Abstract
Oral cancer is a deadly and disfiguring disease which could greatly benefit from new diagnostic approaches enabling early detection. In this pilot study, we describe a nano-bio-chip (NBC) sensor technique for analysis of oral cancer biomarkers in exfoliative cytology specimens, targeting both biochemical and morphologic changes associated with early oral tumorigenesis. Here, oral lesions from 41 dental patients, along with normal epithelium from 11 healthy volunteers, were sampled using a non-invasive brush biopsy technique. Specimens were enriched, immunolabeled, and imaged in the NBC sensor according to previously established assays for the EGFR biomarker and cytomorphometry. A total of 51 measurement parameters were extracted using custom image analysis macros including EGFR labeling intensity, cell and nuclear size, and the nuclear-to-cytoplasmic ratio. Four key parameters were significantly elevated in both dysplastic and malignant lesions relative to healthy oral epithelium, including the nuclear area and diameter (p < 0.0001), the nuclear-to-cytoplasmic ratio (p < 0.0001), and EGFR biomarker expression (p < 0.03). Further examination using logistic regression and receiver operating characteristic (ROC) curve analysis identified morphologic features as the best predictors of disease (AUC ≤ 0.93) individually, while a combination of all features further enhanced discrimination of oral cancer and pre-cancerous conditions (AUC = 0.94) with high sensitivity and specificity. Further clinical trials are necessary to validate the regression model and evaluate other potential biomarkers, but this pilot study supports the NBC sensor technique as a promising new diagnostic tool for early detection of oral cancer, which could enhance patient care and survival.
Keywords: lab-on-a-chip sensor, oral cancer, biomarkers, EGFR, exfoliative cytology
Introduction
Oral cancer, largely oral squamous cell carcinoma (OSCC), is a global health problem afflicting over 300,000 people each year (1). In the United States alone, greater than 35,000 new cases and nearly 8,000 deaths are estimated in 2008, representing approximately 3% of all cancers in men and 2% in women (2). Despite significant advances in surgical procedures and treatment, the long-term prognosis for patients with OSCC remains poor, with a 5-year survival rate at approximately 50% which is among the lowest for all major cancers (2-4). This high mortality rate is often attributed to the advanced disease stage of many OSCCs upon initial identification and biopsy. In contrast, if detected early, the prognosis for oral cancer patients is excellent with 5-year survival rates up to 90% (2, 4). In addition, management of early stage oral cancer is often accomplished with less aggressive methods which can preserve vital organ function and physical appearance, resulting in a better quality of life for patients (3). This trend underscores the need for new diagnostic techniques targeting early tumor progression and molecular transformation which could enable early detection of oral cancer.
The disease OSCC happens to be well suited for development of early detection and screening strategies due to the inherent accessibility of the oral cavity and the presence of pre-malignant lesions (PMLs) which often precede the emergence of invasive OSCC. These lesions typically present clinically as leukoplakia (white lesions) and occasionally as erythroplakia (red lesions); however, only about 5% of these lesions will actually progress to cancer (5-7). While some clinical signs, such as a non-homogeneous surface or a stippled appearance, suggest a higher risk of malignant transformation, there are currently no reliable indicators of which lesions will progress (8). Standard good clinical practice dictates that all suspicious lesions should be subjected to surgical biopsy and pathological evaluation since histological evidence of epithelial dysplasia is regarded as the most reliable indicator of malignant potential (8, 9). However, there are several major limitations of this approach. For one, the low rate of cancerous progression suggests that up to 95% of patients may receive an invasive diagnostic procedure unnecessarily, with associated cost, psychological effects, and physical morbidity. Furthermore, the grading of oral epithelial dysplasia (mild/moderate/severe) is subjective and notoriously unreliable with considerable inter- and intra- examiner variation in the histopathological diagnosis (10, 11).
The use of exfoliative cytology offers a rapid, non-invasive method to obtain a sampling of epithelial cells from PMLs which may be subjected to microscopic or molecular examination for signs of malignant change. Yet, unlike cervical cancer screening which has utilized epithelial brushings or scrapings for decades, conventional oral cytology has long been viewed unfavorably with limited diagnostic ability and low sensitivity. These shortcomings have frequently been attributed to poor or inadequate cell sampling and subjectivity associated with cytologic interpretation and diagnosis (12). Recent improvements in specimen collection and quantitative image analysis techniques, including cytomorphometry and DNA aneuploidy, have stimulated a renewed interest in cytology for detection of oral malignancies (13, 14). In addition, exfoliative cytology has also emerged as an excellent, non-invasive method to obtain cellular material for DNA-, RNA-, and protein-based analysis of tumor biomarkers shifting the clinical impact of oral cytology toward molecular diagnostic techniques (14-18). Bringing these two approaches together on a single nano-bio-chip (NBC) platform for molecular and morphological analysis in oral exfoliative cytology may further enhance the role and utility of oral cytology in clinical diagnostics.
Currently, the only commercially available diagnostic adjunct employing exfoliative cytology is the OralCDx® Brush Test with computer-assisted analysis from OralScan Laboratories. In a large multi-centre study, the OralCDx® test demonstrated high sensitivity and specificity (100% and 93%, respectively) for detection of atypical oral epithelia based upon morphology, keratinization, and ploidy patterns (19). Unfortunately, in this study scalpel biopsy was not performed for all, or even the majority, of OralCDx® “negative” or “atypical” specimens possibly resulting in over-estimation of the sensitivity/specificity for this test (19, 20). Despite promising results of the OralCDx® test and initial interest in this new technique, widespread clinical use by dental practitioners is waning due to high rates of false positive test results (personal communication). This trend is supported by Poate et al. reporting a 71% sensitivity and 32% specificity using the OralCDx® system (21). Here, the low specificity may be due to flaws in the retrospective study design or the high occurrence of false positive results associated with benign inflammatory conditions, such as lichen planus (19, 22). The latter suggests the need for additional disease-specific biomarkers, which may be used in conjunction with brush cytology, to improve molecular-level characterization of oral lesions (21, 22).
