Abstract
Objectives:
The aim of this study was to determine the expression of PLUNC proteins in benign and malignant salivary gland tumours and thus their potential use as diagnostic and/or prognostic tools.
Materials and Methods:
A tissue microarray was assembled from 64 salivary gland tumours including adenoid cystic carcinoma, carcinoma ex-pleomorphic adenoma, mucoepidermoid carcinoma, polymorphous low-grade adenocarcinoma, pleomorphic adenoma, acinic cell carcinoma, myoepithelial carcinoma and papillary cystadenocarcinoma. Clinicopathological data were collected retrospectively and immunohistochemical analysis of 3 PLUNC proteins (SPLUNC1, SPLUNC2, and LPLUNC 1) was performed. Immunoreactivity was assessed as positive or negative.
Results:
PLUNC expression was only found in mucoepidermoid carcinomas and papillary cystadenocarcinoma, all other tumours studied were negative. Mucin plugs, mucous and intermediate cells of mucoepidermoid carcinomas were positive for LPLUNC1 and SPLUNC2 but areas composed of epidermoid and clear cells were negative for all PLUNCs. Papillary cystadenocarcinoma was positive for all PLUNCs. No correlation was found with tumour grade or outcome.
Conclusions:
Intense expression of two PLUNC proteins in mucous cells and mucin plugs of mucoepidermoid carcinoma and papillary cystadenocarcinoma indicate they could be used as additional diagnostic tools in some equivocal cases but further studies are needed to understand the biological processes involved in PLUNC expression.
Keywords: PLUNC, Salivary, Tumour, Mucoepidermoid, Papillary, Cystadenocarcinoma
INTRODUCTION
Salivary gland tumours are uncommon and histologically heterogeneous (Speight and Barrett, 2002). They predominantly affect major salivary glands but minor salivary glands, mainly located in the palate, can also be affected (Vargas et al, 2002). The most common salivary gland tumour is pleomorphic adenoma while mucoepidermoid carcinoma and adenoid cystic carcinoma are the most frequent malignant tumours in the major and minor salivary glands (Jones and Franklin, 2006). Salivary gland tumours show a varied and complex pattern of histopathological features and diagnosis may be difficult even for the most experienced diagnostic pathologist. The latest WHO classification (Barnes et al, 2005) contains over 40 named neoplasms, many of which show significant morphological diversity resulting in overlapping features which make differentiation between tumour types difficult. There is therefore a constant need to evaluate potential new biomarkers that may be useful in diagnosis and prognosis.
Palate Lung Nasal Clone (PLUNC) was first described in the nasal epithelium of the mouse embryo and the trachea and bronchi of adult mouse lung (Weston et al, 1999). We have subsequently shown that human PLUNC belongs to a family of 10 genes located on chromosome 20q11 (Bingle and Bingle, 2000; Bingle and Craven, 2002; Bingle et al, 2004) that are expressed in the upper airways, oro- and nasopharynx. Due to structural similarities with a family of lipid binding/transfer proteins that includes bactericidal/permeability-increasing protein (BPI) and lipopolysaccharide binding protein (LBP), we have suggested that these proteins may play a role in host defence (Bingle and Craven, 2002). PLUNC proteins subdivide into short (SPLUNC) and long (LPLUNC) proteins, which contain domains structurally similar to one or both domains of BPI respectively (Bingle and Craven, 2002). Although many PLUNCs remain poorly characterised, our analysis suggests that they are expressed in the major salivary glands, the minor mucosal glands of the oral cavity and the respiratory epithelium of the nasal, tracheal and bronchial passages (Bingle and Bingle, 2000; Bingle and Craven, 2002). Limited studies have been performed at the protein level but some PLUNCs have been identified in saliva, nasal fluid and pulmonary secretions. For example SPLUNC1, SPLUNC2, and LPLUNC1 have been identified in saliva (Vitorino et al, 2004; Ramachandran et al, 2006; Guo et al, 2006) suggesting that these proteins are produced by the salivary glands. Although human PLUNCs have only recently been identified, a number of rodent orthologues including parotid secretory protein (psp) have been studied extensively (Ball et al, 2003). psp was identified as an abundant component of rat and mouse saliva (Owerbach and Hjorth, 1980; Mirels and Ball, 1992) and is highly expressed in salivary gland tissue. A second psp related protein, submandibular gland protein B (smgb) was also identified as being highly expressed in the salivary glands of rats (Mirels and Ball, 1992). Our recent analysis has shown that SPLUNC2 is the orthologue of rodent psp (Bingle et al, 2004) and consistent with this observation SPLUNC2 is predominantly expressed in the major salivary glands (Bingle and Craven, 2002; Geetha et al, 2005).
