Abstract
Purpose
To explore the association between CYP3A4 and CYP3A5 gene polymorphisms and blood pressure response to amlodipine among participants from the African-American Study of Kidney Disease and Hypertension Trial randomized to amlodipine (n = 164).
Methods
Cox proportional hazards models were used to determine the risk of reaching a target mean arterial pressure (MAP) of ≤107 mm Hg by CYP3A4 (A–392G and T16090C) and CYP3A5 (A6986G) gene polymorphisms, stratified by MAP randomization group (low or usual) and controlling for other predictors for blood pressure response.
Results
Women randomized to a usual MAP goal with an A allele at CYP3A4 A–392G were more likely to reach a target MAP of 107 mm Hg. The adjusted hazard ratio (AA/AG compared to GG) with 95% confidence interval was 3.41 (1.20–9.64; p = 0.020). Among participants randomized to a lower MAP goal, those with the C allele at CYP3A4 T16090C were more likely to reach target MAP: The adjusted hazard ratio was 2.04 (1.17–3.56; p = 0.010). After adjustment for multiple testing using a threshold significance level of p = 0.016, only the CYP3A4 T16090C SNP remained significant. CYP3A5 A6986G was not associated with blood pressure response.
Conclusions
Our findings suggest that blood pressure response to amlodipine among high-risk African-Americans appears to be determined by CYP3A4 genotypes, and sex specificity may be an important consideration. Clinical applications of CYP3A4 genotype testing for individualized treatment regimens warrant further study.
Key Words: Pharmacogenetics, Hypertension, Amlodipine, Renal failure, CYP3A polymorphisms, AASK, African-Americans
Introduction
Approximately 16.8% of the adult US population is diagnosed with chronic kidney disease, and those with preexisting hypertension are almost twice as likely to develop chronic renal disease [1]. African-Americans are at highest risk for end-stage renal disease from hypertension [1], and this disparity remains despite adjustment for age, gender, and co-morbid illness [2,3,4,5]. Genetic factors may influence response to antihypertensive drug treatment, resulting in variability between ethnic groups or races.
CYP3A is a subfamily of the cytochrome P450 superfamily of genes involved in the metabolism of endogenous compounds and over 50% of pharmaceutical agents; CYP3A4is largely responsible for adult drug metabolism [6,7,8], including antihypertensive drug disposition [9,10,11]. CYP3A4 and CYP3A5 polymorphisms have been shown to vary between ethnic (racial) groups, and CYP3A4 polymorphisms may account for much of the observed variability in drug efficacy and toxicity. While CYP3A5 is highly polymorphic, variation at this gene does not appear to result in significant differences in hepatic adult drug metabolism [7,8]. Hepatic P450 gene regulation is also influenced by endogenous steroid hormones, resulting in gender-specific differences in CYP3A expression and drug handling [7,12].
Amlodipine is a long-acting antihypertensive dihydropyridine calcium channel blocker (CCB) that prevents the influx of calcium ions in vascular smooth muscle cells, thereby acting primarily as a peripheral vasodilator. It is slowly absorbed and is extensively metabolized in the liver, mainly by CYP3A4 [11,13]. CCBs have been seen to be more effective in African-Americans than in other ethnic/ancestral groups, suggesting that variation in CYP3A4 and possibly CYP3A5 function or expression might account for differences in CCB drug response between such ethnic groups [14,15,16]. However, CCB pharmacokinetics and efficacy have not as yet been conclusively shown to vary by CYP3A4 and CYP3A5 polymorphisms (table 1) [17,18,19]. In this study, we explored the association of polymorphisms on the CYP3A4 and CYP3A5 genes with blood pressure response to amlodipine in African-American men and women with early hypertensive nephrosclerosis based on randomized trial data from the NIDDK African-American Study of Kidney Disease and Hypertension Study (AASK) [20].
Table 1.
CYP3A polymorphisms: previously observed effects
| Polymorphism | Effects on gene transcription or antihypertensive drug pharmacokinetics |
|---|---|
| CYP3A5 A6986G | G-allele associations (also referenced as CYP3A5∗3C) |
| Ref SNP 776746 | No differences in felodipine pharmacokinetics [17] |
| Intron-3 | No significant differences in nifedipine metabolism [18] |
| CYP3A4 A–392G | G-allele associations (also referenced as CYP3A4∗1B) |
| Ref SNP 2740574 | Increased gene transcription [30] |
| Promoter region | No significant effect on nifedipine metabolism [19] |
| CYP3A4 T16090C | C-allele associations |
| Ref SNP 2246709 | Interaction with CYP2D6; higher metabolism of [26] and lower sympathetic activity from yohimbine [43] |
| Intron-7 | No significant association with CYP3A4 enzyme activity [37, 44] |
Methods
Participants and Study Design
Details of the AASK study [21,22] and details of the AASK Genetics Study [23,24] have been previously published. Informed consent was received from AASK participants at the time of the original study and the study protocol approved by institutional human subjects review boards. Briefly, the original trial included of 1,094 self-identified African-American men and women between the ages of 18 and 70 years and diagnosed with hypertensive kidney disease (glomerular filtration rate between 20 and 65 ml/min per 1.73 m2), randomized to either an angiotensin-converting enzyme inhibitor, a β-adrenergic receptor blocker or a dihydropyridine CCB. Participants were also randomized to either a low mean arterial pressure (MAP) (≤92 mm Hg) or a usual MAP (102–107 mm Hg) group. Unlike the drug randomization group that was double blinded, the MAP group was known by both investigators and research subjects, and medications were titrated based on the MAP treatment group. Of those eligible and consented to participate in the AASK Genetics Study (n = 850), 159 (19.04%) were randomized to the CCB amlodipine. Ten of these observations were removed due to incomplete or missing genotype data; 149 observations were used in the present analysis.