The cyto-analysis solution presented here based on the nano-bio-chip (NBC), synergizes components and achievements from nanotechnology, molecular diagnostic biomarker discovery, and microfluidics to create a powerful new measurement approach condensing all working components into a small device footprint. The NBC boasts of an extremely flexible assay design and has a diverse group of validated analyte subtypes, including nucleic acids, proteins, cells, and bacteria (23-26). Previously, we established the biochip sensor system for rapid detection and quantitation of the epidermal growth factor receptor (EGFR) biomarker in OSCC cell lines (27). The EGF-receptor is a well-characterized biomarker associated with early oral tumorigenesis and aggressive cancer phenotypes that is over-expressed in up to 90% of all OSCCs (28, 29). In the current study, the integration of the NBC sensor system for concurrent and quantitative analysis of cellular biomarkers, using EGFR as an example, and cytomorphology is demonstrated as a new technique for multi-functional cyto-analysis. In addition, this NBC sensor and methodology is tested in a clinical pilot group in order to secure an initial understanding of the diagnostic utility of such biosensor systems in clinical settings.
Experimental Methods
Sensor Design and Instrumentation
The design and fabrication of the membrane-based NBC platform has been described previously (27). Briefly, the cell-based NBC microfluidic sensor is a multi-layered structure built upon a 22 × 30 × 8.6 mm poly-methyl methacrylate (PMMA) base containing a 1mm diameter, round fluid inlet and outlet port. A polycarbonate track-etched membrane, or screen filter, with 0.4μm pores (Isopore™ Millipore, Billerica, MA) and underlying support were embedded within the base, sealed with laminate adhesives containing a precision cut fluid delivery channel and topped with a glass coverslip. Laminate cut-out structures were created using SolidWorks® 3D CAD software and cut at 25 μm resolution using a SummaCut D-60 vinyl plotter cutter. Dimensions of the fluidic channel are 1 mm wide by 125 μm high by 8.2 mm long generating a channel volume of 1.1 μl. The circular membrane capture area and imaging window is 5 mm in diameter by 200 μm high resulting in a reaction volume of 3.9 μl above the membrane surface. Fluid and sample delivery was facilitated by a peristaltic pump with 6-port injection valve at flow rates between 250μl/min-725μL/min. Cells retained on the surface of the membrane using this straightforward filtration mechanism were analyzed for protein and/or nucleic acids using assay-specific fluorescent labeling techniques as described below. Efficiency of membrane capture is dependent upon the ratio of particle size, or in this instance cell size (typically 10 μm), to membrane pore size (0.4 μm). According to the manufacturer, greater than 99% capture efficiency is obtained for particles exceeding the membrane pore size of 0.4 μm.
Sample Collection and Processing
Exfoliative cytology specimens were collected using the OralCDx® cytology brush by placing the nylon brush firmly against the epithelial surface and rotating 10-15 times, while applying moderate pressure, until pinpoint bleeding was attained. Cells were then released from the biopsy brush and suspended in cold Eagle’s Minimum Essential Media (EMEM) supplemented with 2 mM L-glutamine and 10% fetal bovine serum using vigorous agitation for 15-30 seconds. Media was removed by centrifugation at 1200 rpm for 5 minutes. Cells were then washed twice in PBS buffer (BupH™ Modified Dulbecco’s Phosphate Buffered Saline Packs #28374, Pierce/Thermo Fisher Scientific, St. Louis, MO) by centrifugation at 1200 rpm for 5 minutes. Following the final wash, the supernatant was discarded and the cell pellet was resuspended in fetal bovine serum with 10% dimethyl sulfoxide and frozen at −80°C for long-term storage and transport. In preparation for analysis, frozen samples were thawed rapidly in a 37°C water bath, washed twice in PBS, and fixed in 0.5% methanol-free formaldehyde (#18814 Polysciences Inc., Warrington, PA) in PBS buffer at 4°C for 20 minutes to 1 hour. Following fixation, cells were washed twice in PBS and stored in PBS with 0.1% BSA (PBSA) at 4°C for up to 1 week. Healthy brush biopsy samples were all collected from the buccal mucosa, while clinical samples were collected from the lesion site or surgically excised tissue using similar protocols. When multi-focal regions of irregularity were present, multiple brush biopsies from the same lesions were taken sampling clinically distinct lesion areas.
Cell Staining
Approximately 2,500 – 5,000 cells were suspended in 50% glycerol in PBSA, then delivered/captured on the NBC sensor membrane at a flow rate of 2 mL/min for 30 seconds followed by a 2.5 minute PBS buffer wash at 1 mL/min. Next, fluorescent labeling was carried out through sequential delivery of immunoreagents, consisting of primary anti-EGFR antibody (#MS-378-P, LabVision) at 10 μg/mL in PBSA containing 0.1% Tween-20 (PBSAT) followed by a secondary antibody cocktail containing 20 μg/mL goat anti-mouse IgG/AlexaFluor®-488 antibody fragment (#A11017, Molecular Probes), 33 μM phalloidin/AlexaFluor®-647 (#A22287, Molecular Probes), and 5 μM DAPI (#D3571, Molecular Probes) in PBSAT. All reagents were delivered to the membrane-captured cells at 250 μL/min for 2.5 minutes with intermittent buffer washes at 1 mL/min for 2.5 minutes. Total assay time (without imaging and analysis) was 13 minutes.
Imaging and Analysis
Digital micrographs of stained cells were obtained in an (X, Y, Z) membrane scan using a 10X (0.3 NA) objective on an automated Olympus BX-61 modified epifluorescent microscope with motorized stage and 12-bit monochrome CCD camera (Q-Imaging, British Columbia) controlled via Simple PCI software (Compix Inc., Sewickley, PA). Monochrome images of Phalloidin, EGFR, and DAPI fluorescent labels were collected sequentially using appropriate filter cubes (Chroma Technology Corp., Rockingham, VT) then merged into red, green, and blue (RGB) spectral channels, respectively. Multiple Z-focal planes were collected at +5 μm intervals and recombined using a z-stack focusing algorithm in ImageJ (30) in order to accommodate adherent epithelial cell populations with both individual and aggregated cell clusters. Automated image analysis routines utilized ImageJ and/or Cell Profiler (31) open-source software with custom written macros for quantitative intensity standardization and cell/nuclear contouring to define the region-of-interest (ROI) for each object/cell (27). Measurement parameters included whole cell and nuclear area, perimeter, circularity, Feret diameter, minimum and maximum intensity, mean, standard deviation, mode, median, and integrated intensity. All together, 51 parameters, including nine morphological and forty-two intensity parameters were extracted or calculated for each cell with an average of 1420 cells measured in each cytology sample (range 50 – 3624). For statistical analysis, data was exported to Microsoft® EXCEL® with Analyse-It® (Analyse-It Software Ltd., Leeds, UK) software and SigmaPlot 9.0 (Systat Software Inc., San Jose, CA) for graphical representation.
