Abstract
Phospholipid transfer protein (PLTP) belongs to the lipid transfer/lipopolysaccharide-binding protein gene family. Expression of PLTP has been implicated in the development of atherosclerosis. We evaluated the effects of PLTP region tagging single nucleotide polymorphisms (SNPs) on the prediction of both carotid artery disease (CAAD) and PLTP activity. CAAD effects were evaluated in 442 Caucasian male subjects with severe CAAD and 497 vascular disease-free controls. SNP prediction of PLTP transfer activity was evaluated in both a subsample of 87 subjects enriched for an allele of interest and in a confirmation sample of 210 Caucasian males and females. Hemoglobin A1c or insulin level predicted 11–14% of age- and sex-adjusted PLTP activity. PLTP SNPs that predicted ∼11–30% of adjusted PLTP activity variance were identified in the two cohorts. For rs6065904, the allele that was associated with CAAD was also associated with elevated PLTP activity in both cohorts. SNPs associated with PLTP activity also predicted variation in LDL-cholesterol and LDL-B level only in the replication cohort. These results demonstrate that PLTP activity is strongly influenced by PLTP region polymorphisms and metabolic factors.
Keywords: lipids, atherosclerosis, polymorphism
Phospholipid transfer protein (PLTP) belongs to the lipid transfer/lipopolysacharide binding protein gene family. It is expressed in the liver and in macrophages (1). PLTP is responsible for most of the transfer of phospholipids from plasma VLDL to HDL (2–4). Mouse studies support the hypothesis that increased PLTP level is positively associated with atherosclerotic risk. Increased PLTP expression in mice has been associated with atherosclerosis (5–7), impaired reverse cholesterol transport (7), and decreased HDL, LDL, and VLDL levels (5). Elevated PLTP may lower HDL by increased hapatic uptake in mice (8). Mice with PLTP deficiency also absorbed less dietary cholesterol (9). Macrophage cholesterol efflux is impaired in PLTP transgenic mice, which may promote atherosclerosis (7). Acute elevation of PLTP was found to increase both atherosclerotic lesions size and macrophage content in transgenic mice (6). Also, transplantation of bone marrow from PTLP-deficient mice into irradiated LDL receptor-deficient mice reduced atherosclerosis compared with mice possessing functional macrophage PLTP (10). In contrast, macrophage PLTP expression introduced through bone marrow transplantation in double LDL receptor and PLTP-deficient mice decreased atherosclerotic lesions and total cholesterol, while increasing HDL (11). Similarly, bone marrow knockouts of PLTP have also been reported to have lower atherosclerosis on an LDL receptor background (11).
In humans, PLTP has been reported to influence the size, density, and levels of VLDL and HDL (12–15) as well as LDL (15, 16). We recently described the relationship between PLTP mass, activity, and lipid levels. PLTP mass and activity were poorly correlated, but PLTP mass was correlated with HDL size and concentration, while PLTP activity was correlated with VLDL size, total cholesterol, and triglyceride levels. These correlations differ by gender (17).
The relationship of PLTP activity and expression to vascular disease in humans requires further investigation. While studies have associated elevated PLTP with coronary artery disease (18), left ventricular dysfunction (19), and coronary artery events in subjects on statins (20), the opposite result was reported for peripheral artery disease. Peripheral artery vascular disease has been associated with lower PLTP (21), while carotida intima-media thickness (22) has been associated with raised PLTP in humans. PLTP expression in macrophages in the vessel wall has been proposed to be induced in the conversion of macrophages to foam cells, thus impacting atherosclerotic plaque progression (1, 23, 24).
Prior investigation identified genetic variation associated with PLTP level and with lipids. PLTP single nucleotide polymorphism (SNP) rs7679 was found to predict serum triglyceride level as well as RNA expression of PLTP in liver (25). In that report, the rs7679 T allele was associated with increased PLTP expression, increased HDL level, and decreased triglyceride level. SNP rs2294213 has been reported to be associated with raised HDL, lowered triglycerides, and higher PLTP activity (26). The lipid profiles would suggest that increased expression might be cardioprotective. This is inconsistent with elevated PLTP predicting atherosclerosis in humans.
Our goal was to evaluate the relationship of common genetic variation in PLTP with both carotid artery disease (CAAD) and with PLTP activity using a tagging SNP (tagSNP) approach. A cohort of subjects with and without severe carotid artery disease was considered, and a subset was selected for the labor intensive PLTP activity measure. An additional replication cohort was available for evaluation of PLTP activity.
METHODS
Sample
The sample population included 939 unrelated Caucasian males from the CLEAR study of carotid artery disease risk. This study has been previously described in detail elsewhere (27). Briefly, the study gains power per sampled individual from using a tails-of-the-distribution sampling scheme where cases (n = 442) had >80% stenosis of one or both internal carotid arteries, and controls (n = 497) had <15% stenosis bilaterally on duplex ultrasound. Additionally, controls had no known atherosclerotic vascular disease, had normal ankle-arm indices measures for lower extremity vascular disease, and were age distribution matched with the cases based on the age of onset of disease (censored age). Use of medications was ascertained by report and reconciled with review of pharmacy and medical records. Self-reported race was confirmed by analysis with STRUCTURE (28), incorporating HapMap ethnicity groups. One inconsistent subject was removed from the analysis. Height and weight were measured, with medical records or self-report used to complete missing data. The PLTP activity assay has not been adapted to high throughput; hence, 87 genotyped subjects were selected for phenotyping. To minimize confounding with disease status and medication, these subjects were all controls who were not on statin medications. To maximize power to detect effects on PLTP activity, the selection of subjects was enriched for the rare genotype for a SNP associated with CAAD. As shown on Table 1, this strategy raised the minor allele frequency (MAF) of the SNP-predicting CAAD from 0.02 to 0.12. The study was approved by the University of Washington, the Veterans Affairs Puget Sound Health Care System, and the Virginia Mason Medical Center human subject review processes. Subjects gave written informed consent. Demographics are given in supplementary Table A.
TABLE 1.
