Abstract
Fibroblast growth factors (FGFs) mediate a vast range of CNS developmental processes including neural induction, proliferation, migration, and cell survival. Despite the critical role of FGF signaling for normal CNS development, few reports describe the mechanisms that regulate FGF receptor gene expression in the brain. We tested whether FGF8 could autoregulate two of its cognate receptors, Fgfr1 and Fgfr3, in three murine cell lines with different lineages: fibroblast-derived cells (3T3 cells), neuronal cells derived from hippocampus (HT-22 cells), and neuroendocrine cells derived from hypothalamic gonadotropin-releasing hormone (GnRH) neurons (GT1-7 cells). GnRH is produced by neurons in the hypothalamus and is absolutely required for reproductive competence in vertebrate animals. Several lines of evidence strongly suggest that Fgf8 is critical for normal development of the GnRH system, therefore, the GT1-7 cells provided us with an additional endpoint, Gnrh gene expression and promoter activity, to assess potential downstream consequences of FGF8-induced modulation of FGF receptor levels. Results from this study suggest that the autoregulation of its cognate receptor represents a common downstream effect of FGF8. Further, we show that Fgfr1 and Fgfr3 are differentially regulated within the same cell type, implicating these two receptors in different biological roles. Moreover, Fgfr1 and Fgfr3 are differentially regulated among different cell types, suggesting such autoregulation occurs in a cell type-specific fashion. Lastly, we demonstrate that FGF8b decreases Gnrh promoter activity and gene expression, possibly reflecting a downstream consequence of altered FGF receptor populations. Together, our data bring forth the possibility that, in addition to the FGF synexpression group, autoregulation of FGFR expression by FGF8 represents a mechanism by which FGF8 could fine-tune its regulatory actions.
Introduction
Fibroblast growth factors (FGFs) mediate a vast range of CNS developmental processes including neural induction, proliferation, migration, and cell survival. The FGF family consists of four receptors (FGFR1, 2, 3, 4), 22 ligands, and their splice variants that vary in expression patterns both temporally and spatially [1]. The structural components of FGF receptors consist of three extracellular Ig-like domains, a transmembrane domain, and two intracellular tyrosine kinase domains [2]. Despite the critical role of FGF signaling in CNS development, there are few reports to date describing the mechanisms that regulate FGF receptor gene expression in the brain.
Receptor expression is often controlled by autoregulation, where binding of the cognate ligand leads to changes that affect protein turnover, internalization, primary transcript stability, and gene promoter activity [3], [4], [5]. Interestingly, FGFR1 was reported to have a synexpression pattern with its cognate ligand FGF8 [6]. Synexpression is an interesting feature associated with FGF and a few other signaling pathways that involves the coexpression of a set of genes termed the synexpression group [7], [8], [9]. The products of the FGF synexpression group are then capable of modulating the intracellular signaling cascades of several FGF ligands, in particular FGF8, to curtail or achieve specific spatial patterns of FGF signaling [10]. This raises the possibility that FGF8 may control its own activity level via the autoregulation of its own receptors. The upregulation of FGFR1 by FGF8 could represent a positive feedback mechanism that adds another layer of regulatory complexity, further fine-tuning the spatial and temporal specificity of FGF8 actions during development.
Until now, the possibility that FGF8 could add to the modular regulation of its activity in neurons by autoregulating its own receptor has not been adequately explored. Further, it is unclear if FGF8 could autoregulate all cognate receptors in a similar fashion. In this study, we examined if FGF8 autoregulated two of its cognate receptors, Fgfr1 and Fgfr3, in three murine cell lines with different lineages: fibroblast-derived cells (3T3), neuronal cells derived from hippocampus (HT-22), and neuroendocrine cells derived from hypothalamic gonadotropin-releasing hormone neurons (GT1-7). The GT1-7 cells were particularly useful since the in vivo specification of GnRH neuronal fate was shown to be highly dependent on FGF8 signaling and, the expression level of FGF receptors in these cells could be correlated with a hallmark of GnRH neuronal differentiation: the expression of Gnrh gene [11]. Therefore, these cells provided us with an additional endpoint, Gnrh gene expression and promoter activity, to assess potential downstream consequences of FGF8-induced modulation of FGF receptor levels.
Results
Endogenous expression of FGF8 in 3T3, HT-22, and GT1-7 cell lines
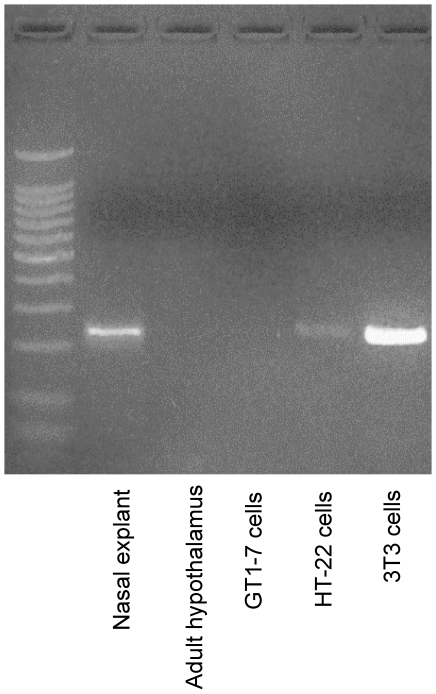
First, we characterized the endogenous expression of FGF8 in the 3T3, HT-22, and GT1-7 and compared it with mouse tissue taken from embryonic nasal explants and adult hypothalamus. Consistent with the widely accepted role of FGF8 during development, mouse nasal explants had high expression levels of endogenous FGF8 (Fig. 1, lane 2). Also, 3T3 cells had high endogenous levels of FGF8 (Fig. 1, lane 6) which was expected due to their fibroblast cell lineage. By contrast, endogenous FGF8 expression was low in the neuronal-derived HT-22 cells (Fig. 1, lane 5) and completely absent in the GT1-7 cells and hypothalamus (Fig. 1, lanes 3 and 4, respectively).