Study Participants and Demographics
A total of 52 participants were enrolled in this study from May 2007 to March 2009. Eleven of these volunteers were healthy individuals with no known oral diseases or signs of epithelial abnormality within the oral cavity. An additional 41 clinical participants were identified by collaborating dentists and physicians located at the UTHSCs at San Antonio (n = 22) and Houston (n = 19). Eligibility criteria included patients presenting with a visible oral lesion(s), leukoplakia or erythroplakia, who were referred to specialists in secondary or tertiary care centers for surgical biopsy or removal. Informed consent was obtained from all participants and the study guidelines were approved by the Institutional Review Board at each institution.
The demographic data and pathological diagnosis available from clinical participants is provided in Table 1. Of the 41 clinical cases, 66% were male, 85% had a history of tobacco use, and 42% also reported moderate to heavy alcohol consumption. The average age in this clinical group was 56 (range 31 – 79 years) and the majority of lesions occurred in the tongue or floor of the mouth. Diagnosis was established for each patient by surgical biopsy of the lesion site and standard histopathology by board certified oral and maxillofacial pathologists at their respective institutions. The group of 11 healthy participants consisted of 55% males with an average age of 33 (range 21 – 53 years). Only one of the healthy participants reported a history of smoking.
Table 1.
Demographic data and pathological diagnosis for study participants enrolled at UTHSC San Antonio and Houston clinics
| Patient ID | Age | Sex | Lesion NBCation | Pathological Diagnosis |
|---|---|---|---|---|
| UTSA-001 | 48 | M | Floor of mouth | SCC, moderately differentiated |
| UTSA-002 | 56 | M | Tongue, base | SCC, poorly differentiated |
| UTSA-003 | 57 | M | Soft palate | Dysplasia, mild-moderate |
| UTSA-004 | 44 | M | Soft palate and tonsil | SCC, moderately differentiated |
| UTSA-005 | 40 | M | Tongue, lateral | SCC |
| UTSA-006 | 48 | M | Hard palate | Benign hyperkeratosis and oral submucous fibrosis |
| UTSA-007 | 45 | F | Tongue, ventral | SCC |
| UTSA-008* | 44 | M | Alveolar ridge | Lymphoma |
| UTSA-009 | 64 | M | Retromolar trigone | SCC |
| UTSA-010 | 59 | M | Tongue, ventrolateral | SCC, well differentiated |
| UTSA-011 | 62 | M | Soft palate | SCC, moderately differentiated |
| UTSA-012 | 60 | M | Tongue, lateral | SCC, moderately differentiated |
| UTSA-013 | 42 | F | Tongue, lateral | SCC, moderate to poorly differentiated |
| UTSA-014 | 59 | F | Floor of mouth/tongue | SCC, moderately differentiated |
| UTSA-015 | 64 | M | Palate | SCC, moderately differentiated |
| UTSA-016 | 72 | F | Hard palate | SCC, well to moderately differentiated |
| UTSA-017 | 68 | M | Hard palate | SCC, verrucous carcinoma |
| UTSA-018 | 50 | F | Soft palate | SCC, poorly differentiated |
| UTSA-019 | 70 | F | Pharynx | SCC |
| UTSA-020 | 30 | M | Tongue, lateral | SCC, well differentiated |
| UTSA-021 | 48 | M | Tongue/floor of mouth | SCC, moderately differentiated |
| UTSA-022 | 55 | M | Retromolar trigone | SCC, moderately differentiated |
| UTH-001 | 56 | M | Hard palate | SCC, moderately differentiated |
| UTH-002 | 68 | M | Hard palate | SCC, moderately differentiated |
| UTH-003 | 53 | M | Tongue | SCC, moderately differentiated |
| UTH-004 | 52 | M | Tongue/floor of mouth | SCC, moderately differentiated |
| UTH-005 | 74 | F | Tongue | SCC, residual ulcerated |
| UTH-006 | 47 | M | Tongue/floor of mouth | SCC, moderately differentiated |
| UTH-007 | 75 | F | Tongue | Dysplasia, moderate |
| UTH-008 | 50 | M | Tongue | SCC, moderately differentiated |
| UTH-009 | 79 | F | Tongue | SCC, moderately differentiated |
| UTH-010 | 69 | M | Tongue | SCC, moderately differentiated, superficially invasive |
| UTH-011 | 62 | F | Gingiva | Dysplasia, severe |
| UTH-012 | 51 | F | Palate | Benign lichen planus |
| UTH-013 | 31 | M | Tongue, lateral | Dysplasia, moderate |
| UTH-014 | 60 | M | Tongue, ventral | Dysplasia, moderate |
| UTH-015 | 65 | F | Tongue/floor of mouth | Dysplasia, moderate |
| UTH-016 | 41 | M | Buccal mucosa | Dysplasia, mild with submucosal fibrosis |
| UTH-017 | 52 | F | Tongue, lateral | Dysplasia, moderate-severe |
| UTH-018 | 72 | M | Tongue | SCC, verrucous carcinoma |
| UTH-019 | 60 | F | Soft palate | Dysplasia, moderate in atypical verrucous/papillary hyperplasia |
Patient was excluded from the study due to non-epithelial nature of malignancy
Statistical Analysis
A Student’s t-test (Analyze-It® for MS Excel®) was utilized to assess statistically significant differences in the measured cellular parameters within each study group, including benign lesions, dysplasia, and invasive SCC, when compared to the healthy controls (p < 0.05). Logistic regression and ROC curve analysis was performed using MedCalc® (MedCalc Software, Mariakerke, Belgium) statistical software. This method requires the input of parameter values measured for each sample (i.e. EGFR intensity, nuclear and cellular area, etc.) according to a binary classification of disease status where non-diseased samples received a value of zero and diseased samples one (“Non-diseased” = 0; “Diseased” = 1). The logistic function, with general equation of f(z) = 1/(1+exp(−z)) serves to predict the probability of an outcome (disease occurrence) between 0 and 1 based upon one or more of these input parameters or factors. The variable (z), is a measure of the total contribution of all the factors used in the model, according to the logit equation (z = a0+ a1xBM1+a2xBM2+.....anxBMn) where a0 is the intercept and (a1....an) are the regression coefficients for each biomarker (BM1....BMn). Predicted values using the linear regression model were utilized to build receiver operating characteristics (ROC) curves plotting the projected true positive and false positive rates (sensitivity vs. 1 – specificity) for disease classification based upon the model.