At the same time as we identified the human SPLUNC1 gene, it was also identified (and named LUNX) as a potential marker for micrometastasis for non-small cell lung cancer (NSCLC) (Iwao et al, 2001). LUNX expression was also detected in colorectal cancer, esophageal cancer, hepatocellular carcinoma and breast cancer (Iwao et al, 2001). The value of SPLUNC1/LUNX as a diagnostic marker in NSCLC has recently been confirmed in two additional studies (Mitas et al, 2003a; Mitas et al, 2003b) and we have also shown that SPLUNC1 protein is highly expressed in lung cancers with a glandular phenotype (Bingle et al, 2005). Differential expression of PLUNC family members has also been demonstrated in head and neck squamous cell carcinoma (Lemaire et al, 2003), nasopharyngeal carcinoma (Zhang et al, 2003) and breast tumours (Egland et al, 2003). Taken together these studies suggest that differential expression of PLUNC genes may be a characteristic of cancers and the study of these genes may prove to be of diagnostic and prognostic value. PLUNC proteins have not been investigated in salivary gland tumours, but their expression in other glandular neoplasms and in normal salivary glands suggests that they should be expressed and may be useful as diagnostic markers.
The aim of this study therefore was to determine the expression of PLUNC family members in benign and malignant salivary gland tumours in an attempt to detect their potential usefulness in the diagnosis and prognosis of these lesions.
PATIENTS AND METHODS
Tissue samples
Archived formalin-fixed, paraffin-embedded tissue blocks of salivary gland tumours were obtained from the Department of Oral Pathology, School of Clinical Dentistry, University of Sheffield. All the samples were examined on haematoxylin and eosin (H&E) stained sections and classified according to the latest World Health Organization (WHO) guidelines (Barnes et al, 2005). The study was approved by the South Sheffield Research Ethics Committee.
Sixty-four cases of eight different types of salivary gland tumours were assembled in a tissue microarray format. There were thirteen adenoid cystic carcinomas (ACC), 10 carcinoma ex pleomorphic adenomas (CEPA), 10 pleomorphic adenomas (PA), 10 mucoepidermoid carcinomas (MEC), 10 polymorphous low-grade adenocarcinomas (PLGA), 9 acinic cell carcinomas (AcCC), 1 myoepithelial carcinoma (MC), 1 papillary cystadenocarcinoma (PC). We assessed recurrent tumours from 3 ACC patients and 1 MC patient. Clinical data including gender, age, tumour size, location of lesion and patient outcome (i.e. recurrence or metastasis) were collected by reviewing the pathology records (Table 1). We have previously reported DNA ploidy findings in part of this group of tumours (Vargas et al, 2007).
Table 1.
