1739 | 202.8 ± 1.2 | 220.7 ± 1.4 | 0.005 | 133.6 ± 1.0 | 143.8 ± 1.3 | 0.002 | 108.0 ± 0.8 | 113.1 ± 0.9 | 0.03 |
| A>G | GA | 828 | 934 | 202.3 ± 1.4 | 221.6 ± 1.6 | 133.4 ± 1.2 | 145.1 ± 1.4 | 107.5 ± 0.9 | 113.4 ± 1.0 | |||
| GG | 104 | 113 | 212.1 ± 3.1 | 227.6 ± 3.4 | 143.8 ± 2.8 | 148.9 ± 3.0 | 114.8 ± 2.1 | 115.3 ± 2.2 | ||||
P values using the three genotypes, men and women combined; adjusted for sex, body mass index, age, alcohol, smoking, diabetes, apoE phenotype, and country.
Significant associations between the -133A>G variant and lipid levels were most pronounced in women, with -133GG female carriers having the lowest TC and LDL-C levels compared with -133AA and -133AG carriers (P for trend = 0.0032 for TC and 0.023 for LDL-C; Table 2). Similar trends were noted in men, but did not reach statistical significance (P for trend = 0.46 for TC and 0.09 for LDL; Table 2).
Haplotype analysis detected two LD blocks with two SNPs each and one SNP between the blocks. In order to test allelic interactions and preserve power, we constructed haplotypes that included one SNP from each haplotype block and the SNP outside of either haplotype block (-18A>C, L272L, and U3_28650A>G) (Fig. 1; Table 3), resulting in four common haplotypes (frequencies >5%). The tests that compared a common haplotype (-18A>C[A]L272L[C]U3_28650A>G[A]) present in 68% of the participants to the others five haplotypes showed that carriers of the common haplotype had 1.8 mg/dl lower baseline LDL-C levels for each copy of this haplotype compared with the alternative haplotype (-18[C]L272L[G]U3_28650G), which was present in 10% of participants (Table 3). This is consistent with the data shown in Table 2, where individual variations in these SNPs were significantly associated with LDL-C levels with homozygous carriers for the minor alleles having the highest levels. These data indicate that most of the variation in TC and LDL-C levels could be explained by individual SNPs and that haplotype analysis did not add significantly to the model performance. The proportion of variation explained by multivariable models with individual SNPs was on average 15.5% for baseline LDL-C levels and 17.4% for baseline TC, while multivariate models with haplotypes explained 15.6% and 17.4%, respectively (P for model comparison >0.05).
TABLE 3.
Incremental effect of rare NPC1L1 haplotypes on baseline lipid levels
| Haplotype |
TC |
LDL-C |
ApoB |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NPC1L1 Haplotype | Frequency | -18A>C | L272L | U3_28650A>G | Estimate, mg/dla | Pb | Estimate, mg/dla | Pb | Estimate, mg/dla | Pb |
| 1 | 0.681 | A | C | A | –c | – | – | – | – | – |
| 2 | 0.102 | C | G | G | 0.18 | 0.0006 | 0.17 | 0.0004 | 0.04 | 0.007 |
| 3 | 0.097 | A | C | G | 0.06 | 0.29 | 0.08 | 0.14 | −0.12 | 0.05 |
| 4 | 0.068 | A | G | A | 0.13 | 0.05 | 0.10 | 0.10 | 0.03 | 0.09 |
| 5 | 0.043 | C | G | A | 0.14 | 0.11 | 0.10 | 0.20 | −0.07 | 0.56 |
| 6 | 0.004 | C | C | A | −0.36 | 0.14 | −0.29 | 0.18 | 0.01 | 0.52 |
Incremental effect of each copy of haplotype.
Compared to the common haplotype.
Referent haplotype.
Associations with response to treatment
To assess differential responsiveness to pravastatin in carriers of various NPC1L1 genotypes, we tested the association between the SNPs under study and the 6 month change in lipid levels in treated individuals with good compliance. The -133A>G variant was associated with LDL-C lowering in the sex-pooled analysis (P = 0.02). However, while -133GG male carriers showed the greatest reduction in LDL-C following pravastatin treatment compared with carriers of one or two major alleles, -133GG female carriers had the lowest reduction compared with carriers of one or two major alleles (P value for SNP-sex interaction is 0.006; Fig. 2). Other SNPs were not related to LDL-C lowering response to pravastatin (see supplementary Table I).