Genomics
Details of DNA extraction and handling have been previously published [23,24]. Two CYP3A4 and one CYP3A5 single nucleotide gene polymorphisms (SNPs) located on chromosome 7q21 [25] known to influence the metabolism of several medications were selected (table 1): (1) CYP3A4 T16090C, a T to C polymorphism within intron-7, 16,090 base pairs from the CYP3A4 cap site (RefSNP rs2246709) [26], (2) CYP3A4 A–392G, a functional A to G polymorphism, and also the most frequently studied genetic variant of CYP3A4 [27], located in the 5′-flanking region (also known as CYP3A4 * 1B; RefSNP rs2740574) [8], and (3) CYP3A5 A6986G, a functional A to G polymorphism located in intron-3 of CYP3A5, responsible for alternative splicing and protein truncation (also known as CYP3A5*3; RefSNP rs776746) [6,10,25]. Genotype assignments were made by capturing images with a flatbed scanner and using proprietary research software developed by Roche Molecular Systems (Pleasanton, Calif., USA). Hardy-Weinberg equilibrium was assessed by using the Pearson goodness of fit (χ2) test statistic.
Statistical Analysis
Because of multiple comparison testing (3 genetic variants), statistical significance was determined by an α-level of 0.016. Descriptive statistics were performed to assess baseline characteristics and preliminary data among CCB genotypes, between males and females, and between low and usual target MAP groups using one-way ANOVAs or t tests. To eliminate any potential for obscuring the genotype-drug response relationship as a result of progressive renal disease, analyses were restricted to the first year after randomization. Haplotypes could not be inferred with reasonable certainty for many individuals; therefore, analyses were limited to individual genotypes.
As described in our previous studies, the main outcome was number of days to reach a target MAP of ≤107 mm Hg. Cox proportional hazards models were used to determine the hazard (relative risk) of reaching target MAP of 107 mm Hg. Cox proportional hazards assumptions were tested and, if violated, the model was stratified by a third variable. A stepwise approach was used to explore the effects of other potential predictors of blood pressure response and variables found to contribute significantly to the overall model were included in the adjusted models. These variables included baseline demographics (i.e., age, insurance status, education), anthropometrics (body mass index, baseline MAP, heart rate), renal function (glomerular filtration rate), other biochemical laboratory measures (serum lipids, creatinine, blood urea nitrogen, liver function tests, etc.) and co-morbid disease such as coronary artery disease. It should be noted that other antihypertensive medications (other than one of the three study drugs) were used to manage blood pressure and were systematically added according to a standardized protocol across study sites. Therefore, the average number of daily medications during the first year of follow-up was used as a proxy measure to control for antihypertensive medication use.
With blood pressure randomization (low or usual MAP) and gender stratification, this study had 80% power to detect a hazard ratio of 2.6. Statistical analyses were performed using SAS statistical software, version 9.1, programming package (SAS Institute, Inc., Cary, N.C., USA) and STATA statistical software, release 9.2 and 10.0 (StataCorp LP, College Station, Tex., USA).
Drug Specificity and Population Stratification
The metabolism and mechanism of action for amlodipine on blood pressure is different from angiotensin enzyme inhibitors. Therefore, participants randomized to ramipril (n = 302) were used as specificity controls. Previously, we had also shown that population stratification did not contribute to blood pressure response in this dataset using a generalized analysis of molecular variance (GAMOVA) [23,28].
Results
Genomics
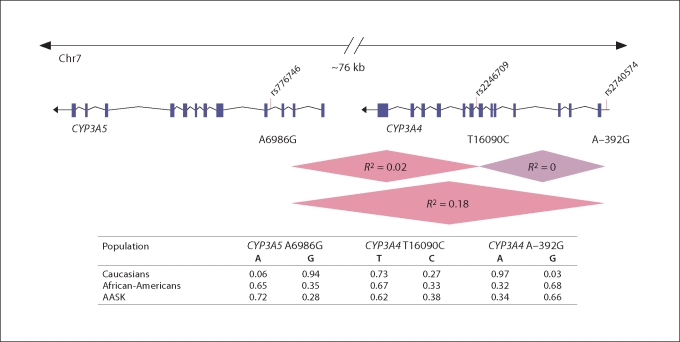
Of the 164 participants randomized to amlodipine, 137 (84%) were genotyped at CYP3A4 A–392G, 143 (87%) at CYP3A4 T16090C and 145 (88%) at CYP3A5 A6986G. All three polymorphisms were in Hardy-Weinberg equilibrium and allele frequencies were similar to frequencies listed on dbSNP (http://www.ncbi.nlm.nih.gov/SNP/; fig. 1). However, there were too few A/A homozygotes at A–392G (n = 10) for a meaningful analysis; homozygous A/A were combined with heterozygous A/G individuals, resulting in an A-dominant model for analysis. Similarly, there were too few homozygous C/C individuals at T16090C (n = 13) and too few G/G individuals at A6986G (n = 8), resulting in C- and G-dominant models for analyses at these sites, respectively. Some linkage disequilibrium was detected between CYP3A5 A6986G and CYP3A4 A–392G (R2 = 0.18) and between CYP3A5 A6986G and CYP3A4 T16090C (R2 = 0.02); however, CYP3A4 A–392G and CYP3A4 T16090C were not in linkage disequilibrium in this population (R2 = 0) (fig. 1).
Fig. 1.
CYP3A4 and CYP3A5 polymorphisms population frequencies. CYP3A4 and CYP3A5 are located in tandem on chromosome 7q21. Two CYP3A4 and one CYP3A5 SNP were selected: (1) CYP3A4 T16090C, a T to C polymorphism within intron-7, 16,090 base pairs from the CYP3A4 cap site (rs2246709); (2) CYP3A4 A–392G, an A to G polymorphism, and also the most common variant of CYP3A4, located in the 5′-flanking region (rs2740574), and (3) CYP3A5 A6986G, an A to G polymorphism located in intron-3, responsible for alternative splicing and protein truncation (rs776746). The extent of linkage disequilibrium between loci is represented as R-squared (R2). Allele frequencies in Caucasian and African-Americans made available dbSNP (http://www.ncbi.nlm.nih.gov/SNP) or the Pharmacogenomics Knowledge Base (http://www.pharmgkb.org) and in the AASK population are also shown.
