Abstract
Helicases are ubiquitous enzymes found in all organisms that are necessary for all (or virtually all) aspects of nucleic acid metabolism. The Pif1 helicase family is a group of 5′→3′ directed, ATP-dependent, super-family IB helicases found in nearly all eukaryotes. Here, we review the discovery, evolution, and what is currently known about these enzymes in Saccharomyces cerevisiae (ScPif1 and ScRrm3), Schizosaccharomyces pombe (SpPfh1), Trypanosoma brucei (TbPIF1, 2, 5, and 8), mice (mPif1), and humans (hPif1). Pif1 helicases variously affect telomeric, ribosomal, and mitochondrial DNA replication, as well as Okazaki fragment maturation, and in at least some cases affect these processes by using their helicase activity to disrupt stable nucleoprotein complexes. While the functions of these enzymes vary within and between organisms, it is evident that Pif1 family helicases are crucial for both nuclear and mitochondrial genome maintenance.
Keywords: Pif1, Rrm3, Pfh1p, helicase, DNA replication
1. Introduction
Since the isolation of the Saccharomyces cerevisiae PIF1 gene more than 25 years ago [1] and the eventual characterization of the S. cerevisiae Pif1 protein (ScPif1) as a helicase [2], work on the Pif1 family of helicases has begun to elucidate the roles of these enzymes in both nuclear and mitochondrial genome stability. ScPif1 and its homologs have been studied in yeasts, parasites, and mammals. The goal of this review is to summarize what is known about the Pif1 helicases, compare their activities in different model systems, and cast an eye towards the future of Pif1 research.
2. History and evolution of the Pif1 helicases
The S. cerevisiae PIF1 gene was isolated in a genetic screen for genes that affect the frequency of mitochondrial DNA (mtDNA) recombination between wild type and cytoplasmic petite mutant strains (i.e., petite integration frequency) [1]. A decade later, the same group succeeded in partially purifying ScPif1 from mitochondria and demonstrated that it possesses single-stranded DNA (ssDNA)-dependent ATPase and 5′→3′ directed helicase activities [2]. Soon after, the study of Pif1 helicases as proteins affecting the nuclear genome began with the “rediscovery” of ScPif1 as an enzyme that inhibits telomere elongation and de novo telomere formation [3] (see Section 3 for an in-depth discussion of ScPif1).
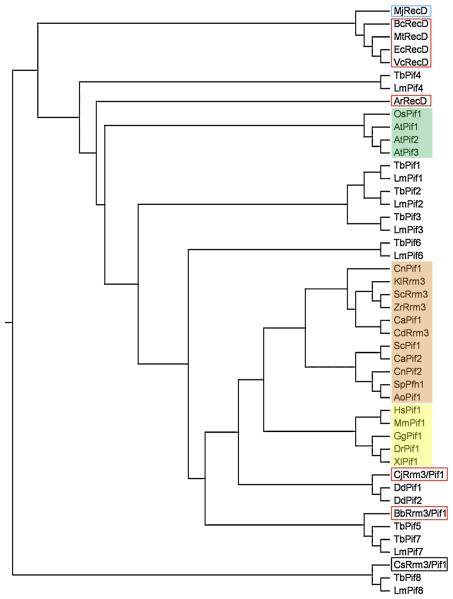
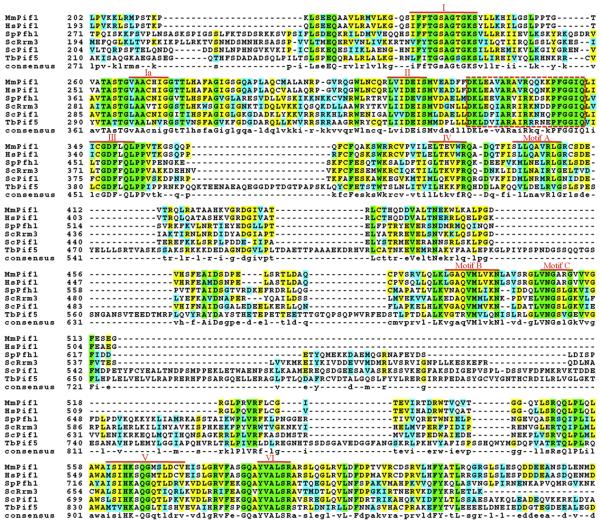
A second PIF1-like gene, now called RRM3, was found in S. cerevisiae by two different groups. In one case, the gene was identified by searching the database for genes with similarity to ScPIF1 [4]. In the second, it was identified in a screen for ribosomal DNA (rDNA) recombination mutants [5], where it was named ScRRM3 [5]. As more genome data became available, it became apparent that ScPif1 was the founding member of a family of helicases conserved in essentially all eukaryotes (Figure 1). However, while several fungal genomes, such as Candida albicans and Cryptococcus neoformans, are like S. cerevisiae in encoding two Pif1 family helicases, most higher eukaryotes and all metazoans contain only one. Two notable exceptions to this rule are Arabidopsis thaliana with three Pifs (Figure 1) and kinetoplastid parasites with seven to eight Pif1 helicases (see Section 7). Regardless of the number of Pif1-like proteins in an organism's genome, all of the Pif1 family proteins are comprised of a conserved 400-500 amino acid ATPase/helicase domain but have divergent N- and C-termini (Figure 2).
Figure 1.
Evolutionary relationship among Pif1 family and RecD helicases. The indicated sequences were aligned using ClustalX [88], and the phylogenetic relationship among them was drawn as a rooted tree (using the unrelated human beta actin protein (NP_001092) as an outgroup (not shown) with TreeView v. 1.6.6. software (http://taxonomy.zoology.gla.ac.uk/rod/rod.html). Prokaryotic proteins are outlined in red, and the archaeal protein is outlined in blue. Fungal proteins are shaded pink, plant proteins are shaded green, and metazoan proteins are shaded yellow. Proteins from the following organisms were aligned: Agrobacterium radiobacter (Ar), Arabidopsis thaliana (At), Aspergillus oryzae (Ao), Bacillus cereus (Bc), Bdellovibrio bacteriovorus (Bb), Campylobacter jejuni (Cj), Candida albicans (Ca), Candida dubliensis (Cd), Clostridium sporogenes (Cs), Cryptococcus neoformans (Cn), Danio rerio (Dr), Dictyostelium discoidium (Dd), Escherichia coli (Ec), Gallus gallus (Gg), Homo sapiens (Hs), Kluveromyces lactis (Kl), Leishmania major (Lm), Methanocaldococcus jannaschii (Mj), Mus musculus (Mm), Mycobacterium tuberculosis (Mt), Oryza sativa (Os), Saccharomyces cerevisiae (Sc), Schizosaccharomyces pombe (Sp), Trypanosoma brucei (Tb), Xenopus laevis (Xl), Vibrio cholerae (Vc), and Zygosaccharomyces rouxii (Zr). The GenBank accession numbers are as follows: AoPif1, XP_001824182; ArRecD, YP_002544895; AtPif1, CAB91581; AtPif2, NP_190738; AtPif3, CAB63155; BbRrm3/Pif1, BcRecD, YP_085716; CaPif1, XP_718694; CaPif2, XP_712340; CdRrm3, XP_002421612; CjRrm3/Pif1, YP_002344343.1; CnPif1, XP_572423; CnPif2, XP_569577; CsRrm3/Pif1, ZP_02995968.1; DdPif1, XP_642006; DdPif2, XP_647539; DrPif1, NP_942102; EcRecD, AAB40466.1; GgPif1, XP_426648; HsPif1, NP_079325; KlRrm3, XP_453658; LmPif1, XP_001681501; LmPif2, XP_001681500; LmPif3, XP_001684538; LmPif4, XP_001685476; LmPif6, XP_001683071; LmPif7, XP_001681013; LmPif8, XP_001684097; MjRecD, NP_248527; MmPif1, EDL26099; MtRecD, NP_215143; OsPif1, ABB47755; ScPif1, NP_013650; ScRrm3, NP_011896; SpPfh1, NP_596488; TbPif1, XP_828762; TbPif2, XP_828763; TbPif3, XP_829242; TbPif4, XP_829537; TbPif5, XP_847187; TbPif6, XP_822349; TbPif7, XP_846907; TbPif8, XP_845724; XlPif1, Q0R4F1; VcRecD, NP_231950; and ZrRrm3, XP_002498680.