Figure 1. FGF expression in mouse brain and representative cell lines.
Photomicrograph of RT-PCR product for FGF8 mRNA stained with ethidium bromide and resolved on a 2% agarose gel. Total RNA was isolated from mouse nasal explant (embryonic day 11.5; lane 2) adult hypothalamus (lane 3), hypothalamic-derived GT1-7 cells (lane 4), hippocampus-derived HT-22 cells (lane 5), and fibroblast-derived 3T3 cells (lane 6). Presence of band indicates FGF8 primary transcripts in representative tissue type or cell line.
Differential effects of FGF8b on the expression of FGF receptors 1 (Fgfr1) and 3 (Fgfr3)
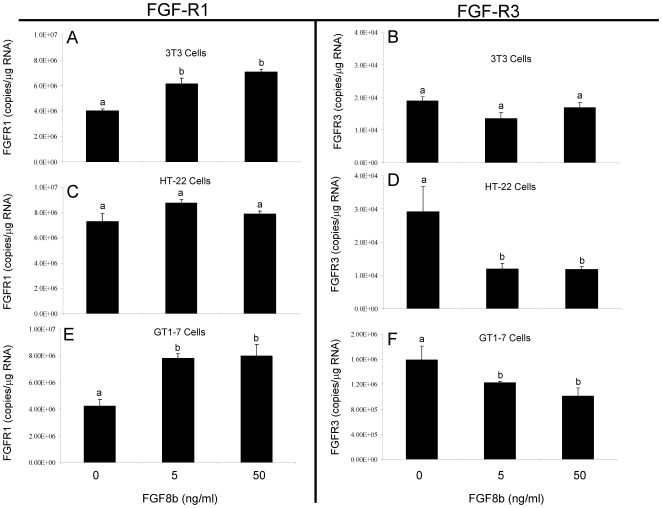
In these experiments, 3T3, HT-22, and GT1-7 cells were treated with 5 or 50 ng/ml of FGF8b for 4 hours in order to determine if FGF8b regulated the expression of its cognate receptors Fgfr1 and Fgfr3 in these cell types. Overall, our data revealed that FGF8b differentially altered the expression of Fgfr1 and Fgfr3 mRNA depending on the cell type. For instance, in 3T3 cells, which express high endogenous levels of FGF8, FGF8b treatment for 4 hours significantly increased Fgfr1 mRNA (Fig. 2A), yet had no effect on Fgfr3 (Fig. 2B). By contrast, in HT-22 cells, which express low endogenous levels of FGF8, FGF8b treatment had no effect on Fgfr1 (Fig. 2C), yet significantly decreased Fgfr3 mRNA (Fig. 2D). Most notably, in the GT1-7 cells, which do not express endogenous FGF8, FGF8b treatment significantly increased Fgfr1 expression (Fig. 2E) while simultaneously decreasing Fgfr3 expression (Fig. 2F). This differential effect of FGF8b on Fgfr1 and Fgfr3 receptor expression in GT1-7 cells did not occur in the HT-22 or 3T3 cells. Therefore, in a subsequent experiment, GT1-7 cells were used to determine whether the differential effects of FGF8b on Fgfr1 and Fgfr3 expression are mediated through the classical membrane FGF receptors.
Figure 2. Comparative effects of FGF8b on Fgfr1 and Fgfr3 mRNA in 3T3, HT-22 and GT1-7 cells.
Fgfr1 and Fgfr3 mRNA levels in 3T3 cells (A, B), HT-22 cells (C, D), or GT1-7 cells (E,F) following treatment with vehicle or FGF8b at 5 or 50 ng/ml. for 4 hours. Data are expressed as mean copies of Fgfr1 or Fgfr3 transcript/µg total RNA ± SEM. Dissimilar letters indicate statistically significant difference among groups, P<0.05.