Effects of genotyped PLTP tagSNPs on prediction of severe carotid artery disease and on PLTP activity level
| n = 939 |
n = 87 Discovery |
n = 210 Confirmation |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs | Chr 20. BP | Location | MAF-All | Minor Allele | OR | p-CAAD | MAF-Discovery | β-PLTPa | p-PLTPa | MAF-Conf | β-PLTPa | p-PLTPa |
| rs1736493 | 43964041 | Intron 4 | 0.069 | G | 1.52 | 1.53E-02* | 0.138 | 0.09 | 1.11E-02* | 0.093 | −0.03 | 2.76E-01 |
| rs553359 | 43964245 | Intron 4 | 0.394 | C | 0.86 | 1.65E-01 | 0.465 | −0.05 | 8.29E-03* | 0.370 | −0.03 | 4.61E-02* |
| rs11569649 | 43964824 | Intron 6 | 0.024 | C | 0.66 | 2.12E-01 | 0.023 | −0.07 | 3.59E-01 | 0.045 | −0.07 | 7.93E-02 |
| rs11086985 | 43965662 | Intron 6 | 0.381 | A | 0.98 | 9.95E-01 | 0.408 | 0.07 | 1.14E-03* | 0.325 | 0.02 | 3.18E-01 |
| rs11569671 | 43965887 | Intron 6 | 0.027 | A | 1.50 | 3.76E-01 | 0.042 | −0.04 | 5.07E-01 | 0.039 | −0.03 | 4.73E-01 |
| rs6073950 | 43966517 | Intron 6 | 0.333 | T | 1.00 | 6.23E-01 | 0.321 | 0.06 | 3.02E-03* | 0.239 | 0.01 | 4.87E-01 |
| rs6065904 | 43968058 | Intron 8 | 0.214 | A | 0.76 | 2.30E-02* | 0.339 | −0.08 | 2.55E-05* | 0.279 | −0.08 | 1.80E-06* |
| rs6073952 | 43970339 | Intron 11 | 0.194 | A | 0.85 | 1.16E-01 | 0.264 | −0.10 | 3.39E-06* | 0.275 | −0.09 | 5.51E-07* |
| rs378114 | 43971834 | Intron 12 | 0.272 | A | 0.86 | 2.71E-01 | 0.218 | 0.10 | 5.29E-05* | 0.157 | 0.04 | 7.22E-02 |
| rs11569633 | 43973142 | Intron 13 | 0.012 | A | 1.48 | 7.83E-01 | 0.017 | 0.01 | 9.28E-01 | 0.005 | 0.08 | 4.73E-01 |
| rs394643 | 43973585 | Intron 15 | 0.461 | A | 0.79 | 2.40E-02* | 0.471 | −0.02 | 2.85E-01 | 0.445 | −0.05 | 1.58E-03* |
| rs11569668 | 43976239 | 5′ (-2046) | 0.024 | A | 0.30 | 8.70E-03* | 0.121 | 0.10 | 1.56E-02* | 0.022 | 0.01 | 8.16E-01 |
| rs4810479 | 43978455 | 5′ (-4262) | 0.249 | G | 0.88 | 4.15E-01 | 0.391 | −0.07 | 4.93E-04* | 0.331 | −0.09 | 8.51E-08* |
| rs7679 | 44009909 | 5′ (-35716) | 0.182 | C | 0.81 | 1.20E-01 | 0.242 | −0.11 | 5.39E-07* | 0.261 | −0.09 | 3.62E-07* |
rs is the number that uniquely identifies the polymorphism; BP is the base pair position on chromosome 20 for the SNP consistent with dbSNP for the 3′ strand; location refers to the placement of the SNP, either in the listed PLTP intron or 5′ of PLTP at the number of listed base pairs (−) from the initiation site per dbSNP build 130; MAF-all is the minor allele frequency for the case control cohort; OR is the odds ratio for the two less common genotype groups pooled relative to the most common group in the prediction of CAAD; p-CAAD is the P value for the test of the SNP effect on CAAD prediction (see Methods); MAF-discovery is the minor allele frequency in the subsample phenotyped for PLTP activity, β-PLTP is the coefficient for the minor allele's regression on PLTP activity; and p-PLTPα is the P value for that test. * P < 0.05.
A separate cohort from a family study was evaluated for confirmation of the SNP PLTP activity effects. This cohort consisted of 210 adult Caucasian subjects, 100 male and 110 female, ages 11–89 (mean age 47 years, eight minors) in four large families ascertained for familial combined hyperlipidemia (29). The families were originally ascertained in the 1980s and recollected from 2003–2009. All data included here are from recent collection. Lipid and PLTP measures are as described. Height and weight were from survey data. Body mass indexes (BMIs) for 17 subjects were imputed based on the regression of age, sex, age by sex interaction, and natural log-transformed C-reactive protein level on BMI. The family study was approved by the University of Washington human subject review processes. Subjects gave written informed consent or assent with parental consent.
Genotyping
DNA was prepared from buffy coat preparations by a modification of the procedure of Miller, Dykes, and Polesky (30) using Puregene reagents (Gentra, Minneapolis, MN). Genotyping (except rs7679) used the Illumina CVD chip of ∼2,100 loci with possible effects in vascular disease, including inflammation, lipid, and oxidation pathways. The effects of PLTP were prespecified as a locus of interest based on our longstanding investigation of this trait. TagSNPs use the correlations among SNPs to type SNPs that represent most common variation in the gene. As the tag SNPs were selected for a multiethnic cohort, some were uncommon and others redundant in Caucasians. Only one SNP's call rate was <99%: rs4810482 had a call rate of 97.4%. Genotype distributions were tested for departure from Hardy-Weinberg equilibrium (HWE) proportions. SNP rs4810482 and rs6073950 genotype distributions were weakly inconsistent with HWE (P values 0.01 to 0.05). Rs7679 was not included on the Illumina chip. It was genotyped by Taqman using Assay by Design. The Taqman genotypes were contrasted with imputed genotypes.
While all SNPs listed in Table 1 were genotyped, we also imputed all chromosome 20 regional SNPs identified in the HapMap spanning base pair 43,907,883 to 44,027,635 for the unrelated discovery sample in an effort to determine which is the best predictor of PLTP activity. SNPs were imputed in BIMBAM, based on linkage disequilibrium (LD) in the HapMap phased genotypes (build 36; Caucasian population; www.hapmap.org), were analyzed both by a Bayesian procedure that allowed weighting by imputed genotype confidence as well as analyzed using the most likely genotype as estimated by PLINK. SNP rs7679 was both imputed and genotyped. Ten of 932 rs7679 genotypes were discordant. Repeat reading by a second blinded technician supported the molecular versus the inferred genotype. SNP rs7679 was consistent with HWE and had a 100% call rate in both samples. All genotyping was blinded to all phenotypes.