Ancillary AASK Study Participants: Characteristics and Preliminary Outcomes
Baseline characteristics were similar between our amlodipine group and the original AASK study cohort [29]. There was complete follow-up in the first year and 11,811 days at risk for analysis. There were 76 men (mean age 52 ± 11) and 61 women (mean age 53 ± 10). Of the 135 participants, only 11 did not reach a target MAP of 107 mm Hg.
There were no significant baseline differences by genotype (appendix 1). There were more men who did not reach target MAP compared to females (12 vs. 1.5%; p = 0.04). Those randomized to lower MAP had lower total cholesterol (206 vs. 222 mg/dl; p = 0.04), required more daily medications (3.89 vs. 3.16 medications; p = 0.0003) and had a lower average MAP in the first year after randomization (96 vs. 104 mm Hg; p < 0.0001). Men had higher average MAPs in the first year after randomization (102 vs. 98 mm Hg; p = 0.003) had higher measures of serum creatinine (2.39 vs. 1.70 mg/dl; p < 0.0001), and on average used more daily medications (3.81 vs. 3.20 medications; p = 0.05; appendix 2).
Survival Analyses: Time to Reach Target MAP (≤107 mm Hg)
Hazard ratios (‘relative risk’ of reaching target MAP ≤107 mm Hg) and 95% confidence intervals were estimated using a Cox proportional hazards model (table 2). Inclusion of low and usual MAP groups violated the Cox proportional hazards assumption; hence, these groups were analyzed in separate models. The effects of gender were also explored by gender stratification.
Table 2.
Cox proportional hazards model in amlodipine-treated subjects, stratified by CYP3A4 and CYP3A5 genotypes
| Model |
CYP3A4 A–392G (A/A or A/G vs. G/G) (n = 135) |
CYP3A4 T16090C (C/C or T/C vs. T/T) (n = 145) |
CYP3A5 A6986G (G/G or A/G vs. A/A) (n = 146) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| hazard ratio | 95% CI | p∗ | hazard ratio | 95% CI | p∗ | hazard ratio | 95% CI | p∗ | |
| Low MAP | |||||||||
| Overall | |||||||||
| Unadjusted | 0.92 | 0.56, 1.54 | 0.76 | 2.18 | 1.27, 3.76 | 0.005 | 0.81 | 0.50, 1.29 | 0.37 |
| Adjusted∗ | 0.81 | 0.48, 1.38 | 0.44 | 2.04 | 1.17, 3.56 | 0.01 | 0.99 | 0.60, 1.61 | 0.95 |
| Males | |||||||||
| Unadjusted | 1.01 | 0.45, 2.30 | 0.98 | 1.94 | 0.95, 4.20 | 0.07 | 0.84 | 0.42, 1.68 | 0.61 |
| Adjusted∗ | 1.10 | 0.46, 2.65 | 0.832 | 2.22 | 1.03, 4.79 | 0.04 | 0.85 | 0.42, 1.73 | 0.66 |
| Females | |||||||||
| Unadjusted | 0.87 | 0.43, 1.78 | 0.70 | 2.22 | 1.01, 5.01 | 0.05 | 1.06 | 0.50, 2.24 | 0.88 |
| Adjusted∗ | 0.67 | 0.31, 1.42 | 0.29 | 2.33 | 1.01, 5.40 | 0.05 | 1.20 | 0.55, 2.59 | 0.649 |
| Usual MAP | |||||||||
| Overall | |||||||||
| Unadjusted | 0.91 | 0.53, 1.56 | 0.72 | 0.81 | 0.49, 1.32 | 0.39 | 0.85 | 0.53, 1.37 | 0.51 |
| Adjusted∗ | 1.48 | 0.82, 2.69 | 0.19 | 0.77 | 0.46, 1.31 | 0.34 | 0.60 | 0.35, 1.02 | 0.06 |
| Males | |||||||||
| Unadjusted | 0.97 | 0.45, 2.10 | 0.94 | 0.77 | 0.39, 1.49 | 0.43 | 0.75 | 0.39, 1.42 | 0.38 |
| Adjusted∗ | 1.31 | 0.58, 2.93 | 0.51 | 0.82 | 0.46, 1.85 | 0.92 | 0.73 | 0.38, 1.38 | 0.32 |
| Females | |||||||||
| Unadjusted | 2.81 | 0.93, 5.59 | 0.07 | 0.95 | 0.46, 2.09 | 0.95 | 1.44 | 0.64, 3.21 | 0.38 |
| Adjusted∗ | 3.41 | 1.20, 9.64 | 0.02 | 1.39 | 0.59, 3.27 | 0.46 | 1.16 | 0.52, 2.60 | 0.72 |
p ≤ 0.05 bolded, marginal significance; p ≤ 0.016 significant after adjustment for three multiple comparisons.
Adjustment for baseline MAP, average number of daily antihypertensive medications, age, gender (for combined male and female models) and/or serum creatinine.
