Abstract
Summary: This article reports the development of SDOP-DB, which can provide definite, detailed and easy comparison of experimental protocols used in mouse phenotypic analyses among institutes or laboratories. Because SDOP-DB is fully compliant with international standards, it can act as a practical foundation for international sharing and integration of mouse phenotypic information.
Availability: SDOP-DB (http://www.brc.riken.jp/lab/bpmp/SDOP/)
Contact: hmasuya@brc.riken.jp
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Recent progress in functional genomics has resulted in an enormous volume of experimental data in the biomedical field. In this context, standardized data formats to describe laboratory workflows are essential for effective archiving and sharing of experimental results. For example, the Functional Genomics Experiment (FuGE) models the components of an experimental activity that are common across different technologies including protocols, samples and data (Jones et al., 2007). Also, the Ontology for Biomedical Investigations (OBIs) provide an integrated ontology for the description of life science and clinical investigations (http://obi-ontology.org/page/Main_Page). Although both the FuGE and OBI provide general components to describe experimental protocols, there has been little effort to provide domain-specific components to directly compare differences between protocols for specific experimental assays.
For that purpose, we have developed a new data format termed Standardized Description of Operating Procedures (SDOPs), which provides an assay-specific descriptive framework and enables direct and detailed comparison of procedural parameters. Using this data format, this work presents a comparative standardized-protocol database, SDOP-DB (http://www.brc.riken.jp/lab/bpmp/SDOP/), as a practical application to share, compare and evaluate the contents of protocols to enable data comparisons and analyses in the field of large-scale mouse phenotyping.
2 DEVELOPMENT OF SDOP-DB
2.1 Development of the SDOP format
There is a long history of and extensive evidentiary support for using the mouse to study function within the mammalian genome and to model human disease. To contribute to comprehensive unraveling of the relationship between gene and phenotype as well as gene and disease, we focused on large-scale mouse phenotypic assessments such as the Japan Mouse Clinic (JMC, Wakana et al., 2009) and the European Mouse Disease Clinic (EUMODIC, http://www.eumodic.org/), which have produced a large volume of phenotyping data for characteristics such as morphology, behavior and pathology. We developed the SDOP format for each of the phenotyping analyses from these projects to provide a foundation for interpretation of the phenotype data. The SDOP targets 16 mouse phenotypic analyses that are common between JMC and EUMODIC.
To provide users with a better understanding of detailed protocol descriptions, we first developed the common framework of the SDOP format for any mouse phenotypic analyses by using elements defined in the XML schemata, ‘Phenotyping Procedures Markup Language (PPML, http://www.interphenome.org/ppxml/ppml_v1_3.html)’ and ‘Standard Operating Procedure Markup Language (SOPML, Green et al., 2005)’. Based on the common framework, ‘assay-specific SDOPs’ for the 16 phenotypic analyses were then developed by adding appropriate elements according to each detailed description of standardized protocols used in JMC and EUMODIC through thorough consultation with analysis experts from JMC (refer to Fig. 1). In principle, each of assay-specific SDOPs covers all contents of its standardized protocols.
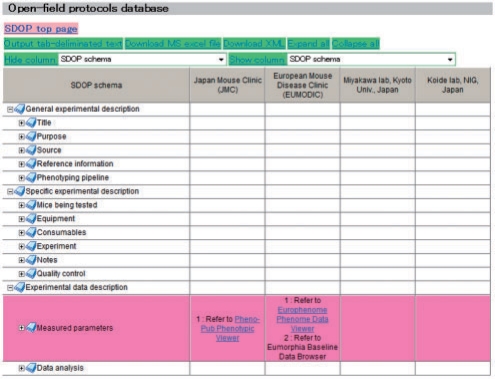
Fig. 1.
Screen shot of the SDOP display in the SDOP-DB browser. The tree-like display in the left-most column enables easy navigation of protocol contents. The tree view shown here is used for all 16 phenotypic analyses. Clicking on an item in the tree view takes users to a more detailed view of the selected item. Users can easily access phenotype databases such as ‘Pheno-Pub’ in JMC and ‘EuroPhenome’ in EUMODIC through hyperlinks. If the parameter descriptions within a row differ among institutes/laboratories, the row is represented in pink, as shown above. Clicking on ‘Expand all’ tab, users can see all the differences. The example shown is for the SDOP of open-field tests.