PLTP activity
Plasma phospholipid transfer activity mediated by PLTP was determined on fasting whole plasma by measuring the transfer of 14C-phosphatidycholine from phospholipid liposomes to HDL using an established radioassay (31). Briefly, each assay tube contained 50 µl HDL3 with 150 nmol phospholipids, 50 µl 14C-phosphatidycholine-labeled liposomes containing 50 nmol phosphatidylcholine, 50 µl of plasma samples prediluted 1:50 with Tris buffer (0.01 mM Tris, 150 mM NaCl, 1 mM EDTA, and 0.01% sodium azide), and 250 µl Tris buffer to bring the total assay volume to 400 µl. The HDL3(d 1.125–1.21) was isolated from pooled plasma from fasting individuals by sequential ultracentrifugation and subsequently passed over a heparin Sepharose column to remove apolipoprotein E (apoE) and residual PLTP activity. Three separate dilutions of each plasma sample were assayed. All samples were incubated at 37°C for 15 min, a condition when the rate of transfer of phospholipids from liposomes to HDL is linear. Donor and acceptor particles were separated by precipitation with dextran sulfate and magnesium chloride (32). Plasma PLTP activity was calculated as the percentage of total radioactivity per assay tube transferred to HDL minus background transfer (tubes without plasma). Three plasma samples stored at −70°C until use were included in each assay to control for interassay variation. PLTP activity phenotypes in both the CLEAR and family cohorts were determined in the Albers lab, blinded to all other subject data.
Lipids, hemoglobin A1c, and insulin
Lipid measurements were performed on fasting whole plasma, as detailed elsewhere (33). Hemoglobin A1c (HbA1c) was measured using the standard HPLC methods. Insulin was measured by double antibody radioimmunoassay. Each assay includes appropriate reference samples.
Analysis
Logistic regression considered an additive genetic model to predict CAAD status, with genotypes coded linearly as 0, 1, and 2. Age and current smoking were considered as covariates for CAAD prediction. Based on the SNPs that predicted CAAD, a subsample was evaluated for SNP prediction of ln-PLTP activity, considering age, BMI, and HbA1c as covariates. These covariates were selected due to prior repeats of PLTP associations with the metabolic syndrome and that glucose has been shown to regulate PLTP expression (34), as discussed below. SNPS that predicted PLTP activity were then further considered for their independent effects on PLTP activity in multi-SNP models including the same covariates. Linkage disequilibrium of the SNPs was evaluated by considering the squared correlation between alleles at two sites, r2. PLTP activity was also measured on the second cohort of 210 subjects in four families ascertained for familial combined hyperlipidemia. In order to use the largest possible sample, we chose to ignore relatedness in the family cohort. This does not influence the estimated effect sizes of genotype effects but could affect the statistical tests by underestimating the variance (35, 36). Single SNP tests of SNPs associated with PLTP activity for effects on other phenotypes, including lipids, used the same models and covariates. Lipid tests performed on the entire cases control cohort of 939 subjects and were repeated on only 511 the subjects who were not taking lipid lowering medications. Due to correlations among SNPs and among phenotypes, it is not possible to quantitate the number of independent tests made; thus, adjustments for multiple contrasts were not made. HbA1c was not available for the family, and serum insulin levels served as a covariate instead, along with age, sex, BMI, and an age by sex interaction term. Insulin level was imputed for 20 subjects, using the regression of insulin on age, sex, BMI, and ln-triglyceride level. Individuals with isolated missing data for single SNPs or lipid phenotypes of interest were dropped from that analysis. Imputation of untyped SNPs was performed using the program BIMBAM (37, 38). Other statistical tests utilized program packages PLINK, R, and SPSS.
RESULTS
Demographics for the CLEAR cohort used for these analyses are reported in supplementary Table A. The PLTP region SNP effects on case status and PLTP activity in the CLEAR subsample and replication families are shown in Table 1. Four SNPs, rs6065904 (P = 0.023), rs11569668 (P = 0.009), rs1736493 (P = 0.015), and rs394643 (P = 0.024), were associated with CAAD status at the 0.05 level, without adjusted for multiple (correlated) comparisons. These four SNPs show only weak evidence for LD. However, SNP rs6065904 was in LD with both SNPs rs6065952 and rs4810479 at r2 > 0.8, neither of which predicted CAAD. LD is summarized in supplementary Table B. The largest effect on CAAD risk was seen for rs11569668, whose minor allele (A, MAF 0.02) was protective. Genotype frequencies for cases and controls are shown in supplementary Table C. Each SNP associated with CAAD was also associated with PLTP activity in at least one cohort. Rs6065904 was associated with PLTP activity in both cohorts. For this SNP, the elevated activity allele predicted CAAD.
PLTP activity was evaluated for its relationship with potential covariates and inflammatory and lipid traits in the 87 control subjects who were not taking statins. Ln-PLTP activity modestly declined with age (P = 0.025, r2 = 0.058; Table 2). Addition of BMI accounted for 2.9% of additional variance but did not contribute significantly to ln-PLTP activity prediction. Age, BMI, and HbA1c predict 20.1% of variance, with only HbA1c (P < 0.001) being significant at the 0.05 level. When age, BMI, and HbA1c are included in the model predicting ln-PLTP activity, addition of the following traits determined that none were significant at the 0.05 level: diabetes status, ln-triglycerides, HDL-cholesterol, LDL-cholesterol (LDL-C), and c-reactive protein. Therefore, age, BMI, and HbA1c were included as covariates in the genetic models predicting ln-PLTP activity. The residuals of the regression did not differ from a normal distribution.
TABLE 2.
Sources of PLTP activity variance
| Percentage of Variance in PLTP Activity Predicted by the Model |
||
|---|---|---|
| Predictors in Model | n = 85*, Discovery | n = 203*, Confirmation |
| Age and sex | 4.8 (male only) | 6.2 (6.2 female, 6.0 male) |
| Age, sex, and BMI | 7.6 | 12.3 (18.1 female, 6.0 male) |
| Age, sex, BMI, and HbA1C or insulin (covariates) | 21.4 | 19.8 (21.9 female, 20.9 male) |
| Covariates and rs7679 | 42.7 | 28.0 (31.1 female, 27.1 male) |
| Covariates and rs4810479 | 34.4 | 20.7 (23.2 female, 21.4 male) |
| Covariates and rs6073952 | 40.9 | 19.8 (21.9 female, 21.7 male) |
| Covariates and rs11569668 | 28.0 | 20.2 (24.0 female, 20.9 male) |
| Covariates and rs378114 | 35.3 | 21.1 (22.9 female, 22.6 male) |
| Covariates and rs6065904 | 37.2 | 23.0 (28.0 female, 21.4 male) |
| Covariates and rs7679, rs11569668 | 47.5 | 28.1 (32.1 female, 27.2 male) |
| Covariates and rs7679, rs378114 | 49.3 | 28.2 (31.1 female, 27.8 male) |
| Covariates and rs7679, rs11569668, and rs1736493 | 50.1 | 28.3 (32.2 female, 28.9 male) |
| Covariates and rs6073952, rs553359, and rs4810479 | 40.9 | 20.9 (23.5 female, 22.2 male) |
| Covariates and rs6073952, rs11569668, and rs1736493 | 48.9 | 20.2 (24.6 female, 22.2 male) |
| Covariates and rs6073952, rs11569668, and rs4810479 | 46.6 | 21.1 (25.2 female, 22.1 male) |
| Covariates and rs11569668, rs1736493, and rs4810479 | 43.7 | 21.2 (26.0 female, 22.0 male) |
| Covariates and rs7679, 6073952, rs11569668, and rs1736493 | 50.2 | 28.3 (32.2 female, 29.6 male) |
| Covariates and rs7679 rs11569668, rs1736493, and rs4810479 | 50.6 | 28.9 (33.3 female, 29.1 male) |
Other PLTP activity tests reported in the article use 87 subjects in discovery and 210 in confirmation cohort; however, this table only uses 85 in discovery cohort and 203 (106 female, 97 male) in confirmation cohort. These individuals do not have any isolated missing genotypes, thus allowing for perfectly nested models to be compared.