FGF8b effects on FGF receptor expression in GT1-7 cells are mediated by FGF receptors
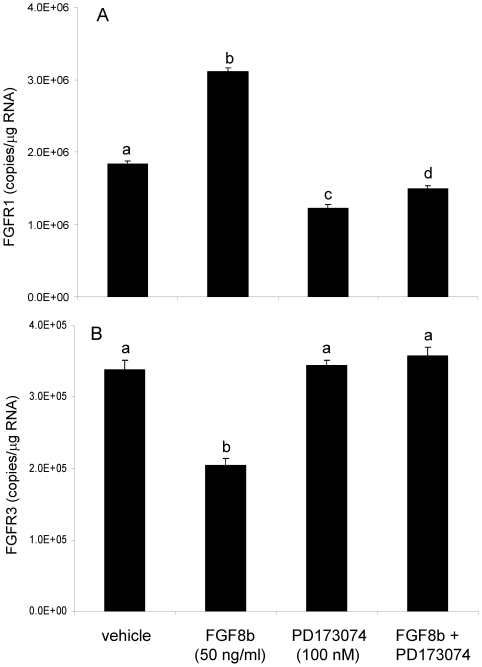
In these experiments, GT1-7 cells were treated with FGF8b (50 ng/ml), the FGF receptor antagonist PD173074 (100 nM), or combined FGF8b + PD173074 for 8 hours. Consistent with our earlier observation, treatment with FGF8b alone significantly increased Fgfr1 (Fig. 3A). Moreover, the FGF receptor anatagonist PD173074, alone or in combination with FGF8b, significantly inhibited Fgfr1 mRNA expression compared to the vehicle group (Fig. 3A). Also consistent with our earlier observation in GT1-7 cells, FGF8b treatment significantly decreased Fgfr3 mRNA (Fig. 3B), but there was no effect of PD173074 alone on the expression of Fgfr3. The inhibitory effect of FGF8b on Fgfr3 was completely abolished in the presence of the antagonist (Fig. 3B).
Figure 3. Differential effects of FGF8b on Fgfr1 and Fgfr3 mRNA in GT1-7 cells.
Fgfr1 (A) and Fgfr3 (B) mRNA levels in GT1-7 cells following treatment with vehicle, FGF8b (50 ng/ml), PD173074 (100 nM), or FGF8b + PD173074 for 8 hours. Data are expressed as mean copies of Fgfr1 or Fgfr3/µg total RNA ± SEM. Dissimilar letters indicate statistically significant difference among groups, P<0.05.
FGF8b decreased gonadotropin-releasing hormone promoter activity and mRNA in GT1-7 cells
Previous work demonstrated that targeted disruption of Fgfr1 signaling in GnRH neurons decreased the numbers of detectable GnRH neurons in the hypothalamus of adult mouse brain, suggesting that FGF is critical for normal GnRH neuronal development [12] Based on this observation, we hypothesized that that the expression level of the Gnrh gene, a hallmark of GnRH neuronal differentiation, could vary according to Fgfr1, and possibly Fgfr3, levels [11]. Further, in the previous experiment we determined that FGF8b differentially altered the expression of its two cognate receptors, Fgfr1 and Fgfr3 in a GnRH-expressing cell line (GT1-7 cells; see Fig. 2E, F). Therefore, we measured Gnrh promoter activity and mRNA levels in GT1-7 cells following treatment with FGF8b to determine whether changes in the FGF receptor population (i.e increased Fgfr1 and decreased Fgfr3) corresponded to changes in Gnrh gene activity.
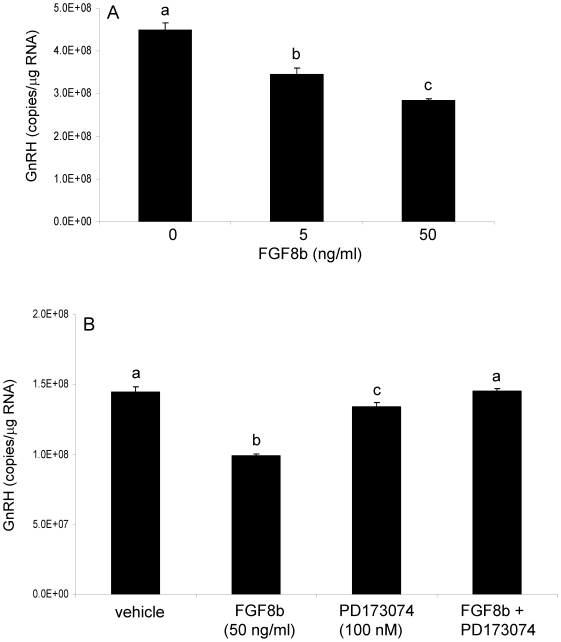
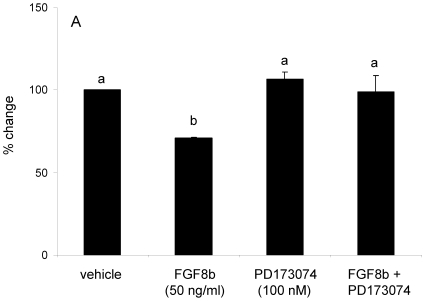
GT1-7 cells were treated with 5 or 50 ng/ml of FGF8b for 4 or 8 hours. Treatment with either 5 or 50 ng/ml of FGF8b significantly reduced Gnrh mRNA in GT1-7 cells after 8, but not 4 (data not shown), hours of FGF8b exposure (Fig. 4A). To determine whether the effects of FGF8b on Gnrh expression in GT1-7 cells were mediated through classical FGF receptors, cells were treated with the broad FGF receptor antagonist PD173074. As expected, FGF8b treatment concomitant with the receptor antagonist had no effect on Gnrh mRNA levels (Fig. 4B), indicating that the FGF8b-induced decrease in Gnrh mRNA in GT1-7 cells was dependent upon its cognate membrane receptors. Interestingly, treatment with PD173074 alone induced a modest, yet significant, decrease in Gnrh mRNA levels, similar that observed previously with Fgfr1 (compare to Fig. 3A). Next, we measured Gnrh promoter activity following FGF8b treatment in GT1-7 cells. Treatment with FGF8b for 8 hours significantly reduced Gnrh promoter activity in GT1-7 cells (Fig. 5) in parallel to the observed reduction in Gnrh mRNA levels (compare to Fig. 4A). Further, the concomitant treatment with PD17074 abolished the FGF8b-induced reduction in promoter activity in GT1-7 cells.