Clinical data and histopathological types of the eight salivary gland tumours (n=64).
| Tumours | number of cases |
Gender M/F |
Age (years) mean(range) |
Site | Size (cm) mean(range) |
|---|---|---|---|---|---|
| ACC | 13 | 4M/9F | 57.1 (37-84) | 2SMG, 11 IOSG | 1.8 (0.8-3.0) |
| MEC | 10 | 2M/8F | 43.4 (16-73) | 2PG, 1SMG, 7 IOSG | 2.4 (0.9-6.5) |
| PA | 10 | 4M/6F | 44.5 (22-68) | 7 PG, 3 IOSG | 3.1 (1.5-5.5) |
| CEPA | 10 | 3M/7F | 54.2 (42-67) | 6 PG, 1 SMG, 3 IOSG | 3.7 (1.3-9.0) |
| PLGA | 10 | 4M/6F | 71.6 (39-91) | 10 IOSG | 2.6 (1.4-3.8) |
| AcCC | 9 | 4M/5F | 36.8 (17-77) | 8 PG, 1 SMG | 3.1 (1.4-8.5) |
| MC | 1 | M | 83 | 1 IOSG | 4.0 |
| PC | 1 | M | 74 | 1 IOSG | na |
|
| |||||
| Total | 64 | 23M/41F | 23PG; 5SMG; 36IOSG | ||
na: not available; M: male; F: female; PG: parotid gland; SMG: submandibular gland; IOSG: intraoral salivary gland; for abbreviation of tumour types see text; ACC: 9 cases were cribriform/tubular, 4 solid variant, and 3 cases recurred (cribriform variant). MEC: 7 low-grade, 2 intermediate-grade and one high-grade cases. AcCC: 1 case metastasised to cervical lymph node. MC: recurrent case.
Tissue Microarray (TMA)
Representative tumour areas were selected and marked on H&E stained sections by using an objective marker (1.8mm, Nikon Corporation, Japan). TMA were constructed using a manual tissue arrayer (Mitogen, Beecher instruments, Inc.). Three representative cylindrical cores of 1.0mm diameter and 3mm depth were taken from each tissue block. The donor cores were then arrayed sequentially into a recipient paraffin block. Three recipient blocks were constructed for immunohistochemical analysis. An oral squamous cell carcinoma (OSCC) was inserted into the left upper corner of each block for orientation.
Generation of SPLUNC2 and LPLUNC1 antibodies
Custom antibodies were generated against SPLUNC2 and LPLUNC1 by Eurogentec (Seraing, Belgium) using established methods. Specifically we generated two antibodies by injection of two synthetic peptides into a pair of rabbits. For SPLUNC2 the peptide sequence corresponded to amino acids, 236-249-COOH: (VDNPQHKTQLQTLI) which represents the extreme C-terminal region of the protein. For LPLUNC1 the peptide sequences corresponded to amino acids, 139-154: (TIRMDTSASGPTRLV) which is located within the first BPI domain of LPLUNC1. Pooled final serum was used for affinity purification against each of the individual peptides. Antibodies were validated by western blotting using recombinant SPLUNC2 and LPLUNC1 as previously described (Bingle et al, 2005).
Immunohistochemical analysis
Immunostaining was performed on 4μm paraffin sections from the TMA blocks, mounted onto adhesive-coated glass slides (Surgipath, Richmond, USA). Slides were dried in the oven for one and a half hours to ensure the sections were firmly attached. All reactions followed standard protocols. Sections were deparaffinized and rehydrated. Endogenous peroxidase activity was blocked by quenching the sections in 3% H2O2 for 20 minutes. The sections were then rinsed in PBS and where necessary (antibodies SPLUNC2, LPLUNC1) antigen retrieval was performed; tri-sodium citrate buffer for 8 minutes in a microwave oven followed by rinsing in PBS. Sections were incubated in 100% normal serum (corresponding to species of secondary antibody) for 30 minutes at room temperature in a humidified chamber. The serum was drained and replaced with primary antibody diluted in 100% normal serum (as used for blocking). The antibody dilutions used were 1:300 SPLUNC1, 1:250 SPLUNC2 and 1:100 LPLUNC1 and left on the sections for an overnight incubation at 4°C in a humidified chamber. A Vectastain kit (Vector Labs) was used for secondary antibodies and enhancement according to manufacturers instructions and a VectorRed kit (Vector Labs) was used for colour development. Expression of PLUNC family members was classified as positive or negative.