A sample of 87 control subjects was constructed maximizing the frequency of rs11569668 GA genotype to improve power to detect allelic effects on PLTP activity. The marginal effect of each SNP on PLTP activity is shown in Table 1, and the percentage of variance in PLTP activity explained by selected models is shown in Table 2. Ten PLTP SNPs predicted ln-PLTP activity at P < 0.05, including three of the four SNPs that predicted CAAD status at the P < 0.05.
We examined the activity effects of untyped SNPs in LD with our tagSNPs in Caucasian male population. No regional SNPs are found in LD (no r2 > 0.35) with rs11569668 or r2 > 0.43 with rs1736493 using Caucasian population from International HapMap. However, SNP rs6073952 was in high LD (r2 = 0.96) with a group of SNPs in complete LD with each other, including rs7679, rs17447545, rs6065906, rs4465830, and rs6073972. Additionally, as noted above, rs6073952 is in LD with tagSNPs rs6065904 (r2 = 0.81) and rs4810479 (r2 = 0.63). Rs378114 is in LD with untyped SNP rs435306; both are in LD with rs6073950 (r2 = 0.78 and 0.79, respectively). The P values for prediction of PLTP activity for the group of SNPs in complete LD with rs7679 were identical, P = 1.16E-06, compared with P = 3.39E-06 for rs6073952. This led us to genotype rs7679, most predictive of the SNPs (P = 5.39E-07; Table 1).
While all other SNPs in the results were genotyped, we investigated the imputed SNP rs22494213 in the discovery cohort based on prior reports associated with HDL level (26). This SNP was not in strong LD with genotyped SNPs. Thus, imputation was poor, with 87% accuracy estimated by doing the same imputation in HAPMAP2 data. Nonetheless, rs2294213 was associated with PLTP activity in the discovery cohort (P = 0.0052, b = 0.17), with its minor allele, G, associated with elevated PLTP as well as reduced LDL-B (P = 0.037) and a trend to LDL-C (P = 0.069). The imputed SNP predicts 4.3% of PLTP activity variance once age, BMI, and HbA1c are considered. This SNP was not associated with CAAD status; however, power for this test is limited by the poor imputation accuracy. It was not associated with other lipid phenotypes, including HDLC. Due to relatedness, it was not imputed in the replication cohort.
We considered a joint model to evaluate the independent effects of the genotyped SNPs that predicted ln-PLTP activity with age, BMI, and HbA1c as covariates to evaluate SNPs with independent effects. Three independent SNPs remained in the model to predict ln-PLTP activity. These included rs7679 (P < 0.0001) SNP, rs11569668 (P = 0.003), and rs1736493 (P = 0.021). This model predicted 50.1% of PLTP activity variance (Table 2). Note that this sample is enriched for the minor alleles of rs11569668 and rs1736493; thus, the variation is expected to overestimate that of the general population. The alleles associated with increased PLTP activity for these C/G SNPs are rs7679: C, rs11569668:G, and rs1736493:G.
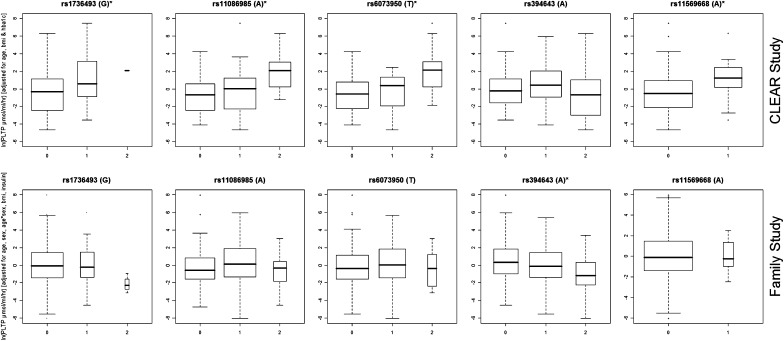
Analysis in the second, confirmatory cohort identified similar covariable effects. Sex, age, and sex*age interaction were all significant predictors of ln-PLTP activity at the 0.001 significance level, together accounting for 6.2% of the PLTP activity variance (Table 2). Addition of BMI was also significant, P = 0.001. HbA1c was not available in this cohort, but ln-insulin level additionally predicted ln-PLTP activity (P = 7.1E-05). Age, sex, age*sex interaction, BMI, and ln-insulin level accounted for 19.8% of ln-PLTP activity variance and were retained as covariates for the genotype effect tests. Covariate effects were similar in males versus females, except for a larger effect of BMI in females (Table 2). The prediction of PLTP activity by rs7679 (P = 3.62E-07), considering covariates, was significant, explaining another 8.2% of PLTP activity variance (Table 1). The effect of rs4810479, which is in an LD (r2 = 0.67) with rs7679, was more highly significant (8.51E-08, r2 = 0.207) than the effect of rs7679 in this replication cohort. The best joint model for PLTP activity prediction in the CLEAR discovery cohort was evaluated, considering covariates plus rs7679, rs11569668, rs1736493, and rs4810479; both rs7679 (P = 0.002) and rs11569668 (P = 0.001) effects on PLTP activity were again significant, while rs1736493 (P = 0.09) and rs4810479 (P = 0.36) effects were not significant. Figure 1 shows the PLTP activity effects of SNPs whose effects are supported in both cohorts and that the genotypic patterns of effects are very similar in the two cohorts. The effects of SNPs supported in only 1 cohort are shown in supplementary Figure A.
Fig. 1.
Box plots of adjusted PLTP activity effects of SNPs that were predictive of PLTP in both the unrelated (CLEAR) and replication (Family) cohorts. The x axis is genotype minor allele number and the y axis is the natural log of PLTP activity measured in μmol/ml/h and adjusted for age, BMI and HbA1c or insulin level. The box shows the 25 to 75 percentile, with a line at the median value. The range excluding outliers is marked by the “whiskers” and the diamonds mark the outliers.