Figure 2.
Conserved motifs in the Pif1 family helicases. The sequences of the Hs-, Mm-, and ScPif1, ScRrm3, SpPfh1, and TbPif5 helicases used to generate Figure 1 were aligned using ClustalW [88], and the BOXSHADE program in the Biology WorkBench suite (http://workbench.sdsc.edu) was used to color-code conserved residues. The completely conserved residues are in green, conserved similarities are in cyan, and identical residues are yellow; the amino acid similarity groups were defined as FYW, IVLM, RK, DE, GA, TS, and NQ. Due to spatial constraints, the divergent N- and C-termini are not shown, leaving only the highly conserved core ATPase/helicase domain. The seven conserved SFI helicase motifs (red Roman numerals; [89]) and three additional motifs with high homology to E. coli RecD (red A, B, and C; [90]) are shown. A putative Pif1 family signature motif is indicated with the dashed red box (see Section 2. for details).
Originally, proteins with high similarity to Pif1 helicases were found only in eukaryotes, although BLAST searching [6] for homologues of ScPif1 revealed that it is distantly related (16.4% identity) to the Escherichia coli RecD helicase [7,8]. A current search of the NCBI protein database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=Protein&itool=toolbar) reveals that several putative prokaryotic proteins are annotated as Rrm3-/Pif1-like (see Figure 1 and legend). To determine if these sequences are evolutionarily related or simply related by being ATPases/helicases, we constructed a phylogenetic tree of prokaryotic RecD (and Rrm3-/Pif1-like) proteins and eukaryotic Pif1 and Rrm3 proteins. As shown in Figure 1, the prokaryotic Bdellovibrio bacteriovorus, Campylobacter jejuni, and Clostridium sporogenes Rrm3/Pif1 proteins do cluster with eukaryotic Pif1 family members as opposed to RecD proteins. It should be noted, however, that these Rrm3-/Pif1-like sequences are grouped with some of the most divergent eukaryotic Pif1 family helicases (e.g., C. sporogenes Rrm3/Pif1 forms a clade with the Pif8 homologues from parasites, which are believed to have lost their helicase activities (see Section 7.4). Also, the Agrobacterium radiobacter RecD appears to be closely related to the Pif1 helicases from plants. In all, these results suggest that RecD, Rrm3, and Pif1 may have evolved from a common proto-helicase.
Regardless of ancestry, the evolution of Pif1 helicases is intriguing. At some point in the past, there existed a single progenitor helicase that evolved into two helicases with distinct functions in fungi (and seven or more in parasites), the most-widely characterized duo being ScPif1 and ScRrm3 (it should be noted that the presence of two Pif1 helicases does not appear to be due to the ancient genome duplication that occurred in S. cerevisiae (J. Bessler, unpublished)). Then, in most eukaryotes, one of these enzymes was lost, resulting in the single Pif1 family helicase found in metazoans. The current situation leaves us with several questions, including whether ScRrm3 or ScPif1 is more closely related to the ancestral form and if metazoan Pif1 helicases are more similar to ScPif1 or ScRrm3. Based on the phylogeny presented in Figure 1, it is difficult to address either of these questions definitively, but the branch lengths of the fungal Pif1 and Rrm3 clades indicate that ScRrm3 may have evolved first.
Questions of evolution aside, the Pif1 family belongs to the super-family IB helicases, which are comprised of mostly monomeric, 5′→3′ directed, P-loop (a conserved nucleotide binding motif, also known as “the Walker A box” or “motif I”) helicases [9]. This view comes from an analysis of Pif1 sequence alignments (Figure 2) that shows the seven conserved SFI motifs (I, Ia, II, III, IV, V, and VI) and the three motifs shared with E. coli RecD (A, B, and C). There is also a putative 21-residue Pif1 family signature sequence located between motifs II and III. This motif is highly conserved in 24 of the 33 (72.7%) eukaryotic Pif1 family proteins used to generate the phylogenetic tree in Figure 1 (M. Bochman, observations) but is degenerate or absent in the plant Pif1 helicases and the parasitic Pif1, 2, 4, and 8 homologues, which are the most divergent sequences in the Pif1 family alignment. This motif is also absent in RecD homologues. Finally, using the consensus sequence for this motif (DKLeXvARaiRkqXkPFGGIQ) to query the NCBI protein database via BLAST searching [6] returns only Pif1 homologues (M. Bochman, observations).
3. ScPif1
As mentioned above, the founding member of the Pif1 family helicases, ScPif1, was first discovered in a genetic screen as a gene affecting mitochondrial DNA [1]. This study identified ScPIF1 as a gene whose mutation results in reduced recombination between the mtDNA of certain rho− (respiratory deficient) and rho+ (respiration proficient) strains [1].
ScPIF1 was rediscovered in a screen for mutations that affect telomeres [3], suggesting that ScPif1 also functions in the nucleus. Both the mitochondrial and nuclear isoforms are expressed from the single ScPIF1 open reading frame but use different translational start sites. A mitochondrial targeting signal (MTS) is located between the first and the second translational start site [10], and translation from the first start site targets ScPif1 to mitochondria (Figure 3). Two isoforms are detected by western blot analysis: a larger isoform, corresponding to the nuclear isoform; and a slightly smaller isoform, corresponding to the mitochondrial isoform, due to the cleavage at the MTS site upon import into the mitochondria [11]. By altering the first (pif1-m1) or the second (pif1-m2) AUG site in the ScPIF1 open reading frame, the functions of the mitochondrial ScPif1 or nuclear ScPif1 can be distinguished [3]. Cells mutated for the second AUG (pif1-m2) express an intact mitochondrial isoform of ScPif1 but not the nuclear isoform. Although pif1-m2 cells are mitochondrial proficient, like pif1Δ cells, they display several telomere defects, as described in Section 3.1 below. However, the telomere phenotypes are more severe in pif1Δ than in pif1-m2 cells [3], suggesting that a fraction of the mitochondrial ScPif1 leaks into the nucleus. Below, both the nuclear and mitochondrial functions of ScPif1 are discussed.
Figure 3.
Mitochondrial and nuclear isoforms of ScPif1. Schematic of the wild type, mitochondrial (m2), and nuclear (m1) PIF1 alleles with the predicted localization in the cell. M1 denotes the position of the first AUG site, and M2 marks the position of the second AUG site. Mutated M2 to alanine is represented with an asterisk. The picture is drawn to scale. MTS, mitochondrial target signal: aa, amino acids.
3.1 Nuclear function
3.1.1. Adding telomeric repeats to double strand breaks
One critical function of telomeres is to enable the cell to distinguish true ends (i.e., telomeres) from double strand breaks (DSBs). Broken chromosomes are most often repaired by one of several recombination pathways. However, telomerase can hinder DSB repair by adding telomeric repeats to a broken chromosome [12], which prevents the break from engaging in recombination. Adding a telomere to a DSB generates a terminally deleting chromosome that lacks the genetic information from the site of the break to the normal end of the chromosome. Thus, telomerase-mediated repair of DSB is normally deleterious because it generates a partially aneuploid cell. In yeast, telomere addition to DSBs is rare, largely because ScPif1 inhibits these events. Thus, the most dramatic telomere defect in pif1Δ and pif1-m2 cells is an increase of up to 1000-fold in telomere addition to DSBs [3,12,13], and this inhibition is telomerase dependent [12].
In addition to the effects of ScPif1 at DSBs, Pif1-depleted cells have longer telomeres than wild type cells [3]. The telomere lengthening in pif1-m2 and pif1Δ cells is also telomerase-dependent [11], showing that ScPif1 regulates telomerase at both telomeres and at DSBs. Finally, the C-terminus of ScPif1 is post-translationally phosphorylated in response to DNA damage [14]. Phosphorylation of ScPif1 is required for its inhibition of de novo telomere addition at DSBs but not for addition of telomeric repeats at telomeres by telomerase [14], suggesting that ScPif1 can distinguish between telomeres and DSBs.