Figure 4. FGF8b decreased GnRH mRNA in GT1-7 cells.
Panel A: GnRH mRNA levels in GT1-7 cells following 8 hours of vehicle or FGF8b treatment at 5 or 50 ng/ml. Panel B: GnRH mRNA levels in GT1-7 cells following treatment with vehicle, FGF8b (50 ng/ml), PD173074 (100 nM), or FGF8b + PD173074 for 8 hours. Data are expressed as mean copies of GnRH transcript/µg total RNA ± SEM. Dissimilar letters indicate statistically significant difference among groups, P<0.05.
Figure 5. Effects of FGF8b on GnRH promoter activity.
Transient transfection of GT1-7 cells with 0.15 µg/well of mouse full-length GnRH-luciferase reporter construct. Following transfection, cells were treated with vehicle, FGF8b (50 ng/ml), PD173074 (100 nM), or FGF8b + PD173074 for 8 hours. Data are represented as mean percent change in RLU's from vehicle-treated controls ± SEM. Dissimilar letters indicate statistically significant difference among groups, P<0.05.
Discussion
Precisely timed and coordinated FGF signaling events are critical for proper CNS development, yet the mechanisms regulating the expression of specific membrane FGF receptors in neurons have not been thoroughly investigated. From our data, the following general conclusions can be drawn. First, FGF8b autoregulates the gene expression of its two cognate receptors, Fgfr1 and Fgfr3, in a cell-type specific manner; second, this autoregulation is mediated by FGF receptors as opposed to a non-classical signaling pathway; third, receptor specific autoregulation might be dependent upon the levels of endogenous FGF8 present in a given cell type; and finally, FGF8b decreases Gnrh promoter activity and gene expression, possibly reflecting a downstream consequence of altered FGF receptor populations.
Ligand-mediated receptor autoregulation is a common feature of many types of receptors, but there are few reports documenting this as a mechanism for regulating FGF receptor gene expression. FGF receptors belong to a large class of cell surface receptors called receptor tyrosine kinases (RTKs) which encompasses multiple receptor families, including the epidermal growth factor (EGF), vascular endothelial growth factor (VEGF), RET, and Eph receptor families [1], [2], [13]. The initial reports characterizing the discovery of an FGF receptor noted that basic FGF (FGF2) downregulated the number of available FGF binding sites in baby hamster kidney cells [14]. These results were later confirmed in the fibroblast-derived 3T3 cell line [15] and in pancreatic-derived AR4-2J cells [16]. Interestingly, while FGF2 downregulated Fgfr1 in AR4-2J cells, an FGF2 protein isoform (22.5 kDa FGF2) upregulated Fgfr1 levels in those same cells by increasing the half-life of the Fgfr1 transcript [16]. For our studies, FGF8b, was chosen over other FGF8 isoforms based on the greater requirement of FGF8b during CNS development [17], [18]. In this respect, the earlier findings are consistent with our current data in supporting the autoregulatory effects of an FGF ligand on its own receptors, at the level of transcription.
The four FGF receptors and their associated splice variants share a considerable amount of overlap in their tissue distribution, ability to bind multiple FGF ligands, and their intracellular signaling pathways. Despite this redundancy, there is mounting evidence that each receptor confers distinct downstream cellular functions. Studies using the tumorigenic pro-B cell line, BaF3, showed that a majority of the FGF ligands are more effective at inducing mitogenic activity through Fgfr1 than the other FGF receptor types [1]. Moreover, abnormally high levels of Fgfr1 gene expression have been observed in prostate, colorectal, bladder, and a subset of breast carcinomas [13], [19], [20], [21]. Together, these studies suggest that Fgfr1 is important for maintaining or inducing a less differentiated cell phenotype. On the other hand, it has been shown that Fgfr3 can inhibit cellular proliferation in multiple cell types including bone, pancreas, and brain [22], [23], [24], [25]. Further, a reciprocal relationship between Fgfr1 and Fgfr3 was observed in colorectal carcinoma cells [20]. In that study, the transcriptional silencing of Fgfr1 with siRNA decreased cellular proliferation and increased Fgfr3 expression, suggesting that Fgfr3 was important for limiting the progression of tumorigenesis. The only cell line in the present study that recapitulated the reciprocal relationship between Fgfr1 and Fgfr3 was GT1-7 cells (Fig. 2). As such, we hypothesize that Fgfr1 and Fgfr3 both mediate biologically important, but opposing, effects in these cells. FGF8 could favor effects mediated by Fgfr1 by upregulating Fgfr1 and downregulating the antagonizing Fgfr3. These data put forth a novel mechanism by which a ligand with multiple receptors could preferentially activate pathways associated with one receptor subtype. This regulatory mechanism would offer both flexibility and selectivity during development, when multiple ligands and their receptors are present at the same time.