Fig. 2.
LDL-C response to pravastatin in the treatment group by gender and -133A>G genotype (shown as mean percentage change from baseline and SE). Men AA = 438, AG = 432, GG = 93; women AA = 402, AG = 429, GG = 113. Adjusted P for genotype-sex interaction = 0.006.
Associations with history and incidence of cardiovascular disease
At baseline, U3_28650 G carriers had significantly lower prevalence of claudication [total number of affected individuals, n = 365; adjusted odds ratio (OR) (95% confidence interval) 0.76 (0.60–0.96); P = 0.02]. No other SNPs were associated with a history of vascular disease at baseline (see supplementary Table II).
Hazard ratios for incident CHD death or nonfatal MI (total number of new cases n = 597) stratified by gender and genotype are shown in Table 4. All the SNPs that were significantly associated with higher TC, LDL-C, and apoB were also found to be associated with a greater risk of CHD death or nonfatal MI in homozygous carriers of the minor alleles compared with heterozygote and homozygous carriers of the common alleles (P < 0.05) regardless of the treatment regimen received over the course of the trial (Table 4). However, statistical significance was achieved only when pravastatin and placebo groups were combined and adjusted for treatment, likely due to reduced statistical power in the stratified analysis (Table 4).
TABLE 4.
Analysis of incidence of CHD death or nonfatal MI on trial by SNP carrier status: new cases/total subjects (597/5,418)
| Adjusteda | Placebob | Pravastatinb | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Number of New Cases/ Total Subjects (%) | HRc | P | Number of new case/ Total subjects (%) | HRc | P | Number of new case/ Total subjects (%) | HRc | P | |
| −133 A>G | AA + AG | 534/4819 (11.1) | 1 | 0.99 | 291/2429 (12.0) | 1 | 0.8 | 243/2390 (10.2) | 1 | 0.8 |
| GG | 63/573 (11.0) | 1.00 (0.77-1.30) | 35/291 (12.0) | 1.04 (0.73-1.48) | 28/282 (9.9) | 0.96 (0.65-1.42) | ||||
| −18 A>C | AA +AC | 574/5272 (10.9) | 1 | 0.04 | 314/2658 (11.8) | 1 | 0.2 | 260/2614 (9.9) | 1 | 0.06 |
| CC | 23/126 (18.3) | 1.67 (1.10-2.54) | 12/67 (17.9) | 1.51 (0.84-2.69) | 11/59 (18.6) | 1.78 (0.97-3.27) | ||||
| L272L | CC + CG | 557/5136 (10.8) | 1 | 0.04 | 301/2587 (11.6) | 1 | 0.05 | 256/2549 (10.0) | 1 | 0.5 |
| GG | 41/259 (15.8) | 1.50 (1.09-2.06) | 26/136 (19.1) | 1.66 (1.11-2.49) | 15/123 (12.2) | 1.23 (0.73-2.06) | ||||
| V1296V | CC + CT | 561/5178 (10.8) | 1 | 0.03 | 306/2602 (11.8) | 1 | 0.1 | 255/2576 (9.9) | 1 | 0.05 |
| TT | 36/216 (16.7) | 1.56 (1.11-2.18) | 21/120 (17.5) | 1.46 (0.94-2.27) | 15/96 (15.6) | 1.68 (0.99-2.83) | ||||
| U3_28650 A>G | AA + AG | 562/5178 (10.9) | 1 | 0.04 | 305/2599 (11.7) | 1 | 0.1 | 257/2579 (10.0) | 1 | 0.1 |
| GG | 35/217 (16.1) | 1.50 (1.07-2.11) | 21/122 (17.2) | 1.43 (0.92-2.23) | 14/95 (14.7) | 1.58 (0.92-2.71) |
P values for men and women combined; adjusted for gender, body mass index, age, alcohol, smoking, diabetes, hypertension, apoE phenotype, randomized treatment, and country. No significant differences were noted when men and women were separated.
P values for men and women combined; adjusted for gender, body mass index, age, alcohol, smoking, diabetes, hypertension, apoE phenotype, and country. No significant differences were noted when men and women were separated.
Hazards ratio (95% confidence intervals).
DISCUSSION
The purpose of this study was to evaluate associations of SNPs at the NPC1L1 locus with baseline lipid levels, history of vascular disease, incident CHD, and LDL-C lowering response to pravastatin in a large population of elderly subjects at increased risk of developing CHD. The NPC1L1 transporter has been shown to be critical for cholesterol absorption and is the site of the action of ezetimibe, a cholesterol lowering agent that inhibits cholesterol absorption (8, 9, 13).