3.1.2. Inhibition of telomerase
Lengthening of telomeres occurs through two different pathways, recombination-dependent elongation (called alternative lengthening of telomeres), which is RAD52 dependent, or the major pathway, which is telomerase-dependent [15]. In the absence of ScPif1, telomeres are at least ~100 bp longer; when ScPif1 is overexpressed, telomeres are modestly shorter [11]. pif1 rad52 cells also have longer telomeres, suggesting that ScPif1 does not inhibit the recombinational pathway of telomere lengthening [11]. Lengthening does not occur in a pif1 strain that is also telomerase deficient. Thus, genetic analysis indicates that ScPif1 acts on the telomerase pathway. Since chromatin immunoprecipitation (ChIP) shows that ScPif1 is enriched at chromosome ends, the helicase likely acts directly to inhibit telomerase [11,14]. This interpretation is supported by the isolation of mutations in the catalytic subunit of telomerase in which telomere length is no longer sensitive to ScPif1 [16].
Biochemical assays also show that ScPif1 uses its helicase activity to reduce telomerase processivity by releasing it from telomeric DNA [17]. Consistent with the biochemical data, ChIP assays demonstrate that overexpression of ScPif1 decreases the association of the telomerase subunits, Est1 and Est2, with telomeric DNA without affecting the binding of telomere structural proteins [17]. In addition, western blot analysis shows that the abundance of nuclear ScPif1 is cell cycle regulated, peaking in late S/G2 phase. This cell cycle regulation of ScPif1 is dependent on the anaphase promoting complex (APC) [18]. Since yeast telomerase lengthens telomeres only in late S/G2 phase [19,20], ScPif1 is maximally expressed at the same time in the cell cycle that telomerase acts [18]
These data support a model in which negative regulation of telomere length by ScPif1 occurs by removal of telomerase from telomeres, either by releasing the telomerase RNA from the telomere ends (which would abolish the association of the telomerase holoenzyme from the telomeres [21]) or by disrupting Est2 from the telomeric ends. The fact that ScPif1 preferentially unwinds forked RNA-DNA hybrids in vitro [22] makes the first model attractive.
3.1.3. G-quadruplex forming DNA
In almost all eukaryotes, telomeric DNA is G-rich and repetitive. G-rich oligonucleotides, including but not limited to telomeric repeats, can form G-quadruplex structures by self-assembly of four guanine bases into a four stranded DNA structure stabilized by monovalent cations [23]. Genome-wide sequence analysis to identify naturally occurring tracts with the potential to form G-quadruplex structures have been conducted in both prokaryotic and eukaryotic genomes [24,25]. Apart from telomeres, these sequences are also found at promoters [23], suggesting a role for these structures in transcriptional regulation. In addition, in S. cerevisiae, such sequences are enriched at sites of spontaneous DSBs (T. Capra, K. Paeschke, M.Singh, and V.A. Z., manuscript under review). G-quadruplex structures are very stable, and both their folding and unfolding may need assistance in vivo.
Inactivation of nuclear ScPif1 causes frequent rearrangements in the G-rich human minisatellite CEB1 when it is inserted into the yeast genome [26]. However, mutation of other helicases does not affect the stability of the CEB1 sequence. These rearrangements are dependent on homologous recombination [26]. The same study reported that the CEB sequence forms G-quadruplex structures in vitro. Moreover, recombinant ScPif1 can unwind the CEB1 G-quadruplex structure, and this unwinding occurs with a faster rate than the unwinding of other dsDNA substrates [26]. The helicase activity of ScPif1 is required for unwinding, as purified ScPif1 with a point mutation in motif I of the helicase domain (pif1-K264A) is unable to unwind the CEB1 G-quadruplex structure [26].
This study was the first evidence that ScPif1 participates in resolving G-quadruplex structures. Other helicases, such as the human RecQ family helicases (WRN and BLM) and the S. cerevisiae RecQ helicase Sgs1, also unwind G-quadruplex structures in vitro [27-29]. Although G-quadruplex structures may have important functions, they are also likely to be an obstacle for the replication machinery if they are present during DNA replication. A possible model is that G-quadruplex formation during S phase stalls the replication fork, and ScPif is recruited to the paused forks to resolve these structures, allowing fork progression. In fact, in contrast to ScRrm3, which is a component of the replisome (see Section 4.1.1 for details), ScPif1 does not travel with the replication fork but rather is recruited to a subset of genomic loci [30] (K. Paeschke and V.A. Z., unpublished results)
3.1.4. Okazaki fragment maturation
Nuclear ScPif1 is implicated in processing Okazaki fragments during semi-conservative DNA replication. Deletion of ScPif1 suppresses the lethal effects of dna2 cells [31]. Dna2 has both helicase and endonuclease activities and functions in removal of long flaps (~30 bp) bound by the ssDNA binding protein RPA during the maturation of Okazaki fragments. In S. cerevisiae, during semi-conservative DNA replication, the lagging strand DNA polymerase δ (Pol δ) [32] produces around 100,000 Okazaki fragments. DNA polymerase α (Polα), a low fidelity DNA polymerase, uses its associated RNA primase activity to initiate and then elongate each Okazaki fragment, which are then extended by the high fidelity Pol δ. To produce a continuous DNA strand on the lagging strand, each of the Pol α generated RNA-DNA segments is displaced and filled in by Pol δ. In vitro experiments demonstrate very efficient strand displacement by Pol δ and the flap endonuclease FEN1. When Pol δ arrives at the 5′ end of the downstream Okazaki fragment, it displace 2-3 nt of the downstream primer at a time, and by the action of FEN1, these flaps are cleaved, eventually leaving a ligatable nick for DNA ligase I [33]. However, in some cases, during lagging strand synthesis, longer flaps are generated. These flaps are bound by RPA, which inhibits FEN1 cleavage but promotes cleavage by Dna2 [34]. The discovery that the lethal effects of dna2 are suppressed by pif1, suggests a model in which ScPif1 is involved in the creation of these long flaps [31]. Long flaps are believed to cause chromosomal instability. By deleting POL32 (the smallest subunit of Pol δ) in dna2 pif1 cells, these cells survive even better at permissive temperatures than a dna2 pif1 double mutant [31]. These genetic results are also supported by elegant biochemical experiments [35,36].
In summary, genetic and biochemical studies suggest that ScPif1 and Pol δ together create a long flap, which requires cleavage by Dna2. The occurrence and processing of long flaps may occur only during the replication of a subset of the genome. In these cases, ScPif1 could be recruited to the lagging strand to facilitate Pol δ displacement of the downstream Okazaki fragment. Perhaps the G-rich repetitive sequences that are capable of forming G-quadruplex DNA structures are particularly prone to forming long flaps (see Section 3.1.3 for details).
3.1.5. Blocking the replication of rDNA
A replication fork barrier (RFB) is found at every rDNA repeat in S. cerevisiae, where it acts as a polar barrier to replication fork progression. By blocking the movement of replication forks in a directional manner, the RFB ensures that replication moves through the rDNA in the same direction as transcription. Thus, the RFB prevents collision of the transcription and replication machineries. Using two-dimensional (2D) gel electrophoresis, the absence of ScPif1 results in less efficient arrest of replication forks at the RFB compared to wild type cells, suggesting that full RFB function requires ScPif1 [4]. Since, by the criterion of ChIP, ScPif1 is RFB-associated, it likely affects rDNA replication directly. The absence of ScRrm3 (see Section 4) leads to the opposite result, suggesting that these homologous helicases affect rDNA replication in opposite ways: ScPif1 inhibits and ScRrm3 promotes fork progression at the RFB.
3.2. Mitochondrial function
Although ScPIF1 was first discovered in a genetic screen for mutations affecting recombination between mutant mtDNA genomes, it also has an important role in the maintenance of wild type mtDNA [37-41]. In S. cerevisiae pif1Δ or pif1-m1 cells, mtDNA is lost more quickly than in wild type cells, and this effect is exacerbated at high temperatures [3,38]. Aging and several human diseases are correlated with mutations in the mitochondrial genome. Endogenous reactive oxygen species, which are the by-product of respiration, cause oxidative DNA damage and are likely a common mtDNA damaging agent. ScPif1 is suggested to be involved in mtDNA recombination and replication [37]. In addition, ScPif1 is likely also involved in a recombination-independent pathway affecting mtDNA [39]. Investigation of replication fork progression by 2D gels shows that pif1 deletion causes mtDNA breakage, suggesting a role for ScPif1 in the repair or prevention of mtDNA breakage [41]. ChIP assays indicate that ScPif1 binds throughout the mtDNA genome, suggesting that it may be part of the mitochondrial replisome [41].