Strong evidence suggests that FGF8 signaling, through its cognate receptor Fgfr1, is critical for normal development of the GnRH system. Mice hypomorphic for Fgf8 or Fgfr1 possessed virtually no GnRH neurons in their forebrains [11]. By contrast, Fgfr3-null mice showed no developmental deficiencies in GnRH neurons (13). In this respect, Fgfr1 and Fgfr3 are clearly not functionally equivalent in driving GnRH neuronal system development, although both are expressed in GnRH neurons [26]. In GT1-7 cells treated with FGF8b, an increase in Fgfr1/Fgfr3 ratio was accompanied by a concomitant decrease in Gnrh mRNA. Extrapolating these results to the endogenous GnRH neurons, FGF8b could induce a general suppression of Gnrh gene expression during early development via the preferential activation of Fgfr1, a phenomenon consistent with low levels of Gnrh gene expression in GnRH neurons before birth [27]. However, the physiological significance of this finding in the endogenous GnRH system requires further exploration.
Overall, our data demonstrate that autoregulation of FGFRs is a cell-type specific process that leads to altered downstream consequences due to individual receptor signaling events. At present, the molecular mechanisms regulating FGF ligand-induced receptor autoregulation and the resulting downstream effects are unclear. However, the data herein provide novel insights into understanding how FGF signaling, with 22 ligands, 4 transmembrane receptors, and their splice variants, could fine-tune their regulatory roles by differentially autoregulating FGF receptor transcription. Such a mechanism could be broadly applicable to the regulation of normal cellular processes, such as neural development, as well as pathological processes, such as cancer.
Materials and Methods
Cell culture
All cell lines used in these studies were verified to be free of mycoplasma contamination (MycoSensor QPCR, Stratagene/Agilent Technologies). The following murine tumorigenic cell lines were used: fibroblast-derived (3T3, American Tissue Type Culture Collection), neuronal derived from hippocampus (HT-22, a subclone of the HT4 cell line [28], generously provided by Dr. Dave Schubert, Salk Institute, San Diego, CA), and neuroendocrine derived from hypothalamic gonadotropin-releasing hormone neurons (GT1-7, generously provided by Dr. Pamela Mellon, University of California, San Diego, CA). Cells were maintained in 50/50 F12/Dulbecco's modified essential medium (DMEM) containing 4.5% glucose and L-glutamine (Invitrogen Inc., Carlsbad, CA), supplemented with 1x non-essential amino acids and 10% fetal bovine serum (Gemini Bioproducts, Woodland, CA). Cells were grown to 70% confluency and used within 10 passages for all experiments.
Peptides
Recombinant mouse fibroblast growth factor-8b carrier-free (FGF8b, R&D Systems, Minneapolis, MN) was reconstituted in sterile phosphate buffered saline (PBS) and diluted further to final concentrations. The FGF/VEGF receptor tyrosine kinase inhibitor PD173074 (Calbiochem, Gibbstown, NJ) was reconstituted in 100% dimethlysulfoxide (DMSO) and used at a final concentration of 100 nM in 0.01% DMSO. PD173074 is an ATP-competitive reversible inhibitor of FGF and VEGF receptors (IC50 = 21.5 nM for FGFR1, Calbiochem) and has been extensively characterized [29], [30], [31], [32].
Reporter plasmid constructs
The full-length mouse (-3446 to +24) GnRH promoter was subcloned into the promoterless firefly luciferase vector (pXP2) and has been extensively characterized [33], [34]. The renilla luciferase pGL4 reporter construct (Invitrogen Inc., Carlsbad, CA) was used as an internal control for calculating plasmid transfection efficiency.
Transient Transfections
GT1-7 cells were plated at a density of 0.2×105 cells/well in 96-well plates for 48 hours prior to transfection to achieve a final confluency of 70–80%. All constructs were transfected in replicates of six wells within each assay, and each transfection assay was repeated a minimum of 3 times. Further, each experiment was performed using a minimum of 3 different preparations for each plasmid reporter construct. Transfections were carried out using a lipid-mediated transfection reagent according to manufacturer's instructions (Fugene6, Roche Molecular Biomedical, Indianapolis, IN). Cells were incubated with transfection media complex overnight followed by replacement with phenol red-free 50/50 F12/DMEM containing 1.0% stripped fetal bovine serum (Hyclone Laboratories, Logan, UT) to minimize the presence of exogenous growth factors in the cell culture media. Notably, all experiments were replicated using media containing 10%, 1%, and dextran-charcoal stripped serum, and no differences were observed. Therefore, all data reported herein are taken from experiments where cells were kept in media containing 1% FBS. Thirty-six hours after transfection, cells were incubated with media containing 0.01% PBS, 50 ng/ml FGF8b, or 100 nM PD173074 for eight hours and then lysed for dual luciferase analysis. Luciferase activity was measured using the Dual Luciferase Reporter Assay system (Promega Corp., Madison, WI) according to manufacturer's instructions. Relative light units were measured using the Synergy HT multimode plate reader (BioTek Instruments Corp., Winooski, VT). Luciferase substrates (100 µl/well) were added to cells using automatic injectors attached to the plate reader.