Results
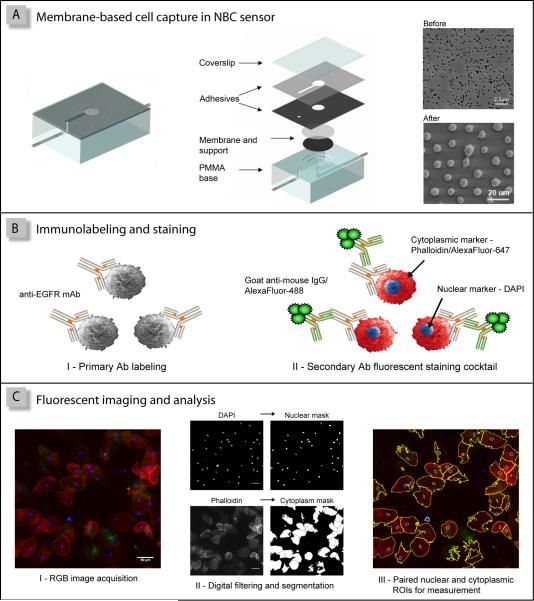
As illustrated in Figure 1, the nano-bio-chip sensor system for OSCC assessment integrates multiple laboratory processes onto a microfluidic platform in 3 primary steps consisting of: (i) cell separation/capture on the membrane filter, (ii) biomarker immunolabeling and cytochemical staining, and (iii) fluorescent imaging and analysis. During cell capture, an oral cytology suspension is delivered to the NBC sensor, using pressure-driven flow, whereupon any particles or cells larger than the membrane pore size are retained on the membrane surface (Figure 1A). Once captured, a series of biomarker-specific immunoreagents and cellular stains (Figure 1B, panels I and II respectively) are utilized to tag the epithelial cells of interest, which are then detected using fluorescence microscopy in an X, Y, Z scan of the membrane surface (Figure 1C, panel I). Subsequent automated image analysis routines employ digital filtering and image segmentation of the DAPI and Phalloidin signals, blue and red spectral channels, respectively, to generate binary masks and contours of the nuclear and cytoplasmic ROIs for measurement in all fluorescent channels (Figure 1C, panels II and III). This semi-automated NBC methodology permits concurrent analysis of EGFR surface biomarker expression and cellular/nuclear morphology using over 50 ROI intensity and shape parameters with particular attention focused on the cellular and nuclear area, the N/C ratio and mean cellular EGFR intensity as early indicators of malignancy.
Figure 1.
The NBC oral cytology assay consists of three primary steps: (A) cell capture on the membrane filter; (B) EGFR/AlexaFluor®488 immunolabeling and staining with Phalloidin/AlexaFluor®647 and DAPI; and (C) fluorescent imaging and analysis, where paired cytoplasmic/nuclear regions of interest (ROIs) are defined for each cell.
Cytomorphometry and EGFR Biomarker Expression Analysis
A total of 56 oral brush cytology specimens from healthy and disease participants were examined using the NBC sensor method. Eleven specimens were obtained from healthy individuals while 45 were from clinically visible lesions identified in 41 patients (Table 1). Histopathological diagnosis identified 3 benign lesions , 8 moderate or severely dysplastic lesions, and 34 invasive squamous cell carcinomas (SCC) (Table 1). Two of the SCC samples were expended during the initial assay development stages, while another SCC and one dysplasia specimen were excluded due to inadequate cell sampling for a sampling error rate of approximately 4%.
Table 2 summarizes the NBC sensor results obtained from the remaining 52 brush cytology specimens analyzed for cytomorphometry and EGFR biomarker expression. Here, the nuclear area was significantly increased in dysplastic and invasive SCC cytospecimens (149 μm2 and 165 μm2, respectively) versus healthy control epithelium (63.4 μm2, p < 0.0001). Similarly, the nuclear diameter was also elevated in dysplastic and SCC lesions (16.1 μm and 17.3 μm, respectively) relative to healthy mucosa (11.5 μm, p < 0.0001). Concurrent with this nuclear enlargement was a decrease in the cellular area from 1040 μm2 in healthy mucosa to 697 μm2 in SCC (p < 0.0001) as well as a decrease in cellular diameter from 51.5 μm in healthy specimens to 41.1 μm in SCC (p < 0.001). This inverse relationship, with an overall increase in nuclear size and decrease in cellular size, yielded a significant elevation in the nuclear-to-cytoplasmic ratio (N/C ratio). Interestingly, this rise in the N/C ratio appeared to be progressive from healthy mucosa (0.063) through pre-cancerous lesions with moderate and/or severe dysplasia (0.223, p < 0.0001) to invasive SCC (0.323, p < 0.001) though the difference in N/C ratio between dysplastic and SCC lesions was not significant (p < 0.08). The EGFR labeling intensity was also demonstrated to increase significantly in diagnosed dysplasia and OSCC relative to healthy epithelium (9.5 au, 11.8 au, and 6.0 au, respectively) (Table 2). Cytology specimens from the three benign lesions exhibited a rise in nuclear area/diameter and N/C ratio over controls (p < 0.005), but their EGFR intensity remained similar to normal oral mucosa. There were no statistically significant differences between patient samples collected at UTHSC San Antonio and Houston facilities for any of the parameters measured (data not shown).
Table 2.
Results of NBC sensor analysis of EGFR biomarker expression and cytomorphometry in oral lesions according to histopathological diagnosis, reported as mean ± standard deviation
|
Healthy n = 11 |
Benign n = 3 |
Dysplasia n = 7 |
SCC n = 31 |
|
|---|---|---|---|---|
| Nuclear Area (μm2) | 63.4 ± 11 | 122 ± 57* p < 0.004 |
149 ± 23* p < 0.001 |
165 ± 46* p < 0.001 |
| Nuclear Diameter (μm) | 11.5 ± 0.70 | 14.8 ± 3.2* p < 0.005 |
16.1 ± 1.2* p < 0.001 |
17.3 ± 2.6* p < 0.001 |
| Cellular Area (μm2) | 1040 ± 160 | 1300 ± 500 p < 0.15 |
1040 ± 260 p < 0.99 |
697 ± 230* p < 0.001 |
| Cellular Diameter (μm) | 51.5 ± 4.4 | 55.1 ± 12 p < 0.4 |
49.9 ± 6.3 p < 0.6 |
41.1 ± 7.1* p < 0.001 |
| N/C Ratio | 0.063 ± 0.02 | 0.206 ± 0.17* p < 0.01 |
0.223 ± 0.10* p < 0.001 |
0.323 ± 0.14* p < 0.001 |
| EGFR Intensity (au) | 5.99 ± 1.4 | 7.10 ± 1.8 p < 0.3 |
9.53 ± 4.3* p < 0.03 |
11.8 ± 6.5* p < 0.006 |
Asteric denotes statistically significant differences relative to healthy controls using paired t-test (p < 0.05).