CYP3A4 A–392G
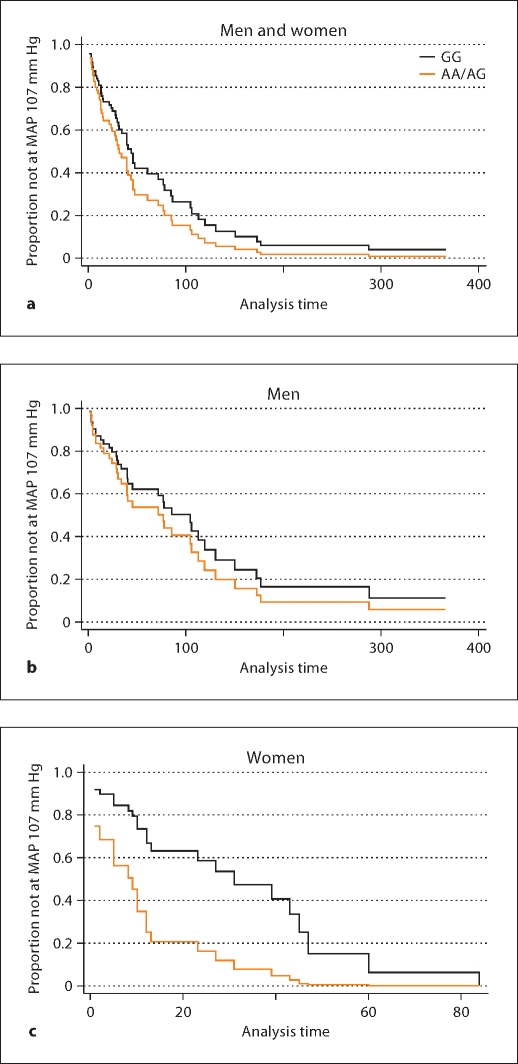
There were no significant differences in the risk of reaching a target MAP ≤107 mm Hg among men and women (combined) randomized to a low MAP goal by CYP3A4 A–392G genotypes (table 2). However, gender stratification suggested that women with an A/A or A/G randomized to a usual MAP genotype were more likely to reach a target MAP of 107 mm Hg. The hazard ratio (A/A and A/G compared to G/G), with 95% confidence interval, was 2.81 (0.93–5.59; p = 0.07). After adjusting for average number of daily medications, this association was stronger, though of borderline significance with correction for multiple comparisons: 3.41 (1.20–9.64; p = 0.02) (fig. 2).
Fig. 2.
Adjusted Cox regression survival curves by CYP3A4 promoter A–392G (usual MAP goal 102–107 mm Hg treatment group). Among men and women randomized to usual MAP treatment goal, there was not a significant difference in rate of reaching a target MAP of 107 mm Hg by A–392G genotypes (a; adjusted p = 0.77). Upon gender stratification, there was no difference among men (b; adjusted p = 0.82); however, women with A/A or A/G (compared to G/G) genotype were over 3 times more likely to reach a target MAP of 107 mm Hg (c; adjusted p = 0.02).
CYP3A4 T16090C
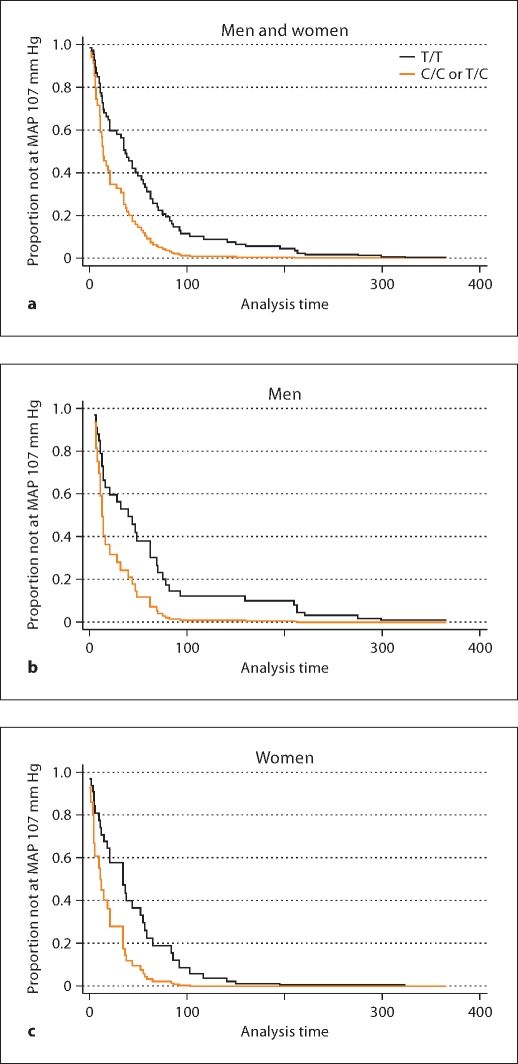
No significant differences were found by CYP3A4 T16090C genotypes among those randomized to the usual MAP group (table 2). However, among those randomized to the low MAP group, individuals with a C/C or T/C genotype were twice as likely to reach a target MAP 107 mm Hg compared to T/T individuals: 2.04 (1.17–3.56; adjusted p = 0.01). Similar results were found among men and women: 2.22 (1.03–4.79; adjusted p = 0.04) and 2.33 (1.01–5.40; adjusted p = 0.048), respectively (fig. 3).
Fig. 3.
Adjusted Cox regression survival curves by CYP3A4 T16090C (low MAP goal <92 mm Hg). Among more aggressively managed men and women, those with a C/C or T/C compared to a T/T genotype were twice as likely to reach a target MAP of 107 mm Hg (a; adjusted p = 0.01). Stratified analyses suggested similar results among men (b; adjusted p = 0.05) and women (c; adjusted p = 0.05).
CYP3A5 A6986G
There were no associations between CYP3A5 A6986G genotypes and risk of reaching target MAP of 107 mm Hg in this dataset.
Drug Specificity
Among those randomized to ramipril, there were no associations between CYP3A4 A–392G, CYP3A4 T16090C, or CYP3A5 A6986G genotypes and time to reach target MAP among men or women randomized to a low or usual MAP.