ScRrm3 deletion suppresses, to some degree, the respiratory deficiencies (petite induction phenotype) of pif1Δ cells [40]. Indeed, by examining replication fork progression by 2D gels, mtDNA replication forks are maintained in pif1 rrm3 cells, suggesting a role for ScPif1 in sustaining the integrity of mtDNA that is counteracted by ScRrm3. However, a synergistic effect of increased point mutations in the genome was also detected [40]. Over-expression of Ribonucleotide Reductase 1 (Rnr1), the large subunit of the RNR enzyme that catalyzes de novo synthesis of dNTPs, also rescues the mtDNA maintenance phenotypes of pif1 cells and does so to the same degree as an rrm3 deletion [40]. This result suggests that ScRrm3 and ScPif1 are both involved in mtDNA repair or maintenance, but they act by two different pathways. In addition, ScRrm3 may be involved in a pathway that involves the elevation of dNTP pools during DNA damage. Elevation of dNTP pools in the absence of ScRrm3 would explain detection of increased point mutations in mtDNA in these cells. In fact, in the presence of nuclear DNA damage, dNTP pools are elevated, resulting in an improved tolerance to DNA damage in the nucleus but at the cost of increased mutations [42]. For example, primer extension assays have shown that elevated dNTP pools engage the leading strand DNA polymerase ε [43] to bypass the oxidative base damage, 8-oxoG by inserting dATP opposite the damaged base [44]. When pif1Δ cells are grown in ethidium bromide (EtBr), especially in the presence of a nonfermentable carbon source such as glycerol [41], their mtDNA is fragmented and eventually lost [41]. Deletion of ScRRM3 does not rescue the mtDNA instability of pif1Δ EtBr-treated cells [45], as it does under non-genotoxic conditions [40].
3.3. Biochemical experiments
ScPif1 has a molecular weight of ~ 98kDa and unwinds DNA with 5′→3′ polarity in an ATP- and Mg2+-dependent manner [11,46]. The ScPif1 used in biochemical studies has been purified from various sources, including baculovirus infected Sf9 insect cells [11], E. coli [22] or yeast mitochondrial membranes from cells overexpressing the protein [46]. Unlike other Pif1 family helicases, ScPif1 is soluble and fairly easy to purify [47].
Purified nuclear ScPif1 unwinds RNA-DNA substrates at rates at least 2-fold higher than its unwinding of DNA-DNA substrates. It also prefers unwinding forked DNA-DNA substrates, and these two preferences are synergistic [22]. Preference for forked DNA-DNA substrates is also seen with the mitochondrial ScPif1 isoform [46]. In addition, nuclear ScPif1 has higher unwinding activity on G-quadruplex structures compared to random double stranded DNA [26]. Glycerol gradient analysis suggests that both the nuclear and mitochondrial isoforms are monomers in solution [22,46]. Finally, recombinant nuclear ScPif1 reduces the processivity of telomerase in a primer extension telomerase assay by its ability to release telomerase from telomeric oligonucleotides. Telomerase release, which requires the helicase activity of ScPif1, is not due to competition between ScPif1 and telomerase for binding to telomeric oligonucleotides [17].
4. ScRrm3
S. cerevisiae RRM3 was first identified in a screen to identify genes that affect the recombination of rDNA [5]. Mutation of ScRRM3 results in the stimulation of mitotic recombination not only in the rDNA but also at other (but not all) tandemly repeated loci (i.e., CUP1 genes but not Ty elements). However, mutation of ScRRM3 was later found to increase Ty1 mobility >100-fold when the element is located upstream of a tRNA gene [48]. At the time, the reason for this selectivity was not known, but recent research suggests that this phenomenon is due to specific, stable protein complexes that affect fork progression (see Section 4.1.2) [5]. The known roles and activities of ScRrm3 are reviewed below.
4.1. Role in nuclear genome stability
4.1.1. ScRrm3 as a component of the replisome
There has been some debate over whether ScRrm3 is a component of the replisome (the protein super-complex found at all eukaryotic replication forks). A ChIP study dissecting the molecular anatomy of stable, paused replisomes concluded that ScRrm3 is not a member of the replisome but rather is specifically recruited to paused replication forks [49]. However, our lab found that ScRrm3 moves with replication forks globally and interacts with Pol2, the catalytic subunit of DNA Pol ε (a replisome component) [30]. These data suggest that ScRrm3 is a stable component of the replisome.
The discrepancy between these two reports could be due to the different experimental systems used and the fact that there may be two pools of ScRrm3 in the nucleus. In the initial study [49], the authors used two ectopic Fob1/RFB blocks on Chromosome III to induce fork pausing, while the second monitored ScRrm3 via ChIP on unmodified chromosomes [30]. If there are two pools of ScRrm3, one associated with replisomes and one free in the nucleus, then the free ScRrm3 may be heavily recruited to exceptionally strong sites of fork pausing such as the RFB. According to this scenario, the amount of ScRrm3 recruited to the ectopic RFB sites likely swamped out the replisome-level ScRrm3 signal associated with unperturbed forks [49]. This interpretation is also supported by ChIP experiments showing higher levels of ScRrm3 binding at natural rDNA repeats than at other genomic sites [50]. Two different pools of ScPif1 have recently been discovered that affect telomeres and DSBs, respectively, that are differentiated by C-terminal phosphorylation [14]. However, post-translationally modified forms of ScRrm3 have not been reported.
Other data also suggest that ScRrm3 is a replisome component. By yeast two-hybrid analysis, ScRrm3 interacts with Orc5 [51], a subunit of the origin recognition complex. This interaction may mediate the loading of ScRrm3 into replisomes at origins of replication. In fact, by ChIP, ScRrm3 binds origins early in S phase, but not in G1 phase, an origin binding pattern similar to that of other replisomal components but different from that of ORC itself [30]. Further, ScRrm3 contains a proliferating cell nuclear antigen (PCNA) interaction motif (i.e., a PIP-box) at its extreme N-terminus [52,53], and ScRrm3 physically interacts with PCNA in vitro [53]. PCNA is the eukaryotic sliding clamp that functions as a processivity factor for DNA polymerases and as a “tool belt” for the attachment of various other replication factors [54]. Perhaps an interaction with PCNA tethers ScRrm3 to the replisome.
4.1.2. The disruption of protein-DNA complexes
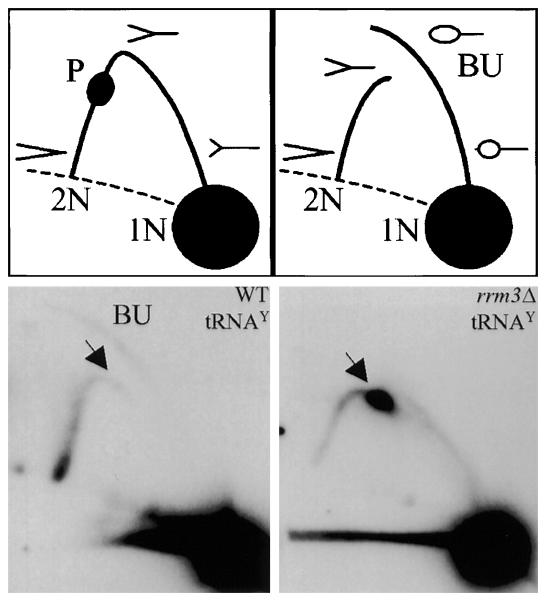
The major nuclear function of ScRrm3 appears to be the disruption of particularly stable non-nucleosomal protein-DNA complexes at what are referred to as Rrm3-sensitive sites. These sites are defined as loci where replication forks pause (or where pausing is exacerbated) in the absence of ScRrm3. These pause sites were first identified in S. cerevisiae by 2D gel analysis of replication fork intermediates [55] and later by microarray studies that monitored DNA Pol2 association genome-wide in rrm3Δ cells [56]. Together, these studies suggest that there are ~1400 Rrm3-sensitve sites in the S. cerevisiae genome, including telomeres [57], rDNA [4], tRNA genes, centromeres, inactive replication origins, and transcriptional silencers [58]. For example, in the absence of ScRrm3, fork pausing dramatically increases at tRNA genes (e.g., ~180-fold at tRNAY in rrm3Δ cells relative to wild type; see Figure 4) [58]. However, when the non-nucleosomal protein-DNA complexes at Rrm3-sensitive loci are artificially disassembled (e.g., at the RFB by deleting FOB1 [59]), fork progression in rrm3 cells largely mirrors that of wild type S. cerevisiae cells.
