Abstract
Apolipoprotein E (apoE), one of the major protein components of lipoproteins in the peripheral and central nervous systems, regulates cholesterol metabolism through its interaction with members of the low density lipoprotein receptor family. One key to understanding apoE function is determining the structure of lipid-bound forms of apoE. Negative-staining (NS) electron microscopy (EM) is an easy and rapid approach for studying the structure and morphology of lipid-bound forms of apoE. However, an artifact of using the conventional NS protocol is that the apoE•phospholipid particles form rouleaux. In this study, we used cryo-electron microscopy (cryo-EM) to examine apoE4•palmitoyl-oleoylphosphatidylcholine (POPC) particles in a frozen-hydrated native state. By comparing the particle sizes and shapes produced by different NS protocols to those produced by cryo-EM, we propose an optimized protocol to examine apoE4•POPC particles. Statistical analysis demonstrated that the particle sizes differ by less than 5% between the optimized protocol and the cryo-EM method, with similar shapes. The high contrast and fine detail of particle images produced using this optimized protocol lend themselves to the structural study of lipid-bound forms of apoE.
Keywords: apolipoprotein E, cryo-EM, electron microscopy, phospholipid
Apolipoprotein E (apoE), 299 amino acids, is a protein component of lipoproteins in plasma and the central nervous system (1, 2). By associating with phospholipids, apoE is critical for lipid transport and cholesterol homeostasis in the central nervous system and plasma and is involved in neuronal plasticity (3). ApoE has three common isoforms (apoE2, apoE3, and apoE4) distinguished by cysteine/arginine content, and each form has different effects on plasma cholesterol levels (4). ApoE4 is associated with the highest plasma cholesterol and low density lipoprotein (LDL) concentrations and is thus associated with an increased risk for cardiovascular disease (5, 6), and is also the major genetic risk factor for sporadic, late-onset Alzheimer's disease (7–10).
To understand the mechanisms of biologically active apoE, its structure must be unraveled. The crystal structure of apoE in the lipid-free state shows that the N-terminal domain is a globular four-helix bundle (11). In the biologically active, lipid-bound state, the four-helix bundle of apoE undergoes a conformational rearrangement (12, 13). Lipid association is required for activities such as high-affinity binding to LDL receptors and modulating neuron maintenance and regeneration (9). The lipid-bound form of apoE has been studied by electron microscopy (EM), neutron scattering, and X-ray scattering for more than two decades. Because of the heterogeneity and dynamic nature of the apoE particles, particle shape and radial distribution of electron density have been difficult to determine (14, 15). So far, two structural models of apoE•phospholipid particles have been proposed. One model is a discoidal model, which is mainly supported by negative-staining (NS) EM. In this model, the phospholipid is arranged in a bilayer with the protein wrapped around the edge of the disc, covering the exposed hydrophobic chains of the phospholipids (16–24). The second, a spherical or ellipsoidal model, was recently proposed based on X-ray crystallography and X-ray small-angle scattering data. In this model, the phospholipids of the apoE particle are arranged in a micelle-like shape, with the protein portion embedded in the micelle surface (14, 19, 25).
EM is the only technology capable of directly visualizing apoE•phospholipid particles for structural study. NS is an excellent tool for examining lipid-bound forms of apoE: it is extremely quick, easy, and qualitative for examining the structure of isolated organelles and individual macromolecules at the nanometer resolution level (26–28). The advantage of cryo-electron microscopy (cryo-EM) is that the “native” structure of biological macromolecules can be examined in a defined physiological buffer without introducing potential artifacts commonly associated with NS preparation methods including chemical fixation, dehydration, embedding, and staining (29). However, the contrast in unstained, vitrified specimens by cryo-EM is very weak (30–32). In comparison, NS provides extremely good contrast because the protein is fixed by a layer of heavy metal ions in an emulsion of high ionic strength (26, 28). Unfortunately, some artifacts in NS are found in many experiments (33–36). For instance, liposomes or lipid tubes tend to collapse in NS (34) but not in cryo-EM (37, 38). The rouleau formation of lipid-bound forms of apoE rendered by conventional NS has been suspected as an artifact because rouleaux are not observed in serum or solutions of phosphate-buffered or Tris-buffered saline (19, 23, 39).
Given the benefits of NS (26–28), we set out to determine if NS artifacts could be eliminated by modifying the NS protocol. We examined apoE4•POPC in physiological buffer using cryo-EM and statistically compared the particle size and shape to particles produced by different NS protocols. Based on these results, we propose an optimized NS protocol to produce “near native state” particle images.
MATERIALS AND METHODS
Recombinant apoE and apoE4•POPC particle production
Sodium cholate in methanol and POPC in chloroform (Avanti Polar Lipids 850457) (wt. ratio 2.5:1) were dried in glass tubes under a stream of nitrogen. A vacuum was applied to remove all traces of the chloroform before reconstitution to 10 mg/ml in Tris-buffered saline (TBS) (10 mM Tris HCl, pH 7.4, 150 mM NaCl, 0.25 mM EDTA, 0.0005% NaN3). ApoE4 in 100 mm ammonium bicarbonate was added to the POPC at a molar ratio of 1:40 (apoE:POPC) as previously described (14). After incubation at room temperature for 3 h, the particles were extensively dialyzed against cold TBS (6 changes over three days). The density of apoE•POPC was raised to d = 1.21 by adding solid KBr, and the particles were isolated by density gradient ultracentrifugation (d = 1.006–1.21) at 55 K (18 h, 15°C), and followed by fast-protein liquid chromatography (FPLC) isolation. The fractionated particles were dialyzed against TBS, analyzed by native gel electrophoresis (4–20% gradient gels), and the major fraction was taken for analysis (Fig. 1).
Fig. 1.
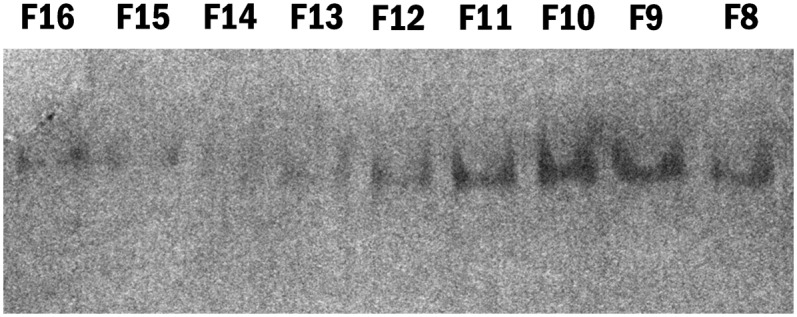
Native gel electrophoresis of apoE4•POPC FPLC fractions. The apoE4•POPC particles were isolated by density gradient ultracentrifugation (d = 1.006–1.21). The fractionated particles were dialyzed against TBS and analyzed by native gel electrophoresis (4–20% gradient gels). The native gel was used to assess the relative particle size and size homogeneity. Fraction 10 (F10) was selected for EM analysis because it was the most homogeneous. apoE4•POPC, POPC-bound form of apoE4; EM, electron microscopy; FPLC, fast-protein liquid chromatography; POPC, palmitoyl-oleoylphosphatidylcholine.