Discussion
Study Overview and Main Results
The CYP3A4 A–392G promoter polymorphism was predictive of blood pressure response among African-American women with early hypertensive nephrosclerosis randomized to a usual MAP group (102–107 mm Hg); though of marginal significance after correction for multiple comparisons, women with an A allele were over 3 times more likely to reach a target MAP of 107 mm Hg (p = 0.02). A less studied CYP3A4 T16090C intronic SNP [26] was significantly associated with blood pressure response to amlodipine among those randomized to a lower MAP group (≤92 mm Hg). Men and women with a C allele at T16090C were twice as likely to reach target MAP 107 mm Hg (p = 0.01). CYP3A4 genotype and blood pressure response appeared to be specific to amlodipine since there were no associations among men or women randomized to ramipril. As shown in our previous studies, population stratification did not appear to be contributing to the relationship between genotype and blood pressure response in this dataset [23,24]. CYP3A5 A6986G was not associated with blood pressure response in this study.
CYP3A4 G–392A
G–392A is a functional promoter polymorphism [27] and is a likely candidate for the regulation of gene transcription [30]. Based on this study, women with an A-allele were more likely to respond to amlodipine, suggesting that the G-allele carriers were more rapid metabolizers of amlodipine. The G-allele at A–392G has been associated with higher drug dose requirements in some [31,32,33] but not all studies [19,34]. However, most studies did not examine the relationship in a sex-specific manner. We found that the effects of CYP3A4 A–392G genotype on therapeutic response to amlodipine differed in men and women, suggesting differential CYP activity in the male versus female hormonal milieu. P-glycoprotein levels, a multidrug transporter that mediates cellular transport of several drugs, have been shown to be 2–3 times higher in men in comparison to women. P-glycoprotein-mediated efflux of drugs such as amlodipine may result in higher intracellular concentrations in women, resulting in more variation in metabolism according to CYP3A4 genotype [35,36]. Another CYP3A4 variant (rs464637, in close proximity to T16090C) also affected CYP expression and activity in a sex-dependent way [37], and CYP3A4 gene polymorphisms have been associated with early onset of puberty in females [38,39,40] and hormone-dependent cancers (e.g. endometrial, breast, prostate) [27,41,42].
CYP3A4 T16090C
Men and women with a C-allele who were more aggressively treated (i.e., randomized to the low MAP of ≤92 mm Hg) were more likely to respond to amlodipine (fig. 3), suggesting that carriers of the C-allele may be poorer metabolizers of amlodipine. Associations for men and women were of similar magnitude and direction, suggesting that the effect of T16090C genotypes is not sex-specific. The intronic CYP3A4 T16090C SNP was first studied by Le Corre et al. [26]. In this original study, the metabolism of yohimbine (α2-adrenergic antagonist that results in increased sympathetic activity) was shown to be dependent on both CYP3A4 T16090C and CYP2D6. Among those in the fast CYP2D6 metabolizer group, the C-allele was associated with faster metabolism of yohimbine in comparison to the T-allele, suggesting functionality at (or in close proximity to) CYP3A4 T16090C. In concordance with these findings, Bharucha et al. [43] found higher sympathetic activity with yohimbine among those classified as poor metabolizers (in comparison to fast metabolizers) according to CYP2D6 and CYP3A4 T16090C status. While our results suggest decreased metabolic activity with the C-allele at CYP3A4 T16090C, epistatic effects or interactions with CYPD26 were not explored here. Other studies have not found significant effects of the T16090C polymorphism on gene expression [37,44] or N-dealkylation of midazolam or verapamil [37,45].
CYP3A5 A6986G
While CCBs are also CYP3A5 substrates, the lack of an association between blood pressure response to amlodipine and CYP3A5 A6986G is consistent with previous studies suggesting no difference in the pharmacokinetics of nifedipine or felodopine (dihydropyridine CCBs, similar to amlodipine) by CYP3A5 A6986G genotypes [17,18]. CYP3A5 is highly polymorphic and CYP3A5 variants result in decreased CYP3A5 protein expression. However, CYP3A5 does not appear to contribute significantly to adult hepatic metabolism in comparison to CYP3A4. Furthermore, the CYP3A5 A6986G allele is not common among African-Americans (28% minor allele frequency in our study population), resulting in limited power to study the effects of this polymorphism with this dataset [46].
Study Advantages and Limitations
The AASK Genomics Study presented a unique opportunity to explore associations between genotype and antihypertensive blood pressure responses in the powerful setting of a large randomized clinical trial using a novel application of survival methodology to the analysis of blood pressure and to explore gene-by-gender interactions with respect to blood pressure response to amlodipine. While we considered adjustment for multiple comparison testing by the three genotypes (significant p value 0.016), we did not adjust for data stratification by MAP randomization group (low and usual MAP) and gender. Results from this study must also be confirmed in independent populations, in other clinical trials or other ethnic/racial groups. TagSNPs (markers for a haplotype block) could also be used in future studies to more comprehensively cover the region of the genome around these sites. Given the modest number of AASK subjects randomized to amlodipine, this study was not designed to detect small differences in genotype-dependent blood pressure response, nor was it adequately powered to study the effects of rare alleles.
Conclusions
Blood pressure response to amlodipine among high-risk African-American patients appears to be partly determined by CYP3A4 genotypes, and sex specificity may be an important consideration. These initial results are provocative and suggest that CYP3A4 genotype testing may be clinically predictive of blood pressure response to amlodipine and perhaps other CCBs. The clinical use of CYP3A4 genotype testing for individualized treatment regimens should be further explored in larger hypertension clinical studies.
Acknowledgements
The authors appreciate the support of the NIH/NCMHD-sponsored (MD000220) EXPORT/CRCHD Minority Health Center, as well as the NIH/NCRR-sponsored (RR00827) General Clinical Research Center. We appreciate the assistance of the UCSD General Clinical Research Center (RR00827), and the UCSD Comprehensive Research Center in Health Disparities (CRCHD, MD000220).
Funding: Satellite Research, NIH (K23 RR020822-01A1, DK048689, RR000071, DK057867, DK60702, RR11145) and the Department of Veterans Affairs.