Figure 4.
Replication fork movement through the tRNAY gene. Top) Cartoon of the 2D gel technique: 1N, non-replicating fragment; 2N, the nearly fully replicated fragment before sister chromatids separate; P, replication pause; BU, bubble-shaped replication intermediates. Bottom) Southern blots were probed to detect the tRNAY gene (tY[GUA]F1; EcoRV fragment, YFR012W). These images are reproduced from [58] and are reprinted following the guidelines of Cell Press's Authors' Rights statement.
Recently, using a ChIP-on-chip approach, the sites of fork pausing in wild type and rrm3 S. cerevisiae cells were mapped genome-wide [60], confirming the existence of the same classes of pause sites [58] as seen earlier by 2D gels in rrm3 cells. The data also demonstrate that previously identified pause sites have a major impact on Pol2 occupancy only in the absence of ScRrm3. In addition, the ORFs of highly transcribed RNA Pol II genes are impediments to fork progression in wild type cells, but this slowing of replication is not exacerbated in the absence of ScRrm3 [60]. Essentially, these data indicate that most natural pause sites are likely due to molecular traffic jams between the replication and transcription machinery, and only in the absence of ScRrm3 does one encounter significant fork pausing at Rrm3-sensitive sites.
Since replication fork progression at Rrm3-sensitive sites is also slowed in helicase-dead rrm3 alleles, ScRrm3 likely uses its catalytic activity to promote fork movement. However, even in regions like the rDNA, which is particularly dependent on ScRrm3 during DNA replication [50], the Mcm2-7 complex, which is the replicative helicase in eukaryotes [61], is still required for fork progression (J. Bessler and VAZ, unpublished results). These data suggest that ScRrm3 is an accessory helicase that acts by disrupting stable protein-DNA complexes at the replication fork that would otherwise slow the translocation of the ScMcm2-7 complex. Such an activity was recently demonstrated for the Rep helicase in E. coli [62]. In addition, the actions of ScRrm3 at stable protein complexes is reminiscent of ScPif1's ability to remove telomerase from telomeres in vitro [17]. Indeed, perhaps a defining characteristic of the non-processive Pif1 family helicases is their ability to generate considerable force while translocating over short stretches of DNA.
4.1.3. The function of ScRrm3 at telomeres
In the absence of ScRrm3, telomeres show modest lengthening, less than in pif1 cells, as well as a very modest decrease in telomeric silencing [57]. Using ChIP, ScRrm3 is associated with telomeric DNA in vivo, and by the criteria of 2D gel analysis, it is also necessary for normal rates of replication through subtelomeric and telomeric DNA. Two-dimensional gel electrophoresis and microarray studies show that, even in wild type cells, replication forks slow as they near telomeres, and this pausing is increased in the absence of ScRrm3 [57,60]. Further, the catalytic activity of the helicase is necessary for efficient replication fork progression, because cells encoding a helicase-dead ScRrm3 (K260–>A) display the same replication pauses by 2D gel analysis as rrm3Δ cells. Fork slowing is not due to the altered chromatin structure associated with telomeric transcriptional silencing because deleting SIR genes, whose products are required for silencing, does not eliminate fork pausing [63]. Likewise, Rif proteins are not required for replication fork pausing within telomeric DNA [64]. The effects of telomeric DNA on fork progression also do not require that the repeats be positioned at a telomere since internal stretches of yeast telomeric DNA also cause fork pausing in wild type cells. As at telomeres, pausing at internal tracts of telomeric DNA is exacerbated in rrm3Δ cells [55]. These findings suggest that ScRrm3 uses its disruption activity (see Section 4.1.2) to dissociate proteins that are bound to telomeric DNA [64]. This finding was the first demonstration that the semi-conservative replication of telomeric DNA, not just replication of the very end of chromosomes, is also a problem for the replication machinery. More recently, telomeric DNA was also shown to impede fork progression in S. pombe [65] and human [66] cells.
4.2. Role in mitochondrial genome stability
Like many of the Pif1 family helicases discussed in this review, ScRRM3 is predicted to encode both nuclear and mitochondrial isoforms [10]. Indeed, an analysis of the S. cerevisiae mitochondrial proteome suggests that ScRrm3 is present in mitochondria [67], and deletion of RRM3 partially rescues the mitochondrial defects of pif1Δ yeast [40,45,68]. This partial rescue may be related to the increase of dNTPs in vivo [40] caused by activation of the intra-S-phase checkpoint [68], which results from increased replication fork pausing in rrm3Δ cells [58]. While these facts may account for the partial rescue, the absence of ScRrm3 has no effect on EtBr-induced mtDNA damage in pif1Δ cells [45]. It remains to be determined if ScRrm3 has a direct effect on mtDNA replication.
4.3. ScRrm3 biochemistry
Regrettably, the in vitro examination of ScRrm3 has lagged behind in vivo studies, largely due to difficulties in purifying the protein. Full-length ScRrm3 expresses poorly and/or forms insoluble aggregates in E. coli, yeast, and baculovirus infected Sf9 insect cells ([57]; M. Bochman and V. Zakian, unpublished data). At least one explanation for these problems is that the N-terminal ~200 amino acids of ScRrm3 are predicted to be natively disordered (M. Bochman, observations) by the criteria of the DISOPRED2 program [69]. Indeed, sequence analyses using DISOPRED2 indicate that an unstructured N-terminus is a hallmark of Pif1 family helicases (M. Bochman, observations). Consistent with this finding, truncating the N-terminal 193 residues of ScRrm3 (Rrm3ΔN) and expressing the protein with a C-terminal GST fusion allows for its purification as a soluble, active helicase [57]. Rrm3ΔN is a Mg+2-dependent ATPase that is stimulated by ssDNA. The enzyme also displays 5′→3′ helicase activity in the presence of Mg+2-ATP. Unfortunately, the Rrm3ΔN protein makes a poor in vitro model of ScRrm3 because the N-terminus is essential for in vivo function [52]. Thus, the biochemical characterization of ScRrm3 (and other Pif1 helicases, see Sections 6 and 7) awaits the development of a robust expression and purification procedure for soluble full-length protein.
5. Pif1 family helicases in other organisms
As discussed in Section 1, it is unclear whether Pif1 family helicases in organisms that encode a single family member more closely resemble ScPif1 or ScRrm3 in their in vivo functions, whether they do the work of both ScPif1 and ScRrm3, or whether they have evolved completely novel activities not displayed by their S. cerevisiae homologues. The following three sections seek to shed light on this issue.
6. SpPfh1
Pif1 family helicases have been most extensively studied in two yeasts, S. cerevisiae and S. pombe, which diverged from each other about 1.1 billion years ago [70]. Unlike S. cerevisiae, but analogous to mammalian cells, S. pombe contains only one Pif1family helicase called Pfh1 (Pif1 family homolog; formerly known as rph1, RRM3/ PIF1 homolog 1). Thus, it is possible that the role of SpPfh1 will be more similar to that of the human Pif1 homolog (hPif1).
Sequence analysis of SpPfh1 reveals equal similarities to both ScPif1 and ScRrm3 [10]. Therefore, the in vivo functions of this helicase cannot be predicted by sequence comparisons. In contrast to ScRrm3, ScPif1 and murine Pif1 (mPif1), pfh1+ is an essential gene [10,71]. This result is not easily explained as S. cerevisiae pif1 rrm3 double mutants are viable, although cell cycle progression and growth rate are perturbed in these double mutants [4]. SpPfh1 deleted cells are able to go through S phase once or twice, but they eventually arrest in G2 phase with an elongated cell shape phenotype characteristic of a defect in nuclear DNA replication [10].