RESULTS
As previous studies have identified SPLUNC1, SPLUNC2 and LPLUNC1 in saliva, we first validated our antibodies by immunohistochemical analysis using sections of disease-free submandibular gland. This confirmed that the three proteins exhibited a different distribution in this tissue. LPLUNC1 was expressed within the ducts, with no staining found in either the mucous or the serous acini (Figure 1). SPLUNC1 staining was also seen within the ducts as well as in the cells of the mucous acini but the serous cells were negative (Figure 1). In contrast to the localisation of the other two proteins, SPLUNC2 was highly expressed in the serous cells of the gland but was negative in both the ducts and the mucous cells of the acini (Figure 1).
Figure 1.
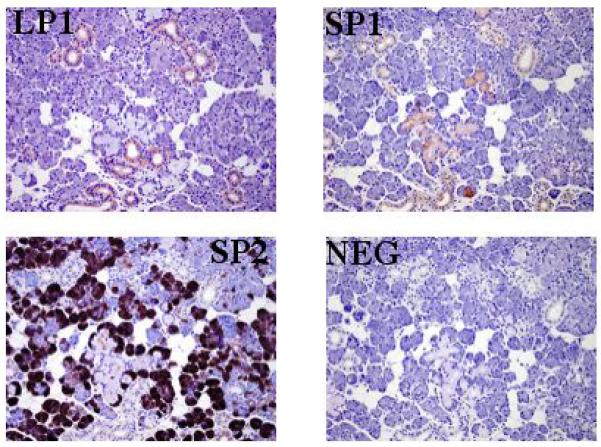
The expression of PLUNC proteins in normal, submandibular gland tissue. LPLUNC1 was localised to the cells of the ducts (LP1), SPLUNC1 was localised to the mucous acini and ducts (SP1) and SPLUNC2 was highly expressed in the serous cells of the acini (SP2). A negative section is included for comparison (NEG). The original magnification for all sections was 400.
Having validated the antibodies on normal salivary gland tissue we applied them to the salivary gland TMAs. An oral squamous cell carcinoma (OSCC) was used as an orientation marker for TMA construction and was negative for all three PLUNC proteins.
The vast majority of the salivary gland tumours were negative for all three PLUNC proteins assessed. No immunoreactivity was seen in any cases of ACC, CEPA, PLGA, PA, AcCC or MC.
In mucoepidermoid carcinomas, LPLUNC1 (Figure 2A) displayed strong cytoplasmic positivity in the mucous cells and the mucin plugs (Figure 2B). Intermediate cells were moderately positive for LPLUNC1 (Figure 2C) and SPLUNC2 (Figure 2D). Epidermoid (Figure 2E) and clear cells (Figure 2F) within these samples were completely negative in all cases. No positive staining for SPLUNC1 was found in any case of MEC. All 10 cases exhibited a similar expression pattern. There were no correlations between PLUNC expression and tumour grade, size, age, gender or site.
Figure 2.
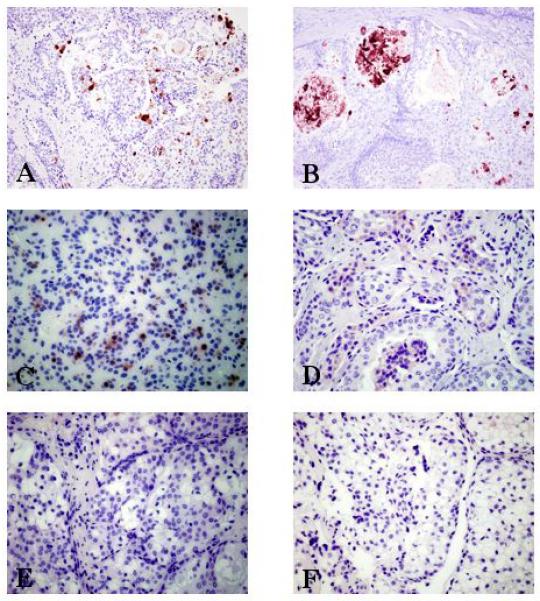
Photomicrographs of mucoepidermoid carcinomas displaying cytoplasmic immunoreactivity for PLUNC family members. MEC exhibiting strong cytoplasmic positivity in the mucous cells for LPLUNC1 (A) (original magnification was x200). (B) Mucin plugs within cystic neoplastic areas positive for LPLUNC1 (original magnification x200). Intermediate cells showing moderate cytoplasmic reaction for LPLUNC1 (C) and SPLUNC2 (D) (original magnification for both LPLUNC1 and SPLUNC2 was x400). Epidermoid (E) and clear cells (F) were negative in all cases for LPLUNC1 (original magnification x400).