Appendix 1
Baseline characteristics and preliminary outcomes stratified by CYP3A genotypes
|
CYP3A4 A–392G (n = 135) |
CYP3A4 T16090C (n = 145) |
CYP3A5 A6986G (n = 146) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/A (n = 10) | A/G (n = 70) | G/G (n = 55) | p | C/C (n = 13) | T/C (n = 79) | T/T (n = 53) | p | A/A (n = 74) | A/G (n = 64) | G/G (n = 8) | p | |
| Number randomized to low MAP (%) | 4 (40) | 31 (44) | 32 (58) | 7 (53) | 40 (49) | 28 (53) | 39 (52) | 31 (47) | 5 (63) | |||
| Number of men (%) | 6 (60) | 48 (69) | 20 (37) | 8 (61) | 45 (55) | 29 (55) | 33 (44) | 41 (62) | 8 (100) | |||
| Number who did not reach target (%) | 0 | 9 (13) | 2 (4) | 0.26 | 1 (8) | 3 (4) | 4 (8) | 0.66 | 1 (1) | 6 (9) | 1 (12) | 0.07 |
| Age, years | 53 ± 10 | 53 ± 11 | 52 ± 11 | 0.96 | 53 ± 12 | 52 ± 11 | 53 ± 10 | 0.90 | 53 ± 11 | 53 ± 11 | 49 ± 13 | 0.74 |
| Baseline MAP, mm Hg | 106 ± 21 | 114 ± 19 | 115 ± 15 | 0.38 | 117 ± 22 | 115 ± 17 | 114 ± 18 | 0.69 | 114 ± 16 | 115 ± 20 | 120 ± 18 | 0.58 |
| Body mass index, kg/m2 | 31 ± 7 | 30 ± 7 | 32 ± 6 | 0.52 | 29 ± 7 | 31 ± 7 | 31 ± 6 | 0.36 | 31 ± 7 | 30 ± 6 | 30 ± 5 | 0.30 |
| Serum creatinine, mg/dl | 2.24 ± 0.82 | 2.21 ± 0.90 | 1.88 ± 0.72 | 0.07 | 2.03 ± 1.0 | 2.01 ± 0.74 | 2.11 ± 0.88 | 0.56 | 1.99 ± 0.78 | 2.01 ± 0.83 | 2.8 ± 0.66 | 0.09 |
| Glomerular filtration rate ml/min per 1.73 m2 | 42 ± 15 | 45 ± 13 | 47 ± 12 | 0.39 | 46 ± 12 | 47 ± 13 | 45 ± 12 | 0.54 | 46 ± 12 | 47 ± 13 | 39 ± 14 | 0.55 |
| Total cholesterol, mg/dl | 227 ± 54 | 207 ± 50 | 219 ± 46 | 0.26 | 202 ± 29 | 212 ± 50 | 218 ± 48 | 0.28 | 216 ± 47 | 211 ± 47 | 216 ± 57 | 0.69 |
| Average number of daily medications | 2.89 ± 1.17 | 3.76 ± 1.34 | 3.41 ± 1.12 | 0.07 | 3.3 ± 1.1 | 3.5 ± 1.3 | 3.6 ± 1.3 | 0.36 | 3.5 ± 1.2 | 3.6 ± 1.3 | 3.3 ± 1.4 | 0.78 |
| Average MAP in first year after randomization | 99 ± 6 | 101 ± 7 | 100 ± 6 | 0.55 | 101 ± 10 | 100 ± 6 | 100 ± 7 | 0.94 | 100 ± 6 | 101 ± 7 | 98 ± 9 | 0.94 |
Appendix 2
Baseline characteristics and outcomes stratified by gender or MAP group
| AASK participant characteristic | All (n = 149) | Gender |
MAP group |
||||
|---|---|---|---|---|---|---|---|
| males (n = 82) | females (n = 67) | p | low MAP (≤92 mm Hg) (n = 75) | usual MAP (≤107 mm Hg) (n = 74) | p | ||
| Number randomized to low MAP (%) | 75 (50) | 39 (52) | 36 (48) | – | – | ||
| Number of men (%) | 82 (55) | – | – | 39 (52) | 43 (58) | ||
| Number who did not reach target (%) | 11 (7) | 10 (12) | 1 (1.5) | 0.04 | 4 (5) | 7 (10) | 0.46 |
| Age at randomization, years | 53 ± 11 | 52 ± 11 | 54 ± 11 | 0.27 | 53 ± 11 | 53 ± 11 | 0.93 |
| Baseline MAP, mm Hg | 115 ± 18 | 117 ± 20 | 112 ± 15 | 0.05 | 116 ± 20 | 112 ± 15 | 0.13 |
| Body mass index, kg/m2 | 31 ± 7 | 31 ± 6 | 30 ± 7 | 0.63 | 31 ± 6 | 31 ± 7 | 0.82 |
| Serum creatinine, mg/dl | 2.05 ± 0.82 | 2.33 ± 0.89 | 1.70 ± 0.55 | <0.0001 | 1.94 ± 0.73 | 2.15 ± 0.89 | 0.13 |
| Glomerular filtration rate, ml/min per 1.73 m2 | 46 ± 13 | 47 ± 13 | 45 ± 12 | 0.17 | 46 ± 12 | 46 ± 13 | 0.82 |
| Total cholesterol, mg/dl | 214 ± 47 | 211 ± 48 | 217 ± 47 | 0.46 | 206 ± 39 | 222 ± 54 | 0.04 |
| Average number of daily medications | 3.53 ± 1.25 | 3.81 ± 1.24 | 3.20 ± 1.19 | 0.005 | 3.89 ± 1.15 | 3.16 ± 1.25 | 0.0003 |
| Average MAP in first year after randomization | 100 ± 7 | 102 ± 7 | 98 ± 5 | 0.003 | 96 ± 6 | 104 ± 5 | <0.0001 |
p < 0.05 bolded.