RNA isolation
3T3, HT-22, and GT1-7 cells were plated at a density of 2.0×105 cells/well in a six-well plate. Cells were allowed to grow in regular media containing 10% FBS for 24–48 hours until 70–80% confluent. Twenty-four hours prior to treatment, cells were washed once with PBS followed by the addition of phenol red-free 50/50 F12/DMEM containing 1% FBS (Hyclone Laboratories, Logan, UT) to minimize the presence of exogenous growth factors in the cell culture media. Notably, all experiments were replicated using media containing 10%, 1%, and dextran-charcoal stripped serum and no differences were observed. All data reported herein are from cells kept in media containing 1% FBS. On the day of treatment, cells were treated with vehicle, 5 or 50 ng/ml FGF8b, 100 nM PD173074, or a combination (FGF8b + PD173074) for 4 or 8 hours. All treatments were done in replicates of 6 wells. Cells were washed once with cold PBS, lysed with Trizol reagent, and total RNA isolated according to manufacturer's instructions (Invitrogen Inc., Carlsbad, CA). Following isolation, genomic DNA contamination was removed using DNAfree reagents (Stratagene, a division of Agilent Corp., La Jolla, CA) according to manufacturer's instructions. Quantification of total RNA was performed using a Nanodrop spectrophotometer, and samples with an OD 260∶280 of 1.7 – 1.9 were used for subsequent reverse transcription assays.
Reverse Transcription
Total RNA (1 µg from cell culture experiments for quantitative real-time RT-PCR; 0.5 µg for FGF8 detection RT-PCR in embryonic nasal explants at E11.5, adult hypothalamus, 3T3 cells, HT-22 cells, and GT1-7 cells) was combined with 0.5 µg oligo d(T), heated to 65°C and rapidly cooled on ice. The RNA-primer mix was combined with M-MLV buffer (50 mM Tris-HCl pH 8.3, 75 mM KCl, 3 mM MgCl2), 10 mM DTT, 0.5 mM dNTP and 0.5 mM M-MLV reverse transcriptase (Invitrogen Inc., Carlsbad, CA). Reverse transcriptase reaction was performed by incubating for 10 minutes at room temp, 50 minutes at 42°C, then 95°C for 5 minutes to terminate the reaction.
RT-PCR
FGF8 Detection. 2 µl of cDNA template, prepared by reverse transcription reaction as described above, was added to a master mix containing 1x Go Taq flexi buffer (Promega Corp.), 1.5 mM MgCl, 200 µM dNTP mix, 0.5 µM forward and reverse primer (see primer sequences below), and 2 U Go Taq flexi DNA polymerase (Promega Corp.,). RT-PCR was performed using the Eppendorf Realplex thermocycler with the following reaction conditions: 95°C for 10 min., 40 repeated cycles including denature (95°C), annealing (62°C), and extension (72°C), final extension for 5 min. at 72°C. PCR products were resolved on a 2% agarose gel and compared with a DNA ladder of known size (Fisher Scientific, Exactgene) to verify product size.
Quantitative Polymerase Chain Reaction
qPCR was performed using FastStart DNA Master SYBR Green I according to manufacturer's instructions (Roche Molecular Biomedical, Indianapolis, IN). Master mix containing MgCL2, SYBR Green, and primer pairs (0.25 µM) were aliquotted into 96-well plates followed by the addition of 1/20th of the reverse transcription reaction (cDNA). No template controls received DNA-free water of the same volume. All cDNA samples were tested in triplicate within an assay and each experiment was repeated three times. Real-time PCR reactions were carried out using the Eppendorf Realplex thermocycler with the following conditions: 95°C for 10 min., 40 repeated cycles including denature (95°C), annealing (60°C), and extension (72°C) with fluorescence detection at the end of each 72°C step, and then melted with continuous fluorescence detection to 95°C. PCR products were resolved on a 2% agarose gel and compared with a DNA ladder of known size (Fisher Scientific, Exactgene 50bp ladder) to confirm product size, and to verify specificity, the products were subjected to a thermal melting curve analysis to determine if the Tm of the product was consistent with the calculated theoretical Tm based on sequence. Primer sequences are as follows: GnRH: forward - 5′CTGCTGACTGTGTGTTTGGAAGG 3′; reverse – 5′CCTGGCTTCCTCTTCAATCA 3′. FGFR1: forward – 5′ATGGTTGACCGTTCTGGAAG 3′; reverse – TGGCTATGGAAGTCGCTCTT 3′; FGFR3: forward – 5′GAGACTTGGCTGCCAGAAAC 3′; reverse – 5′GGAGGACACCAAAAGACCA 3′. FGF8: forward – 5′ GAGCAACGGCAAAGGCAAGG 3′; reverse – 5′ CTCAACTACCCGCCCTTCAC 3′. The FGF sequence targets exon 5 which is present in all FGF8 splice variants.
