Abstract
Motivation: Intriguingly, sequence analysis of genomes reveals that a large number of genes are unique to each organism. The origin of these genes, termed ORFans, is not known. Here, we explore the origin of ORFan genes by defining a simple measure called ‘composition bias’, based on the deviation of the amino acid composition of a given sequence from the average composition of all proteins of a given genome.
Results: For a set of 47 prokaryotic genomes, we show that the amino acid composition bias of real proteins, random ‘proteins’ (created by using the nucleotide frequencies of each genome) and ‘proteins’ translated from intergenic regions are distinct. For ORFans, we observed a correlation between their composition bias and their relative evolutionary age. Recent ORFan proteins have compositions more similar to those of random ‘proteins’, while the compositions of more ancient ORFan proteins are more similar to those of the set of all proteins of the organism. This observation is consistent with an evolutionary scenario wherein ORFan genes emerged and underwent a large number of random mutations and selection, eventually adapting to the composition preference of their organism over time.
Contact: ron@biocoml.ls.biu.ac.il
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
One of the consistent and intriguing observations that emerged from the extensive availability of whole genome sequences is the large number of genes that seem to encode unique proteins that do not exist in other organisms or exist only in very closely related organisms. This appears to be the case even when using sophisticated sequence comparison methods like psi-blast. These genes are commonly called ORFan genes (Fischer and Eisenberg, 1999) and the resulting proteins are called ORFan proteins. It was estimated (Siew and Fischer, 2004) that 20–30% of the open reading frames in a given genome are ORFans. These observations were made early in the history of genome analysis, when only the first organisms had been sequenced. At that time, the common explanation was that these ‘unique’ genes were not unique at all, but that not enough organisms had been sequenced to follow the evolution of these genes. However, while the fraction of ORFan genes has somewhat decreased as more genomes became available, it also became clear that the phenomenon is not a mere artifact of a small sample size; rather, even with the availability of the complete sequence of close to a thousand genomes, there remain a large number of genes whose evolutionary history is not accounted for.
Several possible explanations were given over the years for this phenomenon (for a review see Daubin and Ochman, 2004; Long et al., 2003). One explanation is that those sequences are not real genes; rather they may represent open reading frames that are never expressed. However, several studies have shown (Siew and Fischer, 2003) that these genes are expressed, and some of the resulting proteins have even been subjected to 3D structure analysis (Siew and Fischer, 2004). Another possible explanation is that these genes came from lateral gene transfer (LGT). In order for this explanation to be logically relevant, the transfer should have come from genomes whose sampling is sparse and thus can serve as a reservoir for the unique genes. Viral and phage genomes have been suggested as such a reservoir (Cortez et al., 2009), although other recent studies (Yin and Fischer, 2006) have indicated that LGT cannot be the source for most of these genes. Another possibility that has been suggested (Long et al., 2003) is that ORFan genes originated from ancestral genes, but because of fast evolutionary rate, these genes have mutated their sequence to such an extent that their ancestors are no longer recognizable. Yet another possibility is that some ORFan genes emerged de novo from non-coding regions of each genome without being inherited in the regular evolutionary path, for example by shifting the reading frame, a phenomenon called overprinting (see e.g. Delaye et al., 2008) or by mutations that change non-coding regions to open reading frames (Long et al., 2003).
It is well known that protein sequences have different amino acids compositions, i.e. not all of the 20 amino acids appear in proteins with the same frequency of 5%. The composition is different for different organisms (Pe'er et al., 2004) and has both evolutionary and functional origin and consequences. Furthermore, within genomes, different sequences have different compositions, and we term the deviation of each sequence from the average composition of the organism as composition bias. The composition of sequences has been used as one of the main considerations in predicting the sub-cellular localization of proteins (Nair and Rost, 2003). Furthermore, it was observed (Ofran and Margalit, 2006) that proteins of the same fold but with unrelated sequences have similar amino acid composition, and thus it was suggested that amino acid composition can help to predict structural folds.
In an attempt to shed light on the evolutionary history of ORFan proteins, we explored the composition bias of 47 prokaryotic organisms. Using a simple measure, we compared the composition bias of the set of all proteins, of random proteins (Section 2) and of ORFan proteins in each genome. We show that the tendency of ORFan proteins to behave like the rest of the proteins increases with the evolutionary age of the ORFans, and we discuss the evolutionary implications of this observation.