Cryo-EM specimen preparation
ApoE4•POPC particles were diluted to 0.01 mg/ml with 1× Dulbecco's phosphate-buffered saline (Invitrogen, La Jolla, CA, 2.7 mM KCl, 1.46 mM KH2PO4, 136.9 mM NaCl, and 8.1 mM Na2HPO4). Samples (∼4 μl) were incubated for 1 min at room temperature on Quantifoil holey carbon-film-coated 400-mesh copper grids (Quantifoil Micro Tools, Jena, Germany), which were rendered hydrophilic by glow-discharge for 20 s. Specimens were flash-frozen with a Vitrobot rapid-plunging device (FEI, Eindhoven, The Netherlands) at 100% humidity and 4°C and blotted for 2.5 s on filter paper (#595, Schleicher and Schuell, Dassel, Germany) as described (29, 40). The frozen, hydrated particles embedded in vitreous ice over the holes in the carbon film were stored in liquid nitrogen until used for cryo-EM observation.
Preparation of NS specimens by conventional protocol
The most popular NS protocol for lipoprotein preparation was reported by Forte et al. (41, 42). Using this protocol, lipoprotein solution (0.1 mg/ml) and 2% sodium phosphotungstate (pH 7.4) were mixed at 1:1 ratio by volume before sonification. A droplet (∼4 μl) of the mixture was placed on a glow-discharged carbon-coated grid and allowed to sit for 60 s. Excess solution was removed by touching the grid with filter paper placed onto the backside of the grid.
Preparation of NS specimens by a washing protocol
To reduce the effects of salt, we modified a second protocol (43–45). Aliquots (∼2.5 μl, 0.01 mg/ml) of apoE4•POPC particles were adhered to glow-discharged thin carbon-coated 300-mesh copper grids (Cu-300CN, Pacific Grid-Tech, San Francisco, CA) and incubated for 1 min at room temperature. Excess sample solution was removed by blotting with filter paper touched to the edge of the grid. After removing excess fluid, three droplets of deionized water were adhered to the grid consequently for 1–2 s. Immediately after removal of excess fluid, the grid was stained with 2% (w/v) sodium phosphotungstate (pH 7.4) for 30 s.
Preparation of NS specimens by our optimized protocol
The optimized protocol was modified from a third protocol (28, 46). The apoE4•POPC sample was diluted to 0.01 mg/ml with Dulbecco's PBS (Invitrogen Corporation, Carlsbad, CA). Aliquots (∼3 μl) were adhered to thin pure-carbon-coated 300-mesh copper grids, which had been rendered hydrophilic by glow discharge for 20 s. After incubating for ∼2 min at 4°C, the grids were rapidly washed with three successive drops of deionized water (∼35 µl each) and then exposed to three successive drops of 2% (w/v) uranyl formate (UF) pH 4.6. UF solution was filtered through 0.02 μm inorganic membrane filters (Whatman, Maidstone, UK) before use. After incubation for ∼2 min in a dark chamber on an ice bed, excess solution was removed with filter paper from the backside of the grids, after which the specimens were dried under a heating light (at a distance of ∼20 cm under a lamp with a 60 W tungsten filament light bulb).
EM data collection
ApoE4•POPC particles were examined at –183°C with an FEI Polara Cryo-EM microscope operated at a 300 kV high tension. Micrographs were acquired under the low-dose mode (maximal dose, 30 e/Å2) at a magnification of 77,000 with a Gatan lens-coupled 4k × 4k high-resolution charge-coupled device (UltraCam) and a 10 eV postcolumn energy filter (GIF). Negatively stained specimens were examined with the FEI Tecnai T20 microscope operated at 200 kV. Data were collected at a magnification of 80,000 with the bottom mounted four-quadrant 4k × 4k Gatan UltraScan CCD.
Statistical analysis
For morphometric analysis, individual particle images were selected using the program e2boxer.py in the EMAN2 software package (47). Particles were picked automatically and manually checked to remove overlapping or damaged particles using boxer in the EMAN software package (48). A total of 554 particle images from cryo-EM micrographs, 826 from conventional NS, 97 from NS with washing, and 598 from our optimized NS protocol were used for statistical analysis. The size of each particle was determined by measuring diameters along two orthogonal directions, one of which was the longest dimension of the particle. Histograms of diameter size were generated based on a sampling step of 5 Å. After normalization, the histograms were fitted with ninth-degree polynomial functions in Matlab for data analysis.
Reference free classification and averaging
Isolated particles (∼17,844 particles) were selected and extracted as 160 × 160 pixel images with the program boxer in the EMAN package (48, 49). The particle images were normalized after filtering out the X-ray sparkles. A Gaussian boundary circular shaped mask was applied on all images before classification. A total of 1,374 classes were generated and averaged by refine2d.py in the EMAN software suite for four iterations.