All samples were first normalized to the constitutively expressed hypoxanthine phosphoribosyl transferase 1 (HPRT) housekeeping gene followed by absolute quantification extrapolated from known quantities in a standard curve. The Eppendorf Realplex software plots a standard curve of the crossing line intercepts of the standard vs. the known concentrations of these standards. The crossing line intercept is parallel to the x-axis on a graph of fluorescent intensity vs. cycle number and occurs at a point where the template amplification enters the logarithmic phase of the curve. Samples with higher concentrations of starting material enter the logarithmic phase earlier than samples with a lower concentration of starting material and consequently, have a smaller crossing point value. The crossing line intercept of unknowns is then compared with that of known values to calculate the actual amount. Data are represented as mRNA copies/µg total RNA.
Statistics
Data were analyzed by one-way ANOVA followed by Tukey's HSD test. Differences were considered significant when P<0.05. All transfection data are represented as percent change compared to vehicle-treated, empty vector controls.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
References
- 1.Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, et al. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reuss B, von Bohlen und Halbach O. Fibroblast growth factors and their receptors in the central nervous system. Cell Tissue Res. 2003;313:139–157. doi: 10.1007/s00441-003-0756-7. [DOI] [PubMed] [Google Scholar]
- 3.Alarid ET. Lives and times of nuclear receptors. Mol Endocrinol. 2006;20:1972–1981. doi: 10.1210/me.2005-0481. [DOI] [PubMed] [Google Scholar]
- 4.Bridges TM, Lindsley CW. G-protein-coupled receptors: from classical modes of modulation to allosteric mechanisms. ACS Chem Biol. 2008;3:530–541. doi: 10.1021/cb800116f. [DOI] [PubMed] [Google Scholar]
- 5.Marchese A, Chen C, Kim YM, Benovic JL. The ins and outs of G protein-coupled receptor trafficking. Trends Biochem Sci. 2003;28:369–376. doi: 10.1016/S0968-0004(03)00134-8. [DOI] [PubMed] [Google Scholar]
- 6.Scholpp S, Groth C, Lohs C, Lardelli M, Brand M. Zebrafish fgfr1 is a member of the fgf8 synexpression group and is required for fgf8 signalling at the midbrain-hindbrain boundary. Dev Genes Evol. 2004;214:285–295. doi: 10.1007/s00427-004-0409-1. [DOI] [PubMed] [Google Scholar]
- 7.Jen WC, Gawantka V, Pollet N, Niehrs C, Kintner C. Periodic repression of Notch pathway genes governs the segmentation of Xenopus embryos. Genes Dev. 1999;13:1486–1499. doi: 10.1101/gad.13.11.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamar E, Deblandre G, Wettstein D, Gawantka V, Pollet N, et al. Nrarp is a novel intracellular component of the Notch signaling pathway. Genes Dev. 2001;15:1885–1899. doi: 10.1101/gad.908101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Onichtchouk D, Chen YG, Dosch R, Gawantka V, Delius H, et al. Silencing of TGF-beta signalling by the pseudoreceptor BAMBI. Nature. 1999;401:480–485. doi: 10.1038/46794. [DOI] [PubMed] [Google Scholar]
- 10.Niehrs C, Meinhardt H. Modular feedback. Nature. 2002;417:35–36. doi: 10.1038/417035a. [DOI] [PubMed] [Google Scholar]
- 11.Chung WC, Moyle SS, Tsai PS. Fibroblast growth factor 8 signaling through fibroblast growth factor receptor 1 is required for the emergence of gonadotropin-releasing hormone neurons. Endocrinology. 2008;149:4997–5003. doi: 10.1210/en.2007-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai PS, Moenter SM, Postigo HR, El Majdoubi M, Pak TR, et al. Targeted expression of a dominant-negative fibroblast growth factor (FGF) receptor in gonadotropin-releasing hormone (GnRH) neurons reduces FGF responsiveness and the size of GnRH neuronal population. Mol Endocrinol. 2005;19:225–236. doi: 10.1210/me.2004-0330. [DOI] [PubMed] [Google Scholar]
- 13.Kwabi-Addo B, Ozen M, Ittmann M. The role of fibroblast growth factors and their receptors in prostate cancer. Endocr Relat Cancer. 2004;11:709–724. doi: 10.1677/erc.1.00535. [DOI] [PubMed] [Google Scholar]
- 14.Neufeld G, Gospodarowicz D. The identification and partial characterization of the fibroblast growth factor receptor of baby hamster kidney cells. J Biol Chem. 1985;260:13860–13868. [PubMed] [Google Scholar]
- 15.Moscatelli D, Quarto N. Transformation of NIH 3T3 cells with basic fibroblast growth factor or the hst/K-fgf oncogene causes downregulation of the fibroblast growth factor receptor: reversal of morphological transformation and restoration of receptor number by suramin. J Cell Biol. 1989;109:2519–2527. doi: 10.1083/jcb.109.5.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estival A, Monzat V, Miquel K, Gaubert F, Hollande E, et al. Differential regulation of fibroblast growth factor (FGF) receptor-1 mRNA and protein by two molecular forms of basic FGF. Modulation of FGFR-1 mRNA stability. J Biol Chem. 1996;271:5663–5670. doi: 10.1074/jbc.271.10.5663. [DOI] [PubMed] [Google Scholar]
