Abstract
Although genetic studies have been critically important for the identification of therapeutic targets in Mendelian disorders, genetic approaches aiming to identify targets for common, complex diseases have traditionally had much more limited success. However, during the past year, a novel genetic approach — genome-wide association (GWA) — has demonstrated its potential to identify common genetic variants associated with complex diseases such as diabetes, inflammatory bowel disease and cancer. Here, we highlight some of these recent successes, and discuss the potential for GWA studies to identify novel therapeutic targets and genetic biomarkers that will be useful for drug discovery, patient selection and stratification in common diseases.
Genetic factors are known to have an important role in many common diseases, and the identification of genetic determinants for such diseases has the potential to provide insights into disease pathogenesis, revealing novel therapeutic targets or strategies. Genetic factors could also provide useful biomarkers for diagnosis, patient stratification and prognostic or therapeutic categorization. In addition, given that inherited genetic factors are present at birth, knowledge of these factors could facilitate timely preventative or ameliorative interventions.
During the past 25 years, genetic linkage-based studies have proved very effective in identifying causal genetic factors in Mendelian (single gene) disorders; causal genes for more than 1,300 dominant and recessive Mendelian diseases have been identified1. Most common diseases and endophenotypes, however, do not exhibit Mendelian inheritance, but rather feature complex, multifactorial expression and inheritance. Although linkage-based methods have been broadly applied, these studies have had little success in identifying the allelic determinants of common disorders2. In particular, there has been poor replication among studies, whereby an initial study identifies an allele (genotype) with large estimated genetic effects (relative risk) but subsequent studies fail to corroborate the results3,4. In part, this reflects the dependence of linkage-based studies on unusually informative families (with multiple affected and unaffected individuals), which induce a bias toward rare, semi-Mendelian disease subsets in subpopulations. Reports of successful identification of genetic variants in common diseases using an approach that circumvents this limitation — genome-wide association (GWA) studies — have therefore generated considerable excitement.
Human GWA studies are based on three hypotheses: First, the common trait/common variant hypothesis proposes that the genetic architecture of complex traits consists of a limited number of common alleles, each conferring a small increase in risk to the individual5,6; second, the brief history of most human populations precludes sufficient generations (or meioses) to create recombination events (or mutations) between closely located, common (ancient) variants; and, third, suppression of meiotic recombination (coldspots) occurs very frequently. Thus, approximately 80% of the human genome is comprised of around 10 kb regions that exhibit reduced recombination in human populations (haplotypes)7. Genetic variants (alleles) within haplotypes are in linkage disequilibrium (LD). This phenomenon enables much of the recombination history in a population to be ascertained by genotyping a large set of well-spaced, common (ancient) variants throughout the genome, especially if variant selection is informed by knowledge of haplotypes. During the last 10 years, more than 10 million single nucleotide polymorphisms (SNPs) have been identified8. Furthermore, the International HapMap project has genotyped approximately 4 million common SNPs (occurring with a minor-allele frequency of more than 5%) in human populations and has assembled these genotypes computationally into a genome-wide map of SNP-tagged haplotypes7. These resources, together with array technologies for massively parallel SNP genotyping and the well-established epidemiological case-control association studies have rendered GWA feasible (BOX 1, FIG. 1).
Box 1. Useful resources and databases for genetic-based studies.
Genetic Association Database: An archive of human genetic association studies of complex diseases. http://geneticassociationdb.nih.gov/
Schizophrenia Gene Database: An archive of genetic association studies performed on schizophrenia phenotypes. http://www.schizophreniaforum.org/res/sczgene/default.asp
Online Mendelian Inheritance in Man: A catalogue of human genes and genetic disorders. http://www.ncbi.nlm.nih.gov/sites/entrez?db=OMIM&itool=toolbar
Human Gene Mutation Database. A catalogue of published gene lesions responsible for human inherited disease. http://www.hgmd.cf.ac.uk/ac/index.php
Human Genome Variation Database: A catalogue of normal human gene and genome variation. http://www.hgvbase.org/
dbSNP: A catalogue of human single nucleotide polymorphisms. http://www.ncbi.nlm.nih.gov/projects/SNP/
GeneSNPs: A database of polymorphisms in human genes that are thought to have a role in susceptibility to environmental exposure. http://www.genome.utah.edu/genesnps/
PharmGKB: A database of pharmacogenomics research. http://www.pharmgkb.org/index.jsp
GeneCards: A database of human genes that includes genomic, proteomic and transcriptomic information, as well as orthologies, disease relationships, SNPs, gene expression and gene function. http://www.genecards.org/
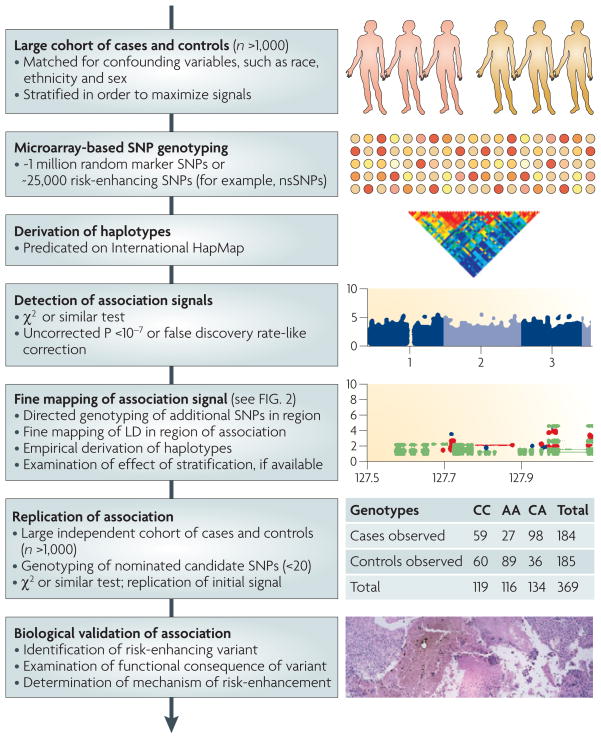
Figure 1. Overview of the general design and workflow of a genome-wide association (GWA) study.
The discovery phase entails genotyping many case and control DNA samples and evaluation for significant associations. The replication phase involves fine mapping of association signals and independent confirmation in a second cohort. Biological validation is important for translation of GWA findings into diagnostic or therapeutic discoveries.
Initial genetic association studies focused on candidate loci and exhibited a lack of replication among studies9,10. There were biological explanations for inconsistent results: unobserved, confounding biological sources of heterogeneity, including inconsistent or poorly defined measurements of the phenotype, heterogeneous genetic sources for the phenotype (genocopies), population stratification (ethnic ancestry), population-specific LD, heterogeneous genetic and epigenetic backgrounds or heterogeneous environmental influences (phenocopies). In addition, there were statistical reasons for irreproducibility, including failure to control the rate of false discoveries, model misspecification and heterogeneous bias in estimated effects among studies11–14. Also, a frequent source of non-replication was lack of power due to the limited number of individuals genotyped and phenotyped15,16.