The single case of PC studied showed cytoplasmic positivity for all three antibodies but staining intensity varied. LPLUNC1 showed strong cytoplasmic immunoreactivity in the mucous, cuboidal, columnar and clear cells and was also expressed in the luminal mucin (Figure 3A). SPLUNC1 showed strong immunoreactivity in the mucous, cuboidal and columnar cells (Figure 3B) while SPLUNC2 exhibited moderate positivity in these cells (Figure 3C).
Figure 3.
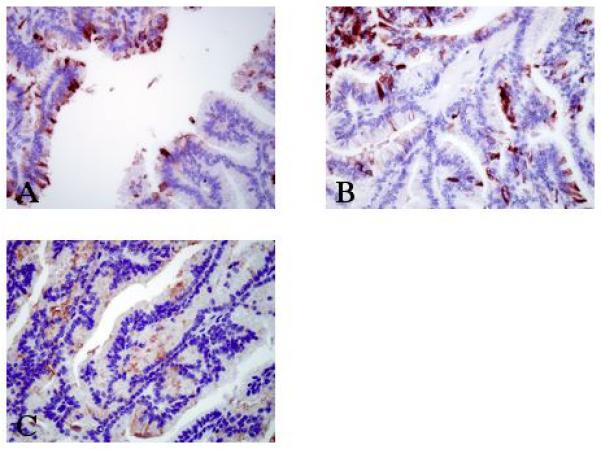
Photomicrographs showing immunoreactivity for PLUNC family members in one case of papillary cystadenocarcinoma. (A) Intraluminal papillary tumour cells showing a strong cytoplasmic positivity for LPLUNC1 in the luminal mucin, mucous, cuboidal, columnar and clear cells (original magnification x400). (B) Intense cytoplasmic immunoreactivity was seen in the mucous, cuboidal and columnar cells with SPLUNC1 antibody (original magnification x400) whilst only moderate cytoplasmic positivity was seen in these cells with the SPLUNC2 antibody (C, original magnification x400).
DISCUSSION
This study examined the expression of three members of the PLUNC family of proteins in a range of salivary gland tumours in order to determine their potential role as diagnostic and/or prognostic tools. We focused on these particular PLUNC proteins as it has previously been reported that they are secreted into saliva (Vitorino et al, 2004; Ramachandran et al, 2006; Guo et al, 2006) and their corresponding mRNAs have been identified in salivary glands (Bingle and Bingle, 2000; Bingle and Craven, 2002; Geetha et al, 2005). We showed that in normal submandibular gland the three proteins were readily detectable. Interestingly they also exhibited a different expression pattern where at least one family member was found in each of the three cellular compartments, namely ducts (LPLUNC1, SPLUNC1) or mucous (SPLUNC1) and serous acini (SPLUNC2).
Surprisingly only 2 of the 8 types of salivary gland tumours on our TMA demonstrated immunopositivity for any of the PLUNC family members studied. The mucin or mucous cells detected in the MECs and our single PC case were positive showing a strong relation between PLUNC expression and mucus-secreting salivary gland tumours. We have made a similar observation for SPLUNC1 in lung cancer (Bingle et al, 2005). The lack of immunoreactivity for PLUNC proteins in ACC, AcCC, CEPA, PA, PLGA and MC might be related to their histogenesis as these tumours do not produce or secrete mucin.