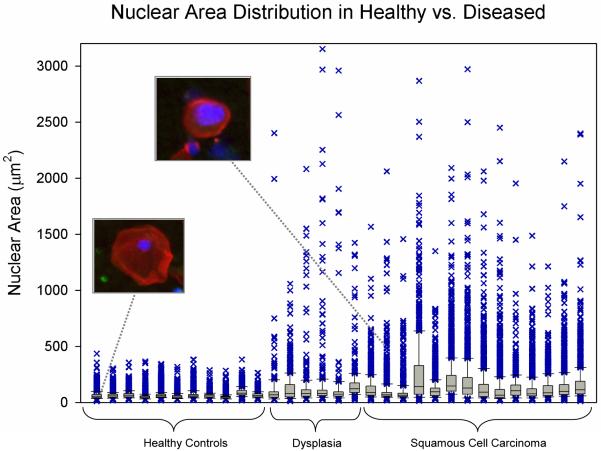
Further examination of the population distribution within individual samples revealed additional features which could potentially be exploited for disease characterization. Box and whisker plots presented in Figure 2 depict the median, interquartile range and outliers of nuclear area measurements from a subset of patients. Here, the median nuclear area for healthy control specimens ranged from 47 μm2 to 63 μm2, while the SCC and dysplasia samples exhibited median values from 66 μm2 to 141 μm2. Interestingly, it is the spread of outliers that appeared to be the most distinguishing characteristic from which to differentiate normal cytology specimens from dysplasia and invasive SCC. As such, none of the healthy specimens possess outliers over 500 μm2 while all of the SCC/dysplasia possessed numerous events above this level.
Figure 2.
Box and whisker plot of the median, interquartile range, and the distribution of outliers in the nuclear area data for each cell population according to disease status. Two representative cells from a healthy participant and SCC patient are shown (inset).
Case Study: Comparison with Histology and Immunohistochemistry
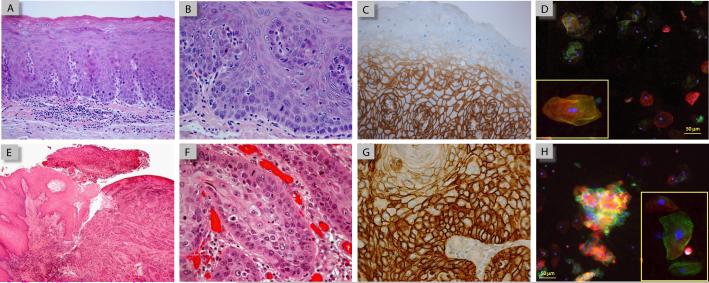
While histology and EGFR immunohistochemistry (IHC) was performed for all surgically biopsied oral tissue, two cases were selected to demonstrate the similarities and difference in the type and range of cellular features examined in traditional diagnostic methods versus the NBC sensor system using brush cytology (Figure 3). One case was diagnosed as mild to moderate dysplasia in the soft palate (Figure 3A) and the other an invasive squamous cell carcinoma located on the ventral tongue (Figure 3E). In the hematoxylin and eosin (H&E) stained tissue shown in Figure 3A, a thin keratin layer is seen at the surface of the epithelium with the various stratified layers beneath, relatively normal for mucosa at this oral cavity site. Upon higher magnification (Figure 3B), several dysplastic features are seen including enlarged and pleomorphic nuclei found above the basal cell layer, with loss of basal cell polarity and maturation sequence. Corresponding EGFR IHC revealed EGFR (+) positive membrane labeling in the lower layers of the epithelium, including the basal cells, which dissipated in the more superficial layers and in the surface epithelium (Figure 3C). Cytologic analysis using the NBC sensor assay from this patient displayed a heterogeneous mixture of EGFR (+) positive and (−) negatively labeled epithelial cells (Figure 3D), derived from the differential EGFR-expressing epithelial layers as depicted in IHC, along with an elevated nuclear area (~120 μm2). This pattern suggests that the NBC cytologic method may effectively reflect biochemical and morphological changes in neoplastic tissue, even at these early pre-malignant stages.
Figure 3.
Comparison of histology, IHC, and NBC sensor micrographs in case studies of mild to moderate dysplasia (A-D) and invasive SCC (E-H). H&E sections at low magnification (A and E) demonstrate the overall tissue architecture while at higher mag (B and F) abnormal cellular alterations, such as nuclear enlargement, loss of polarity, and an increase in the number mitotic figures, are visible. EGFR IHC using peroxidase-DAB staining (C and G) identifies the differential expression of EGFR within the dysplastic/malignant tissue. Micrographs from NBC sensor assay (D and H) demonstrate a corresponding heterogeneous mixture of normal and “abnormal” cells with elevated EGFR biomarker expression and enlarged nuclei (arrow).
Similar pathological features were also seen in the OSCC tissue, albeit more pronounced, with dyskeratosis , cellular and nuclear pleomorphism and mitotic figures visible in H&E stained sections (Figure 3E-F) while IHC demonstrated strong EGFR (+) labeling throughout the tumor (Figure 3G). In the NBC sensor cytology assay, extremely intense EGFR (+) labeling was also detected and localized in the cell membranes of adherent epithelial cell clusters as well as some individual cells (Figure 3H, arrow). In the particular cluster shown, a number of the nuclei present also appear irregular and/or enlarged, indicating that these cells exhibit “cancer-like” nuclear characteristics reflective of what is seen in tumor tissue.