In order to ameliorate poor replication, GWA experiments employ multi-tiered experimental designs with discovery, replication and biological validation stages17 (FIG. 1). Tiered designs are critical for cost-effective detection of meaningful, hypothesis-generating, genotype–phenotype associations given the large number of comparisons involved, prior probability estimates of association, sample sizes, resampling procedures and statistical significance thresholds. GWA studies also owe their statistical power to their large cohort size and high rate of SNP detection. Currently, a respected threshold for uncorrected, significant associations is P <5 × 10−7 (REFS 18,19). Alleles with moderately less significant associations, however, are often also reported, as they might indicate loci that reach the aforementioned threshold in subsequent studies.
Results of initial GWA studies
The first GWA study, published in 2002, evaluated acute myocardial infarction (AMI)20. The discovery, or nomination, phase comprised the examination of genotype–phenotype association signals in 65,671 coding domain SNPs (cSNPs) in 752 cases and controls (TABLE 1). Although subsequent studies have used up to 20 times this number of non-coding SNPs, gene-tagging SNPs are more informative, as the majority of true-positive associations are expected to be with genes1. Even more informative are screens that employ functional cSNPs, such as nonsynonymous SNPs (nsSNPs), that are candidate, causal (risk-enhancing) gene alleles1,21–28. The replication, or confirmatory, phase examined associations of 26 SNPs in 2,137 individuals and confirmed association of AMI with a 50 kb region containing lymphotoxin-α (LTA), nuclear factor of kappa light polypeptide gene enhancer in B cells (also known as RELA), nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor-like 1 (NFKBIL1) and human leukocyte antigen (HLA)-B associated transcript 1 (BAT1) genes. Additional replication studies have been undertaken, some of which have confirmed an association of this region with AMI-related phenotypes and, in particular, one nsSNP in LTA29–35. The association of LTA with AMI was an unexpected finding, suggesting a novel therapeutic target.
Table 1.
Discovery and replication designs of recent GWA studies
| Disease | Discovery Phase | Replication Phase | Refs | ||||
|---|---|---|---|---|---|---|---|
| Number of individuals examined | Number of SNPs | Population | Number of individuals examined | Number of SNPs validated/tested | Population | ||
| AMD | 146 | 105,980 | Caucasian | 96 | 2/50 | Same | 36 |
| Asthma | 2,642 | 307,328 | UK/German | 2,320 | 0/9 | German | 92 |
| Atrial fibrillation | 5,026 | 316,515 | Icelandic | 17,810 | 2/18 | Icelandic/European | 94 |
| Bipolar disorder | 1,024 (pooled) | 555,235 | Western European | 1,648 | 1/37 | Same | 91 |
| Breast cancer | 754 | 227,876 | European | 45,426 | 7/30 | Same | 85 |
| 2,287 | 528,173 | European | 3,848 | 1/8 | Same | 19 | |
| 13,163 | 311,524 | Icelandic | 7,968 | 2/9 | Various | 86 | |
| Celiac disease | 2,200 | 310,605 | UK | 2,480 | 5/27 | Dutch/Irish | 97 |
| Colorectal cancer | 1,890 | 547,647 | Caucasian | 23,121 | 2/18 | Same | 111 |
| 2,593 | 99,632 | Canadian | 23,325 | 2/1,143 | Same | 112 | |
| Crohn’s disease | 1,923 | 304,413 | European | 2,150 | 4/37 | Same | 45 |
| 1,103 | 16,360 nsSNP | German | 2,670 | 3/72 | Same | 24 | |
| 1,475 | 302,451 | Belgian | 2,236 | 7/10 | Same | 44 | |
| IBD | 1,095 + 834 | 308,332 | European | 2,885 | 10/27 | Same | 43 |
| LOAD | 1,086 | 502,627 | Caucasian | ND | ND | ND | 90 |
| Lung cancer | 673 | 116,204 | Italian | 621 | 0/1 | Caucasian/Norwegian | 98 |
| Memory | 341 (pooled) | 502,627 | Swiss | 680 | ½ | Several | 87 |
| AMI | 752 | 65,671 cSNPs | Japanese | 2,137 | 4/26 | Same | 20 |
| Nicotine dependence | 548 (pooled) | 2,427,357 | European | 1,929 | 0/31,960 | European | 93 |
| Obesity | 4,862 | 490,032 | British/Irish | 29,596 | 1/1 | Same | 71 |
| Prolonged QT interval | 3,966 | 88,500 | German | 4,451 | 1/7 | European | 96 |
| Prostate cancer | 4,517 | 316,515 & 243,957 haplotypes | Icelandic | 3,655 | 2/2 | Several | 53 |
| 12,791 | 310,520 | Icelandic | 5,050 | 2/5 | European | 78 | |
| 2,339 | 550,000 | European | 6,266 | 2/2 | Several | 79 | |
| RLS | 2,045 | 236,758 | European | 2,336 | 9/13 | Same | 101 |
| 15,970 | 306,937 | Icelandic | 2,206 | 1/70 | Icelandic/US | 100 | |
| T2DM | 2,335 | 315,635 | Finnish | 2,473 | 10/80 | Same | 55 |
| 4,900 | 393,453 | European | 9,103 | 10/77 | Same | 63 | |
| 7,805 | 313,179 & 339,846 haplotypes | Icelandic/Danish | 3,382 | 2/47 | Same | 57 | |
| 1,316 | 392,935 | French | 5,511 | 8/57 | Same | 56 | |
| T2DM and Triglyceride levels | 2,931 | 386,731 & 284,968 haplotypes | Finnish/Swedish | 10,850 | 3/107 | Several | 54 |
| T1DM | 3,388 | 6,500 nsSNPs | European | 12,229 | 1/1 | Same | 28 |
| Bipolar disorder, Crohn’s disease, T2DM, T1DM, HT, RA, CAD* | 4,868 | 392,575 | UK | ND | ND | ND | 18 |
AMI, acute myocardial infarction; AMD, age-related macular degeneration; CAD, coronary artery disease; HT, hypertension; IBD, inflammatory bowel disease; LOAD, late-onset Alzheimer’s disease; ND, not determined; nsSNP, non-synonymous SNP; RLS, restless leg syndrome; RA, rheumatoid arthritis; SNP, single nucleotide polymorphism; T1DM, type 1 diabetes mellitus; T2DM, type 2 diabetes mellitus.
A second, pioneering GWA study examined age-related macular degeneration (AMD)36 (TABLE 1). The discovery phase sought associations of 105,980 SNPs with AMD in 96 cases and 50 control individuals. Despite the small cohort size, SNPs in the complement factor H (CFH) gene, including an nsSNP, showed significant association with AMD. Replication was not performed, but subsequent studies have replicated associations of CFH alleles with AMD37–40. Of all common diseases examined by GWA to date, AMD is unique in that a single haplotype explains 61% of the genetic variance, conferring a homozygous odds ratio of 7.4. To put this in perspective, this is of a similar magnitude to the classic associations of HLA-B27 with anterior uveitis/ankylosing spondylitis and HLA alleles with type 1 diabetes mellitus (T1DM). Complement pathway dysregulation was a novel, unexpected association with AMD. Subsequent studies have shown an association of AMD with two additional members of the alternative complement pathway (factor B (CFB) and C3)41,42. These findings, together with biological validation studies, have led to the initial development of new AMD therapies, based upon complement inhibition.