RESULTS
Cryo-EM of apoE4•POPC particles
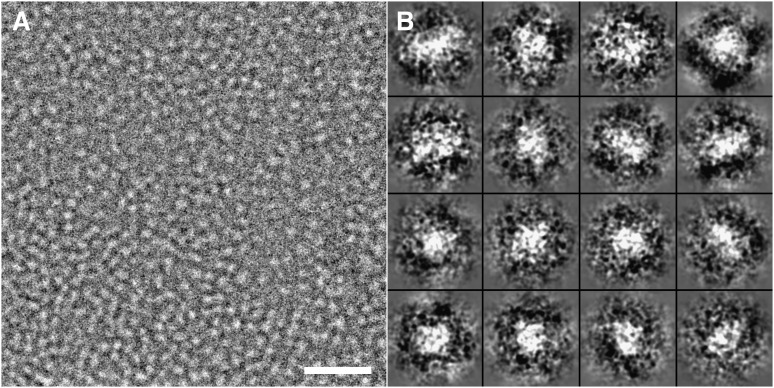
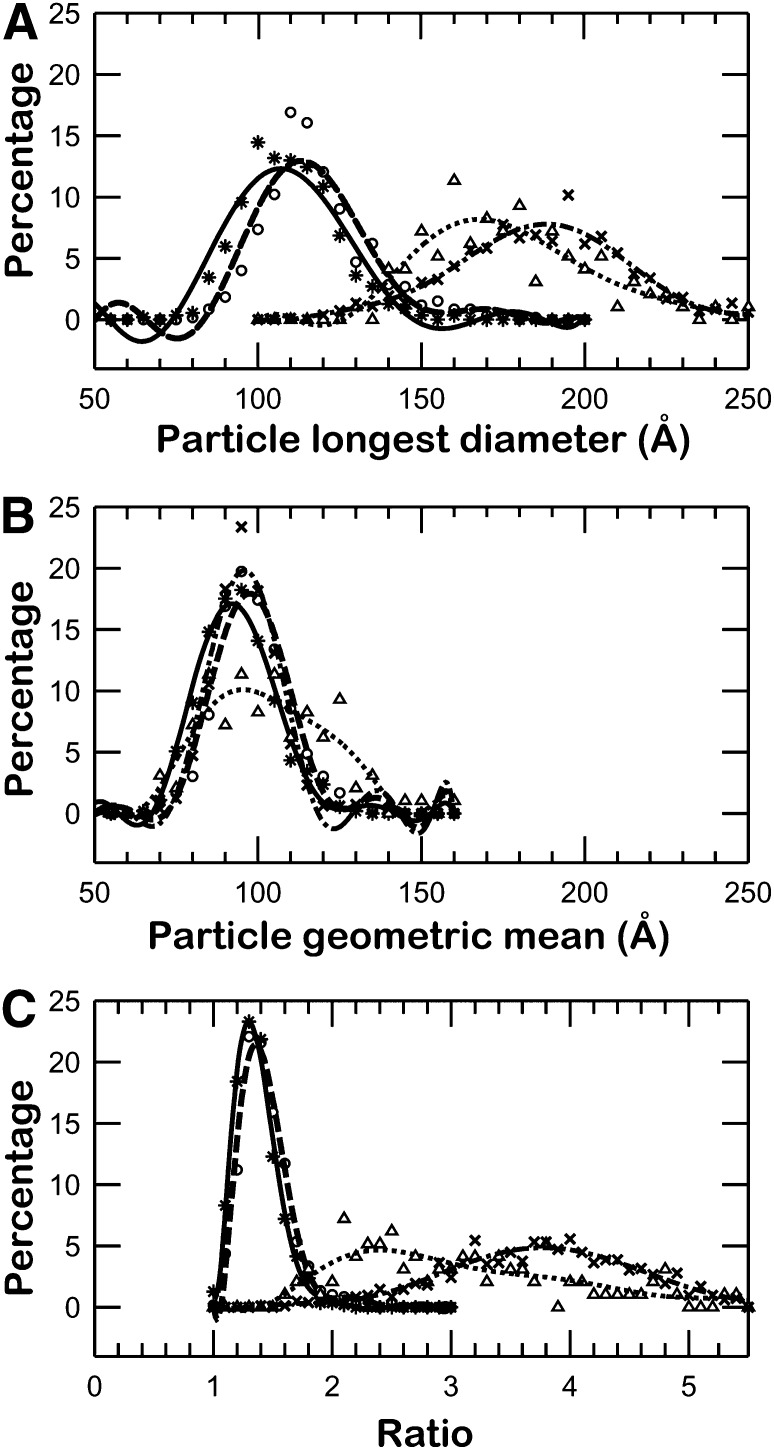
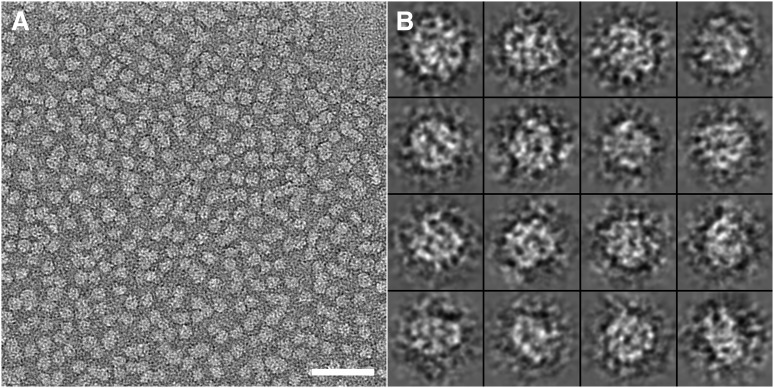
To visualize apoE4•POPC particles in their hydrated native state, the particles were prepared for ultrastructural analysis by cryo-EM. Samples were embedded in vitreous ice and imaged using the low-dose mode. ApoE4•POPC particles in cryo-EM micrographs appeared as isolated spherical and ellipsoidal particles (Fig. 2A). To quantitatively investigate the size and shape of particles, all individual apoE4•POPC particle images (total of 554) were selected and windowed from a CCD frame with no apparent astigmatism or drift (Fig. 2B). The size of each particle was determined by measuring diameters along two perpendicular directions, one of which was the longest dimension. The morphology of the particles was statistically analyzed using three methods. The first method analyzed the distribution of particle sizes based on the longest diameter of each particle (Fig. 3A, asterisks and solid line). The distribution showed ∼90% of particles had maximum diameters of 80–150 Å, and the peak population (∼12.3% of total particles) occurred at 107.0 ± 2.5 Å. Because the longest diameter is not an accurate measurement of size for nonspherical particles, the second method calculated the geometric mean of two diameters, the longest diameter and its perpendicular diameter, to represent particle size. As the geometric mean is the square root of the product of two diameters, it reflects the in-plane area of a particle on supporting film. The distribution of geometric means showed that more than 90% of particles were between 70 Å and 120 Å, and the peak population (∼17% of particles) occurred at 92.0 ± 2.5 Å (Fig. 3B, asterisks and solid line). To quantitatively identify particle shape, we used a third method that statistically analyzed the ratio between the particle's longest diameter and its perpendicular diameter. The distribution of diameter ratios showed that more than 90% of the particles had an oval shape (with the peak at 1.3), suggesting an ellipsoidal shape for apoE particles in physiological buffer (Fig. 3C, asterisks and solid line).
Fig. 2.
Cryo-EM micrograph of apoE4•POPC particles. (A) A representative area of apoE4•POPC particles suspended in vitrified buffer over a hole in a carbon film. Particles are individual and circular or oval in shape. Bar = 500 Å. (B) Enlarged views of windowed raw apoE4•POPC particles selected from the cryo-EM micrographs. Box = 200 Å. apoE4•POPC, POPC-bound form of apoE4; Cryo-EM, cryo-electron microscopy; POPC, palmitoyl-oleoylphosphatidylcholine.
Fig. 3.