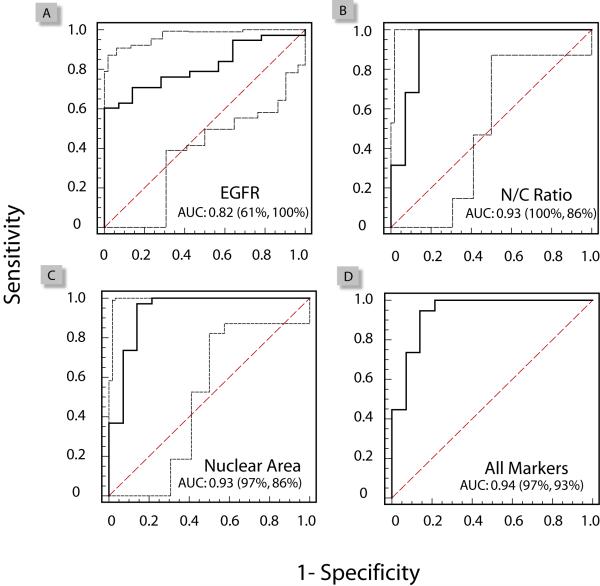
ROC Curve Analysis and Logistic Regression
Logistic regression and ROC curve analysis enabled further exploration into which of these cellular markers, and in what combinations, could be most effective toward diagnosis of OSCC. Each individual parameter was evaluated by a ROC curve, as shown in Figure 4 (panels A-C) for EGFR, N/C ratio, and nuclear area. The ROC curve is a plot of a diagnostic test’s sensitivity, or true positive rate (TPR) versus 1-specificity, or the false positive rate (FPR) at various discrimination thresholds depicting the trade-offs between the true positives (benefits) and the false positives (costs) in diagnostic accuracy. All of the parameters tested exhibited significant capacity for disease classification and discrimination between patients with OSCC or dysplasia, versus healthy controls or benign conditions, as demonstrated by the area under the curve (AUC) values greater than 0.5. Of these parameters, the N/C ratio and nuclear area exhibited the best performance characteristics with AUC of 0.93, followed by the EGFR biomarker (0.82) (Figure 4A-C). While the N/C ratio or nuclear areas, alone, are excellent diagnostic markers, the added value of these markers in a combined panel was further examined using logistic regression. Here, the ROC curve generated from the predicted values in the combined biomarker panel, exhibited AUC of 0.94 with a projected 97% sensitivity and 93% specificity for detection and classification of malignant and pre-malignant oral lesions (Figure 4D).
Figure 4.
Receiver operating characteristic (ROC) curves for evaluation of the diagnostic performance characteristics for the individual markers: EGFR (A), nuclear area (B), and N/C ratio (C). Using a logistic regression model the combined EGFR and cytomorphometry marker panel (D) is predicted to exhibit an AUC value of 0.94 with 97% sensitivity and 93% specificity for characterization of oral malignant and pre-malignant lesions.
Discussion
In this pilot study, a new nano-bio-chip cellular analysis technique for characterizing oral malignant and pre-malignant lesions was evaluated. Six parameters were found to be significantly altered in OSCC cytospecimens versus healthy mucosa including: (1) the nuclear area, (2) nuclear diameter, (3) cellular area, (4) cellular diameter, (5) nuclear-to-cytoplasmic ratio, and (6) EGFR biomarker immunolabeling (Table 2). The nuclear area, nuclear diameter, N/C ratio and EGFR expression were also found to be significantly altered in oral lesions with diagnosed dysplasia, supporting use of these markers as diagnostic indicators of early cancer development and pre-malignancy. These findings are in line with earlier reports by Ramesh, et.al. and Grandis et.al. identifying significant changes in cellular and nuclear morphology, and EGFR expression associated with oral tumorigenesis (32-34), while providing a new method for automated cytoanalysis that is rapid, quantitative, and requires small sample volumes. Using the mean EGFR intensity values from healthy control subjects (Table 2) plus two standard deviations (6.0 ± 2.8), a rough threshold for EGFR over-expression can be established. According to this criterion, 63% (20 out of 32) of SCC tumors and 67% (4/6) of pre-cancerous dysplastic lesions over-express the EGFR biomarker while none (0/3) of the benign specimens exhibit biomarker over-expression. Interestingly, these benign lesions demonstrated a significantly elevated nuclear area and N/C ratio, but not EGFR expression, relative to normal controls (Table 2). Though the number of benign oral lesions examined in this study is small, this finding suggests that a both morphological and biochemical markers are important for successful characterization of oral lesions, particularly when discriminating them from other non-malignant conditions and inflammation (35). This issue will be explored more thoroughly in future studies.
In addition to the mean values, we examined the population distribution, with particular attention toward outliers in the data. These outliers likely represent the “rare events” important for early disease detection which may be obscured or masked in mean measurements over the entire population. As shown in Figure 2, the spread of outliers in the nuclear area measurements visibly distinguishes healthy controls from dysplasia and SCC. Particularly for dysplastic lesions this makes sense, where the top of the mucosa is mature and pathologically similar to healthy normal epithelium, while it is the smaller portion of deep cells that contain the information as to the presence and extent of abnormality. As such, many of the cells in a cytology suspension would be expected to possess N/C ratios in the normal range and only a fraction of the cells would be expected to exhibit dysplastic/malignant changes, (i.e. an elevation in the N/C ratio, represented here as the N/C ratio outliers). A new calculated parameter, such as a “polymorphic index”, representing the range in this distribution or ratio of identifiable cell types may provide an addition handle from which to classify disease with enhanced accuracy over mean values in future assays.
Comparison of the NBC cellular analysis methods with standard histopathology and immunohistochemistry, as shown in Figure 3, offers insight into the types of malignant features available with each method and their associated benefits and trade-offs. Based upon standard histopathology, case one was diagnosed with mild to moderate dysplasia and exhibits several abnormal/enlarged nuclei along with a loss of basal cell polarity in H&E stained tissue sections, but with the overall epithelial structure and stratification maintained (Figure 3A-B). The IHC revealed EGFR expression restricted to the lower layers of epithelium (Figure 3C). These alterations in nuclear morphology and EGFR expression were successfully detected and quantitated in the NBC sensor assay (Figure 3D), suggesting that the sensor method adequately reflects cellular alterations in tumor tissue, even at early premalignant stages. A similar comparison was seen in the case of invasive SCC (Figure 3E-H) where intense EGFR staining was apparent throughout the epithelium in IHC stained histologic sections as well as in a cluster of “cancer-like” cells with EGFR (+) membrane labeling and enlarged nuclei in the NBC sensor (Figure 3H, arrow). When using the bio-chip sensor system a trade-off does exist between the loss of information regarding cellular architecture and cell-cell or cell-matrix interactions and the added benefit of a rapid, non-invasive automated cellular analysis technique.