In the past year, technical challenges associated with GWA were largely overcome, genotyping costs were decreased and a significant number of studies have used SNP genotyping arrays in larger population groups to produce replicated associations between individual SNP alleles and common diseases.
Inflammatory bowel disease
Five large GWA studies have examined Crohn’s disease and ulcerative colitis, two histologically distinct types of inflammatory bowel disease (IBD) (TABLE 1). Four of the studies used micro-arrays featuring between 300,000 and 400,000 SNPs18,43–45, whereas the fifth study genotyped approximately 16,000 nsSNPs24. Two follow-up studies sought to replicate the most significant signals from the Wellcome Trust case control consortium (WTCCC) study18, one in a European population and another in a Japanese population46,47. The European study replicated significant signals of the WTCCC study, but some of the alleles failed to reach significance in the Japanese study and others were not detected. The failure to replicate signals in different studies might reflect true differences between populations, differences in phenotype ascertainment or a lack of power.
Considering the six studies of European populations, there was significant replication of specific allele associations with Crohn’s disease (TABLES 1,2). Three associations were concordant in four out of five studies (representing the genes caspase recruitment domain 15 protein (CARD15, also known as NOD2), interleukin 23 receptor (IIL23R) and ATG16 autophagy related 16-like 1 (ATG16L1)). Of note, CARD15 had previously been identified as a susceptibility gene by linkage-based approaches48,49. One gene, prostaglandin E receptor 4 (PTGER4) showed association in two out of five studies. In addition, several disease-associated intergenic segments have been replicated. IBD susceptibility genes that have been identified to date appear to coalesce into biological networks involving innate immunity, autophagy and phagocytosis50. In addition, alleles of two genes associated with Crohn’s disease (IL23R and PTPN2) have shown association with other autoimmune disorders21,51, suggesting the existence of autoimmune susceptibility ‘supergenes’. There is great interest in alleles that exhibit pleiotropic associations, as they potentially represent blockbuster targets that cross-over therapeutic categories (TABLE 2).
Table 2.
Loci and variants associated with multiple diseases in GWA studies
| Locus | Variant (rs) | Disease | Refs |
|---|---|---|---|
| PTPN22 | 6679677 | RA | 18 |
| T1DM | 18 | ||
| IL2RA | 2104286 | RA | 18 |
| T1DM | 18 | ||
| PTPN2 | 2542151 | T1DM | 51 |
| CD | 18, 46 | ||
| TCF2 | 4430796 | T2DM | 53 |
| PC | 53 | ||
| FTO | 9939609 | T2DM | 18 |
| Obesity | 71 | ||
| APOE | 4420638 | Triglyceride level | 54 |
| Alzheimer’s disease | 90 | ||
| 8q24 | 6983267 | PC | 79, 81, 113 |
| CC | 111–113 | ||
| IL23R | 11209026 | CD | 43 |
| Psoriasis | 21 |
Above variants are associated P <5 × 10−7. CC, colorectal cancer; CD, Crohn’s disease; PC, prostate cancer; RA, rheumatoid arthritis; T1DM, type 1 diabetes mellitus; T2DM, type 2 diabetes mellitus.
In common with most GWA studies to date, estimated genetic effects (relative risks) of IBD-associated loci are small18,46. However, as many of these variants were common, the population attributable risk — an estimate of the percentage of cases of disease that would be avoided if the allele(s) were absent — was substantial. Of several studies that looked for epistatic interactions between IBD association signals, two found suggestive evidence of epistasis involving two different pairs of genes24,52.
Diabetes mellitus
A good example of the capabilities and limitations of GWA studies is type 2 diabetes mellitus18,53–57 (T2DM; TABLE 1). Two studies examined association both with SNPs and haplotypes in the discovery phase54,57. Haplotype-based analysis can be more powerful than marker-by-marker analysis in association studies22,58–61. For example, haplotypes can correlate a specific phenotype with a specific gene in a small population sample even when individual SNPs cannot62. Case-control and family-based association studies were employed in several studies of T2DM.
The replication phases of these studies were impressive; two of them included over 9,000 replication individuals53,54. One study sought to replicate signals identified by the WTCCC study18 by genotyping the most significant SNPs; 9 of 77 candidate SNPs reached a P <5 × 10−7 significance level63. The eight genes represented by these SNPs were replicated in at least one other independent study (TABLES 1–3). The concordance of T2DM-associated genes between GWA studies is striking: of 10 novel associations, only two were unique to a single study.
Table 3.
Loci and variants exhibiting association with type 2 diabetes mellitus in GWA studies
| T2DM phenotype | Locus | Variants (rs) | Refs |
|---|---|---|---|
| Susceptibility | TCF2 | 4430796, 7501939 | 78 |
| TCF7L2 | 4506565, 7903146, 7901695 | 18,54–57,63 | |
| PPARG | 1801282 | 55,63 | |
| KCNJ11 | 5219, 5212 | 54,55,63 | |
| SLC30A8 | 13266634, rs118253964 | 55,56,63 | |
| HHEX | 1111875 | 55,63 | |
| IGF2BP2 | 4402960 | 54,55,63 | |
| CDKAL1 | 9456871, 7754840, 10946398, 7756992 | 18,55,57,63 | |
| CDKN2A/B | 10811661, 564398 | 54,55,63 | |
| Chromosome 11, intergenic | 9300039 | 55 | |
| FTO | 9939609, 7193144, 8050136 | 18,55,63 | |
| Low-density lipoprotein | APOE | 4420638 | 54 |
| High-density lipoprotein | CETP | 1800775 | 54 |
| High-triglyceride level | LPL | 17482753 | 54 |
| GCKR | 780094 | 54 |
Above variants are associated P <5 × 10−7. T2DM, type 2 diabetes mellitus.
Reassuringly, some of the genes identified by GWA in studies of TD2M have previously been associated with the disease in other types of genetic studies. For example, transcription factor 7-like 2 (TCF7L2) had previously shown linkage to T2DM in the Icelandic population, and significant association in a candidate gene association study64. Heterozygous and homozygous carriers of TCF7L2 risk alleles had relative risks of 1.45 and 2.41, respectively. TCF7L2 is a transcription factor that regulates the pro-glucagon gene in entero-endocrine cells65. TCF7L2 alleles have also shown associations with endophenotypes such as a lower likelihood of response to the oral hypoglycaemic drug sulphonylurea66 and increased risk of progression to T2DM among persons with impaired glucose tolerance67.
In common with IBD, T2DM associations exhibited small estimated-effect sizes. Some of the candidate genes from GWA studies were consistent with biological processes that have previously been implicated in the pathogenesis of T2DM, such as pancreatic islet beta-cell function and insulin biosynthesis. However, these studies also suggested new components of these processes, such as zinc transport and Wnt-signalling56,64,68. Validation of T2DM candidate genes as therapeutic targets will require additional studies to identify causal susceptibility alleles and to determine their precise effect on cell biology.