2 METHODS
Dataset: our dataset started with a collection of 66 representative prokaryotic genomes (Yan and Moult, 2005). For these genomes, the sequences and annotations were taken from NCBI. In each organism, ORFan genes were defined as genes that appear only in their genome-of-origin, and do not have any similar genes based on a Blast run against the entire NCBI-NR database. The parameters used to define a hit were E-value <0.05, and match-length that covers at least 50% of the ORFan length. Three organisms were found to have another related organism with which they share many proteins (Escherichia coli with Shigella, Methanococcus jannaschii with Methanocaldococcus and Nostoc sp PCC 7120 with Anabaena). For these organisms, we considered genes as ORFans if they appeared only in their original genome and in the very close relative.
The analysis presented here included the 47 genomes (out of the 66) that have at least 25 ORFan genes each. The list includes 38 bacteria and 9 archaea (see Supplementary Table S1). All together, we identified 8812 ORFan genes out of 123 444 genes (∼7%) in our ensemble (Supplementary Table S2).
Real and random proteins: we called the set of all proteins in an organism the set of ‘real proteins’. For each organism, three sets of random sequences were created. Each set was matched to the set of real proteins in terms of the number of proteins and the length of each protein. The three sets of random sequences were created based on the nucleotide frequency (i.e. the A/C/G/T ratios) of (i) the entire genome, (ii) of only the coding regions and (iii) only of the non-coding regions. The nucleotide sequences were translated to amino acid sequences. All sequences started with ATG, and to maintain protein length, stop codons, when generated, were replaced by other random codons.
Translating proteins from intergenic regions: nucleotide sequences that came from intergenic regions of the genome (i.e. regions that are between genes and do not reside on the opposite strand of coding regions) were translated into proteins. Stop codons were skipped over and the subsequent nucleotides were used to create additional codons such that the lengths of these ‘proteins’ match those of the real proteins. Since the number of intergenic regions in prokaryotic genomes is limited, the set sampled was 1/3 the number of proteins in each genome.
Translating antisense proteins: for each protein, the antisense sequence (i.e. its reverse complement sequence) was also translated. Thus, the size of this set of proteins was the same as that of the real proteins in each genome. Stop codons were skipped over.
Calculating composition bias: for each organism, a reference composition vector was calculated by averaging the percentage of each of the 20 amino acids in each protein over all real proteins of the genome according to NCBI annotation. For each amino acid, the SD about the average composition was also determined. For each amino acid sequence s, the composition bias cs was calculated by comparing its composition vector to the reference composition vector according to:
| (1) |
Where i ranges over the 20 amino acids, fis is the i-th component of the composition vector of the given sequence, fir is the i-th component of the reference composition vector and SDir is the standard deviation of the reference composition of the i-th amino acid about its average. Thus, each ‘protein’ is assigned a composition bias, and for a set of ‘proteins’ in a given organism, we created a histogram of these composition biases, showing for each composition bias bin (in the range of 0–60), the fraction of the proteins in this bin. The histogram is presented as a continuous line. For the ORFan proteins, the histogram was scaled up by a factor based on the fraction of ORFan proteins. For example, if an organism has 4000 proteins of which 400 are ORFans, then the values in the ORFan histogram were scaled up by a factor of 10 (4000/400).
We have also compared the frequency vector of the given sequence to that of the reference vector using a root mean square (RMS) measure. The RMS measure square the difference in the frequency of corresponding amino acids without normalization to the SD weight that appear in Equation (1). The results of using these two measures were similar and thus in this article we show only the results of the first measure.
Calculating the difference between histograms of composition biases: the difference between the histograms was calculated as the difference between the average values of each histogram. We also measured the difference by computing the overlap between the two histograms. We then calculated the ratio between the overlap of the ORFans and real protein and the overlap of the ORFans and the random proteins. This ratio reflects the relatedness between the ORFan proteins to either the real proteins (low ratio values) or the random proteins (high ratio values).
Phylogenetic tree construction and measuring the relative age of ORFans: since ORFan genes are found in only a single branch of the phylogenetic tree, they must have emerged subsequent to the split of that branch. The maximum age of the ORFan genes must be smaller than the age of the organism, and thus it assumed to be proportional to the relative length of their terminal branch (Supplementary Figure S1). This length was used to estimate the approximate relative age of the ORFan.