Histograms of particle size and shape. (A) Histogram of longest particle diameter. The apoE4•POPC particles in each method were selected and measured by their longest diameter. The histograms have a sampling step of 5 Å. After normalization, the histograms were fitted with a ninth-degree polynomial function in Matlab for statistical analysis. The peak populations for cryo-EM (asterisks and solid line), PTA (crosses and dash-dot line), PTA with washing (triangles and dotted line), and UF (circles and dashed line) occurred at 107.0 Å (12.3%), 188.2 Å (7.8%), 167.6 Å (8.2%), and 113.2 Å (13.0%), respectively. (B) Histogram of particle diameter geometric mean. The geometric mean of each particle was calculated based on measurement of two perpendicular diameters on each particle. One of these diameters was the longest diameter. For statistical analysis, the histograms were normalized and fitted with a ninth-degree polynomial function. The peak populations for cryo-EM, PTA, PTA with washing, and UF occurred at 92.0 Å (17.1%), 95.8 Å (19.8%), 95.4 Å (10.1%), and 97.4 Å (17.9%) respectively. (C) Histogram of particle diameter ratio. To quantitatively identify the shapes of particles, the ratio of the two diameters was calculated for each particle (longest diameter/perpendicular diameter). The data for each method was normalized and fitted with a ninth-degree polynomial function. The peak population for cryo-EM occured at 1.30 (23.3%), which is similar to that of the optimized protocol, 1.36 (21.4%), but it is much different from that of conventional NS (3.80, 4.9%) and PTA with washing (2.40, 4.7%). apoE4•POPC, POPC-bound form of apoE4; Cryo-EM, cryo-electron microscopy; POPC, palmitoyl-oleoylphosphatidylcholine; PTA, phosphotungstic acid; UF, uranyl formate.
Conventional NS of apoE4•POPC particles
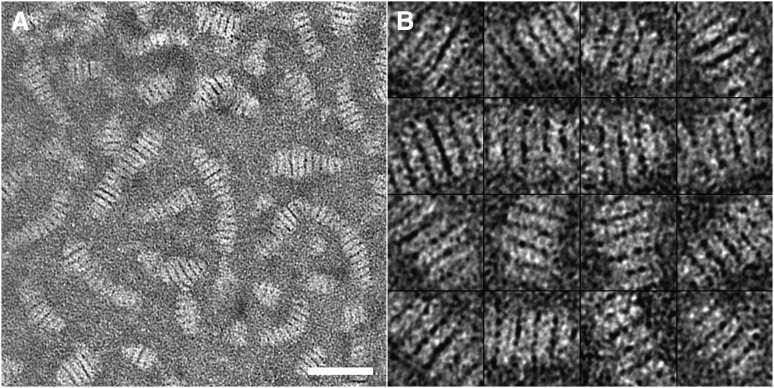
High-contrast EM imaging of proteins processed by NS has been widely used for examining protein structure because of its ease of use and rapidity in a regular EM laboratory (26–28). A conventional NS protocol has been widely used for apoE•phospholipid particles (41, 42). With this protocol, the particles tend to stack together in rouleaux (18–23, 50). To confirm this observation, we used this conventional NS protocol to examine the same apoE4•POPC sample used in the above cryo-EM experiment. The NS micrograph showed that the apoE4•POPC particles formed rouleaux (Fig. 4) similar to other apoE•phospholipid particles (19, 22, 50, 51). In the rouleaux, particles could be distinguished based on dark boundaries between them. The surfaces of the particles were mostly parallel to each other. The longest diameter of each particle was nearly perpendicular to the rouleau central axis.
Fig. 4.
Electron micrograph of negatively stained apoE4•POPC particles prepared using the conventional protocol. (A) A representative area of apoE4•POPC particles stained by PTA. Particles are in the form of rouleaux. Bar = 500 Å. (B) Enlarged views of windowed raw apoE4•POPC particles in rouleaux selected from the NS micrographs. Box = 300 Å. apoE4•POPC, POPC-bound form of apoE4; NS, negative staining; POPC, palmitoyl-oleoylphosphatidylcholine; PTA, phosphotungstic acid.
To quantitatively describe particle size and shape, pairs of perpendicular diameters were measured from 826 particles. As with the particle measurement in cryo-EM, one diameter was the longest. Analysis of the size distribution of the longest diameter showed that ∼80% particles were between 140 Å and 240 Å. The particle size distribution peaked at 188.2 ± 2.5 Å (Fig. 3A, dash-dot line), which was about twice the particle size of the peak population from cryo-EM data (Fig. 3A, solid line).
Although the PTA-stained particles had a much wider size distribution and much larger peak than the cryo-EM particles, this was insufficient to conclude that the particles in PTA-stain images were more heterogeneous in size than those of cryo-EM images. This is because the longest diameter is only a one-dimensional measurement that is not sufficient or accurate enough to represent the actual particle size/volume (three-dimensional). To present a more accurate representation of particle size, we used the geometric mean to represent the particle size because the measurement is related to the projected area of the particle. Thus, the geometric mean more accurately represents the particle size/volume than any one dimensional measurement.
The distribution of the geometric means suggested that the in-plane area of particles was similar to that of cryo-EM. More than 90% of particles had average diameters between 75 and 115 Å, and the peak population (∼20%) had a size of 95.8 ± 2.5 Å (Fig. 3B, dash-dot line), a difference of less than 5% from the cryo-EM results. The distribution of the geometric mean of the particles in the PTA-stained images was not significantly different from the cryo-EM images (Fig. 3B), indicating that particle fusion or size heterogeneity was introduced by the PTA sample preparation. Without any obvious fusion of particles in the micrographs, the particle volume distribution should be the same as that of cryo-EM. The similar distribution of the geometric mean of diameters suggested that the third dimension, perpendicular to the supporting film, might be similar to that of cryo-EM. Considering that the longest diameters were more than twice as long as those in cryo-EM, its perpendicular diameter had to lie along the rouleau center axis and had to be no more than half that of cryo-EM. This revealed that the particles were compressed into discs along the center axis of rouleaux.
To quantitatively investigate the shape of the particles, we generated a distribution for the ratios of these pairs of diameters. The ratio distribution shows that the particle shape was dramatically elongated (Fig. 3C, dash-dot line). More than 90% of the particles had a ratio between 2.5 and 5.5, while the peak population occurred at a ratio of 3.8, about three times that of cryo-EM. In the images of windowed particles, the particles tended to dramatically flatten their surfaces parallel to each other and form into rouleaux. The difference in particle shapes between cryo-EM and conventional NS suggested that the artifact in apoE4•POPC particle shape was caused by the use of the conventional NS protocol.
Reducing the size of rouleaux by washing the sample before staining
Salt concentration affects lipoprotein structure (52). To determine if the salt concentration in the sample buffer contributed to the artifacts in NS, we washed the sample on the supporting film with three successive drops of deionized water immediately before exposing the sample to a drop of phosphotungstic acid (PTA). The micrographs showed the particles still formed rouleaux (Fig. 5). The selected and windowed images of rouleaux were similar to those from conventional NS, but the rouleaux were shorter, and the discs looked thicker. In addition to the rouleaux, some isolated particles were also found in the micrographs. The particles varied in size, but most of them were circular or oval.
Fig. 5.
Electron micrograph of negatively stained apoE4•POPC particles prepared by washing the sample before staining. (A) A representative area of apoE4•POPC particles stained with PTA. Particles are mostly in the form of rouleaux with some individual particles. Bar = 500 Å. (B) Enlarged views of windowed rouleaux and individual apoE4•POPC particles selected from the NS micrographs. Box = 300 Å. apoE4•POPC, POPC-bound form of apoE4; NS, negative staining; POPC, palmitoyl-oleoylphosphatidylcholine; PTA, phosphotungstic acid.