Three studies performed initial modelling of how loci combine to affect susceptibility to T2DM56,63,69. One study found evidence of epistatic interactions between two genes. Otherwise, T2DM appeared to fit a polygenic threshold model with additive/multiplicative effects of individual loci. However, until the causal alleles that underpin these association signals have been found, it is not possible to make categorical statements about the allelic architecture of T2DM.
Frequencies of T2DM associated alleles showed considerable variation between ethnic and racial groups. Despite these differences, however, T2DM-associated risk alleles were conserved between independent populations, implying an ancient origin of these polymorphisms70.
Expansion of an initial association of an allele with a categorical trait (such as the presence of a disease) with quantitative component phenotypes (endophenotypes) is an approach pioneered with apolipoprotein E (APOE) alleles in Alzheimer’s disease. It appears to be highly instructive in elucidating the mechanism of action of alleles in disease pathogenesis. One T2DM GWA extended its analysis to a quantitative endophenotype: T2DM-related obesity (measured by body-mass index (BMI); TABLE 1)54,71. Alleles associated with T2DM in the fat-mass and obesity-associated gene (FTO)18,55,63 also showed an association with BMI (TABLES 2,3). Association of FTO with obesity has since been confirmed72.
Two GWA studies examined T1DM. One examined 6,500 nsSNPs28 and the other evaluated 392,575 SNPs18. Four T1DM, susceptibility loci had previously been identified by linkage-based methods (class II MHC alleles, CTLA4, PTPN22 and insulin). GWA studies replicated the association with PTPN22 and identified several novel loci, including C12orf30, KIAA0350 (also known as CLEC16A) and IFIH1 (each replicated in two studies). Twenty-one T1DM candidate genes that have previously shown linkage or association are currently undergoing replication studies73.
T1DM, like rheumatoid arthritis and IBD, is an autoimmune disorder. Medical practitioners have long noted familial aggregation of autoimmune diseases. One study showed association of both rheumatoid arthritis and T1DM with specific polymorphisms (IL2RA-rs2104286 and PTPN22-rs6679677; TABLE 2)18. T1DM, rheumatoid arthritis and IBD also show association with MHC alleles74–76. These findings suggest common underlying aetiological pathways (and therapeutic targets) for several, common autoimmune disorders77.
Cancer
GWA studies of cancer based on common, inherited SNPs are useful for the identification of germ-line risk alleles, but not somatic mutations. Three GWA studies sought inherited association signals in prostate cancer53,78,79; FIG. 2 shows details of the discovery phase of one of these studies. An association signal at chromosome 8q24 that had previously been identified by linkage analysis80 was replicated in two GWA studies53,79. In addition, these studies identified a second 8q24 association, approximately 300 kb upstream from the first. As yet, the functional basis of these associations is unclear. Although individual 8q24 alleles showed modest estimated genetic effects, the cumulative effect of several loci fit a multiplicative model that conferred a population-attributable risk (PAR), that is, an expected reduction in prostate-cancer incidence if the risk alleles did not exist in the population, of up to 68%81. As noted above, PAR values are strongly affected by allele frequency and represent only an approximate measure of the contribution of those alleles to disease incidence.
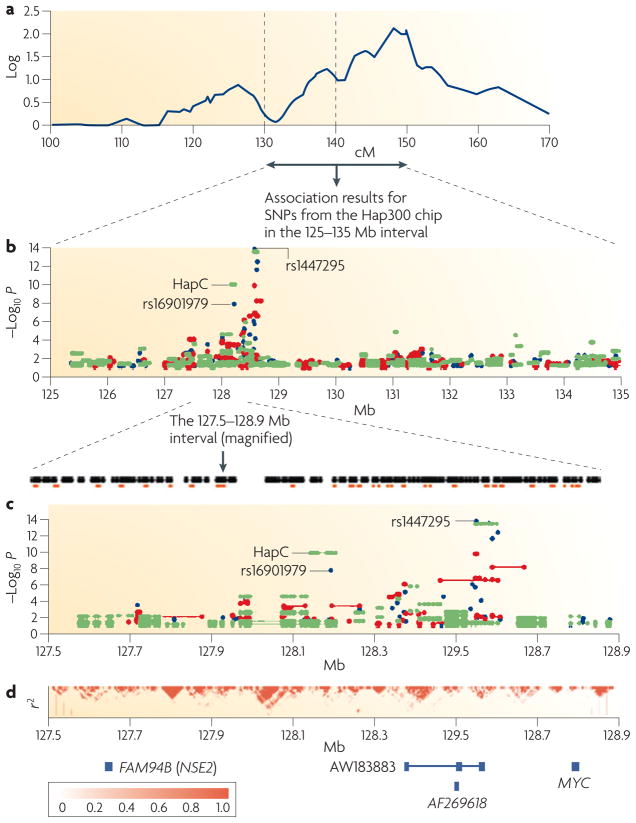
Figure 2. Schematic view of genetic linkage, GWA results, fine mapping and linkage disequilibrium structure in a region of chromosome 8q24.21 that demonstrates an association of rs1447295 and rs16901979 with prostate cancer.
a | Previously reported genetic linkage scan results for chromosome 8, centiMorgans (cM) 100–170 (that is, 8q) from 871 Icelandic individuals with prostate cancer in 323 extended families. A quantitative trait locus (QTL) for prostate cancer susceptibility with log of the odds (lod) score of ~2 is shown. The interval between the two dashed horizontal lines corresponds to a previously reported admixture signal that is associated with prostate cancer. b | Genome-wide association (GWA) results for 1,660 single nucleotide polymorphisms (SNPs) mapping to chromosome 8 Mb 125–135 in 1,453 Icelandic individuals with prostate cancer and 3,064 controls. Association testing P values smaller than 0.1, corrected for relatedness and population stratification, are shown for single SNPs (blue circles), two SNPs (red circles) and linkage disequilibrium (LD)-block haplotypes (green circles). Four SNPs (including rs1447295) and three haplotype blocks (including Hap C, defined by 14 SNPs) show significant association signals (P <1.58 × 10−7). Single SNP association two-sided P values were derived using Fisher’s exact test and were unadjusted for multiple comparisons. Association testing of haplotype block P values were carried out using the expectation-maximization (EM) algorithm directly for the observed data. c | Association results from b, shown in greater detail, for a 1.4 Mb interval on 8q24.21. Filled black circles represent 225 SNPs and the orange boxes represent recombination hotspots (calculated from the HapMap using the likelihood ratio test). d | LD between SNPs, measured by the square of the correlation coefficient calculated for each pairwise comparison of SNPs (r2) from the Centre d’Etude du Polymorphisme Humain from Utah (CEU) HapMap population for the 225 SNPs in c; the blue boxes at the bottom indicate the location of the FAM84B, AF268618 and MYC genes and the AW183883 expressed sequence tag. Figure modified, with permission, from Nature Genetics REF. 53 © 2007 Macmillan Publishers Ltd.