Footnotes
V.B. and E.P.G. contributed equally to this work.
References
- 1.Rizzo JA, Simons WR. Variations in compliance among hypertensive patients by drug class: implications for health care costs. Clin Ther. 1997;19:1446–1457. doi: 10.1016/s0149-2918(97)80018-5. [DOI] [PubMed] [Google Scholar]
- 2.Appel LJ, et al. The rationale and design of the AASK cohort study. J Am Soc Nephrol. 2003;14(suppl 2):S166–S172. doi: 10.1097/01.asn.0000070081.15137.c0. [DOI] [PubMed] [Google Scholar]
- 3.Chobanian AV, et al. The Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: the JNC 7 Report. JAMA. 2003;289:2560–2572. doi: 10.1001/jama.289.19.2560. [DOI] [PubMed] [Google Scholar]
- 4.McClellan W, Tuttle E, Issa A. Racial differences in the incidence of hypertensive end-stage renal disease are not entirely explained by differences in the prevalence of hypertension. Am J Kidney Dis. 1988;12:285–290. doi: 10.1016/s0272-6386(88)80221-x. [DOI] [PubMed] [Google Scholar]
- 5.Safford MM, et al. Understanding racial disparities in hypertension control: intensity of hypertension medication treatment in the REGARDS study. Ethn Dis. 2007;17:421–426. [PubMed] [Google Scholar]
- 6.Hustert E, et al. The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics. 2001;11:773–779. doi: 10.1097/00008571-200112000-00005. [DOI] [PubMed] [Google Scholar]
- 7.Kreutz R, et al. The role of the cytochrome P450 3A5 enzyme for blood pressure regulation in the general Caucasian population. Pharmacogenet Genomics. 2005;15:831–837. doi: 10.1097/01213011-200512000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Lamba JK, et al. Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev. 2002;54:1271–1294. doi: 10.1016/s0169-409x(02)00066-2. [DOI] [PubMed] [Google Scholar]
- 9.Park JY, et al. Effect of CYP3A5*3 genotype on the pharmacokinetics and pharmacodynamics of alprazolam in healthy subjects. Clin Pharmacol Ther. 2006;79:590–599. doi: 10.1016/j.clpt.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 10.Kim KA, et al. Effect of polymorphic CYP3A5 genotype on the single-dose simvastatin pharmacokinetics in healthy subjects. J Clin Pharmacol. 2007;47:87–93. doi: 10.1177/0091270006295063. [DOI] [PubMed] [Google Scholar]
- 11.Kim KA, et al. Effect of CYP3A5*3 genotype on the pharmacokinetics and pharmacodynamics of amlodipine in healthy Korean subjects. Clin Pharmacol Ther. 2006;80:646–656. doi: 10.1016/j.clpt.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Davis BR, et al. Antihypertensive therapy, the α-adducin polymorphism, and cardiovascular disease in high-risk hypertensive persons: the Genetics of Hypertension-Associated Treatment Study. Pharmacogenomics J. 2007;7:112–122. doi: 10.1038/sj.tpj.6500395. [DOI] [PubMed] [Google Scholar]
- 13.Brooks BA, et al. The gene CYP3 encoding P450pcn1 (nifedipine oxidase) is tightly linked to the gene COL1A2 encoding collagen type 1 α on 7q21-q22.1. Am J Hum Genet. 1988;43:280–284. [PMC free article] [PubMed] [Google Scholar]
- 14.Cushman WC, et al. Regional and racial differences in response to antihypertensive medication use in a randomized controlled trial of men with hypertension in the United States. Department of Veterans Affairs Cooperative Study Group on Antihypertensive Agents. Arch Intern Med. 2000;160:825–831. doi: 10.1001/archinte.160.6.825. [DOI] [PubMed] [Google Scholar]
- 15.Damasceno A, et al. Efficacy of captopril and nifedipine in black and white patients with hypertensive crisis. J Hum Hypertens. 1997;11:471–476. doi: 10.1038/sj.jhh.1000428. [DOI] [PubMed] [Google Scholar]
- 16.Weir MR, et al. Influence of race and dietary salt on the antihypertensive efficacy of an angiotensin-converting enzyme inhibitor or a calcium channel antagonist in salt-sensitive hypertensives. Hypertension. 1998;31:1088–1096. doi: 10.1161/01.hyp.31.5.1088. [DOI] [PubMed] [Google Scholar]
- 17.Guo LQ, et al. Different roles of pummelo furanocoumarin and cytochrome P450 3A5*3 polymorphism in the fate and action of felodipine. Curr Drug Metab. 2007;8:623–630. doi: 10.2174/138920007781368917. [DOI] [PubMed] [Google Scholar]
- 18.Fukuda T, et al. CYP3A5 genotype did not impact on nifedipine disposition in healthy volunteers. Pharmacogenomics J. 2004;4:34–39. doi: 10.1038/sj.tpj.6500218. [DOI] [PubMed] [Google Scholar]
- 19.Ball SE, et al. Population distribution and effects on drug metabolism of a genetic variant in the 5′ promoter region of CYP3A4. Clin Pharmacol Ther. 1999;66:288–294. doi: 10.1016/S0009-9236(99)70037-8. [DOI] [PubMed] [Google Scholar]
- 20.Wright JT, Jr, et al. Effect of blood pressure lowering and antihypertensive drug class on progression of hypertensive kidney disease: results from the AASK trial. JAMA. 2002;288:2421–2431. doi: 10.1001/jama.288.19.2421. [DOI] [PubMed] [Google Scholar]
- 21.Agodoa LY, et al. Effect of ramipril vs. amlodipine on renal outcomes in hypertensive nephrosclerosis: a randomized controlled trial. JAMA. 2001;285:2719–2728. doi: 10.1001/jama.285.21.2719. [DOI] [PubMed] [Google Scholar]