Table 3 summarized the lipid effects for SNPs that did affect PLTP activity in that specific cohort. This table is limited to SNP with LD <0.5 to reduce redundant information. PLTP genotypes did not predict fasting HDL-cholesterol, LDL-C, triglyceride, LDL-B, or HbA1c levels at the P = 0.05 level in either the entire discovery cohort of 939 (not adjusting for medications) or in the 511 subjects not taking lipid lowering medications (adjusted for the PLTP activity covariates, no adjustment for multiple contrasts; data not shown). Only the SNPs in LD with rs7679 were predictive of ln-apoA1 at the 0.05 level (without adjustment for the multiple correlated contrasts), including rs6065904 (P = 0.006), rs6073952 (P = 0.007), rs7679 (P = 0.017), and rs4810479 (P = 0.031). In the replication cohort, these PLTP SNPs did not predict ApoA1 level (Table 3), except for rs6073952 (P = 0.02). Rs394643 predicted HDLC (P = 0.007) and, marginally, apoAI (P = 0.052). However, rs7679 predicted LDL-C (P = 3.69E-04) and LDL-B (P = 1.64E-03) levels, as did all other SNPs that predicted PLTP activity. In the replication cohort, in each case alleles that predicted elevated PLTP also predicted higher LDL-B and LDL-C. This is opposite the trend in the discovery cohort. Rs7679 did not predict lipoprotein(a) [Lp(a)] variation in either cohort. While other SNPs did, the direction of Lp(a) effects of given alleles were generally not consistent between cohorts (except for rs378114) or with the effect on PLTP activity. The PLTP SNPs were not predictive of insulin or HbA1c level (Table 3).
TABLE 3.
PLTP SNP effects on lipid traits
| PLTP_Activity |
LDL-C |
LDL-B |
lnLp(a) |
lnAPOAI |
lnHDLC |
lnTRIG |
Insulin/HbA1C |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Sample | β | P | β | P | β | P | β | P | β | P | β | P | β | P | β | P |
| rs7679 | 1 | −0.11 | 5.93E-07* | 0.17 | 9.26E-01 | 0.25 | 8.24E-01 | 0.07 | 4.34E-01 | 0.02 | 2.83E-02* | 0.02 | 3.42E-01 | 0.01 | 8.40E-01 | 0.02 | 7.85E-01 |
| 2 | −0.09 | 3.62E-07* | −12.63 | 3.69E-04* | −7.94 | 1.64E-03* | 0.05 | 7.70E-01 | 0.03 | 1.20E-01 | 0.03 | 2.80E-01 | 0.09 | 1.80E-01 | 0.02 | 7.21E-01 | |
| rs378114 | 1 | 0.10 | 5.29E-05* | −0.89 | 5.75E-01 | −0.04 | 9.70E-01 | −0.17 | 4.49E-02* | −0.01 | 3.22E-01 | 0.00 | 9.29E-01 | 0.03 | 2.91E-01 | −0.02 | 7.86E-01 |
| 2 | 0.04 | 7.22E-02 | 10.52 | 1.56E-02* | 5.31 | 8.39E-02 | −0.53 | 6.71E-03* | 0.01 | 7.89E-01 | 0.06 | 1.31E-01 | −0.19 | 1.66E-02* | −0.05 | 5.37E-01 | |
| rs553359 | 1 | −0.05 | 8.29E-03* | 0.88 | 5.52E-01 | 0.60 | 5.20E-01 | −0.17 | 3.04E-02* | 0.01 | 5.10E-01 | 0.01 | 4.10E-01 | 0.00 | 9.03E-01 | 0.02 | 7.67E-01 |
| 2 | −0.03 | 4.61E-02* | −7.47 | 1.83E-02* | −4.15 | 6.28E-02* | 0.10 | 4.70E-01 | 0.00 | 9.38E-01 | 0.01 | 6.36E-01 | 0.05 | 3.62E-01 | 0.02 | 7.16E-01 | |
| rs11086985 | 1 | 0.07 | 1.14E-03* | −1.54 | 3.26E-01 | −0.70 | 4.49E-01 | −0.17 | 2.75E-02* | −0.01 | 1.66E-01 | 0.00 | 7.65E-01 | 0.01 | 7.88E-01 | −0.03 | 5.41E-01 |
| 2 | 0.02 | 3.18E-01 | |||||||||||||||
| rs1736493 | 1 | 0.09 | 1.11E-02* | −0.14 | 9.63E-01 | 0.35 | 8.49E-01 | −0.18 | 2.28E-01 | −0.02 | 1.75E-01 | −0.03 | 2.02E-01 | −0.02 | 6.56E-01 | 0.04 | 6.63E-01 |
| 2 | −0.03 | 2.76E-01 | |||||||||||||||
| rs11569668 | 1 | 0.10 | 1.56E-02* | 2.45 | 6.07E-01 | 3.36 | 2.59E-01 | −0.26 | 3.05E-01 | 0.04 | 1.81E-01 | 0.06 | 1.99E-01 | 0.04 | 6.31E-01 | −0.16 | 3.53E-01 |
| 2 | 0.01 | 8.16E-01 | |||||||||||||||
| rs394643 | 1 | −0.02 | 2.85E-01 | ||||||||||||||
| 2 | −0.05 | 1.58E-03* | −5.78 | 7.04E-02 | −4.65 | 3.85E-02* | −0.35 | 1.39E-02* | 0.03 | 5.20E-02 | 0.07 | 7.02E-03* | −0.03 | 5.57E-01 | −0.01 | 8.19E-01 | |
Summary of the regression coefficient, β, and P value for the lipid-related measures considering age, sex, BMI, and insulin or HbA1C. The list of SNPs has been reduced to only SNPs that are not in strong LD with other listed SNPs. Lipid effects were only tested if the SNP was associated with PLTP activity in that sample. Covariates for insulin or HbA1C were age, sex, and BMI. Sample 1 is the full case control cohort (n = 939, expect for PLTP activity, where n = 87). Sample 2 is the confirmation cohort (n = 210).
DISCUSSION
In this study, a large proportion of PLTP activity variation can be accounted for by covariates and PLTP SNPs. Interestingly, the strongest covariate of PLTP activity found was HbA1c, which accounted for 13.8% of PLTP activity, above that of age in the older, male, discovery cohort. Plasma insulin level predicted 7.5% in the younger, mixed-gender confirmation cohort. A total of 50.6% of PLTP activity variance is explained by the covariates and multiple PLTP SNPs in the discovery cohort, while 28.9% is explained in the confirmation cohort.