One study of prostate cancer78 identified a TCF2 (also known as HNF1B) susceptibility allele. Intriguingly, this allele appeared to diminish the risk of T2DM (TABLE 2), possibly representing antagonistic pleiotropy. This is supported by epidemiological evidence which suggests that diabetic men have a slightly lower prostate cancer risk than non-diabetic men82.
Another allele exhibiting association in two diseases is rs6983267 at chromosome 8q24, which has shown replicated associations with prostate and colorectal cancer79,82–84 (TABLE 2).
Three GWA studies sought inherited associations with breast cancer19,85,86. Although each study identified significant novel loci, two genes and one allele were each supported in two studies.
Complex traits
In addition to common diseases, GWA studies are applicable to complex traits. One study undertook GWA with numerous quantitative and categorical memory-associated endophenotypes87. Despite a small discovery cohort (341 individuals), associations with the KIBRA (also known as WWC1) gene have been replicated87–89. A notable innovation in this study was that associations were sought with multi-scale and multi-modality endophenotypes; that is, performance in seven memory-associated tests and functional magnetic resonance image-based measures of the hippocampus during three memory-associated tests. This study provides evidence that progress can be made in the elucidation of the genetic determinants of subjective, qualitative neurologic traits by using objective, quantitative, surrogate endophenotypes.
As well as identifying novel associations, GWA studies have confirmed several susceptibility genes that were previously established by linkage analysis in large pedigrees. For example, a GWA study of late-onset Alzheimer’s disease (LOAD) identified the well-established APOE-susceptibility allele90. This association was also replicated in a study that genotyped 17,343 putative functional cSNPs23.
A remaining problem with large GWA studies is the cost of genotyping, but one study provided evidence that sample pooling strategies might help to overcome this issue. In a GWA study of bipolar disorder, investigators created 39 pools, containing DNA from 2,672 individuals91. These pools were used for both discovery and replication experiments. Pools were individually genotyped for 555,235 SNPs and normalized allele frequencies were inferred from intensity data. Replicates were assayed for each pool. Thirty-seven SNPs showing allele frequency differences in both cohorts were individually genotyped and one SNP retained a significant association. The aforementioned WTCCC study also studied bipolar disorder, identifying an association at 16p12 (REF. 18). One locus, for glutamate receptor, metabotropic 7 (GRM7), showed association in both studies.
The rate of publication of GWA studies continues to increase. Recent studies have investigated asthma92, nicotine dependence93, coronary artery disease19,26, atrial fibrillation94, prolonged QT interval and sudden cardiac death95,96, coeliac disease97, lung cancer98, psoriasis21 and liver cirrhosis25, among others (TABLE 1).
Initial conclusions on the utility of GWA
The utility of GWA studies for the identification of novel genomic associations with complex diseases has unambiguously been established over the past year. In general, GWA studies have employed large case–control cohorts featuring both familial and sporadic cases, categorical trait definitions and up to half a million commonly polymorphic SNPs. To date, with the exception of CFH in AMD, the estimated genetic effects of replicated associations have been uniformly and surprisingly small.
Encouragingly, most associated haplotype intervals identified to date are sufficiently small to feature a single gene. In large measure, this reflects the use of several, outbred populations in confirmatory, fine-mapping studies. Even when the association is within a single gene, the predisposing variant might affect an adjacent gene, as in adult lactose intolerance99. Although some association intervals have been found to contain a single, unequivocally functional gene variant, the causality of alleles has been established in only a minority of cases. Causal alleles identified to date do not yet show much difference in genetic mechanism from those identified in Mendelian disorders; this could reflect ascertainment bias1,83,84.
Many genes identified by GWA were not candidate genes previously, highlighting the hypothesis-informing value of genetic studies. Already, there are examples of potentially tractable therapeutic targets that had not previously been considered in a disease or trait. As yet, the confluence of associated genes into biological networks and pathways is at an early stage. In part, this reflects scant or incorrect annotation of many genes. There appears to be a significant conservation of associations of common alleles between human populations. Thus, to date, it appears that GWA studies are fulfilling expectations with regard to the elucidation of molecular mechanisms underpinning poorly understood, common diseases.
In the few informative studies reported to date, endophenotypes have been highly instructive in dissecting the network or pathway that is perturbed by an individual allele, which affects a complex trait. It is particularly exciting to see the application of multi-mode endophenotypes, such as combinations of psychological testing, brain imaging and gene expression87. This is clearly an area of potential opportunity.
The cost of enrolling the very large cohorts that are needed to discover and validate alleles with small effect sizes has hitherto precluded the collection and integration of rich, accurate clinical metadata. It is likely that future studies will use a much greater stratification of traits than the phenotypically crude studies reported so far. Recent GWA studies of breast cancer provide a good example of the added genetic complexity that can be revealed by trait stratification19,85,86. In addition, following replication of associations with categorical traits, it is anticipated that targeted genotypic examination of many endophenotypes will be highly instructive in the dissection of the role of individual alleles in disease pathogenesis.
GWA studies show significant potential to redefine disease classification. In some cases, GWA studies are identifying molecular factors that enable patient stratification and might prove useful in personalized medicine. Cancers provide the clearest examples of this to date. In other cases, exemplified by IBD, GWA studies are pointing to common molecular underpinnings in diseases that were believed to be distinct. In restless leg syndrome, replicated associations have provided concrete evidence that the phenotype represents a bona fide neurological disorder100,101. In mental illness, there is great anticipation that GWA studies will provide an objective, molecular revision of disease categorization.
Many questions remain concerning the genetic architecture of common diseases. These include the extent of locus and allelic heterogeneity, fit with an additive-threshold model or, alternatively, the extent of epistasis (the relative contributions of rare and common, and high and low, penetrance alleles) and various types of variation, from genome rearrangements to SNPs. GWA studies are not designed to evaluate these questions. Once loci have been identified, however, methods such as deep resequencing can nominate candidate susceptibility alleles and provide data for the evaluation of genetic architecture102. Meaningful, individual risk determinations will require the identification of causal alleles, the development of multiplexed molecular diagnostics and significant modelling.
Future developments and implications
The trends observed in recent GWA studies are anticipated to continue. Chips with 900,000 and 1,000,000 million SNPs were recently launched and genotyping accuracies have improved. Cohort sizes are steadily increasing and biobanks of unparalleled size and phenotype definition are being established. Combinations of genotype- and haplotype-based associations are becoming more prevalent. Experimental designs and statistical methods are also becoming more uniform, enabling more meaningful meta-analysis. In particular, the emergence of adaptive designs and the use of Bayesian inferential methods will produce a probabilistic synthesis from combined analyses83. Importantly, this will provide an intuitive framework for combining information from multiple studies, resulting in more effective detection and replication of weak associations103.