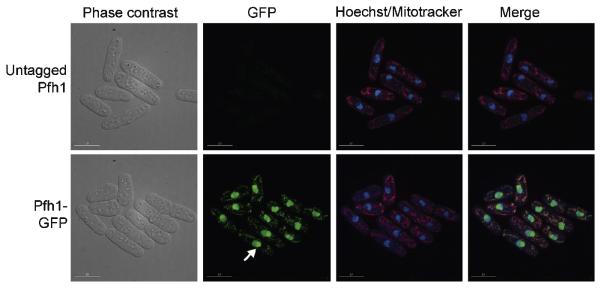
A purified truncated version of Pfh1p has 5′→3′ helicase activity, dependent on ATP and Mg2+ [10] [71], much like ScPif1 and ScRrm3. Since cells expressing a point mutation in the ATP binding domain in motif I (K337A) are not viable, the helicase activity of the SpPfh1 is essential [10,71]. Similar to ScPif1, there are at least two translational start sites in the pfh1+ gene, with a mitochondrial target signal (MTS) site between them [72]. By mutating either the first AUG site or the second AUG site, generating pfh1-m1 and pfh1-m21, respectively, two isoforms of Pfh1p were found. Using fluorescence microscopy, the two isoforms are localized either in mitochondria (pfh1-m21) or in nuclei (pfh1-m1) (Figure 5) [72].
Figure 5.
SpPfh1 is detected in both the nucleus and mitochondria. Wild type cells (untagged Pfh1) and cells expressing Pfh1 fused with GFP at the C-terminus (Pfh1-GFP) are viewed by phase contrast and fluorescence microscopy. Pfh1 is visualized by GFP (green), DNA by Hoechst (blue), and mitochondria by mitotracker (red). The white arrow points out concentrated Pfh1-GFP in the nucleolus. The scale bar indicates 10 μm. This figure is adapted from the journal of Molecular and Cellular Biology, copyright © American Society for Microbiology [Molecular and Cellular Biology , Vol 28, 2008, p.6598, doi:10.1128/MCB.00191-08] [72].
6.1. Replication of chromosomal DNA
6.1.1. Continuation of a discontinuous strand
SpPfh1 interacts genetically with Cdc24 and Dna2. Cdc24 is an essential gene whose exact function is not known, although it is thought to be involved in Okazaki fragment maturation [73,74]. A mutant version of SpPfh1 (pfh1-R20) suppresses the temperature sensitivity of cdc24-M38 and dna2-C2, while overexpression of pfh1+ in either pfh1-R20 cdc24-M38 or pfh1-R20 dna2-C2 double mutants restores the temperature sensitivity [75]. These data led to the proposal that in the absence of SpPfh1, long flaps between Okazaki fragments are not produced (and therefore do not need to be processed), and as a result, the lack of Dna2 is no longer toxic in this background. Thus, like ScPif1, SpPfh1 likely functions in Okazaki fragment maturation [31,75]. In vitro helicase assays demonstrated that SpPfh1 has the ability to unwind both DNA-DNA and RNA-DNA flap structures with equal efficiency [75], suggesting that the RNA-DNA primer on the 5′ end of the downstream Okazaki fragment would not inhibit SpPfh1's unwinding capacity.
6.1.2. Other potential SpPfh1 substrates
Structural elements such as rDNA, tRNA, telomeres, centromeres, and the silent mating type loci are all sites whose replication is promoted by ScRrm3 [4,57,58]. ScRrm3 activity is thought to be important for disrupting non-nucleosomal protein complexes during semi-conservative DNA replication (see section 4.1.2).
The effects of SpPfh1 depletion on telomere length were examined in two studies with different results. In the first, Pfh1 was depleted by sporulating a pfh1+/pfh1Δ heterozygous diploid and examining telomere length in pfh1Δ spore clones, which undergo 1-3 cell divisions before arresting in G2 phase. In this case, telomeres were modestly shorter than in wild type spores [10]. The other study saw no effect on telomere length in cells regulating the expression of SpPfh1 under the thiamine-repressible nmt1 promoter [72]. The lack of effect of SpPfh1 depletion in the second study may be because SpPfh1 is expressed at low levels under the nmt1 promoter, and even low levels are sufficient to supply its telomere function [72]. Although the effects of SpPfh1 on telomere length are not clear, there is no evidence that it inhibits telomerase-mediated telomere lengthening as does ScPif1.
Cytological studies shows that SpPfh1 is in both nuclei and mitochondria, with the highest concentration in nucleoli [72]. Since nucleoli are sites of rDNA transcription and ribosome biogenesis, detection of SpPfh1 in the nucleoli suggests that SpPfh1 may be important for rDNA integrity (Figure 5). Indeed, 2D gel analysis shows increased replication fork arrest and breakage within rDNA in SpPfh1 depleted compared to wild type cells (Sabouri and Zakian, unpublished). The role of SpPfh1 in rDNA replication is similar to what was found for ScRrm3 [58]. This result may explain why ScRrm3, but not ScPif1 or hPif1, can supply the essential nuclear function(s) of SpPfh1 [72]. However, unlike SpPfh1, ScRrm3 is not essential. This difference can be explained if the two proteins perform the same function in their respective organisms, but in S. pombe, replication of one (or more) of the ScRrm3-sensitive substrates is absolutely dependent on SpPfh1. For example, if SpPfh1, like ScRrm3, promotes replication through centromeres, this function, which is not essential in S. cerevisiae, could be critical in S. pombe owing to the much larger size of S. pombe centromeres (S. pombe centromeres are ≥ 35 kb; S. cerevisiae centromeres are only ~125 bp).
6.2. Mitochondrial DNA
SpPfh1 has a mitochondrial targeting signal [10]. Immunoblotting identifies several SpPfh1 isoforms and shows that the mitochondrial version migrates faster than the nuclear isoform, due to cleavage of the MTS as the protein enters the mitochondria [72]. Although, both isoforms are essential, low amounts of the mitochondrial isoform are sufficient to provide the essential nuclear SpPfh1 functions [72]. Cells depleted of SpPfh1 quickly lose mtDNA; by quantifying the amount of mtDNA by real-time PCR, it is 5-fold less abundant after 48 h of SpPfh1 depletion compared to wild type cells [72]. However, no rearrangements of the mitochondrial genome are detected [72]. Thus, SpPfh1 is essential for maintaining mtDNA. Unlike, S. cerevisiae, S. pombe cells are not viable if they lack mtDNA. Therefore, SpPfh1 is essential in both the nuclei and mitochondria. One possibility consistent with these data is that SpPfh1 is the replicative helicase for S. pombe mtDNA. Based on mitochondrial phenotypes, SpPfh1 is more similar to SpPif1, than to ScRrm3.
6.3. DNA repair
In addition to nuclear and mitochondrial isoforms, a third SpPfh1 isoform is detected by immunoblotting [72]. This isoform, which migrates more slowly than either the nuclear or mitochondrial isoforms, increases in abundance in the presence of camptothecin-induced DNA damage [72]. Together these data suggest a post-translationally modified SpPfh1 function in the DNA damage response. Phosphorylated SpPfh1 isoforms are detected by mass spectroscopy (K. McDonald, V.A. Zakian, and I. Cristea, unpublished data). Likewise, ScPif1 is phosphorylated in response to DNA damage [14]. Additionally, sumoylation motifs are found in both SpPfh1 and ScPif1 (S. Pinter and N. Sabouri observations). Furthermore, fluorescence microscopy shows that SpRad22 (the homolog of S. cerevisiae Rad52) and nuclear SpPfh1 co-localize in DNA damaged cells. In addition to supplying the essential nuclear functions of SpPfh1, ScRrm3 also reduces spontaneous, but not induced, DNA damage in the absence of nuclear SpPfh1 [72]. One interpretation of these results is that SpPfh1 has an essential role in DNA replication, the absence of which results in DNA damage, that can be supplied by ScRrm3. However, the function of SpPfh1 in DNA repair cannot be performed by ScRrm3. The ability of ScRrm3 (but not ScPif1) to supply the essential SpPfh1 nuclear functions suggests strong functional similarity between SpPfh1 and ScRrm3.