To quantitatively examine the changes in particle stacking, we statistically analyzed the morphology of the particles as we did in the above experiments. Two diameters were measured in each particle: the longest diameter and the diameter perpendicular to it. Statistical analysis of the longest diameter showed that the particle sizes were mostly 125–230 Å, similar to the results from conventional NS. The peak population (∼8.2%) occurred at a particle size of 167.6 ± 2.5Å, which is ∼20 Å shorter than that of conventional NS, but still significantly longer (∼60 Å) than that of cryo-EM (Fig. 3A, solid line). Analysis of the distribution of the geometric means of diameters suggested that the in-plane area remained similar to that of both conventional NS and cryo-EM. The peak population of particles (∼10%) had an average diameter of 95.4 ± 2.5 Å (Fig. 3B, dotted line). The longest diameter was shorter than that of conventional NS suggesting that the perpendicular diameter was longer than that of conventional NS. The perpendicular diameter was along the direction of the stacking. Thus, the longer perpendicular direction suggested the disc shape of particles tended to be more globular. To quantitatively confirm this change, we analyzed the ratios of these two diameters. The distribution showed the particles within half the height of the peak population had a ratio between 1.7 and 3.5 (with the peak at 2.4), which was obviously smaller than that from conventional NS (2.9–4.8) (Fig. 3C). Although the ratio in the peak population was still higher than that from cryo-EM (2.4 vs. 1.3), the ratio was significantly reduced from that in conventional NS (2.4 vs. 3.8). This result suggested that lowering the salt concentration in the sample buffer reduced PTA-introduced rouleau formation when using the conventional NS protocol.
An optimized NS protocol for apoE4•POPC
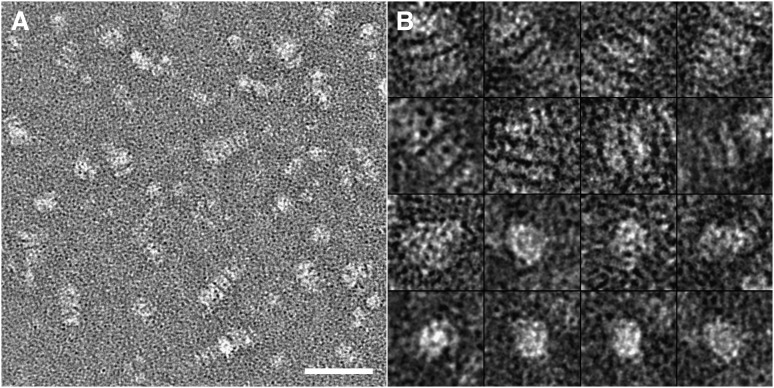
To further improve the NS specimen preparation protocol, we continued to modify the above protocol by using various NS reagents. We found that, when using UF instead of PTA, the particles in the micrographs appeared as well-isolated spheres or ellipsoids (Fig. 6). To quantitatively analyze the particle size and shape, we used analytical methods similar to those used above. We first measured two diameters along each particle's longest axis and its perpendicular direction. The statistical distribution of the longest diameter showed the particles of the peak population had a size of 113.2 ± 2.5 Å, which was only 6 Å larger (∼5%) than the measurement from cryo-EM, but significantly different from the 188.2 ± 2.5 Å in conventional NS (Fig. 3A). Additionally, the distribution of the geometric mean showed the peak population of particles (∼17.9%) had a size of 97.4 ± 2.5 Å, which was less than 6% larger than that from cryo-EM (Fig. 3B). Most importantly, the distribution of the ratio of two diameters demonstrated that the peak population (∼21.4%) of the particles occurred at a ratio of 1.36, which was a less than 5% different from that of cryo-EM, but dramatically different from the 3.8 in conventional NS (Fig. 3C). These results suggested that our optimized NS protocol could produce near native images of the apoE4•POPC particles similar to those produced by cryo-EM. Additionally, the contrast of the particle images, which is significantly higher than with cryo-EM, could be used for monitoring the morphology of the lipid-bound forms of apoE by regular EM laboratories and could even be used for reconstruction of the three-dimensional density map.
Fig. 6.
Electron micrograph of negatively stained apoE4•POPC particles prepared using our optimized protocol. (A) Representative micrographs of apoE4•POPC particles stained by UF by our protocol. The particles show a homogeneous population of compact individual particles with circular or oval shape. Bar = 500 Å. (B) Enlarged views of apoE4•POPC particles selected from the NS micrographs. Box = 200 Å. apoE4•POPC, POPC-bound form of apoE4; NS, negative staining; POPC, palmitoyl-oleoylphosphatidylcholine; UF, uranyl formate.
DISCUSSION
The lipid-bound form of apoE4•phospholipid particles plays a critical role in cholesterol transfer (3). Conventional NS reveals a discoid particle shape (19, 22, 50, 51), which is different from what is seen with small angle scattering (25). In this study, we used different EM methods to examine the same batch of apoE4•POPC sample and proposed an optimized protocol for NS. The statistical analysis showed that the artifacts have been significantly reduced by our NS protocol. The particles rendered by our protocol are similar in size and shape to that of native state particles from cryo-EM. However, the contrast of the particle images is significantly better than that of cryo-EM.
The effects of phosphotungstic acid on lipoprotein interaction
PTA, a soluble and stable heteropolyacid at physiological pH and room temperature, has been widely used for NS of biological samples in regular EM laboratories (26–28). PTA was first used in EM study of lipoproteins about half a century ago (41, 53). Since then, it has been reported to interact with lipoproteins and cause the formation of “myelin figures”—stacked bilayers of amphiphiles (34). In the last two decades, studies on the lipid-bound form of apoE by PTA also showed the particles appeared to be disc-shaped and tended to form rouleaux. Similar stacking behavior has also been observed in other lipid-containing samples, such as PTA-stained phospholipids, phospholipid-cholesterol dispersions, mixtures of phospholipids, cholesterol, and bile acids, as well as lipid vesicles and bilayer membranes (18–23). Use of PTA as a staining reagent was the only link among these different studies, which suggested that the artifacts might be due to PTA. Stacking might occur because PTA has multiple negative charges that can interact with the surface positive charges of phospholipids (on the protonated amino groups of choline) and draw amphiphilic bilayers of different particles together to form rouleaux. The stacked conformation, in which PTA molecules may be located between the discs, allows the system of the particles and PTA molecules to have maximum interaction surface area and minimum electrostatic energy.