As noted above, phenotypes studied to date have been crude. The use of endophenotypes is expected to increase significantly. In particular, biomarker phenotypes are anticipated to become widely used. These will probably include gene expression, proteomic, metabolomic and imaging biomarkers. As determinants of complex traits are identified, genetic stratification will become possible, potentially reducing the genetic complexity of traits and enabling the identification of additional association signals. An example of this was the recent use of periodic limb movements and serum ferritin levels in GWA studies of restless leg syndrome100. An area of substantial future interest for the pharmaceutical industry will be pharmacogenetic GWA studies to identify markers for patient stratification in clinical trials. Comprehensive pharmacogenetic information will, in turn, facilitate the practice of personalized medicine. Pharmacogenetic GWA studies and early adoption of personalized therapy are likely to be used in the selection of expensive or chronic medications in life threatening conditions or where the therapeutic index is narrow or adverse event concerns are high, such as cancer chemotherapy.
Despite the current excitement, GWA studies have only been able to account for a small proportion of the expected genetic variance in complex traits24,102. This is not surprising given current limitations. First, current GWA studies are designed to identify common risk alleles that are predicted to be important in complex disorders under the common disease/common alleles hypothesis5,6. Increasing evidence suggests that some complex disorders and traits, such as schizophrenia, hypercholesterolaemia and body mass, are genetically heterogeneous104–107. The genetic basis of such diseases is more likely to conform to the common trait/rare variant hypothesis, which proposes that many rare variants exist, with substantial allelic heterogeneity at causal loci58,108,109. The GWA approach is unable to detect susceptibility loci that harbour numerous, individually rare (recent), polymorphisms. Instead, a resequencing approach will be needed to identify rare alleles. Encouragingly, massively parallel sequencing methods provide a potential solution102,107,110, suggesting disease-specific rare alleles and recent mutations that provide supplementary genotyping array content. Second, a proportion of the genome cannot effectively be examined on the basis of tag SNP genotypes. Approximately 20% of the genome is comprised of recombination hotspots that are not amenable to LD-based approaches7. Alternatively, at recombination coldspots, haplotype blocks might be too large for unambiguous identification of causal loci. The extent of the effect of genomic copy number variation (CNV) on association signals is not yet clear, although recent genotyping arrays do provide CNV information. Insufficient numbers of cases will be available for GWA studies of many orphan diseases, uncommon disease complications or adverse events. For some common diseases, these considerations could obfuscate a substantial proportion of the genetic variance. Supplementation of genotyping array content reflective of CNV regions should, however, circumvent some of these limitations. Use of adaptive statistical methods and resampling strategies might also circumvent the need for thousands of affected individuals in studies of orphan diseases83.
GWA successes are creating substantial need for downstream genetics, biochemistry and cell biology efforts to confirm the biological relevance of genotype–phenotype associations and to elucidate the underlying mechanisms of disease. This is especially true of association signals in gene deserts or alleles without apparent functional consequence. Translation of the fruits of GWA studies to clinical practice will require the derivation of predictive models of the genetic architecture of complex traits that evaluate with much greater precision the contributions of factors such as epistasis, genocopies, phenocopies and penetrance.
Acknowledgments
A Deo lumen, ab amicis auxilium. This work was partially supported by National Institutes of Health grants N01A000,064 and U01AI066,569, and by National Science Foundation grant 0524,775. The authors thank the reviewers for their helpful suggestions.
- Genetic linkage
Co-segregation (reduced recombination) of a trait and an allele in related subjects (pedigrees) more often than explicable by chance
- Dominant
An allele that confers a trait even when it is heterozygous (present as a single copy in a genome)
- Recessive
An allele that confers a trait only when it is homozygous (present in two copies in a genome, one from each parent)
- Endophenotype
A measurable component of a phenotype
- Multifactorial
Inheritance of a trait that is attributable to two or more genes and their interaction with the environment (also known as polygenic inheritance)
- Allele
The DNA code at a given locus (position) on a chromosome
- Genome-wide association study
A comprehensive search of the human genome for genetic risk factors for a trait by a case-control association study involving comparisons of hundreds of thousands of alleles between unrelated subjects with and without a trait
- Haplotype
A combination of alleles at linked loci (on a single chromatid) that are transmitted together more often than explicable by chance
- Linkage disequilibrium
(LD). Combinations of alleles in a population that differ in frequency from that expected from random formation of haplotypes from alleles based on their frequencies
- Minor-allele frequency
The allele frequency of the less frequently occurring allele of a polymorphism
- Case–control association study
Comparison of the frequency of an allele between unrelated subjects with and without a trait. A difference in allele frequency between the two groups indicates that the allele might change the likelihood of the trait
- Genetic association
Correlation of a trait and an allele in a population more often than explicable by chance
- Genocopy
A genotype at a locus that produces a phenotype that is indistinguishable from that produced by a genotype at another locus
- Phenocopy
An environmentally produced phenotype that simulates the effect of a particular genotype
- Non-synonymous SNP
(nsSNP). A SNP that leads to a change in the amino-acid sequence of the gene’s resulting protein and that might therefore affect its function
- Odds ratio
A measure of risk that compares the probability of occurrence of a disease in a group with a risk allele with the probability in a control group
- Pleiotropy
A single gene that influences multiple phenotypic traits
- Epistasis
Modification of the action of a gene by another gene
- Family-based association study
Evaluation of the frequency of co-transmission of an allele and a trait from parents to offspring. Co-transmission of an allele and trait to offspring more often than expected by chance indicates that the allele might change the likelihood of the trait
- Antagonistic pleiotropy
A single gene that influences multiple competing phenotypes such that beneficial effects of a trait created by the gene are offset by losses in other traits
Footnotes
DATABASES
Entrez Gene: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene APOE | ATG16L1 | BAT1 | C3 | CFB | CFH | CLEC16A | FTO | GRM7 | HLA-B27 | HNF1B | IFIH1 | IL2RA | IL23R | NFKBIL1 | NOD2 | PTGER4 | PTPN2 | PTPN22 | RELA | TCF7L2 | WWC1
OMIM: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM age-related macular degeneration | acute myocardial infarction | Alzheimer’s disease | anterior uveitis/ankylosing spondylitis | breast cancer | colorectal cancer | Crohn’s disease | prostate cancer | restless leg syndrome | type 1 diabetes mellitus | type 2 diabetes mellitus | ulcerative colitis
FURTHER INFORMATION
Stephen Kingsmore’s homepage: http://www.ncgr.org
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nature Genet. 2003;33 (Suppl):228–237. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 2.Freimer N, Sabatti C. The use of pedigree, sib-pair and association studies of common diseases for genetic mapping and epidemiology. Nature Genet. 2004;36:1045–1051. doi: 10.1038/ng1433. [DOI] [PubMed] [Google Scholar]
- 3.Goring HH, Terwilliger JD, Blangero J. Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet. 2001;69:1357–1369. doi: 10.1086/324471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nature Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 5.Chakravarti A. Population genetics — making sense out of sequence. Nature Genet. 1999;21:56–60. doi: 10.1038/4482. [DOI] [PubMed] [Google Scholar]