7. Trypanosome Pif1 helicases
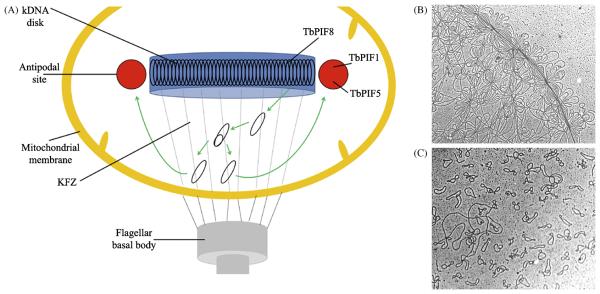
Trypanosoma brucei, and the related kinetoplastid parasites T. cruzi and Leishmania major, have a single, unusual mitochondrion containing a structure known as the kinetoplast (reviewed in [76] and references therein). The kinetoplast contains the mitochondrial genome (kinetoplast DNA or kDNA) that is composed of dozens of maxicircles and thousands of minicircles, all of which are interlocked in a chainmail-like network. The network in vivo is compressed into a disk-shaped structure (Figure 6). In common with the mtDNA from higher eukaryotes, maxicircles encode ribosomal RNAs and proteins necessary for cellular respiration. However, expression of these maxicircle genes requires minicircles because maxicircle transcripts must be edited via the insertion or deletion of uridylate residues, and this editing depends on guide RNAs encoded by minicircle mtDNA.
Figure 6.
The kinetoplast and kDNA. A) In the kinetoplast, the network of maxicircles and minicircles is located near the flagellar basal body. While little is known about maxicircle replication [76], research shows that minicircles (black ovals) detach from the kDNA disk and migrate to the KFZ where they replicate unidirectionally via θ-type replication. Daughter minicircles then move to the antipodal sites where replication continues (including Okazaki fragment maturation) [76]. TbPIF1 (B. Liu and P. Englund, unpublished data) and 5 [79] are located in the antipodal sites, and TbPIF8 localizes to the kDNA disk (B. Liu and P. Englund, unpublished data). Highly expressed GFP-tagged TbPIF2 (not shown) was found throughout the kinetoplast [79]. This figure is based on Figure 3 from [76]. B) Electron micrograph of part of a kDNA network from the kinetoplastid Crithidia fasciculata. C) Topoisomerase II decatenation yields 2.5 kb minicircles and 38 kb maxicircles (left, middle). Micrographs are at approximately the same magnification. The images in B) and C) and legends are adapted from [91] with kind permission of Springer Science+Business Media.
The replication of minicircles and maxicircles differs and is as unusual and complex as the kDNA itself [76]. As such, as many as 100 or more specialized replication factors are necessary for kDNA replication, including six DNA polymerases [77,78]. These factors are located in the kDNA disk itself, in the kinetoflagellar zone (KFZ; a discrete region within the kinetoplast matrix between the kDNA disk and the basal body of the flagellum), and in the antipodal sites (two protein assemblies located on opposite ends of the kDNA disk) (see Figure 6 and [76]). It is thought that the distinct spatial localization of these factors is related to their function in the ordered and tightly regulated replication of the kDNA which occurs once per cell cycle (Figure 6).
Recent genome analysis revealed that T. brucei encodes eight Pif1 family helicases (TbPIF1-8), six of which (TbPIF1, 2, 4, 5, 7, and 8) are mitochondrial, one (TbPIF6) localizes to the nucleus, and one (TbPIF3) is cytoplasmic [79]. However, only TbPIF1, 2, 5, and 8 have been studied in any detail (see below). RNAi shows that three of the mitochondrial TbPIFs (TBPIF1, 2, and 8) are essential, suggesting that one or more of the TbPIFs acts as the mitochondrial replicative helicase. This arsenal of Pif1-like helicases seems to be a common feature of related parasites as the T. cruzi genome contains eight predicted homologs, and the L. major genome contains seven (it lacks a TbPIF5 homolog).
7.1. TbPIF1
Myc-tagged TbPIF1, expressed at near endogenous levels, localizes to antipodal sites at opposite ends of the condensed kDNA disk (B. Liu and P. Englund, unpublished data). Although RNAi eliminates only ~70% of the TbPIF1 mRNA, cells stop growing, and kDNA is lost. The kinetics of minicircle versus maxicircle loss suggest that TbPIF1 likely functions in minicircle replication. Indeed, when TbPIF1 expression is reduced, minicircle replication intermediates are decreased, and there is a concomitant increase in the fraction of multiply-interlocked, covalently-closed minicircle dimers (fraction U). In addition, RNAi against the mitochondrial topoisomerase II results in the appearance of fraction U, suggesting that TbPIF1 is essential for the segregation of newly replicated minicircles. Although it is unclear why a helicase would affect decatenation of minicircle dimers by topoisomerase II, one interesting possibility is that TbPIF1 is needed to dissociate a protein that inhibits topoisomerase activity, and this protein is tightly bound to minicircle DNA.
7.2. TbPIF2
Highly expressed, GFP-tagged TbPIF2 localizes throughout the single tubular T. brucei mitochondrial structure [79]. RNAi against TbPIF2 decreases its mRNA level by 90% in two days and stops growth in six. However, unlike RNAi against many replication factors, reduced TbPIF2 expression does not result in decreased kDNA size. Rather, the relative size of the kDNA disk is maintained because TbPIF2 RNAi causes a severe decrease in maxicircle abundance but a simultaneous 2- to 3-fold increase in minicircle abundance. Overexpression of TbPIF2 drastically increases maxicircle abundance without affecting the number of minicircles. TbPIF2 overexpression also results in kDNA loss in some cells due to defects in kDNA segregation. When a Walker A box mutant (Motif I, K462–>A) TbPIF2 is overexpressed, it acts as a dominant negative, decreasing maxicircles and increasing minicircles, much like TbPIF2 RNAi. These results led the authors to conclude that TbPIF2 uses it catalytic activity for maxicircle replication, perhaps functioning as the maxicircle replicative helicase. Further, previous results showed that RNAi knockdown of the T. brucei HslVU protease causes a striking increase in minicircles and maxicircles [80], which was hypothesized to occur because a positive regulator of kDNA replication was not proteolyzed. It was then demonstrated that RNAi of TbHslVU also causes an increase in TbPIF2 levels [79], suggesting that the helicase is a substrate for the protease and illuminating a possible mechanism for the regulation of maxicircle replication.
7.3. TbPIF5
TbPIF5 was C-terminally Myc-tagged at its endogenous locus [79], and much like TbPIF1 (B. Liu and P. Englund, unpublished data), it localizes to the antipodal sites of the kDNA disk throughout the cell cycle [79]. Reducing TbPIF5 expression (~90%) by a variety of means has no effect on cell growth. However, the authors were unable to delete both TbPIF5 alleles, suggesting that TbPIF5 is essential. Moreover, a 15-fold overexpression of TbPIF5 causes kDNA shrinkage and loss, largely due to a decrease in minicircle abundance. This loss of minicircles is linked to a defect in their replication, specifically the joining of Okazaki fragments. Based on sequence alignments (Figure 1), TbPIF5 appears to be the most closely related to the well-studied fungal Pif1 helicases (see Sections 3-6). If TbPIF5 performs a function similar to that proposed for ScPif1 and SpPfh1 in Okazaki fragment processing (see Section 3.1.4, 6.1.1 and [31,35,36,75,81]), it may unwind RNA primers on the lagging strand, creating flaps that are then degraded by a mitochondrial nuclease [79]. Because Okazaki fragments are not joined in T. brucei until after minicircle segregation and migration to the antipodal sites, overexpression of TbPIF5 likely exerts its negative effects on mtDNA by misregulating the timing of Okazaki fragment processing.
7.4. TbPIF8
Highly expressed, GFP-tagged TbPIF8 localizes largely to the kDNA disk, and as stated above, is essential for growth [79]. TbPIF8 is the smallest and most divergent of the T. brucei Pif1 family helicases (Figure 1; J. Wang and P. Englund, unpublished data). Notably, TbPIF8 and L. major PIF8 are missing two ATPase/helicase motifs (Ia and V; Figure 2) and contain non-canonical Walker A and B boxes (motifs I and II; Figure 2). Thus, TbPIF8 probably does not hydrolyze ATP. Nonetheless, preliminary work suggests that TbPIF8 is required for growth, and maintenance of kDNA ([79]; J. Wang and P. Englund, unpublished data).