Uranyl formate is an ideal stain for apoE4•POPC
Uranyl stains, including uranyl acetate, uranyl nitrate, and UF, are good choices for NS of most biological samples (28). Compared with the pH 7.4 of PTA, the pH 4.6 of UF is obviously different from the pH of apoE•POPC solution (pH 7.4); however, the UF-stained images contained no stacking, suggesting that pH contributes minimally to the observed difference in apoE•POPC particle stacking. Unlike PTA, uranyl stains are cationic and have less opportunity to interact with surfacial positive charges of phospholipids. This may be the reason that no reports exist about uranyl stains causing the rouleau formation in samples involving lipids. Additionally, the polyions of PTA have a diameter of 0.8–0.9 nm, and its grain size is ∼1.2 nm. A dried layer of uranyl stain possesses granularity on the order of 0.3 nm (54–56). Among the uranyl stains, UF has finest grain size, which provides the best staining for structural detail (28). This advantage is very important in visualization of molecules that are smaller than 100 kDa (28). With our optimized protocol, the micrographs of the apoE4•POPC particles contain rich structural detail (the high-density portion of the particle). The high contrast of particle images is promising for 3-D reconstruction of apoE structure in its lipid-bound forms. A minor disadvantage of UF is that solutions are light-sensitive and only stable for a few days.
Ellipsoidal shape of the apoE4•POPC particles
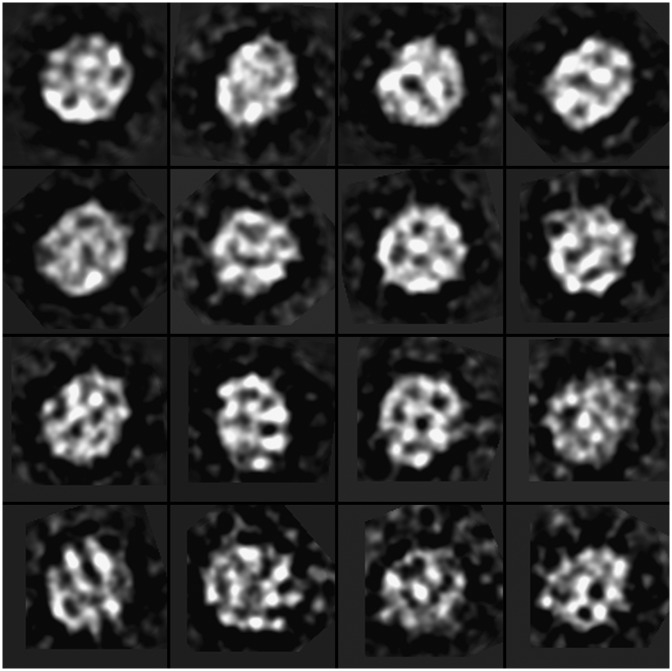
By using our optimized protocol, all particle images on the micrographs displayed a circular or oval shape (Fig. 6). The 2-D images of the particles were, in general, insufficient to identify the 3-D shape of the particles as discoid or ellipsoid. However, considering the significant difference in shape between discoid and ellipsoid, the 3-D shape could still be identified by the following logic: (a) If the particles are discoid, the only possibility for all the particle images appearing as circular or oval is that they all lie on the supporting film on one of their flat disc surfaces as a strongly preferred orientation; and (b) In the discoidal model of apoE•phospholipid particles, the center of disc contains mostly low-density lipid bilayer, while the edge region consists of high-density apoE (24). The only freedom of the particle is the in-plane rotation on the supporting film. Thus, the class averaging of the particle images should have very limited variation between the averages (e.g., the particles should be similar to each other). To test this hypothesis, we used the refine2d.py program in the EMAN software package for classification and averaging to generate very high quality 2-D class averages based on a singular value decomposition and multivariate statistical analysis iterative classification scheme (48). The class averages showed that the averages were significantly different from each other (Fig. 7). Moreover, the high-density portion in the class averages appeared near both the edges and centers of the particle, making the discoidal model of apoE4•POPC less likely. In contrast, the ellipsoidal shape, proposed recently by structural analyses of X-ray crystallography, SAXS, and biophysical studies (14, 19, 23, 25, 57, 58), is more likely supported by the class averages.
Fig. 7.
Selected class averages of negatively stained apoE4•POPC particles. The 17,844 individual particle images were classified and averaged based on a singular value decomposition and multivariate statistical analysis iterative classification scheme. The generated high-quality, two-dimensional class averages show that the high density portion is distributed in the edge as well as the center area. Box = 225 Å (160 pix × 1.406 Å/pix = 225). apoE4•POPC, POPC-bound form of apoE4; POPC, palmitoyl-oleoylphosphatidylcholine.
Geometric mean of diameters is an ideal parameter for demonstrating particle size
As demonstrated here, particle shape depended on the NS protocol. In morphological studies, the particle size could be affected by the arbitrary measurement of the particle diameter. We found that although the particle shapes differed significantly in different experiments, distributions of the geometric means of diameters remained surprisingly similar (Fig. 3B). The peak population of the geometric mean of diameters by cryo-EM was 92.0 Å, which was similar to the 97.4 Å by our optimized protocol and 95.8 Å by the conventional NS protocol. Moreover, the half-widths of the distributions of the geometric means of diameters were similar to each other. These distributions suggest that the geometric mean of diameters was less dependent on the choice of NS protocol than any diameter measurement. This may be because the geometric mean of diameters was measured from 2-D images, which are more precise than any 1-D measurement for presentation of the 3-D volume of particles. Thus, the geometric mean of diameters was a reliable parameter to represent the particle size distribution for statistical analysis of lipid-bound apoE4.
CONCLUSION
Our results suggest an optimized NS protocol, which allowed us to produce images of apoE4•POPC particles similar to those from cryo-EM. In this procedure, the high-contrast images supported the ellipsoid model of apoE•POPC particle determined by X-ray. This optimized protocol may be useful as a general NS protocol for the specimen preparation of other lipoproteins.