- 6.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 7.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nature Genet. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 11.Cardon LR, Bell JI. Association study designs for complex diseases. Nature Rev Genet. 2001;2:91–99. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- 12.Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- 13.Redden DT, Allison DB. Nonreplication in genetic association studies of obesity and diabetes research. J Nutr. 2003;133:3323–3326. doi: 10.1093/jn/133.11.3323. [DOI] [PubMed] [Google Scholar]
- 14.Sillanpaa MJ, Auranen K. Replication in genetic studies of complex traits. Ann Hum Genet. 2004;68:646–657. doi: 10.1046/j.1529-8817.2004.00122.x. [DOI] [PubMed] [Google Scholar]
- 15.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nature Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 16.Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;405:847–856. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- 17.Chanock SJ, et al. Replicating genotype–phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 18.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. The largest GWA study undertaken to date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nature Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ozaki K, et al. Functional SNPs in the lymphotoxin-α gene that are associated with susceptibility to myocardial infarction. Nature Genet. 2002;32:650–654. doi: 10.1038/ng1047. The first large scale association study of a complex human disorder. [DOI] [PubMed] [Google Scholar]
- 21.Cargill M, et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80:273–290. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark AG, Li J. Conjuring SNPs to detect associations. Nature Genet. 2007;39:815–816. doi: 10.1038/ng0707-815. [DOI] [PubMed] [Google Scholar]
- 23.Grupe A, et al. Evidence for novel susceptibility genes for late-onset Alzheimer’s disease from a genome-wide association study of putative functional variants. Hum Mol Genet. 2007;16:865–873. doi: 10.1093/hmg/ddm031. [DOI] [PubMed] [Google Scholar]
- 24.Hampe J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nature Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 25.Huang H, et al. Identification of two gene variants associated with risk of advanced fibrosis in patients with chronic hepatitis C. Gastroenterology. 2006;130:1679–1687. doi: 10.1053/j.gastro.2006.02.032. [DOI] [PubMed] [Google Scholar]
- 26.Luke MM, et al. A polymorphism in the protease-like domain of apolipoprotein(a) is associated with severe coronary artery disease. Arterioscler Thromb Vasc Biol. 2007;27:2030–2036. doi: 10.1161/ATVBAHA.107.141291. [DOI] [PubMed] [Google Scholar]
- 27.Shiffman D, et al. Identification of four gene variants associated with myocardial infarction. Am J Hum Genet. 2005;77:596–605. doi: 10.1086/491674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smyth DJ, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nature Genet. 2006;38:617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 29.Clarke R, et al. Lymphotoxin-α gene and risk of myocardial infarction in 6,928 cases and 2,712 controls in the ISIS case-control study. PLoS Genet. 2006;2:e107. doi: 10.1371/journal.pgen.0020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimura A, et al. Lack of association between LTA and LGALS2 polymorphisms and myocardial infarction in Japanese and Korean populations. Tissue Antigens. 2007;69:265–269. doi: 10.1111/j.1399-0039.2006.00798.x. [DOI] [PubMed] [Google Scholar]
- 31.Koch W, et al. Association of variants in the BAT1–NFKBIL1–LTA genomic region with protection against myocardial infarction in Europeans. Hum Mol Genet. 2007;16:1821–1827. doi: 10.1093/hmg/ddm130. [DOI] [PubMed] [Google Scholar]
- 32.Laxton R, Pearce E, Kyriakou T, Ye S. Association of the lymphotoxin-α gene Thr26Asn polymorphism with severity of coronary atherosclerosis. Genes Immun. 2005;6:539–541. doi: 10.1038/sj.gene.6364236. [DOI] [PubMed] [Google Scholar]
- 33.Mizuno H, et al. Impact of atherosclerosis-related gene polymorphisms on mortality and recurrent events after myocardial infarction. Atherosclerosis. 2006;185:400–405. doi: 10.1016/j.atherosclerosis.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 34.Sedlacek K, et al. Lymphotoxin-α and galectin-2 SNPs are not associated with myocardial infarction in two different German populations. J Mol Med. 2007;85:997–1004. doi: 10.1007/s00109-007-0211-4. [DOI] [PubMed] [Google Scholar]
- 35.Yamada A, et al. Lack of association of polymorphisms of the lymphotoxin α gene with myocardial infarction in Japanese. J Mol Med. 2004;82:477–483. doi: 10.1007/s00109-004-0556-x. [DOI] [PubMed] [Google Scholar]
- 36.Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. Discovery of a single variant that explains a large component of the genetic variance in a common human disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hageman GS, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci USA. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magnusson KP, et al. CFH Y402H confers similar risk of soft drusen and both forms of advanced AMD. PLoS Med. 2006;3:e5. doi: 10.1371/journal.pmed.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Souied EH, et al. Y402H complement factor H polymorphism associated with exudative age-related macular degeneration in the French population. Mol Vis. 2005;11:1135–1140. [PubMed] [Google Scholar]
- 40.Zareparsi S, et al. Strong association of the Y402H variant in complement factor H at 1q32 with susceptibility to age-related macular degeneration. Am J Hum Genet. 2005;77:149–153. doi: 10.1086/431426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gold B, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nature Genet. 2006;38:458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yates JR, et al. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357:553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 43.Duerr RH, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Libioulle C, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rioux JD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nature Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parkes M, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nature Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamazaki K, et al. Association analysis of genetic variants in IL23R, ATG16L1 and 5p13.1 loci with Crohn’s disease in Japanese patients. J Hum Genet. 2007;52:575–583. doi: 10.1007/s10038-007-0156-z. [DOI] [PubMed] [Google Scholar]
- 48.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 49.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 50.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 51.Todd JA, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nature Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raelson JV, et al. Genome-wide association study for Crohn’s disease in the Quebec Founder Population identifies multiple validated disease loci. Proc Natl Acad Sci USA. 2007;104:14747–14752. doi: 10.1073/pnas.0706645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gudmundsson J, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nature Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 54.Saxena R, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 55.Scott LJ, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sladek R, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 57.Steinthorsdottir V, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nature Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 58.Liu PY, et al. A survey of haplotype variants at several disease candidate genes: the importance of rare variants for complex diseases. J Med Genet. 2005;42:221–227. doi: 10.1136/jmg.2004.024752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morris RW, Kaplan NL. On the advantage of haplotype analysis in the presence of multiple disease susceptibility alleles. Genet Epidemiol. 2002;23:221–233. doi: 10.1002/gepi.10200. [DOI] [PubMed] [Google Scholar]