PLTP has been implicated as a factor in the “metabolic syndrome.” Its activity has been correlated with insulin level, glucose, subcutaneous abdominal fat, and BMI (15, 39). PLTP levels are increased in both type 2 (NIDDM) and type 1 (IDDM) diabetes (40, 41). In healthy men, induced hyperglycemia lowered PLTP levels, and PLTP activity was negatively correlated with insulin sensitivity (42). We have previously noted a correlation with insulin in a smaller group of subjects with coronary artery disease (43). Our results of 7–11% of additional variance in PLTP activity accounted for by insulin or HbA1c levels, above that accounted for by age, sex, and BMI, further support a relationship with metabolic syndrome. Glucose has been shown to regulate PLTP expression (34). Thus, it seems unlikely that PLTP affects insulin level or HbA1c, particularly when no SNP effects on insulin or HbA1c have been identified.
These genetic data support the hypothesis that multiple independent PLTP SNPs account for a large proportion of PLTP activity variance. Our results suggested that LD with rs7679 may account for the associations of rs6073952, rs4810479, and rs6065904 with PLTP activity. Rs7679 itself is in complete LD with a group of SNPs in Caucasians, including rs17447545, rs6065906, rs4465830, and rs6073972. However, determination of which of these, if any, is the functional SNP cannot be made from these data. Three independent SNPs affecting PLTP activity are replicated in both samples, rs7679, rs553359, and rs378114, though the latter had a marginal test in the smaller replication cohort (P = 0.07). Other SNPs were associated in only one cohort, including rs11569668, which had significant effects in the CLEAR sample, where it was oversampled, and not in the family sample, where its rarity resulted in an underpowered test. This 5′ SNP is not in LD with other HapMap SNPs in Caucasians.
As none of the SNPs tested impact the amino acid sequence of PLTP or are found to be in LD with coding or splicing site SNPs, all may be expected to influence PLTP gene expression. Despite the poor correlation between mass and activity (17), the increased levels of RNA transcription likely influence the total amount of active PLTP. The regulation of PLTP expression is complex and may differ between liver and macrophages. The nuclear receptor family liver X regulates expression in both liver (44, 45) and macrophages (1). Fibrates are reported to downregulate PLTP expression, dependent on −322 to −299 regulatory regions (46). While the −230 to −72 region contains Sp1 and AP-2 transcription motifs (14), multiple initiation sites have been reported (14). Thus, there are multiple ways in which PLTP expression may be influenced by regulatory SNPs.
Our data are consistent with the recent report of rs7679 (25) being associated with PLTP expression in the liver, as this SNP accounted for the largest percentage of PLTP activity variance. However, we do not detect the reported triglyceride effects of rs7679 (25) in either cohort. Rs7679 was associated with apoAI levels in the discovery cohort only and with LDL-C and LDL-B in the replication cohort only. Using imputed data, we find that SNP rs2294213 predicts PLTP activity. We do not confirm effects of this SNP on HDLC (26) or other lipids; however, power for these tests is limited by the poor imputational accuracy.
The role of PLTP in human atherosclerosis is only beginning to be explored. Schgoer et al. (21) reported lower PLTP activity associated with peripheral artery disease, while de Vries et al. (22) found activity to be positively associated with carotid intima-media thickness. Lacking vascular disease status in the replication cohort, no firm conclusions can be made about the role of PLTP SNPs and vascular disease from this study. The results are suggestive in that each SNP found to be associated with CAAD had a significant PLTP activity effect in at least one of our cohorts. Additionally, there is an excess of CAAD-associated SNPs over random chance, even when LD is considered. Yet, rs7679, with stronger PLTP activity effects in these cohorts, was not predictive of CAAD.
The power to detect the effects on atherosclerosis of SNPs that predict atherosclerotic risk factors is limited by the complex genetic architecture of atherosclerosis. This can be seen in the tests of lipid risk SNPs, which best predict atherosclerotic heart disease in combination (47). Also limiting power is the fact that the largest impact alleles may be expected to be rare, requiring a sequencing approach.
The effects of PLTP SNPs on lipids were not consistent between the two cohorts, and neither cohort replicated the rs7679 association with triglycerides (25). The larger CLEAR cohort suggested possible effects of genotype on apoA1 level, while the family cohort suggested effects on LDL-C and LDL-B. These cohorts differ in age distribution and gender composition. The family cohort also was ascertained for familial combined hyperlipidemia, which is characterized by an excess of apoB. It is possible that the effects of PLTP SNPs on lipids are influenced by a variety of host factors.
We identify multiple PLTP region SNPs that predict PLTP activity and confirm that SNP rs2294213 predicts activity. These are likely regulatory effects. Additionally, we find large magnitude effects of HbA1c or insulin on PLTP activity that may reflect the role of metabolic syndrome in affecting PLTP activity levels. Even among SNPs with a marked PLTP activity effect, we do not find that genetic variation in PLTP has triglyceride effects in these two cohorts. PLTP SNP effects on lipids and CAAD require further study.
Supplementary Material
Acknowledgments
The authors thank the subjects for their participation and the following people for their technical assistance: Tamara Bacus, Loida Erhard, Martha Horike-Pyne, Julieann Marshall, Karen Nakayama, Jane Ranchalis, and Jeff Rodenbaugh.
Footnotes
Abbreviations:
- apo
- apolipoprotein
- BMI
- body mass index
- CAAD
- carotid artery disease
- HbA1c
- hemoglobin A1c
- HWE
- Hardy-Weinberg equilibrium
- Lp(a)
- lipoprotein(a)
- LD
- linkage disequilibrium
- LDL-C
- low density lipoprotein-cholesterol
- MAF
- minor allele frequency
- PLTP
- phospholipid transfer protein
- SNP
- single nucleotide polymorphism
- tagSNP
- tagging single nucleotide polymorphism
This work was funded by the National Institutes of Health (PO1 HL 030086) and the Veteran Affairs Epidemiology Research and Information Center Program (Award CSP 701S). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure and three tables.