- 17.Guo Q, Li JY. Distinct functions of the major Fgf8 spliceform, Fgf8b, before and during mouse gastrulation. Development. 2007;134:2251–2260. doi: 10.1242/dev.004929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Q, Li K, Sunmonu NA, Li JY. Fgf8b-containing spliceforms, but not Fgf8a, are essential for Fgf8 function during development of the midbrain and cerebellum. Dev Biol. 338:183–192. doi: 10.1016/j.ydbio.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andre F, Job B, Dessen P, Tordai A, Michiels S, et al. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res. 2009;15:441–451. doi: 10.1158/1078-0432.CCR-08-1791. [DOI] [PubMed] [Google Scholar]
- 20.Jang JH. Reciprocal relationship in gene expression between FGFR1 and FGFR3: implication for tumorigenesis. Oncogene. 2005;24:945–948. doi: 10.1038/sj.onc.1208254. [DOI] [PubMed] [Google Scholar]
- 21.Tomlinson DC, Lamont FR, Shnyder SD, Knowles MA. Fibroblast growth factor receptor 1 promotes proliferation and survival via activation of the mitogen-activated protein kinase pathway in bladder cancer. Cancer Res. 2009;69:4613–4620. doi: 10.1158/0008-5472.CAN-08-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnaud-Dabernat S, Kritzik M, Kayali AG, Zhang YQ, Liu G, et al. FGFR3 is a negative regulator of the expansion of pancreatic epithelial cells. Diabetes. 2007;56:96–106. doi: 10.2337/db05-1073. [DOI] [PubMed] [Google Scholar]
- 23.Arnaud-Dabernat S, Yadav D, Sarvetnick N. FGFR3 contributes to intestinal crypt cell growth arrest. J Cell Physiol. 2008;216:261–268. doi: 10.1002/jcp.21401. [DOI] [PubMed] [Google Scholar]
- 24.Inglis-Broadgate SL, Thomson RE, Pellicano F, Tartaglia MA, Pontikis CC, et al. FGFR3 regulates brain size by controlling progenitor cell proliferation and apoptosis during embryonic development. Dev Biol. 2005;279:73–85. doi: 10.1016/j.ydbio.2004.11.035. [DOI] [PubMed] [Google Scholar]
- 25.Thomson RE, Pellicano F, Iwata T. Fibroblast growth factor receptor 3 kinase domain mutation increases cortical progenitor proliferation via mitogen-activated protein kinase activation. J Neurochem. 2007;100:1565–1578. doi: 10.1111/j.1471-4159.2006.04285.x. [DOI] [PubMed] [Google Scholar]
- 26.Gill JC, Moenter SM, Tsai PS. Developmental regulation of gonadotropin-releasing hormone neurons by fibroblast growth factor signaling. Endocrinology. 2004;145:3830–3839. doi: 10.1210/en.2004-0214. [DOI] [PubMed] [Google Scholar]
- 27.Walker DM, Juenger TE, Gore AC. Developmental profiles of neuroendocrine gene expression in the preoptic area of male rats. Endocrinology. 2009;150:2308–2316. doi: 10.1210/en.2008-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morimoto BH, Koshland DE., Jr Induction and expression of long- and short-term neurosecretory potentiation in a neural cell line. Neuron. 1990;5:875–880. doi: 10.1016/0896-6273(90)90347-i. [DOI] [PubMed] [Google Scholar]
- 29.Grand EK, Chase AJ, Heath C, Rahemtulla A, Cross NC. Targeting FGFR3 in multiple myeloma: inhibition of t(4;14)-positive cells by SU5402 and PD173074. Leukemia. 2004;18:962–966. doi: 10.1038/sj.leu.2403347. [DOI] [PubMed] [Google Scholar]
- 30.Koziczak M, Holbro T, Hynes NE. Blocking of FGFR signaling inhibits breast cancer cell proliferation through downregulation of D-type cyclins. Oncogene. 2004;23:3501–3508. doi: 10.1038/sj.onc.1207331. [DOI] [PubMed] [Google Scholar]
- 31.Mohammadi M, Froum S, Hamby JM, Schroeder MC, Panek RL, et al. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. Embo J. 1998;17:5896–5904. doi: 10.1093/emboj/17.20.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skaper SD, Kee WJ, Facci L, Macdonald G, Doherty P, et al. The FGFR1 inhibitor PD 173074 selectively and potently antagonizes FGF-2 neurotrophic and neurotropic effects. J Neurochem. 2000;75:1520–1527. doi: 10.1046/j.1471-4159.2000.0751520.x. [DOI] [PubMed] [Google Scholar]
- 33.Chandran UR, Attardi B, Friedman R, Dong KW, Roberts JL, et al. Glucocorticoid receptor-mediated repression of gonadotropin-releasing hormone promoter activity in GT1 hypothalamic cell lines. Endocrinology. 1994;134:1467–1474. doi: 10.1210/endo.134.3.8119188. [DOI] [PubMed] [Google Scholar]
- 34.Pak TR, Chung WC, Roberts JL, Handa RJ. Ligand-independent effects of estrogen receptor beta on mouse gonadotropin-releasing hormone promoter activity. Endocrinology. 2006;147:1924–1931. doi: 10.1210/en.2005-1297. [DOI] [PubMed] [Google Scholar]