Papillary cystadenocarcinoma is a rare malignant neoplasm of the salivary gland mainly located in the parotid and intra-oral minor salivary glands (Harimaya et al, 2006). They are characterized by cystic structures lined by cuboidal, columnar and mucus-secreting cells and arranged in a papillary architecture (Nakagawa et al, 2002). The differential diagnosis includes papillary cystic variant of AcCC, low-grade MEC, salivary duct carcinoma, and PLGA (Foss et al, 1996). PC can usually be distinguished from these tumours using traditional stains such as H&E and PAS. However, some equivocal PC cases remain as diagnostic challenges even for senior pathologists. Slootweg (1993) reported the importance of correctly differentiating papillary PLGAs from PC in the oral cavity. Klijanienko and Vielh (1998) retrospectively compared fine needle aspiration samples from 12 PLGAs and 5 PC, both tumours showed variable proportions of malignant cells, occasionally arranged in pseudopapillary formations, whilst some PLGAs presented hyaline globules. In our study PC was immunoreactive for some PLUNC family members whereas the PLGA and AcCC cases were uniformly negative. Thus PLUNCs could aid in distinguishing PLGA and the papillary variant of AcCC from PC.
Metastases from prostate, gastrointestinal and thyroid tumors can affect the head and neck region and may also show papillary architecture. Established immunomarkers, such as PSA and thyroglobulin, could be useful in distinguishing PC from metastatic lesions but as expression of PLUNCs is limited to the lung, airways and oral cavity (Bingle and Bingle, 2000; Bingle and Craven, 2002; Bingle et al, 2004) they have the potential to give improved distinction.
MEC is the most common malignant salivary gland tumour (Speight and Barrett, 2002). Histopathologically, they are graded as low, intermediate or high-grade malignancy. H&E and other staining methods, notably, PAS and mucicarmine, are used to make a full diagnosis and grade lesions appropriately. High-grade lesions show a small number of mucous cells, which are difficult to recognize using these traditional staining methods. Mucous cells of the MEC studied were strongly positive for LPLUNC1 indicating a potential diagnostic use for this antibody in equivocal cases.
Another interesting finding was the immunoreactivity of some intermediate cells for LPLUNC1 and SPLUNC2. Based on these findings we can hypothesize that intermediate cells that are positive for PLUNC may have the potential to become mucous cells but further studies are needed to confirm this.
In conclusion, the majority of salivary gland tumours did not show immunoreactivity for PLUNC proteins but mucous cells present in MEC and PC did show intense positivity. These immunomarkers might be used as an additional diagnostic tool to identify mucous cells in some equivocal MEC or PC cases.
ACKNOWLEDGEMENTS
This work was supported by the British Lung Foundation and the Wellcome Trust.