The tree was constructed incorporating information from accepted amino acid substitutions per site between species in a large set of protein families, to avoid bias issues encountered in methods where only a small number of families is used. The set of orthologous protein domain families previously constructed (Yan and Moult, 2005) from 66 prokaryotic genomes was used. Multiple sequence alignments for each family were generated using MUSCLE (Edgar, 2004). The estimated accepted amino acid substitutions per site between each pair of domains ‘i’ and ‘j’ in each family ‘u’, S(i, j, u) were then obtained using the PROTDIST module in PHYLIP (Felsenstein, 1989) with the Jones–Taylor–Thornton amino acid substitution matrix (Jones et al., 1992).
The numbers of accepted substitutions per site for each family were placed on the same scale by comparison with the average rates of substitution Sref(i, j) between genomes ‘i’ and ‘j’ in a set of 14 highly conserved families. The rate of sequence change for each family, C(u), relative the reference set was obtained using a robust least median square procedure (Rousseeuw and Leroy, 1987), finding the C(u) which minimizes the median value of the set [r(i, j, u)2], where
and the set includes contributions from all pairs of genomes ‘i’ and ‘j’ with members in family ‘u’ (Yan, 2005). A robust method was necessary to avoid distortions of C(u) arising from anomalous S(i, j, u) values caused by LGT and other factors.
The intergenome distance, D(i, j), between each pair of genomes ‘i’ and ‘j’ was estimated using D(i, j) = <S(i, j, u)/C(u)>u where the average includes contributions from all families with members in genomes ‘i’ and ‘j’. A phylogenetic tree was then built from this distance matrix, using the neighbor joining method (Saitou and Nei, 1987), as implemented in PHYLIP.
Correlations were calculated using the standard Pearson correlation coefficient comparing the two properties of interest (e.g. number of ORFans and relative age) for each of the 47 genomes.
3 RESULTS
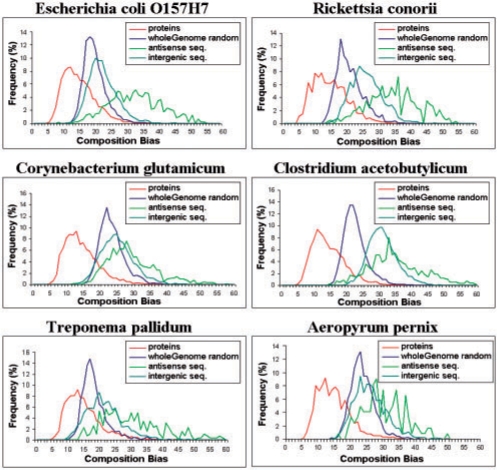
The list of the genomes and the number of proteins and ORFan proteins in each genome is given in Supplementary Table S1. We started by calculating the composition bias of the proteins translated from the coding genes, from random ‘genes’ (based on the nucleotide frequency of the entire genome), from the antisense strands of the coding genes and from the intergenic regions of the genome. The histograms (Section 2) of the composition biases are shown in Figure 1 for six organisms: E.coli. Rickettsia conorii, Treponema pallidum, Corynebacterium glutamicum, Aeropyrum pernix and Clostridium acetobutylicum. As the number of sequences in the intergenic sets is 1/3 of those of the other sets (Section 2), their histograms were normalized by multiplying each value by 3. The real proteins have smaller composition bias (as is evident from the fact that their histogram is the leftmost) than the composition bias of the random proteins. This is expected since the compositions are compared with the average compositions of the real proteins. Surprisingly, the composition bias of the antisense proteins is greater than that of the random proteins. We also note that for all organisms the composition bias histogram of ‘proteins’ translated from intergenic regions are further shifted to the right.
Fig. 1.
Histograms showing the composition bias for six organisms of several sets of proteins. All histograms were computed by using the average composition vector of the real proteins as the reference, and the composition bias of each protein relative to that reference was calculated. As expected, the real proteins have the smallest bias. Surprisingly, the composition bias of intergenic ‘proteins’ is significantly larger than that of random or antisense proteins. For the random genes, very similar results were obtained when using either the genome's coding or non-coding frequencies.