REFERENCES
- 1.Laffitte B. A., Joseph S. B., Chen M., Castrillo A., Repa J., Wilpitz D., Mangelsdorf D., Tontonoz P. 2003. The phospholipid transfer protein gene is a liver X receptor target expressed by macrophages in atherosclerotic lesions. Mol. Cell. Biol. 23: 2182–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tall A. R., Krumholz S., Olivecrona T., Deckelbaum R. J. 1985. Plasma phospholipid transfer protein enhances transfer and exchange of phospholipids between very low density lipoproteins and high density lipoproteins during lipolysis. J. Lipid Res. 26: 842–851. [PubMed] [Google Scholar]
- 3.Tollefson J. H., Ravnik S., Albers J. J. 1988. Isolation and characterization of a phospholipid transfer protein (LTP-II) from human plasma. J. Lipid Res. 29: 1593–1602. [PubMed] [Google Scholar]
- 4.Jiang X. C., Bruce C., Mar J., Lin M., Ji Y., Francone O. L., Tall A. R. 1999. Targeted mutation of plasma phospholipid transfer protein gene markedly reduces high-density lipoprotein levels. J. Clin. Invest. 103: 907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lie J., de Crom R., van Gent T., van Haperen R., Scheek L., Sadeghi-Niaraki F., van Tol A. 2004. Elevation of plasma phospholipid transfer protein increases the risk of atherosclerosis despite lower apolipoprotein B-containing lipoproteins. J. Lipid Res. 45: 805–811. [DOI] [PubMed] [Google Scholar]
- 6.Moerland M., Samyn H., van Gent T., van Haperen R., Dallinga-Thie G., Grosveld F., van Tol A., de Crom R. 2008. Acute elevation of plasma PLTP activity strongly increases pre-existing atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 28: 1277–1282. [DOI] [PubMed] [Google Scholar]
- 7.Samyn H., Moerland M., van Gent T., van Haperen R., Grosveld F., van Tol A., de Crom R. 2009. Elevation of systemic PLTP, but not macrophage-PLTP, impairs macrophage reverse cholesterol transport in transgenic mice. Atherosclerosis. 204: 429–434. [DOI] [PubMed] [Google Scholar]
- 8.van Haperen R., Samyn H., van Gent T., Zonneveld A. J., Moerland M., Grosveld F., Jansen H., Dallinga-Thie G. M., van Tol A., de Crom R. 2009. Novel roles of hepatic lipase and phospholipid transfer protein in VLDL as well as HDL metabolism. Biochim Biophys Acta. 1791: 1031–1036. [DOI] [PubMed] [Google Scholar]
- 9.Liu R., Iqbal J., Yeang C., Wang D. Q., Hussain M. M., Jiang X. C. 2007. Phospholipid transfer protein-deficient mice absorb less cholesterol. Arterioscler. Thromb. Vasc. Biol. 27: 2014–2021. [DOI] [PubMed] [Google Scholar]
- 10.Vikstedt R., Ye D., Metso J., Hildebrand R. B., Van Berkel T. J., Ehnholm C., Jauhiainen M., Van Eck M. 2007. Macrophage phospholipid transfer protein contributes significantly to total plasma phospholipid transfer activity and its deficiency leads to diminished atherosclerotic lesion development. Arterioscler. Thromb. Vasc. Biol. 27: 578–586. [DOI] [PubMed] [Google Scholar]
- 11.Valenta D. T., Bulgrien J. J., Bonnet D. J., Curtiss L. K. 2008. Macrophage PLTP is atheroprotective in LDLr-deficient mice with systemic PLTP deficiency. J. Lipid Res. 49: 24–32. [DOI] [PubMed] [Google Scholar]
- 12.Tu A-Y., Nishida H. I., Nishida T. 1993. High density lipoprotein conversion mediated by human plasma phospholipid transfer protein. J. Biol. Chem. 268: 23098–23105. [PubMed] [Google Scholar]
- 13.Jauhiainen M., Metso J., Pahlman R., Blomqvist S., van Tol A., Ehnholm C. 1993. Human plasma phospholipid transfer protein causes high density lipoprotein conversion. J. Biol. Chem. 268: 4032–4036. [PubMed] [Google Scholar]
- 14.Albers J. J., Wolfbauer J. G., Cheung M. C., Day J. R., Ching A. F. T., Lok S., Tu A-Y. 1995. Functional expression of human and mouse plasma phospholipid transfer protein: effect of recombinant and plasma PLTP on HDL subspecies. Biochim. Biophys. Acta. 1258: 27–34. [DOI] [PubMed] [Google Scholar]
- 15.Murdoch S. J., Carr M. C., Hokanson J. E., Brunzell J. D., Albers J. J. 2000. PLTP activity in premenopausal women: relationship with lipoprotein lipase, HDL, LDL, body fat, and insulin resistance. J. Lipid Res. 41: 237–244. [PubMed] [Google Scholar]
- 16.Murdoch S. J., Carr M. C., Kennedy H., Brunzell J. D., Albers J. J. 2002. Selective and independent associations of phospholipid transfer protein and hepatic lipase with the LDL subfraction distribution. J. Lipid Res. 43: 1256–1263. [PubMed] [Google Scholar]
- 17.Cheung M. C., Wolfbauer G., Deguchi H., Fernandez J. A., Griffin J. H., Albers J. J. 2009. Human plasma phospholipid transfer protein specific activity is correlated with HDL size: implications for lipoprotein physiology. Biochim. Biophys. Acta. 1791: 206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlitt A., Bickel C., Thumma P., Blankenberg S., Rupprecht H. J., Meyer J., Jiang X. C. 2003. High plasma phospholipid transfer protein levels as a risk factor for coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 23: 1857–1862. [DOI] [PubMed] [Google Scholar]
- 19.Cavusoglu E., Marmur J. D., Chhabra S., Chopra V., Eng C., Jiang X. C. 2009. Relation of baseline plasma phospholipid transfer protein (PLTP) activity to left ventricular systolic dysfunction in patients referred for coronary angiography. Atherosclerosis. 207: 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlitt A., Blankenberg S., Bickel C., Lackner K. J., Heine G. H., Buerke M., Werdan K., Maegdefessel L., Raaz U., Rupprecht H. J., et al. 2009. PLTP activity is a risk factor for subsequent cardiovascular events in CAD patients under statin therapy: the AtheroGene study. J. Lipid Res. 50: 723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schgoer W., Mueller T., Jauhiainen M., Wehinger A., Gander R., Tancevski I., Salzmann K., Eller P., Ritsch A., Haltmayer M., et al. 2008. Low phospholipid transfer protein (PLTP) is a risk factor for peripheral atherosclerosis. Atherosclerosis. 196: 219–226. [DOI] [PubMed] [Google Scholar]
- 22.de Vries R., Dallinga-Thie G. M., Smit A. J., Wolffenbuttel B. H., van Tol A., Dullaart R. P. 2006. Elevated plasma phospholipid transfer protein activity is a determinant of carotid intima-media thickness in type 2 diabetes mellitus. Diabetologia. 49: 398–404. [DOI] [PubMed] [Google Scholar]