In this study, we documented that homozygous carriers of the minor alleles at -18A>C, L272L, V1296V, and U3_28650A>G at the NPC1L1 locus had 2–8% higher TC, LDL-C, and apoB levels at baseline compared with their common allele homozygous counterparts. An opposite trend was observed for NPC1L1 -133A>G, primarily in women. Additional support for the effect of the NPC1L1 -133A>G SNP and lipid levels was obtained in a recent meta-analysis of genome-wide association studies involving 8,816 subjects that included controls, diabetics, and myocardial infarction survivors, mainly of European origin (32). In that study, carriers of the NPC1L1 -133A>G common allele had higher LDL-C levels (on average 2.4 mg/dl per copy, P = 0.018, not significant at the genome-wide level) (32). Neither L272L nor V1296V or U3_28650A>G were found to be associated with cross-sectional lipid levels assessed in this meta-analysis (32). It is possible that population characteristics, such as the cardiovascular risk profile, age, and prior history of CHD, as well as differences in covariates controlled for by our studies, can, at least in part, explain these latter differences.
Analyses of associations between genotypes and response to pravastatin were prompted by reports that NPC1L1 variants were related to LDL-C lowering response to ezetimibe (14–16). We found that the NPC1L1 -133A>G SNP, but not other NPC1L1 SNPs, was associated with response to pravastatin.
Simon et al. (15), using two large ethnically diverse study populations derived from the Ezetimibe Add on to Statin for Effectiveness trial and Vytorin (ezetimibe + simvastatin) versus Atorvastatin trial, reported that patients carrying at least one copy of the minor allele of -18A>C had close to 15% greater response following 6 weeks of treatment with ezetimibe added to ongoing statin therapy compared with homozygous carriers of the common allele (mean LDL-C change from baseline, 27.8% versus 24.2%, respectively). Our results show that this NPC1L1 SNP was not associated with response to pravastatin alone.
Simon et al. (15) reported that NPC1L1 -133GG carriers had a poorer LDL-C lowering response to statin-ezetimibe combinations compared with NPC1L1 -133AA carriers, similar to our finding with pravastatin therapy alone. In our study, these differences were only observed in women. Moreover, female homozygotes for -133AA had significantly greater reductions than their male counterparts, while the converse was true for females carrying -133GG (see Fig. 2). It should, however, be noted that the former group had lower LDL-C levels at baseline, which, unless the effect of pravastatin is scale independent, could have accounted for some of these results. Moreover, although we cannot rule out a spurious association, we speculate that this phenomenon can also be explained by differences in binding affinity of transcription factors and their ligands, such as estrogen receptors, whose activity may differ between men and women. We have previously documented that genetic variation at the estrogen receptor α gene locus is associated with gender-specific differences in LDL-C lowering response to atorvastatin (33). While our data suggest sex-specific effects of the NPC1L1 genotype on lipid-lowering response, none of the prior NPC1L1 pharmacogenetic studies have reported testing for gene-gender interactions. Discrepancies between pharmacogenetic studies like the ones discussed above are expected and can be explained by different population baseline characteristics, type of statin, dosage, duration of treatment, and concomitant medications, as well as covariates used in the analyses. In our study, we excluded apoE 2/4 carriers from further analyses and adjusted each model for apoE phenotype, a significant predictor of plasma lipid concentrations not used by other studies (14–16).
A novel feature of our study is that we ascertained whether there was an association of NPC1L1 polymorphisms and vascular disease. More than half of the men and more than a third of the women participating in PROSPER had a prior history of cardiovascular events. However, none of the SNPs were associated with a history of vascular disease at baseline. Of note is that the allele frequencies for the NPC1L1 SNPs that we observed in PROSPER were similar to those reported by other studies (15, 16) and conformed to those expected by Hardy- Weinberg equilibrium. Therefore, no survival bias with regard to these variants was implied. However, the rare alleles -18A>C, L272L, V1296V, and U3_28650A>G at NPC1L1 were associated with a 50–67% increased risk of developing fatal or nonfatal CHD events on trial. Moreover, the higher incidence of new CHD death or nonfatal MI in carriers of minor alleles did not appear to be modified by statin treatment, indicating that pravastatin is not effective in CHD risk reduction in this subgroup of patients. These subjects may be similar to those previously identified by Miettinen et al. (21) that had higher baseline markers of cholesterol absorption and had no significant CHD risk reduction despite simvastatin treatment compared with placebo in the 4S study.