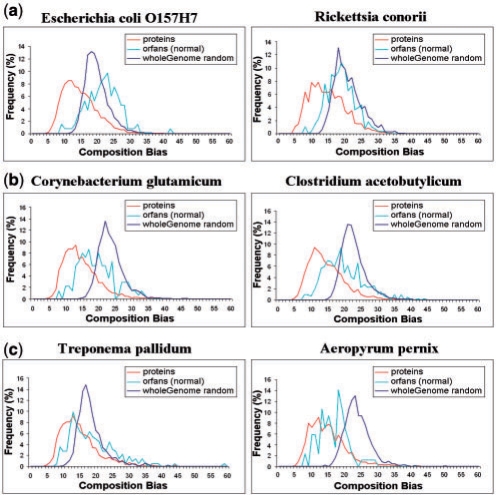
We next compared the composition bias of ORFan proteins to that of the other datasets.Figure 2 shows the composition bias histograms of real proteins, random proteins and ORFan proteins (scaled up to the size of the other groups) for several genomes. We noticed that ORFan proteins from different species behave differently in their similarity to either the coding or the random groups. The ORFans of E.coli and R.conorii look like random proteins (Fig. 2a), the ORFans of Treponema pallidum and Aeropyrum pernix resemble real proteins (Fig. 2c) while the ORFan proteins of Corynebacterium glutamicum and Clostridium acetobutylicum have intermediate assignments (Fig. 2b).
Fig. 2.
Histograms of the composition bias of the set of ORFan proteins are compared with the composition bias of all proteins and of random proteins for six organisms. Since there are fewer ORFan proteins, their histograms were scaled up accordingly (the results were validated to ensure that they are not due to sampling effects). In the two examples in the top panel (a), the ORFan proteins behave like random proteins; in the two examples in the bottom panel (c), the ORFans behave like the real proteins; and the behavior of the examples in the middle panel (b) is intermediate.
From the results of the calculations for all 47 organisms, we noticed that indeed there is a range in the similarity of the composition bias between the ORFan proteins and the real and random proteins. In an effort to understand this range, we looked at the relative age of the ORFans, as determined by the phylogenetic tree (Section 2), as a possible explanation.
First, we checked the correlation between the number of ORFans in each genome and their relative age, and found a weak correlation (0.36). A more significant correlation (0.5) was found between the relative age of the ORFans and the percentage of ORFan genes from the total number of coding genes in the organism (see scatter plots in Supplementary Figure S2).
Next, we found a surprising strong correlation coefficient of 0.59 between the relative age of the ORFans and the distance between the average composition bias of the ORFan and the random proteins. Similarly, the correlation coefficient between the relative age and the distance between the average composition bias of the ORFan and the real proteins is −0.66 (see scatter plots in Supplementary Figures S3a and b). To make sure that these high correlations are not dependent on the particular way of comparing the composition bias, we also calculated the correlation between the relative age and the ratio of the overlaps (Section 2) and got similar results (correlation coefficient of −0.58, see Supplementary Figure S3c).
4 DISCUSSION
The main finding of this study is the correlation between the relative age of the ORFans and the degree of similarity of their composition to that of the real proteins of the organism. We found a significant correlation (correlation coefficients between 0.58 and 0.66) between the relative age of the ORFans and their composition bias, as determined by various measures of the composition distance between the set of the ORFan proteins and the set of real proteins. Thus, the older the ORFans, i.e., the more ancient the organism, the more the amino acid composition of its ORFans resembles that of the rest of the proteins. Young organisms, i.e. organisms that split from their ancestor organisms more recently, tend to have ORFan genes with composition that is more different from that of the rest of the proteins, and more similar to that of the random genes.
We tested to see if there are other factors that correlate with the relative age of the ORFan proteins and with the composition bias. As expected, we found that the fraction of ORFan genes among all coding genes in each organism is correlated with the evolutionary age of the organism (correlation coefficient of 0.5). Older organisms that have, almost by definition, fewer close relatives, tend to have more ORFan genes. No other factors that we tested, including the GC content of the organism, the size of the genome and the ratio of coding to intergenic regions, showed a strong correlation (<0.3) with the ORFan behavior.
Thus, our data are consistent with a model wherein ORFan genes emerged with a composition that was similar to the random composition of the genome. Then, during evolution and due to the selective pressures that shape the composition bias of each organism, the composition of ORFan genes gradually converged to be more similar to the composition of the rest of the proteins of the genome.
We may examine the three possible explanations for the origin of ORFan genes in light of this observation. The first explanation is that ORFan genes originated from bacteriophages (see a review in Daubin and Ochman, 2004). We think that this is unlikely. First, note that bacterial genes that have known homologues in bacteriophage are not considered ORFans by our definition. Second, for six bacteria for which sufficient bacteriophages have been sequenced, we compared the composition of the ORFan genes with the composition of bacteriophage proteins and found that the composition of the ORFan genes of the bacteria is not similar to the composition of the bacteriophage proteins (see Supplementary Figure S4).