REFERENCES
- Ball WD, Mirels L, Hand AR. Psp and Smgb: a model for developmental and functional regulation in the rat major salivary glands. Biochem. Soc. Trans. 2003;31:777–780. doi: 10.1042/bst0310777. [DOI] [PubMed] [Google Scholar]
- Barnes L, Eveson JW, Reichart P, Sidransky D. Pathology and Genetics of Head and Neck Tumours. IARC Press; Lyon: 2005. World Health Organization Classification of Tumours; p. 210. [Google Scholar]
- Bingle CD, Bingle L. Characterisation of the human plunc gene, a gene product with an upper airways and nasopharyngeal restricted expression pattern. Biochem. Biophys. Acta. 2000;1493:363–367. doi: 10.1016/s0167-4781(00)00196-2. [DOI] [PubMed] [Google Scholar]
- Bingle CD, Craven CJ. PLUNC: a novel family of candidate host defence proteins expressed in the upper airways and nasopharynx. Hum. Mol. Genet. 2002;11:937–943. doi: 10.1093/hmg/11.8.937. [DOI] [PubMed] [Google Scholar]
- Bingle CD, LeClair EE, Havard S, Bingle L, Gillingham P, Craven CJ. Phylogenetic and evolutionary analysis of the PLUNC gene family. Protein Science. 2004;13:422–430. doi: 10.1110/ps.03332704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingle L, Cross SS, High AS, Wallace WA, Devine DA, Havard S, Campos MA, Bingle CD. SPLUNC1 (PLUNC) is expressed in glandular tissues of the respiratory tract and in cancers with a glandular phenotype. J. Path. 2005;205:491–497. doi: 10.1002/path.1726. [DOI] [PubMed] [Google Scholar]
- Egland KA, Vincent JJ, Strausberg R, Lee B, Pastan I. Discovery of the breast cancer gene BASE using a molecular approach to enrich for genes encoding membrane and secreted proteins. PNAS. 2003;100:1099–1104. doi: 10.1073/pnas.0337425100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss RD, Ellis GL, Auclair PL. Salivary gland cystadenocarcinomas: a clinicopathologic study of 57 cases. Am. J. Surg. Pathol. 1996;20:1440–1447. doi: 10.1097/00000478-199612000-00002. [DOI] [PubMed] [Google Scholar]
- Geetha C, Venkatesh SG, Bingle L, Bingle CD, Gorr SU. Design and validation of anti-inflammatory peptides from human Parotid Secretory Protein. J. Dent. Res. 2005;84:149–153. doi: 10.1177/154405910508400208. [DOI] [PubMed] [Google Scholar]
- Guo T, Rudnick PA, Wang W, Lee CS, Devoe DL, Balgley BM. Characterization of the human salivary proteome by capillary isoelectric focusing/nanoreversed-phase liquid chromatography coupled with ESI-tandem MS. J. Proteome Res. 2006;5:1469–1478. doi: 10.1021/pr060065m. [DOI] [PubMed] [Google Scholar]
- Harimaya A, Somekawa Y, Sasaki M, Ohuchi T. Cystadenocarcinoma (papillary cystadenocarcinoma) of the submandibular gland. J. Laryngol. Otol. 2006;120:1077–1080. doi: 10.1017/S0022215106003100. [DOI] [PubMed] [Google Scholar]
- Iwao K, Watanabe T, Fujiwara Y, Takami K, Kodama K, Higashiyama M, Yokouchi H, Ozaki K, Monden M, Tanigami A. Isolation of a novel human lung-specific gene, LUNX, a potential molecular marker for detection of micrometastasis in non-small-cell lung cancer. Int. J. Cancer. 2001;91:433–437. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1059>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Jones AV, Franklin CD. An analysis of oral and maxillofacial pathology found in adults over a 30-year period. J. Oral. Pathol. Med. 2006;35:392–401. doi: 10.1111/j.1600-0714.2006.00451.x. [DOI] [PubMed] [Google Scholar]