It is important to note that the design of this study does not allow us to draw definitive conclusions about the exact mechanisms that may lead to genotype-related differences in lipid concentrations. However, there are several biologically plausible explanations for our findings. The promoter region SNPs at positions -18 and -133 could affect alternative splicing and produce NPC1L1 isoforms with properties that may differ from those of the full-length gene product (9). However, further studies are required to determine how these SNPs can potentially affect NPC1L1 gene expression. If such SNPs were associated with decreased gene expression, their presence could result in less efficient intestinal absorption of cholesterol. There is no experimental data at this point to support these concepts. In addition, NPC1L1 L272L and V1296V polymorphisms result in synonymous substitutions that do not produce alterations in the coding sequences and therefore would not be expected to change the function of the protein. Nevertheless, there is accumulating evidence suggesting that synonymous changes introduce rare codons, which may result in altered translation rates and protein folding (34, 35). In addition, the SNP in the untranslated region could be associated with alterations in the posttranscriptional regulation that might affect NPC1L1 gene expression. The next step would be to sequence the genomic region at and near NPC1L1 and search for functional, and as yet unknown, variants that can explain the associations detected in this study.
The limitations of our study include the fact that our cohort is elderly and of European origin, restricting generalization of our findings to those of younger individuals and those of other ethnic backgrounds. However, the consistency and biological relevance of these associations should motivate further research to verify and extend these findings and test their relationship with specific markers of intestinal cholesterol absorption, such as β-sitosterol and campesterol (17).
In summary, our data are consistent with the hypothesis that genetic variation at NPC1L1 is associated with plasma total cholesterol and LDL-C levels and CHD risk due to alterations in cholesterol absorption in an elderly European population. In the future, resequencing of NPC1L1 and precise elucidation of the effects of NPC1L1 variants on NPC1L1 gene expression and function should improve our understanding of how genetic variation at this gene affects lipid concentrations, CHD risk, and response to lipid-lowering treatments.
Supplementary Material
Footnotes
Abbreviations:
- apo
- apolipoprotein
- CHD
- coronary heart disease
- LD
- linkage disequilibrium
- LDL-C
- low density lipoprotein-choleserol
- MI
- myocardial infarction
- NPC1L1
- Niemann-Pick C1-like 1
- PROSPER
- Prospective Study of Pravastatin in the Elderly at Risk
- SNP
- single nucleotide polymorphism
- TC
- total cholesterol
This work was supported by Grant R01 HL-74753 from the National Institutes of Health and contract 53-1950-5-003 from the Agricultural Research Service of the USDA. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
The online version of this article (available at http://www.jlr.org)contains supplementary data in the form of two tables.
REFERENCES
- 1.Ingelsson E., Schaefer E. J., Contois J. H., McNamara J. R., Sullivan L., Keyes M. J., Pencina M. J., Schoonmaker C., Wilson P. W., D'Agostino R. B., et al. 2007. Clinical utility of different lipid measures for prediction of coronary heart disease in men and women. JAMA. 298: 776–785. [DOI] [PubMed] [Google Scholar]
- 2.Wilson P. W., D'Agostino R. B., Levy D., Belanger A. M., Silbershatz H., Kannel W. B. 1998. Prediction of coronary heart disease using risk factor categories. Circulation. 97: 1837–1847. [DOI] [PubMed] [Google Scholar]
- 3.Miettinen T. A., Kesaniemi Y. A. 1989. Cholesterol absorption: regulation of cholesterol synthesis and elimination and within population variations of serum cholesterol levels. Am. J. Clin. Nutr. 49: 629–635. [DOI] [PubMed] [Google Scholar]