- 22.Norris KC, et al. Baseline predictors of renal disease progression in the African-American Study of Hypertension and Kidney Disease. J Am Soc Nephrol. 2006;17:2928–2936. doi: 10.1681/ASN.2005101101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhatnagar V, et al. Angiotensin-converting enzyme gene polymorphism predicts the time-course of blood pressure response to angiotensin-converting enzyme inhibition in the AASK trial. J Hypertens. 2007;25:2082–2092. doi: 10.1097/HJH.0b013e3282b9720e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhatnagar V, et al. G-protein-coupled receptor kinase 4 polymorphisms and blood pressure response to metoprolol among African-Americans: sex specificity and interactions. Am J Hypertens. 2009;22:332–338. doi: 10.1038/ajh.2008.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuehl P, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 26.Le Corre P, et al. Human sympathetic activation by α2-adrenergic blockade with yohimbine: bimodal, epistatic influence of cytochrome P450-mediated drug metabolism. Clin Pharmacol Ther. 2004;76:139–153. doi: 10.1016/j.clpt.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 27.Rebbeck TR, et al. Modification of clinical presentation of prostate tumors by a novel genetic variant in CYP3A4. J Natl Cancer Inst. 1998;90:1225–1229. doi: 10.1093/jnci/90.16.1225. [DOI] [PubMed] [Google Scholar]
- 28.Nievergelt CM, Libiger O, Schork NJ. Generalized analysis of molecular variance. PLoS Genet. 2007;3:e51. doi: 10.1371/journal.pgen.0030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Contreras G, et al. Blood pressure control, drug therapy, and kidney disease. Hypertension. 2005;46:44–50. doi: 10.1161/01.HYP.0000166746.04472.60. [DOI] [PubMed] [Google Scholar]
- 30.Amirimani B, et al. Increased transcriptional activity of the CYP3A4*1B promoter variant. Environ Mol Mutagen. 2003;42:299–305. doi: 10.1002/em.10199. [DOI] [PubMed] [Google Scholar]
- 31.Anglicheau D, et al. Consequences of genetic polymorphisms for sirolimus requirements after renal transplant in patients on primary sirolimus therapy. Am J Transplant. 2005;5:595–603. doi: 10.1111/j.1600-6143.2005.00745.x. [DOI] [PubMed] [Google Scholar]
- 32.Hesselink DA, et al. Genetic polymorphisms of the CYP3A4, CYP3A5, and MDR-1 genes and pharmacokinetics of the calcineurin inhibitors cyclosporine and tacrolimus. Clin Pharmacol Ther. 2003;74:245–254. doi: 10.1016/S0009-9236(03)00168-1. [DOI] [PubMed] [Google Scholar]
- 33.Wei-lin W, et al. Tacrolimus dose requirement in relation to donor and recipient ABCB1 and CYP3A5 gene polymorphisms in Chinese liver transplant patients. Liver Transpl. 2006;12:775–780. doi: 10.1002/lt.20709. [DOI] [PubMed] [Google Scholar]
- 34.Wandel C, et al. CYP3A activity in African-American and European-American men: population differences and functional effect of the CYP3A4*1B5-promoter region polymorphism. Clin Pharmacol Ther. 2000;68:82–91. doi: 10.1067/mcp.2000.108506. [DOI] [PubMed] [Google Scholar]
- 35.Meibohm B, Beierle I, Derendorf H. How important are gender differences in pharmacokinetics? Clin Pharmacokinet. 2002;41:329–342. doi: 10.2165/00003088-200241050-00002. [DOI] [PubMed] [Google Scholar]
- 36.Cummins CL, Wu CY, Benet LZ. Sex-related differences in the clearance of cytochrome P450 3A4 substrates may be caused by P-glycoprotein. Clin Pharmacol Ther. 2002;72:474–489. doi: 10.1067/mcp.2002.128388. [DOI] [PubMed] [Google Scholar]
- 37.Schirmer M, et al. Sex-dependent genetic markers of CYP3A4 expression and activity in human liver microsomes. Pharmacogenomics. 2007;8:443–453. doi: 10.2217/14622416.8.5.443. [DOI] [PubMed] [Google Scholar]
- 38.Kadlubar FF, et al. The CYP3A4*1B variant is related to the onset of puberty, a known risk factor for the development of breast cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:327–331. [PubMed] [Google Scholar]
- 39.Lai J, et al. CYP gene polymorphisms and early menarche. Mol Genet Metab. 2001;74:449–457. doi: 10.1006/mgme.2001.3260. [DOI] [PubMed] [Google Scholar]
- 40.Xin X, et al. Association study of four activity SNPs of CYP3A4 with the precocious puberty in Chinese girls. Neurosci Lett. 2005;381:284–288. doi: 10.1016/j.neulet.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 41.Chu W, et al. Association between CYP3A4 genotype and risk of endometrial cancer following tamoxifen use. Carcinogenesis. 2007;28:2139–2142. doi: 10.1093/carcin/bgm087. [DOI] [PubMed] [Google Scholar]
- 42.Plummer SJ, et al. CYP3A4 and CYP3A5 genotypes, haplotypes, and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:928–932. [PubMed] [Google Scholar]
- 43.Bharucha AE, et al. Relationship of cytochrome P450 pharmacogenetics to the effects of yohimbine on gastrointestinal transit and catecholamines in healthy subjects. Neurogastroenterol Motil. 2008;20:891–899. doi: 10.1111/j.1365-2982.2008.01124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perera MA, et al. Prediction of CYP3A4 enzyme activity using haplotype tag SNPs in African-Americans. Pharmacogenomics J. 2009;9:49–60. doi: 10.1038/tpj.2008.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perera MA, et al. Prediction of CYP3A4 enzyme activity using haplotype tag SNPs in African-Americans. Pharmacogenomics J. 2009;9:49–60. doi: 10.1038/tpj.2008.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ingelman-Sundberg M, et al. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther. 2007;116:496–526. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]