Evaluation of which cellular and biochemical markers, alone or in combination, could be most effective toward diagnosis of malignant and pre-malignant oral lesions was undertaken in a more quantitative manner using logistic regression and ROC curves analysis (Figure 4). Logistic regression serves as a model to predict the outcome of disease, based upon input of one or more variables, such as the N/C ratio, nuclear area, and EGFR biomarker measurements obtained in NBC sensor assays. The predicted values generated from logistic regression were used to build ROC curves, a graphical representation of a diagnostic test’s sensitivity, or true positive rate (TPR) versus 1-specificity, or the false positive rate (FPR) at various discrimination thresholds. In Figure 4, the ROC curves for the EGFR biomarker, nuclear area, and N/C ratio are presented for the combined patient population. Here, the EGFR biomarker exhibited moderate disease discrimination capability with an area under the curve (AUC) value of 0.82, while the nuclear area and N/C ratio both exhibited AUC values of 0.93 indicating excellent disease classification performance. The logistic regression model and ROC curve for a 3 marker panel (Figure 4-D) exhibited a slight increase in AUC value to 0.94 with a predicted sensitivity of 97% and specificity of 93%. These results suggest that the combined cytomorphometry and EGFR panel likely holds the greatest potential for cancer detection and diagnosis. Yet, the true diagnostic need lies not in the identification or oral cancer, but in the identification of premalignant lesions. Analysis of the data from Figure 4 to include dysplasia only (moderate or severe) yielded AUC values of 0.83 for EGFR, 0.90 for the N/C ratio, and 0.92 for the nuclear area (data not shown). Unfortunately, the limited number of dysplastic samples prevented further statistical analysis using logistic regression which typically requires a minimum of 10 cases per independent variable (36). However, the close similarity between AUC values from dysplasia alone and the combined patient population suggests that the parameters that were most predictive in the combined cohort are also predictive of oral premalignancy.
It is important to note that this work represents a development effort for a novel nano-bio-chip system with integrated microfluidic elements and proof of principle study; not a validation of a commercial product. As such, the clinical implications are somewhat limited due to a relatively small sample size, particularly within the benign and dysplasia diagnostic categories, and an uneven distribution of samples between the diagnostic groups that can weigh one category too heavily in comparative analyses. In particular, the SCC group accounted for 60% of all samples. Such imbalances are often found in early-stage diagnostic development studies where clinical collaborators are typically located in regional specialty clinics or research-based cancer centers where the patient population is skewed toward more severe cases of dysplasia and/or SCC (37, 38).
However, the largest challenge for early detection in the oral cavity is not the diagnosis of actual SCC, rather it is determining which “innocuous” lesions may be biologically worrisome. Building upon the promising cytoanalysis techniques presented here, our future research studies target a cohort of ~500 patients presenting with oral leukoplakia, or otherwise “innocuous” lesions, which may harbor early signs of dysplasia and molecular transformation. These prospective studies will target a low-risk population and greatly expand the sample size in order to improve statistical power of NBC sensor results. In addition, we will examine the role of benign inflammation on assay specificity while continuing to integrate and evaluate additional disease-specific biomarkers. We believe that the true potential for this work rests in the ability to extend the rapid assay techniques developed here for examination of multiple disease-specific biomarkers in an effort to improve risk assessment and early detection and specificity of oral diagnostics.
Acknowledgements
The authors would like to thank Dr. Rebecca Richard-Kortum at Rice University for her generous assistance and insight throughout this research. Funding for this project was provided by the National Institute for Dental and Craniofacial Research (U01-DE15017) and the Welch Foundation (F-1193).
Abbreviations
- NBC
nano-bio-chip
- EGFR
epidermal growth factor receptor
- OSCC
oral squamous cell carcinoma
- SCC
squamous cell carcinoma
- IHC
immunohistochemistry
- ROC
receiver operating characteristic curve
- AUC
area under curve
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society . Cancer Facts and Figures 2008. American Cancer Society; Atlanta, GA: 2008. [Google Scholar]
- 3.Seiwert TY, Cohen EE. State-of-the-art management of locally advanced head and neck cancer. Br J Cancer. 2005;92:1341–8. doi: 10.1038/sj.bjc.6602510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ries LAGMD, Krapcho M, Mariotto A, Miller BA, Feuer EJ, Clegg L, Horner MJ, Howlader N, Eisner MP, Reichman M, Edwards BK. SEER Cancer Statistics Review. National Cancer Institute; Bethesda, MD: 1975-2004. [Google Scholar]
- 5.Lee JJ, Hong WK, Hittelman WN, et al. Predicting cancer development in oral leukoplakia: ten years of translational research. Clin Cancer Res. 2000;6:1702–10. [PubMed] [Google Scholar]
- 6.Schepman KP, van der Meij EH, Smeele LE, van der Waal I. Malignant transformation of oral leukoplakia: a follow-up study of a hospital-based population of 166 patients with oral leukoplakia from The Netherlands. Oral Oncol. 1998;34:270–5. [PubMed] [Google Scholar]
- 7.Speight PM, Farthing PM, Bouquot JE. The pathology of oral cancer and precancer. Curr Diag Pathol. 1996;3:165–76. [Google Scholar]
- 8.Reibel J. Prognosis of oral pre-malignant lesions: significance of clinical, histopathological, and molecular biological characteristics. Crit Rev Oral Biol Med. 2003;14:47–62. doi: 10.1177/154411130301400105. [DOI] [PubMed] [Google Scholar]
- 9.van der Waal I, Schepman KP, van der Meij EH, Smeele LE. Oral leukoplakia: a clinicopathological review. Oral Oncol. 1997;33:291–301. doi: 10.1016/s1368-8375(97)00002-x. [DOI] [PubMed] [Google Scholar]
- 10.Karabulut A, Reibel J, Therkildsen MH, Praetorius F, Nielsen HW, Dabelsteen E. Observer variability in the histologic assessment of oral premalignant lesions. J Oral Pathol Med. 1995;24:198–200. doi: 10.1111/j.1600-0714.1995.tb01166.x. [DOI] [PubMed] [Google Scholar]