- 4.Tikkanen M. J. 2005. Plant sterols and stanols. Handbook of Experimental Pharmocology. Springer-Verlag, Berlin: 215–230. [DOI] [PubMed] [Google Scholar]
- 5.Gylling H., Miettinen T. A. 2002. Inheritance of cholesterol metabolism of probands with high or low cholesterol absorption. J. Lipid Res. 43: 1472–1476. [DOI] [PubMed] [Google Scholar]
- 6.Gylling H., Laaksonen D. E., Atalay M., Hallikainen M., Niskanen L., Miettinin T. A. 2007. Markers of absorption and synthesis of cholesterol in men with type 1 diabetes. Diabetes Metab. Res. Rev. 23: 372–377. [DOI] [PubMed] [Google Scholar]
- 7.Davies J. P., Levy B., Ioannou Y. A. 2000. Evidence for a Niemann-pick C (NPC) gene family: identification and characterization of NPC1L1. Genomics. 65: 137–145. [DOI] [PubMed] [Google Scholar]
- 8.Carstea E.D., Morris J.A., Coleman K.G., Loftus S.K., Zhang D., Cummings C., Gu J., Rosenfeld M.A., Pavan W.J., Krizman D.B., et al. 1997. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science. 277: 228–231. [DOI] [PubMed] [Google Scholar]
- 9.Davis H. R., Jr., Altmann S. W. 2009. Niemann-Pick C1 Like 1 (NPC1L1) an intestinal sterol transporter. Biochim. Biophys. Acta. 1791: 679–683. [DOI] [PubMed] [Google Scholar]
- 10.Altmann S. W., Jr. Davis H. R., Zhu L. J., Yao X., Hoos L. M., Tetzloff G., Iyer S. P., Maguire M., Golovko A., Zeng M., et al. 2004. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science. 303: 1201–1204. [DOI] [PubMed] [Google Scholar]
- 11.Temel R. E., Tang W., Ma Y., Rudel L. L., Willingham M. C., Ioannou Y. A., Davies J. P., Nilsson L. M., Yu L. 2007. Hepatic Niemann-Pick C1-like 1 regulates biliary cholesterol concentration and is a target of ezetimibe. J. Clin. Invest. 117: 1968–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen J. C., Pertsemlidis A., Fahmi S., Esmail S., Vega G. L., Grundy S. M., Hobbs H. H. 2006. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc. Natl. Acad. Sci. USA. 103: 1810–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Calvo M., Lisnock J., Bull H. G., Hawes B. E., Burnett D. A., Braun M. P., Crona J. H., Davis H. R., Jr., Dean D. C., Detmers P. A., et al. 2005. The target of ezetimibe is Niemann-Pick C1-Like 1 (NPC1L1). Proc. Natl. Acad. Sci. USA. 102: 8132–8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J., Williams C. M., Hegele R. A. 2005. Compound heterozygosity for two non-synonymous polymorphisms in NPC1L1 in a non-responder to ezetimibe. Clin. Genet. 67: 175–177. [DOI] [PubMed] [Google Scholar]
- 15.Simon J. S., Karnoub M. C., Devlin D. J., Arreaza M. G., Qiu P., Monks S. A., Severino M. E., Deutsch P., Palmisano J., Sachs A. B., et al. 2005. Sequence variation in NPC1L1 and association with improved LDL-cholesterol lowering in response to ezetimibe treatment. Genomics. 86: 648–656. [DOI] [PubMed] [Google Scholar]
- 16.Hegele R. A., Guy J., Ban M. R., Wang J. 2005. NPC1L1 haplotype is associated with inter-individual variation in plasma low-density lipoprotein response to ezetimibe. Lipids Health Dis. 4: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Himbergen T. M., Matthan N. R., Resteghini N. A., Otokozawa S., Ai M., Stein E. A., Jones P. H., Schaefer E. J. 2009. Comparison of the effects of maximal dose atorvastatin and rosuvastatin therapy on cholesterol synthesis and absorption markers. J. Lipid Res. 50: 730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miettinen T. A., Gylling H. 2003. Synthesis and absorption markers of cholesterol in serum and lipoproteins during a large dose of statin treatment. Eur. J. Clin. Invest. 33: 976–982. [DOI] [PubMed] [Google Scholar]
- 19.Miettinen T. A., Stranberg T. E., Gylling H. 2000. Noncholesterol sterols and cholesterol lowering by long term simvastatin treatment in coronary patients:realtion to basal serum cholestanol. Arterioscler. Thromb. Vasc. Biol. 20: 1340–1346. [DOI] [PubMed] [Google Scholar]