- Klijanienko J, Vielh P. Salivary carcinomas with papillae: cytology and histology analysis of polymorphous low-grade adenocarcinoma and papillary cystadenocarcinoma. Diagn. Cytopathol. 1998;19:244–249. doi: 10.1002/(sici)1097-0339(199810)19:4<244::aid-dc3>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Lemaire F, Millon R, Young J, Cromer A, Wasylyk C, Schultz I, Muller D, Marchal P, Zhao C, Melle D, Bracco L, Abecassis J, Wasylyk B. Differential expression profiling of head and neck squamous cell carcinoma (HNSCC) Br. J. Cancer. 2003;89:1940–1949. doi: 10.1038/sj.bjc.6601373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirels L, Ball WD. Neonatal rat submandibular gland protein SMG-A and parotid secretory protein are alternatively regulated members of a salivary protein multigene family. J. Biol. Chem. 1992;267:2679–2687. [PubMed] [Google Scholar]
- Mitas M, Cole DJ, Hoover L, Fraig MM, Mikhitarian K, Block MI, Hoffman BJ, Hawes RH, Gillanders WE, Wallace MB. Real-time reverse transcription-PCR detects KS1/4 mRNA in mediastinal lymph nodes from patients with non-small cell lung cancer. Clin. Chem. 2003a;49:312–315. doi: 10.1373/49.2.312. [DOI] [PubMed] [Google Scholar]
- Mitas M, Hoover L, Silvestri G, Reed C, Green M, Turrisi AT, Sherman C, Mikhitarian K, Cole DJ, Block MI, Gillanders WE. Lunx is a superior molecular marker for detection of non-small lung cell cancer in peripheral blood. J. Mol. Diagn. 2003b;5:237–242. doi: 10.1016/s1525-1578(10)60480-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Hattori K, Iwata N, Tsujimura T. Papillary cystadenocarcinoma arising from minor salivary glands in the anterior portion of the tongue: a case report. Auris. Nasus. Larynx. 2002;29:87–90. doi: 10.1016/s0385-8146(01)00121-3. [DOI] [PubMed] [Google Scholar]
- Owerbach D, Hjorth JP. Inheritance of a parotid secretory protein in mice and its use in determining salivary amylase quantitative variants. Genetics. 1980;95:129–141. doi: 10.1093/genetics/95.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P, Boontheung P, Xie Y, Sondej M, Wong DT, Loo JA. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J. Proteome Res. 2006;5:1493–1503. doi: 10.1021/pr050492k. [DOI] [PubMed] [Google Scholar]
- Slootweg PJ. Low-grade adenocarcinoma of the oral cavity: polymorphous or papillary? J. Oral Pathol. Med. 1993;22:327. doi: 10.1111/j.1600-0714.1993.tb01083.x. 330. [DOI] [PubMed] [Google Scholar]
- Speight PM, Barrett AW. Salivary gland tumours. Oral Dis. 2002;8:229–240. doi: 10.1034/j.1601-0825.2002.02870.x. [DOI] [PubMed] [Google Scholar]
- Vargas PA, Gerhard R, Araújo Filho VJ, de Castro IV. Salivary gland tumors in a Brazilian population: a retrospective study of 124 cases. Rev. Hosp. Clín. Fac. Med. S. Paulo. 2002;57:271–276. doi: 10.1590/s0041-87812002000600005. [DOI] [PubMed] [Google Scholar]
- Vargas PA, Torres-Rendon A, Speight PM. DNA ploidy analysis in salivary gland tumours by image cytometry. J. Oral. Pathol. Med. 2007;36(6):371–376. doi: 10.1111/j.1600-0714.2007.00551.x. In press. [DOI] [PubMed] [Google Scholar]
- Vitorino R, Lobo MJ, Ferrer-Correira AJ, Dubin JR, Tomer KB, Domingues PM, Amado FM. Identification of human whole saliva protein components using proteomics. Proteomics. 2004;4:1109–1115. doi: 10.1002/pmic.200300638. [DOI] [PubMed] [Google Scholar]
- Weston WM, LeClair EE, Trzyna W, McHugh KM, Nugent P, Lafferty CM, Ma L, Tuan RS, Greene RM. Differential display identification of plunc, a novel gene expressed in embryonic palate, nasal epithelium, and adult lung. J. Biol. Chem. 1999;274:13698–13703. doi: 10.1074/jbc.274.19.13698. [DOI] [PubMed] [Google Scholar]
- Zhang B, Nie X, Xiao B, Xiang J, Shen S, Gong J, Zhou M, Zhu S, Zhou J, Qian J, Lu H, He X, Li X, Hu G, Li G. Identification of tissue-specific genes in nasopharyngeal epithelial tissue and differentially expressed genes in nasopharyngeal carcinoma by suppression subtractive hybridization and cDNA microarray. Genes Chromosomes Cancer. 2003;38:80–90. doi: 10.1002/gcc.10247. [DOI] [PubMed] [Google Scholar]