2.2 Implementation and user interface
The cross-browser JavaScript user interface of SDOP-DB was constructed using the dhtmlxTreeGrid API, which provides components designed to represent tabular data in hierarchical view (http://dhtmlx.com/docs/products/dhtmlxTreeGrid/). In the hierarchical view, users can click on individual parameters to expand the view to show more detailed breakdowns of the selected parameters. Each parameter of the experimental assays in this view represents the direct comparison of its values in different protocols (Fig. 1, see legend). The data description in each SDOP format is performed with MS-Excel and is then converted to XML format for dhtmlxTreeGrid representation using a custom-developed VBA program embedded in the Excel file. Users can download the Excel, XML and tab-delimited text versions of the SDOP data from the web page.
3 DISCUSSION
To ensure the integration and sharing of experimental data with high reliability, we have developed an unprecedented database, SDOP-DB, enabling users to directly compare detailed protocol differences among institutes/laboratories (refer to Supplementary Figure S1).
The Mouse Phenotype Database Integration Consortium (InterPhenome, The Mouse Phenotype Database Integration Consortium, 2007) has proposed a draft version of Minimal Information to describe Mouse Phenotyping Procedures (MIMPPs, http://mibbi.org/index.php/Projects/MIMPP). As one of the MIMPP data models, the PPML format, which is an XML schema used to describe a phenotyping procedure, is also proposed as the international standard data model. Because the assay-specific SDOPs were developed in complete compliance with the MIMPP and PPML, they are in accordance with international standards for data exchange. All of the SDOP formats are openly available on the web site to facilitate discussion on developing a common format for sharing and comparing detailed experimental protocol information among research communities. We propose that these SDOP formats can be used as a tool for protocol version management both within and between laboratories. Therefore, the SDOP-DB has the potential to function as an information infrastructure for the international sharing and integration of phenotyping protocols in mice, as well as in broader research communities.
The SDOP-DB is also helpful for identifying procedural parameter(s) that can result in differences in data between different protocols. Users can easily access phenotype data through hyperlinks to mouse phenotype databases such as ‘Pheno-Pub’ (http://www.brc.riken.jp/lab/jmc/mouse_clinic/en/m-strain_en.html) in JMC and ‘EuroPhenome’ (Morgan et al., 2010; http://www.europhenome.org/databrowser/baselineViewer.jsp) in EUMODIC, allowing association of the data with each user's own in-house data (refer to Supplementary Figure S2).
We are now preparing the SDOP-DB data in the PPML format, and further plan to expand SDOP-DB to cover all 33 phenotypic analyses that are included in high-throughput phenotyping pipelines of JMC and EUMODIC. For all analyses to be covered in SDOP-DB, users can submit in-house standardized protocols by filling in MS-Excel form downloaded from the web site.
In addition, we are now planning to annotate all the data fields and parameters in SDOP with ontologies such as Experiment ACTions (EXACT), enabling protocol data to be compared and integrated computationally. Therefore, the parameterization in SDOP would be an essential approach for the integration of protocol data with ontologies.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J.M. Hancock and A.M. Mallon for sending us the MIMPP draft and for helpful discussions. We also thank H. Yokoyama and S. Nishimura for valuable advice in developing SDOP. Finally, we thank T. Takatsuki for her help in creating the SDOP-DB web site.
Funding: Japan Society for the Promotion of Science [Grant-in-Aid for Scientific Research (B) (20300148), 2008–2009].
Conflict of Interest: none declared.
REFERENCES
- Green ECJ, et al. EMPReSS: European mouse phenotyping resource for standardized screens. Bioinformatics. 2005;21:2930–2931. doi: 10.1093/bioinformatics/bti441. [DOI] [PubMed] [Google Scholar]
- Jones AR, et al. The Functional Genomics Experiment model (FuGE): an extensible framework for standards in functional genomics. Nat. Biotechnol. 2007;25:1127–1133. doi: 10.1038/nbt1347. [DOI] [PubMed] [Google Scholar]
- Morgan H, et al. EuroPhenome: a repository for high-throughput mouse phenotying data. Nucleic Acids Res. 2010;38:D577–D585. doi: 10.1093/nar/gkp1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Mouse Phenotype Database Integration Consortium. Mouse Phenotype Database Integration Consortium: integration of mouse phenome data resources. Mamm. Genome. 2007;18:157–163. doi: 10.1007/s00335-007-9004-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakana S, et al. Introduction to the Japan Mouse Clinic at the RIKEN BioResource Center. Exp. Anim. 2009;58:443–450. doi: 10.1538/expanim.58.443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.