- 20.Duane W. C. 1993. Effects of lovastatin and dietary cholesterol on sterol homeostasis in healthy human subjects. J. Clin. Invest. 92: 911–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miettinen T. A., Gylling H., Stransberg T., Sarna S. 1998. Baseline serum cholestanol as predictopr of recurrent coronary events in subgroup of Scandinavian simvastatin survival study. BMJ. 316: 1127–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shepherd J., Blauw G. J., Murphy M. B., Cobbe S. M., Bollen E. L., Buckley B. M., Ford I., Jukema J. W., Hyland M., Gaw A., et al. 1999. The design of a prospective study of Pravastatin in the Elderly at Risk (PROSPER). PROSPER Study Group. PROspective Study of Pravastatin in the Elderly at Risk. Am. J. Cardiol. 84: 1192–1197. [DOI] [PubMed] [Google Scholar]
- 23.Shepherd J., Blauw G. J., Murphy M. B., Bollen E. L., Buckley B. M., Cobbe S. M., Ford I., Gaw A., Hyland M., Jukema J. W., et al. 2002. PROSPER study group. PROspective Study of Pravastatin in the Elderly at Risk. Pravastatin in elderly individuals at risk of vascular disease (PROSPER): a randomised controlled trial. Lancet. 360: 1623–1630. [DOI] [PubMed] [Google Scholar]
- 24.Havekes L. M., de Knijff P., Beisiegel U., Havinga J., Smit M., Klasen E. 1987. A rapid micromethod for apolipoprotein E phenotyping directly in serum. J. Lipid Res. 28: 455–463. [PubMed] [Google Scholar]
- 25.Lahoz C., Osgood D., Wilson P. W., Schaefer E. J., Ordovas J. M. 1996. Frequency of phenotype-genotype discrepancies at the apolipoprotein E locus in a large population study. Clin. Chem. 42: 1817–1823. [PubMed] [Google Scholar]
- 26.Ordovas J. M., Lopez-Miranda J., Perez-Jimenez F., Rodriguez C., Park J. S., Cole T., Schaefer E. J. 1995. Effect of apolipoprotein E and A-IV phenotypes on the low density lipoprotein response to HMG CoA reductase inhibitor therapy. Atherosclerosis. 113: 157–166. [DOI] [PubMed] [Google Scholar]
- 27.Pedro-Botet J., Schaefer E. J., Bakker-Arkema R. G., Black D. M., Stein E. M., Corella D., Ordovas J. M. 2001. Apolipoprotein E genotype affects plasma lipid response to atorvastatin in a gender specific manner. Atherosclerosis. 158: 183–194. [DOI] [PubMed] [Google Scholar]
- 28.Wilson P. W. F., Schaefer E. J., Larson M. G., Ordovas J. M. 1996. Apolipoprotein E alleles and risk of coronary disease: a meta-analysis. Arterioscler. Thromb. Vasc. Biol. 16: 1250–1255. [DOI] [PubMed] [Google Scholar]
- 29.Lahoz C., Schaefer E. J., Cupples L. A., Wilson P. W., Levy D., Osgood D., Parpos S., Pedro-Botet J., Daly J. A., Ordovas J. M. 2001. Apolipoprotein E genotype and cardiovascular disease in the Framingham Heart Study. Atherosclerosis. 154: 529–537. [DOI] [PubMed] [Google Scholar]
- 30.Lewontin R. C. 1964. The interaction of selection and linkage. II. Optimum models. Genetics. 50: 757–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Expert Panel. 2001. Executive summary of the third report of the National Cholesterol Education Program (NCEP). Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III). JAMA. 285: 2486–2497. [DOI] [PubMed] [Google Scholar]
- 32.Willer C. J., Sanna S., Jackson A. U., Scuteri A., Bonnycastle L. L., Clarke R., Heath S. C., Timpson N. J., Najjar S. S., Stringham H. M., et al. 2008. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kajinami K., Brousseau M. E., Lamon-Fava S., Ordovas J. M., Schaefer E. J. 2005. Gender specific effects of estrogen receptor alpha gene haplotype on lipid response to atorvastatin: interaction with apolipoprotein AI promoter gene polymorphism. Atherosclerosis. 178: 331–338. [DOI] [PubMed] [Google Scholar]
- 34.Nackley A. G., Shabalina S. A., Tchivileva I. E., Satterfield K., Korchynskyi O., Makarov S. S., Maixner W., Diatchenko L. 2006. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 314: 1930–1933. [DOI] [PubMed] [Google Scholar]
- 35.Kimchi-Sarfaty C., Oh J. M., Kim I. W., Sauna Z. E., Calcagno A. M., Ambudkar S. V., Gottesman M. M. 2007. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 315: 525–528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.