- 11.Abbey LM, Kaugars GE, Gunsolley JC, et al. Intraexaminer and interexaminer reliability in the diagnosis of oral epithelial dysplasia. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1995;80:188–91. doi: 10.1016/s1079-2104(05)80201-x. [DOI] [PubMed] [Google Scholar]
- 12.Sugerman PB, Savage NW. Exfoliative cytology in clinical oral pathology. Aust Dent J. 1996;41:71–4. doi: 10.1111/j.1834-7819.1996.tb05915.x. [DOI] [PubMed] [Google Scholar]
- 13.Ogden GR, Cowpe JG, Green M. Cytobrush and wooden spatula for oral exfoliative cytology. A comparison. Acta Cytol. 1992;36:706–10. [PubMed] [Google Scholar]
- 14.Ogden GR. The future role for oral exfoliative cytology--bleak or bright? Oral Oncol. 1997;33:2–4. doi: 10.1016/s0964-1955(96)00047-4. [DOI] [PubMed] [Google Scholar]
- 15.Driemel O, Dahse R, Hakim SG, et al. Laminin-5 immunocytochemistry: a new tool for identifying dysplastic cells in oral brush biopsies. Cytopathology. 2007;18:348–55. doi: 10.1111/j.1365-2303.2006.00401.x. [DOI] [PubMed] [Google Scholar]
- 16.Spafford MF, Koch WM, Reed AL, et al. Detection of head and neck squamous cell carcinoma among exfoliated oral mucosal cells by microsatellite analysis. Clin Cancer Res. 2001;7:607–12. [PubMed] [Google Scholar]
- 17.Schwartz JL, Panda S, Beam C, Bach LE, Adami GR. RNA from brush oral cytology to measure squamous cell carcinoma gene expression. J Oral Pathol Med. 2008;37:70–7. doi: 10.1111/j.1600-0714.2007.00596.x. [DOI] [PubMed] [Google Scholar]
- 18.Veltman JA, Hopman AH, Bot FJ, Ramaekers FC, Manni JJ. Detection of chromosomal aberrations in cytologic brush specimens from head and neck squamous cell carcinoma. Cancer. 1997;81:309–14. doi: 10.1002/(sici)1097-0142(19971025)81:5<309::aid-cncr9>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 19.Sciubba JJ, U.S. Collaborative OralCDx Study Group Improving detection of precancerous and cancerous oral lesions. Computer-assisted analysis of the oral brush biopsy. J Am Dent Assoc. 1999;130:1445–57. doi: 10.14219/jada.archive.1999.0055. [DOI] [PubMed] [Google Scholar]
- 20.Lingen MW. Critical evaluation of diagnostic aids for the detection of oral cancer. Oral Oncol. 2007 doi: 10.1016/j.oraloncology.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poate TW, Buchanan JA, Hodgson TA, et al. An audit of the efficacy of the oral brush biopsy technique in a specialist Oral Medicine unit. Oral Oncol. 2004;40:829–34. doi: 10.1016/j.oraloncology.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Scheifele C, Schmidt-Westhausen AM, Dietrich T, Reichart PA. The sensitivity and specificity of the OralCDx technique: evaluation of 103 cases. Oral Oncol. 2004;40:824–8. doi: 10.1016/j.oraloncology.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Ali MF, Kirby R, Goodey AP, et al. DNA hybridization and discrimination of single-nucleotide mismatches using chip-based microbead arrays. Anal Chem. 2003;75:4732–9. doi: 10.1021/ac034106z. [DOI] [PubMed] [Google Scholar]
- 24.Christodoulides N, Floriano PN, Miller CS, et al. Lab-on-a-chip methods for point-of-care measurements of salivary biomarkers of periodontitis. Oral-Based Diagnostics. 2007:411–28. doi: 10.1196/annals.1384.035. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez WR, Christodoulides N, Floriano PN, et al. A microchip CD4 counting method for HIV monitoring in resource-poor settings. Plos Medicine. 2005;2:663–72. doi: 10.1371/journal.pmed.0020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Floriano PN, Christodoulides N, Romanovicz DK, et al. Membrane-based on-line optical analysis system for rapid detection of bacteria and spores. Biosensors and Bioelectronics. 2005;20:2079–88. doi: 10.1016/j.bios.2004.08.046. [DOI] [PubMed] [Google Scholar]
- 27.Weigum SE, Floriano PN, Christodoulides N, McDevitt JT. Cell-based sensor for analysis of EGFR biomarker expression in oral cancer. Lab on a Chip. 2007;7:995–1003. doi: 10.1039/b703918b. [DOI] [PubMed] [Google Scholar]
- 28.Grandis JR, Tweardy DJ. Elevated levels of transforming growth factor alpha and epidermal growth factor receptor messenger RNA are early markers of carcinogenesis in head and neck cancer. Cancer Res. 1993;53:3579–84. [PubMed] [Google Scholar]
- 29.Shin DM, Ro JY, Hong WK, Hittelman WN. Dysregulation of epidermal growth factor receptor expression in premalignant lesions during head and neck tumorigenesis. Cancer Res. 1994;54:3153–9. [PubMed] [Google Scholar]
- 30.Rasband WS, Image J, Bethesda MD. USA: U.S. National Institutes of Health. 2006 19997. [Google Scholar]
- 31.Carpenter AE, Jones TR, Lamprecht MR, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pektas ZO, Keskin A, Gunhan O, Karslioglu Y. Evaluation of nuclear morphometry and DNA ploidy status for detection of malignant and premalignant oral lesions: quantitative cytologic assessment and review of methods for cytomorphometric measurements. J Oral Maxillofac Surg. 2006;64:628–35. doi: 10.1016/j.joms.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 33.Ramaesh T, Mendis BR, Ratnatunga N, Thattil RO. Cytomorphometric analysis of squames obtained from normal oral mucosa and lesions of oral leukoplakia and squamous cell carcinoma. J Oral Pathol Med. 1998;27:83–6. doi: 10.1111/j.1600-0714.1998.tb02099.x. [DOI] [PubMed] [Google Scholar]
- 34.Cowpe JG, Longmore RB, Green MW. Quantitative exfoliative cytology of abnormal oral mucosal smears. J R Soc Med. 1988;81:509–13. doi: 10.1177/014107688808100905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anuradha C, Reddy BV, Nandan SR, Kumar SR. Oral lichen planus. A review. N Y State Dent J. 2008;74:66–8. [PubMed] [Google Scholar]
- 36.Peduzzi P, Concato J, Kemper E, Holford TR, Feinstein AR. A simulation study of the number of events per variable in logistic regression analysis. J Clin Epidemiol. 1996;49:1373–9. doi: 10.1016/s0895-4356(96)00236-3. [DOI] [PubMed] [Google Scholar]
- 37.Lane PM, Gilhuly T, Whitehead P, et al. Simple device for the direct visualization of oral-cavity tissue fluorescence. J Biomed Opt. 2006;11 doi: 10.1117/1.2193157. 024006. [DOI] [PubMed] [Google Scholar]
- 38.Ram S, Siar CH. Chemiluminescence as a diagnostic aid in the detection of oral cancer and potentially malignant epithelial lesions. Int J Oral Maxillofac Surg. 2005;34:521–7. doi: 10.1016/j.ijom.2004.10.008. [DOI] [PubMed] [Google Scholar]