The second possible explanation is that ORFan genes emerged de novo from non-coding regions of the genome (see a review in Long et al., 2003). This is also not consistent with our observation that protein created from intergenic sequences are distinct (further to the right in Fig. 1) from the random proteins, while the ORFan proteins fall between the random and the real proteins. If ORFan proteins emerged from intergenic regions, then we would expect the ORFan genes to behave more closely to intergenic non-coding regions of the genome, and not like random sequences.
The third explanation is that ORFan genes result from a very fast evolutionary clock rate of mutations operating on genes that are under positive selection (Long et al., 2003). This explanation is the most consistent with our observations. Random mutations are likely to create nucleotide sequences that have A/C/G/T frequencies that are similar to random sequences, thus creating novel proteins whose amino acid sequences have composition bias similar to the random proteins that we have created. Over time, the sequences underwent further mutations and selection that changed their composition and brought their composition bias to be more similar to that of the rest of the proteins.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge Nuttinee's Fellowship.
Funding: This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Partial support from NIH R01GM081511 to J.M., Fulbright Fellowship for University of Thai Chamber of Commerce, University Staff Development Program to N.T. and Israel Science Foundation 1339/08 to R.U.
Conflict of Interest: none declared.
REFERENCES
- Cortez D, et al. A hidden reservoir of integrative elements is the major source of recently acquired foreign genes and ORFans in archaeal and bacterial genomes. Genome Biol. 2009;10:R65. doi: 10.1186/gb-2009-10-6-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin V, Ochman H. Start-up entities in the origin of new genes. Curr. Opin. Genet. Dev. 2004;14:616–619. doi: 10.1016/j.gde.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Delaye L, et al. The origin of a novel gene through overprinting in Escherichia coli. BMC Evol. Biol. 2008;8:31. doi: 10.1186/1471-2148-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Mathematics vs. evolution: mathematical evolutionary theory. Science. 1989;246:941–942. doi: 10.1126/science.246.4932.941. [DOI] [PubMed] [Google Scholar]
- Fischer D, Eisenberg D. Finding families for genomic ORFans. Bioinformatics. 1999;15:759–762. doi: 10.1093/bioinformatics/15.9.759. [DOI] [PubMed] [Google Scholar]
- Jones DT, et al. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Long M, et al. The origin of new genes: glimpses from the young and old. Nat. Rev. Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- Nair R, Rost B. Better prediction of sub-cellular localization by combining evolutionary and structural information. Proteins. 2003;53:917–930. doi: 10.1002/prot.10507. [DOI] [PubMed] [Google Scholar]
- Ofran Y, Margalit H. Proteins of the same fold and unrelated sequences have similar amino acid composition. Proteins. 2006;64:275–279. doi: 10.1002/prot.20964. [DOI] [PubMed] [Google Scholar]
- Pe'er I, et al. Proteomic signatures: amino acid and oligopeptide compositions differentiate among phyla. Proteins. 2004;54:20–40. doi: 10.1002/prot.10559. [DOI] [PubMed] [Google Scholar]
- Rousseeuw PJ, Leroy MA. Robust Regression and Outlier Detection. New York: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Siew N, Fischer D. Analysis of singleton ORFans in fully sequenced microbial genomes. Proteins. 2003;53:241–251. doi: 10.1002/prot.10423. [DOI] [PubMed] [Google Scholar]
- Siew N, Fischer D. Structural biology sheds light on the puzzle of genomic ORFans. J. Mol. Biol. 2004;342:369–373. doi: 10.1016/j.jmb.2004.06.073. [DOI] [PubMed] [Google Scholar]
- Yan Y. PhD Thesis. MD, USA: University of Maryland, College Park; 2005. ‘Computational analysis of Microbial genomes – operons, protein families and lateral gene transfer’. [Google Scholar]
- Yan Y, Moult J. Protein family clustering for structural genomics. J. Mol. Biol. 2005;353:744–759. doi: 10.1016/j.jmb.2005.08.058. [DOI] [PubMed] [Google Scholar]
- Yin Y, Fischer D. On the origin of microbial ORFans: quantifying the strength of the evidence for viral lateral transfer. BMC Evol. Biol. 2006;6:63. doi: 10.1186/1471-2148-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.