- 23.O'Brien K. D., Vuletic S., McDonald T. O., Wolfbauer G., Lewis K., Tu A. Y., Marcovina S., Wight T. N., Chait A., Albers J. J. 2003. Cell-associated and extracellular phospholipid transfer protein in human coronary atherosclerosis. Circulation. 108: 270–274. [DOI] [PubMed] [Google Scholar]
- 24.Desrumaux C. M., Mak P. A., Boisvert W. A., Masson D., Stupack D., Jauhiainen M., Ehnholm C., Curtiss L. K. 2003. Phospholipid transfer protein is present in human atherosclerotic lesions and is expressed by macrophages and foam cells. J. Lipid Res. 44: 1453–1461. [DOI] [PubMed] [Google Scholar]
- 25.Kathiresan S., Voight B. F., Purcell S., Musunuru K., Ardissino D., Mannucci P. M., Anand S., Engert J. C., Samani N. J., Schunkert H., et al. 2009. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 41: 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engler M. B., Pullinger C. R., Malloy M. J., Natanzon Y., Kulkarni M. V., Song J., Eng C., Huuskonen J., Rivera C., Poon A., et al. 2008. Genetic variation in phospholipid transfer protein modulates lipoprotein profiles in hyperalphalipoproteinemia. Metabolism. 57: 1719–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crawford D. C., Nord A. S., Badzioch M. D., Ranchalis J., McKinstry L. A., Ahearn M., Bertucci C., Shephard C., Wong M., Rieder M. J., et al. 2008. A common VLDLR polymorphism interacts with APOE genotype in the prediction of carotid artery disease risk. J. Lipid Res. 49: 588–596. [DOI] [PubMed] [Google Scholar]
- 28.Pritchard J. K., Stephens M., Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics. 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Badzioch M. D., Igo R. P., Jr., Gagnon F., Brunzell J. D., Krauss R. M., Motulsky A. G., Wijsman E. M., Jarvik G. P. 2004. Low-density lipoprotein particle size loci in familial combined hyperlipidemia: evidence for multiple loci from a genome scan. Arterioscler. Thromb. Vasc. Biol. 24: 1942–1950. [DOI] [PubMed] [Google Scholar]
- 30.Miller S. A., Dykes D. D., Polesky H. F. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16: 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tishkoff S. A., Dietzsch E., Speed W., Pakstis A. J., Kidd J. R., Cheung K., Bonne-Tamir B., Santachiara-Benerecetti A. S., Moral P., Krings M. 1996a. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science. 271: 1380–1387. [DOI] [PubMed] [Google Scholar]
- 32.Warnick G. R., Benderson J., Albers J. J. 1982. Dextran sulfate-Mg2+ precipitation procedure for quantitation of high-density-lipoprotein cholesterol. Clin. Chem. 28: 1379–1388. [PubMed] [Google Scholar]
- 33.Jarvik G. P., Rozek L. S., Brophy V. H., Hatsukami T. S., Richter R. J., Schellenberg G. D., Furlong C. E. 2000. Paraoxonase (PON1) phenotype is a better predictor of vascular disease than is PON1(192) or PON1(55) genotype. Arterioscler. Thromb. Vasc. Biol. 20: 2441–2447. [DOI] [PubMed] [Google Scholar]
- 34.Tu A. Y., Albers J. J. 2001. Glucose regulates the transcription of human genes relevant to HDL metabolism: responsive elements for peroxisome proliferator-activated receptor are involved in the regulation of phospholipid transfer protein. Diabetes. 50: 1851–1856. [DOI] [PubMed] [Google Scholar]
- 35.Durbin J., Watson G. S. 1950. Testing for serial correlation in least squares regression. I. Biometrika. 37: 409–428. [PubMed] [Google Scholar]
- 36.Tregouet D. A., Ducimetiere P., Tiret L. 1997. Testing association between candidate-gene markers and phenotype in related individuals, by use of estimating equations. Am. J. Hum. Genet. 61: 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Servin B., Stephens M. 2007. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 3: e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guan Y., Stephens M. 2008. Practical issues in imputation-based association mapping. PLoS Genet. 4: e1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dullaart R. P. F., Sluiter W. J., Dikkeschei L. D., Hoogenberg K., van Tol A. 1994. Effect of adiposity on plasma lipid transfer protein activities: a possible link between insulin resistance and high density lipoprotein metabolism. Eur. J. Clin. Invest. 24: 188–194. [DOI] [PubMed] [Google Scholar]
- 40.Riemens S., van Tol A., Sluiter W., Dullaart R. 1998. Elevated plasma cholesteryl ester transfer in NIDDM: relationships with apolipoprotein B-containing lipoproteins and phospholipid transfer protein. Atherosclerosis. 140: 71–79. [DOI] [PubMed] [Google Scholar]
- 41.Colhoun H. M., Taskinen M. R., Otvos J. D., Van Den Berg P., O'Conner J., van Tol A. 2002. Relationship of phospholipid transfer protein activity to HDL and apolipoprotein B-containing lipoproteins in subjects with and without type 1 diabetes. Diabetes. 51: 3300–3305. [DOI] [PubMed] [Google Scholar]
- 42.van Tol A., Ligtenberg J. J. M., Riemens S. C., Van Haeften T. W., Reitsma W. D., Dullaart R. P. F. 1997. Lowering of plasma phospholipid transfer protein activity by acute hyperglycaemia-induced hyperinsulinaemia in healthy men. Scand. J. Clin. Lab. Invest. 57: 147–158. [DOI] [PubMed] [Google Scholar]
- 43.Cheung M. C., Brown B. G., Marino Larsen E. K., Frutkin A. D., O'Brien K. D., Albers J. J. 2006. Phospholipid transfer protein activity is associated with inflammatory markers in patients with cardiovascular disease. Biochim. Biophys. Acta. 1762: 131–137. [DOI] [PubMed] [Google Scholar]
- 44.Urizar N. L., Dowhan D. H., Moore D. D. 2000. The farnesoid X-activated receptor mediates bile acid activation of phospholipid transfer protein gene expression. J. Biol. Chem. 275: 39313–39317. [DOI] [PubMed] [Google Scholar]
- 45.Cao G., Beyer T. P., Yang X. P., Schmidt R. J., Zhang Y., Bensch W. R., Kauffman R. F., Gao H., Ryan T. P., Liang Y., et al. 2002. Phospholipid transfer protein is regulated by liver X receptors in vivo. J. Biol. Chem. 277: 39561–39565. [DOI] [PubMed] [Google Scholar]
- 46.Tu A. Y., Albers J. J. 1999. DNA sequences responsible for reduced promoter activity of human phospholipid transfer protein by fibrate. Biochem. Biophys. Res. Commun. 264: 802–807. [DOI] [PubMed] [Google Scholar]
- 47.Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N. P., Rieder M. J., Cooper G. M., Roos C., Voight B. F., Havulinna A. S., et al. 2008. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 40: 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.