Footnotes
Abbreviations:
- apoE
- apolipoprotein E
- apoE4•POPC
- POPC-bound form of apoE4
- Cryo-EM
- cryo-electron microscopy
- EM
- electron microscopy
- FPLC
- fast-protein liquid chromatography
- NS
- negative staining
- POPC
- palmitoyl-oleoylphosphatidylcholine
- PTA
- phosphotungstic acid
- UF
- uranyl formate
This work was supported by the W. M. Keck Foundations (G.R.); in part by the National Pre-doctoral Scholarship of China through the China Scholarship Council (CSC) (File No. 2008628018) (L.Z.); and by RO1 AG028793 (K.W.) from the National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
REFERENCES
- 1.Elshourbagy N. A., Liao W. S., Mahley R. W., Taylor J. M. 1985. Apolipoprotein E mRNA is abundant in the brain and adrenals, as well as in the liver, and is present in other peripheral tissues of rats and marmosets. Proc. Natl. Acad. Sci. USA. 82: 203–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahley R. W., Huang Y. 1999. Apolipoprotein E: from atherosclerosis to Alzheimer's disease and beyond. Curr. Opin. Lipidol. 10: 207–217. [DOI] [PubMed] [Google Scholar]
- 3.Mahley R. W., Rall S. C., Jr 2000. Apolipoprotein E: far more than a lipid transport protein. Annu. Rev. Genomics Hum. Genet. 1: 507–537. [DOI] [PubMed] [Google Scholar]
- 4.Weisgraber K. H., Rall S. C., Jr., Mahley R. W. 1981. Human E apoprotein heterogeneity. Cysteine-arginine interchanges in the amino acid sequence of the apo-E isoforms. J. Biol. Chem. 256: 9077–9083. [PubMed] [Google Scholar]
- 5.Mahley R. W. 1988. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 240: 622–630. [DOI] [PubMed] [Google Scholar]
- 6.Weisgraber K. H. 1994. Apolipoprotein E: structure-function relationships. Adv. Protein Chem. 45: 249–302. [DOI] [PubMed] [Google Scholar]
- 7.Luc G., Bard J. M., Arveiler D., Evans A., Cambou J. P., Bingham A., Amouyel P., Schaffer P., Ruidavets J. B., Cambien F., et al. 1994. Impact of apolipoprotein E polymorphism on lipoproteins and risk of myocardial infarction. The ECTIM Study. Arterioscler. Thromb. 14: 1412–1419. [DOI] [PubMed] [Google Scholar]
- 8.Saunders A. M., Strittmatter W. J., Schmechel D., George-Hyslop P. H., Pericak-Vance M. A., Joo S. H., Rosi B. L., Gusella J. F., Crapper-MacLachlan D. R., Alberts M. J., et al. 1993. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 43: 1467–1472. [DOI] [PubMed] [Google Scholar]
- 9.Weisgraber K. H., Mahley R. W. 1996. Human apolipoprotein E: the Alzheimer's disease connection. FASEB J. 10: 1485–1494. [DOI] [PubMed] [Google Scholar]
- 10.Mahley R. W., Weisgraber K. H., Huang Y. 2009. Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer's disease to AIDS. J. Lipid Res. 50(Suppl.): S183–S188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson C., Wardell M. R., Weisgraber K. H., Mahley R. W., Agard D. A. 1991. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 252: 1817–1822. [DOI] [PubMed] [Google Scholar]
- 12.Pitas R. E., Innerarity T. L., Mahley R. W. 1980. Cell surface receptor binding of phospholipid. protein complexes containing different ratios of receptor-active and -inactive E apoprotein. J. Biol. Chem. 255: 5454–5460. [PubMed] [Google Scholar]
- 13.Koo C., Innerarity T. L., Mahley R. W. 1985. Obligatory role of cholesterol and apolipoprotein E in the formation of large cholesterol-enriched and receptor-active high density lipoproteins. J. Biol. Chem. 260: 11934–11943. [PubMed] [Google Scholar]
- 14.Peters-Libeu C. A., Newhouse Y., Hatters D. M., Weisgraber K. H. 2006. Model of biologically active apolipoprotein E bound to dipalmitoylphosphatidylcholine. J. Biol. Chem. 281: 1073–1079. [DOI] [PubMed] [Google Scholar]
- 15.Newhouse Y., Peters-Libeu C., Weisgraber K. H. 2005. Crystallization and preliminary X-ray diffraction analysis of apolipoprotein E-containing lipoprotein particles. Acta Crystallogr. 61: 981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tall A. R., Small D. M., Deckelbaum R. J., Shipley G. G. 1977. Structure and thermodynamic properties of high density lipoprotein recombinants. J. Biol. Chem. 252: 4701–4711. [PubMed] [Google Scholar]
- 17.Assmann G., Brewer H. B., Jr 1974. A molecular model of high density lipoproteins. Proc. Natl. Acad. Sci. USA. 71: 1534–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong E. L., Nichols A. V., Weisgraber K. H., Forte T. M., Shore V. G., Blanche P. J. 1989. Discoidal complexes containing apolipoprotein E and their transformation by lecithin-cholesterol acyltransferase. Biochim. Biophys. Acta. 1006: 317–328. [DOI] [PubMed] [Google Scholar]
- 19.Schneeweis L. A., Koppaka V., Lund-Katz S., Phillips M. C., Axelsen P. H. 2005. Structural analysis of lipoprotein E particles. Biochemistry. 44: 12525–12534. [DOI] [PubMed] [Google Scholar]
- 20.Raussens V., Drury J., Forte T. M., Choy N., Goormaghtigh E., Ruysschaert J. M., Narayanaswami V. 2005. Orientation and mode of lipid-binding interaction of human apolipoprotein E C-terminal domain. Biochem. J. 387: 747–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narayanaswami V., Maiorano J. N., Dhanasekaran P., Ryan R. O., Phillips M. C., Lund-Katz S., Davidson W. S. 2004. Helix orientation of the functional domains in apolipoprotein e in discoidal high density lipoprotein particles. J. Biol. Chem. 279: 14273–14279. [DOI] [PubMed] [Google Scholar]
- 22.Li X., Kan H. Y., Lavrentiadou S., Krieger M., Zannis V. 2002. Reconstituted discoidal ApoE-phospholipid particles are ligands for the scavenger receptor BI. The amino-terminal 1–165 domain of ApoE suffices for receptor binding. J. Biol. Chem. 277: 21149–21157. [DOI] [PubMed] [Google Scholar]
- 23.Innerarity T. L., Pitas R. E., Mahley R. W. 1979. Binding of arginine-rich (E) apoprotein after recombination with phospholipid vesicles to the low density lipoprotein receptors of fibroblasts. J. Biol. Chem. 254: 4186–4190. [PubMed] [Google Scholar]
- 24.Blanchette C. D., Law R., Benner W. H., Pesavento J. B., Cappuccio J. A., Walsworth V., Kuhn E. A., Corzett M., Chromy B. A., Segelke B. W., et al. 2008. Quantifying size distributions of nanolipoprotein particles with single-particle analysis and molecular dynamic simulations. J. Lipid Res. 49: 1420–1430. [DOI] [PubMed] [Google Scholar]
- 25.Peters-Libeu C. A., Newhouse Y., Hall S. C., Witkowska H. E., Weisgraber K. H. 2007. Apolipoprotein E*dipalmitoylphosph atidylcholine particles are ellipsoidal in solution. J. Lipid Res. 48: 1035–1044. [DOI] [PubMed] [Google Scholar]
- 26.Oliver R. M. 1973. Negative stain electron microscopy of protein macromolecules. Methods Enzymol. 27: 616–672. [DOI] [PubMed] [Google Scholar]