- 60.Zhang K, Calabrese P, Nordborg M, Sun F. Haplotype block structure and its applications to association studies: power and study designs. Am J Hum Genet. 2002;71:1386–1394. doi: 10.1086/344780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang K, Sun F. Assessing the power of tag SNPs in the mapping of quantitative trait loci (QTL) with extremal and random samples. BMC Genet. 2005;6:51. doi: 10.1186/1471-2156-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drysdale CM, et al. Complex promoter and coding region β2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc Natl Acad Sci USA. 2000;97:10483–10488. doi: 10.1073/pnas.97.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeggini E, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grant SF, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nature Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 65.Yi F, Brubaker PL, Jin T. TCF-4 mediates cell type-specific regulation of proglucagon gene expression by β-catenin and glycogen synthase kinase-3β. J Biol Chem. 2005;280:1457–1464. doi: 10.1074/jbc.M411487200. [DOI] [PubMed] [Google Scholar]
- 66.Pearson ER, et al. Variation in TCF7L2 influences therapeutic response to sulfonylureas: a GoDARTs study. Diabetes. 2007;56:2178–2182. doi: 10.2337/db07-0440. [DOI] [PubMed] [Google Scholar]
- 67.Florez JC, et al. Haplotype structure and genotype–phenotype correlations of the sulfonylurea receptor and the islet ATP-sensitive potassium channel gene region. Diabetes. 2004;53:1360–1368. doi: 10.2337/diabetes.53.5.1360. [DOI] [PubMed] [Google Scholar]
- 68.Helgason A, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nature Genet. 2007;39:218–225. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- 69.Weedon MN, et al. Combining information from common type 2 diabetes risk polymorphisms improves disease prediction. PLoS Med. 2006;3:e374. doi: 10.1371/journal.pmed.0030374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stephens JC, et al. Haplotype variation and linkage disequilibrium in 313 human genes. Science. 2001;293:489–493. doi: 10.1126/science.1059431. [DOI] [PubMed] [Google Scholar]
- 71.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dina C, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nature Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 73.Rich SS, et al. The Type 1 Diabetes Genetics Consortium. Ann NY Acad Sci. 2006;1079:1–8. doi: 10.1196/annals.1375.001. [DOI] [PubMed] [Google Scholar]
- 74.Ahmad T, Marshall SE, Jewell D. Genetics of inflammatory bowel disease: the role of the HLA complex. World J Gastroenterol. 2006;12:3628–3635. doi: 10.3748/wjg.v12.i23.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Orozco G, Rueda B, Martin J. Genetic basis of rheumatoid arthritis. Biomed Pharmacother. 2006;60:656–662. doi: 10.1016/j.biopha.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 76.Sia C, Weinem M. The role of HLA class I gene variation in autoimmune diabetes. Rev Diabet Stud. 2005;2:97–109. doi: 10.1900/RDS.2005.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Criswell LA, et al. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet. 2005;76:561–571. doi: 10.1086/429096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gudmundsson J, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nature Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 79.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nature Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 80.Amundadottir LT, et al. A common variant associated with prostate cancer in European and African populations. Nature Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 81.Haiman CA, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nature Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rodriguez C, et al. Diabetes and risk of prostate cancer in a prospective cohort of US men. Am J Epidemiol. 2005;161:147–152. doi: 10.1093/aje/kwh334. [DOI] [PubMed] [Google Scholar]
- 83.Knight JC. Regulatory polymorphisms underlying complex disease traits. J Mol Med. 2005;83:97–109. doi: 10.1007/s00109-004-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thomas PD, Kejariwal A. Coding single-nucleotide polymorphisms associated with complex vs. Mendelian disease: evolutionary evidence for differences in molecular effects. Proc Natl Acad Sci USA. 2004;101:15398–15403. doi: 10.1073/pnas.0404380101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stacey SN, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nature Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 87.Papassotiropoulos A, et al. Common Kibra alleles are associated with human memory performance. Science. 2006;314:475–478. doi: 10.1126/science.1129837. [DOI] [PubMed] [Google Scholar]
- 88.Rodriguez-Rodriguez E, et al. Age-dependent association of KIBRA genetic variation and Alzheimer’s disease risk. Neurobiol Aging. 2007 Aug 16; doi: 10.1016/j.neurobiolaging.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 89.Schaper K, Kolsch H, Popp J, Wagner M, Jessen F. KIBRA gene variants are associated with episodic memory in healthy elderly. Neurobiol Aging. 2007 Mar 10; doi: 10.1016/j.neurobiolaging.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 90.Coon KD, et al. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer’s disease. J Clin Psychiatry. 2007;68:613–618. doi: 10.4088/jcp.v68n0419. [DOI] [PubMed] [Google Scholar]
- 91.Baum AE, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2007 May 8; doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 93.Bierut LJ, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gudbjartsson DF, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 95.Aarnoudse AJ, et al. Common NOS1AP variants are associated with a prolonged QTc interval in the Rotterdam Study. Circulation. 2007;116:10–16. doi: 10.1161/CIRCULATIONAHA.106.676783. [DOI] [PubMed] [Google Scholar]
- 96.Arking DE, et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nature Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 97.van Heel DA, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nature Genet. 2007;39:827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Spinola M, et al. Genome-wide single nucleotide polymorphism analysis of lung cancer risk detects the KLF6 gene. Cancer Lett. 2007;251:311–316. doi: 10.1016/j.canlet.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 99.Olds LC, Sibley E. Lactase persistence DNA variant enhances lactase promoter activity in vitro: functional role as a cis regulatory element. Hum Mol Genet. 2003;12:2333–2340. doi: 10.1093/hmg/ddg244. [DOI] [PubMed] [Google Scholar]
- 100.Stefansson H, et al. A genetic risk factor for periodic limb movements in sleep. N Engl J Med. 2007;357:639–647. doi: 10.1056/NEJMoa072743. [DOI] [PubMed] [Google Scholar]
- 101.Winkelmann J, et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nature Genet. 2007;39:1000–1006. doi: 10.1038/ng2099. [DOI] [PubMed] [Google Scholar]
- 102.Altshuler D, Daly M. Guilt beyond a reasonable doubt. Nature Genet. 2007;39:813–815. doi: 10.1038/ng0707-813. [DOI] [PubMed] [Google Scholar]
- 103.Hunter DJ, Kraft P. Drinking from the fire hose — statistical issues in genomewide association studies. N Engl J Med. 2007;357:436–439. doi: 10.1056/NEJMp078120. [DOI] [PubMed] [Google Scholar]
- 104.Ahituv N, et al. Medical sequencing at the extremes of human body mass. Am J Hum Genet. 2007;80:779–791. doi: 10.1086/513471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cohen JC, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. An example of a phenotype that fits the common disorder: rare alleles hypothesis. [DOI] [PubMed] [Google Scholar]
- 106.Fanous AH, Kendler KS. Genetic heterogeneity, modifier genes, and quantitative phenotypes in psychiatric illness: searching for a framework. Mol Psychiatry. 2005;10:6–13. doi: 10.1038/sj.mp.4001571. [DOI] [PubMed] [Google Scholar]
- 107.McClellan JM, Susser E, King MC. Schizophrenia: a common disease caused by multiple rare alleles. Br J Psychiatry. 2007;190:194–199. doi: 10.1192/bjp.bp.106.025585. [DOI] [PubMed] [Google Scholar]
- 108.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69:124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease–common variant…or not? Hum Mol Genet. 2002;11:2417–2423. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- 110.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tomlinson I, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nature Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 112.Zanke BW, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nature Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 113.Haiman CA, et al. A common genetic risk factor for colorectal and prostate cancer. Nature Genet. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]