Figure 5.
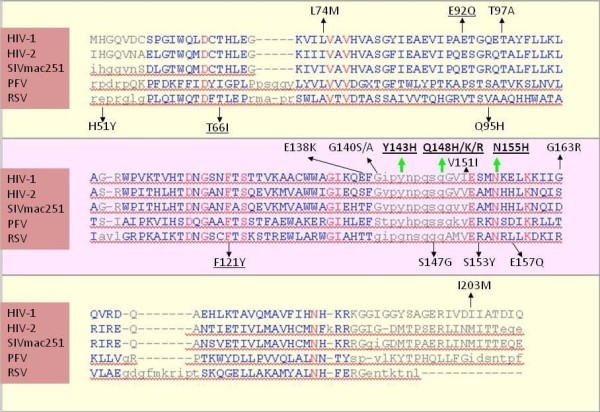
Sequence alignment of the integrase catalytic core domains of HIV-1 subtype B (PDB: 1BL3_C), HIV-2 (PDB: 3F9K_A), SIVmac251 (PDB: 1C6V_A), prototype foamy virus/PFV (PDB: 3DLR_A), and Rous Sarcoma virus/RSV (PDB: 1ASU_A). The sequence alignment is based on a structural alignment performed using the VAST algorithm. Regions showing significant structural alignment are presented in blue, with the highly conserved residues shown in red. Above the alignments are shown the mutations found in HIV-1 infected individuals failing raltegravir-based drug regimens (the green arrows indicate the primary resistance mutations Y143H, Q148H/K/R, and N155H; black arrows indicate secondary resistance mutations). Other drug resistance mutations induced by other integrase strand transfer inhibitors are shown below the alignments. The mutations shown by site-directed mutagenesis to confer resistance to raltegravir are underlined. Note that the structure for HIV-1 subtype B integrase catalytic core domain (PDB: 1BL3_C) presents the secondary drug resistance mutation V151I.