- 27.Woeste S., Demchick P. 1991. New version of the negative stain. Appl. Environ. Microbiol. 57: 1858–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohi M., Li Y., Cheng Y., Walz T. 2004. Negative staining and image classification - powerful tools in modern electron microscopy. Biol. Proced. Online. 6: 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubochet J., Adrian M., Chang J. J., Homo J. C., Lepault J., McDowall A. W., Schultz P. 1988. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21: 129–228. [DOI] [PubMed] [Google Scholar]
- 30.Taylor K. A., Glaeser R. M. 1976. Electron microscopy of frozen hydrated biological specimens. J. Ultrastruct. Res. 55: 448–456. [DOI] [PubMed] [Google Scholar]
- 31.Jaffe J. S., Glaeser R. M. 1984. Preparation of frozen-hydrated specimens for high resolution electron microscopy. Ultramicroscopy. 13: 373–377. [DOI] [PubMed] [Google Scholar]
- 32.Milligan R. A., Brisson A., Unwin P. N. 1984. Molecular structure determination of crystalline specimens in frozen aqueous solutions. Ultramicroscopy. 13: 1–9. [DOI] [PubMed] [Google Scholar]
- 33.Bradley D. E. 1962. A study of the negative staining process. J. Gen. Microbiol. 29: 503–516. [DOI] [PubMed] [Google Scholar]
- 34.Melchior V., Hollingshead C. J., Cahoon M. E. 1980. Stacking in lipid vesicle-tubulin mixtures is an artifact of negative staining. J. Cell Biol. 86: 881–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Egelman E. H., Amos L. A. 2009. Electron microscopy of helical filaments: rediscovering buried treasures in negative stain. Bioessays. 31: 909–911. [DOI] [PubMed] [Google Scholar]
- 36.Cunningham W. P., Staehelin L. A., Rubin R. W., Wilkins R., Bonneville M. 1974. Effects of phosphotungstate negative staining on the morphology of the isolated Golgi apparatus. J. Cell Biol. 62: 491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang Q. X., Chester D. W., Sigworth F. J. 2001. Spherical reconstruction: a method for structure determination of membrane proteins from cryo-EM images. J. Struct. Biol. 133: 119–131. [DOI] [PubMed] [Google Scholar]
- 38.Wang L., Bose P. S., Sigworth F. J. 2006. Using cryo-EM to measure the dipole potential of a lipid membrane. Proc. Natl. Acad. Sci. USA. 103: 18528–18533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng D., Song C., Reardon C. A., Liao S., Getz G. S. 2003. Lipoproteins produced by ApoE−/− astrocytes infected with adenovirus expressing human ApoE. J. Neurochem. 86: 1391–1402. [DOI] [PubMed] [Google Scholar]
- 40.Ren G., Reddy V. S., Cheng A., Melnyk P., Mitra A. K. 2001. Visualization of a water-selective pore by electron crystallography in vitreous ice. Proc. Natl. Acad. Sci. USA. 98: 1398–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forte T., Norum K. R., Glomset J. A., Nichols A. V. 1971. Plasma lipoproteins in familial lecithin: cholesterol acyltransferase deficiency: structure of low and high density lipoproteins as revealed by elctron microscopy. J. Clin. Invest. 50: 1141–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forte T. M., Nordhausen R. W. 1986. Electron microscopy of negatively stained lipoproteins. Methods Enzymol. 128: 442–457. [DOI] [PubMed] [Google Scholar]
- 43.Ren G., Gao K., Bushman F. D., Yeager M. 2007. Single-particle image reconstruction of a tetramer of HIV integrase bound to DNA. J. Mol. Biol. 366: 286–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang Y., Gursky O., Atkinson D. 2003. Lipid-binding studies of human apolipoprotein A-I and its terminally truncated mutants. Biochemistry. 42: 13260–13268. [DOI] [PubMed] [Google Scholar]
- 45.Jayaraman S., Gantz D. L., Gursky O. 2008. Effects of protein oxidation on the structure and stability of model discoidal high-density lipoproteins. Biochemistry. 47: 3875–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen B., Ren X., Neville T., Jerome W. G., Hoyt D. W., Sparks D., Ren G., Wang J. 2009. Apolipoprotein AI tertiary structures determine stability and phospholipid-binding activity of discoidal high-density lipoprotein particles of different sizes. Protein Sci. 18: 921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J. 2007. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157: 38–46. [DOI] [PubMed] [Google Scholar]
- 48.Ludtke S. J., Baldwin P. R., Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128: 82–97. [DOI] [PubMed] [Google Scholar]
- 49.Ludtke S. J., Jakana J., Song J. L., Chuang D. T., Chiu W. 2001. A 11.5 A single particle reconstruction of GroEL using EMAN. J. Mol. Biol. 314: 253–262. [DOI] [PubMed] [Google Scholar]
- 50.Lu B., Morrow J. A., Weisgraber K. H. 2000. Conformational reorganization of the four-helix bundle of human apolipoprotein E in binding to phospholipid. J. Biol. Chem. 275: 20775–20781. [DOI] [PubMed] [Google Scholar]
- 51.Gorshkova I. N., Kypreos K. E., Gantz D. L., Zannis V. I., Atkinson D. 2008. Biophysical properties of apolipoprotein E4 variants: implications in molecular mechanisms of correction of hypertriglyceridemia. Biochemistry. 47: 12644–12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jayaraman S., Gantz D. L., Gursky O. 2006. Effects of salt on the thermal stability of human plasma high-density lipoprotein. Biochemistry. 45: 4620–4628. [DOI] [PubMed] [Google Scholar]
- 53.Nichols A. V., Forte G. M., Coggiola E. L. 1969. Electron microscopy of very low density lipoproteins from egg yolk using negative staining. Biochim. Biophys. Acta. 175: 451–453. [DOI] [PubMed] [Google Scholar]
- 54.Harris J. R., Gebauer W., Guderian F. U., Markl J. 1997. Keyhole limpet hemocyanin (KLH), I: reassociation from Immucothel followed by separation of KLH1 and KLH2. Micron. 28: 31–41. [DOI] [PubMed] [Google Scholar]
- 55.Harris J. R., Gebauer W., Sohngen S. M., Nermut M. V., Markl J. 1997. Keyhole limpet hemocyanin (KLH), II: characteristic reassociation properties of purified KLH1 and KLH2. Micron. 28: 43–56. [DOI] [PubMed] [Google Scholar]
- 56.Hayat M. A. 2000. Negative stains. Principles and Techniques of Electron Microscopy: Biological Applications. Cambridge University Press, Cambridge; 372–377. [Google Scholar]
- 57.Settasatian N., Barter P. J., Rye K. A. 2008. Remodeling of apolipoprotein E-containing spherical reconstituted high density lipoproteins by phospholipid transfer protein. J. Lipid Res. 49: 115–126. [DOI] [PubMed] [Google Scholar]
- 58.Drury J., Narayanaswami V. 2005. Examination of lipid-bound conformation of apolipoprotein E4 by pyrene excimer fluorescence. J. Biol. Chem. 280: 14605–14610. [DOI] [PubMed] [Google Scholar]