7.5. TbPIF biochemistry
Like all Pif1 family helicases, except ScPif1 (see Section 3.3), the TbPIFs are difficult to purify and, additionally, are often unstable in storage (B. Liu and P. Englund, unpublished data). However, preliminary biochemical characterizations have been carried out with TbPIF1, 2, and 5. For instance, recombinant TbPIF1 made in E. coli is a Mg+2-dependent, ssDNA-stimulated ATPase/helicase. Several versions of TbPIF2 have been purified for biochemical analysis: the full-length protein (minus the first 41 amino acids that comprise a putative mitochondrial targeting sequence) N-terminally tagged with GST (GST-TbPIF2) [82]; a His-tagged, N-terminally truncated construct (similar to Rrm3ΔN, see [57] and Section 4.3) missing the first 416 amino acids (ΔNTbPIF2) [79]; and predicted helicase-dead (K462–>A) versions of each. All of the recombinant proteins were expressed in E. coli and recovered with ~90-95% purity, but they are also extremely unstable, requiring immediate use in assays after purification. Regardless, both GST-TbPIF2 and ΔNTbPIF2 are able to unwind M13-based substrates in a Mg+2-ATP-dependent manner, and the K462–>A mutants greatly decrease the in vitro helicase activity of both recombinant forms. Finally, His-tagged TbPIF5 expresses well in E. coli, but much like TbPIF1, is not stable in storage (B. Liu, personal communication). The enzyme hydrolyzes ATP, and this hydrolysis is entirely dependent upon the presence of Mg+2 and ssDNA [79]. As with other Pif1 family helicases, recombinant TbPIF5 unwinds DNA in a 5′–>3′ direction.
8. Mammalian Pif1
All mammalian genomes studied to date encode a single Pif1-like protein of which two, human Pif1 (hPif1) and mouse Pif1 (mPif1), have been examined. Sequence alignments show that hPif1 and mPif1 are 84% identical over their entire open reading frames [83].
8.1. Human Pif1
hPif1 has a predicted molecular weight of ~70 kDa. Alignment of hPif1 to ScRrm3 and ScPif1 shows 24% identity over the helicase domain [83]. Therefore, as with SpPfh1, sequence similarities do not predict the functions of the human protein. Due to difficulties in expressing and purifying full-length hPif1, the first biochemical study of hPif1 was performed on N-terminally truncated hPif1 [8]. This truncated recombinant hPif1 has the expected 5′→3′ helicase activity and unwinds both DNA/DNA [8,84,85] and DNA/RNA substrates [8].
Immunofluorescence analysis shows hPif1 localization to both nuclei [83,86] and mitochondria [86], similar to other Pif1 family members. hPif1 is found in highly proliferating cells [83]. Similar to ScPif1 [18], hPif1 is tightly cell cycle regulated, with peak abundance in G2-phase [83]. As with ScPif1, this cell cycle regulated abundance is APC (anaphase promoting complex) dependent.
Although by the criterion of gel-shift assays, hPif1 binds telomeric DNA with a 100-fold higher affinity compared to random sequence DNA [8], only one of three studies on hPif1 overexpression revealed an effect on telomere length. One study found telomere shortening when overexpressing hPif1 in a telomerase-positive human fibrosarcoma cell line [8], while others saw no effects on telomere length [8,83,86]. The first study also found an inhibition of telomerase when truncated hPif1 is added to an in vitro telomerase assay, and found that this inhibition is due to reduced telomerase processivity [8]. Since FLAG-tagged hPif1 and Myc-tagged hTERT (the catalytic subunit of telomerase) coimmunoprecipitate, [83], together the data suggest a role for hPif1 in telomere biology
8.2. Mouse Pif1
As in humans, mPif1 is found only in highly proliferating cells and interacts with telomerase in mouse extracts, suggesting that mPif1 affects telomeres [87]. However, mouse knockout animals that completely lack mPif1 have no obvious phenotypes such as changes in telomere length or chromosomal abnormalities. Thus, if mPif1 has telomere functions, its role must be fairly subtle or perhaps redundant with that of another helicase. There is no evidence as yet for a mitochondrial function for mPif1. Even if mPif1 localizes to mitochondria, it cannot be required for maintenance of mtDNA since mPif1 knockout mice are viable, and mammals cannot live without mtDNA.
9. Conclusions/Outlook
The studies discussed in this review suggest that Pif1 family helicases share several mutual DNA targets, such as mtDNA, rDNA, and telomeres. However, as summarized below, their functions on these DNA targets are not necessarily the same even in the same organism. In addition, in vitro and in vivo experiments indicate that ScPif1 [31] and SpPfh1 [74] function during Okazaki fragment maturation. These helicases may be important on the lagging strand in situations where the strand displacement activity of Pol δ is not sufficient to dislodge the RNA-DNA primer segment on the downstream Okazaki fragment. Such instances may be more likely to occur at special loci, perhaps where non-nucleosomal protein-DNA complexes are bound and must be dissociated for the continuation of lagging strand synthesis or at special DNA structures, such as G-quadruplex DNA. A role in Okazaki fragment maturation has not yet been described for the other Pif1 family homologues.
The roles of S. cerevisiae Pif1 and Rrm3 in telomere biology are very different. In vivo data support a model where ScRrm3 promotes replication fork progression through telomeres by removing tightly bound protein-DNA complexes that otherwise slow fork movement [55,63,64]. In contrast, ScPif1 is a negative regulator of telomerase that likely acts by unwinding the RNA/DNA hybrid between telomerase RNA and telomeric DNA at the very ends of chromosomes [17]. Although, ScRrm3 and ScPif1 function differently at telomeres, they appear to share a mechanistic property, the ability to disrupt protein complexes bound to DNA. All Pif1 family helicases discussed in this review, except the T. brucei PIFs, are known or suspected to interact with telomeres, but at least in S. pombe, this interaction is more similar to that of ScRrm3 than ScPif1.
ScPif1, SpPfh1, and the T. brucei PIFs are clearly critical for mtDNA replication, perhaps acting as replicative helicases for mtDNA since SpPfh1 [72] and three of the T. brucei homologues (B. Liu, J Wang, and P. Englund, unpublished data; [79]) are essential for maintenance of mtDNA. Although ScPif1 is not essential for mtDNA replication, mtDNA is lost at high rates under normal growth conditions in its absence. Moreover, under stress conditions, such as high temperature, ScPif1 is essential for mtDNA maintenance [3,38]. ScRrm3 is also predicted to localize to mitochondria, but its function seems to be quite different from that of ScPif1 [10,45]. hPif1 is also detected in mitochondria [86], although there is no functional evidence for a role of mammalian Pif1 family proteins in the maintenance of mtDNA. However, it would not be surprising if effects on mtDNA are a general and conserved feature of Pif1 family helicases.
Though they are distantly related to the prokaryotic RecD helicases [7,8], Pif1 helicases clearly have evolved to participate in different DNA transactions at least some of which are eukaryotic specific. However, even the various eukaryotic Pif1 homologues have evolved separate functions. How can such similar proteins have such divergent roles? While it is likely that the basic mechanism of DNA unwinding has been preserved (based on the high similarity between the Pif1 helicases in their internal motor domain), their non-homologous N- and Ctermini may determine their specific activities, either directly or via specific interactions with other proteins.
In summary, it appears that most of the studied Pif1 family helicases have a more ScRrm3-like activity in the nucleus and a more ScPif1-like activity in the mitochondria. However, additional studies are needed to clarify this family's role in the cell, both in organisms where work has been performed and in those not yet studied. The combination of genetics and biochemistry used to date is well suited to elucidate the functions and mechanisms of these enzymes. Given the important roles of Pif1 family helicases in lower eukaryotes, it will not be surprising if eventually mutations in hPif1 are found to be associated with a predisposition to genome instability and hence human disease
Acknowledgements
We thank Beiyu Liu, Jiangyang Wang, and Paul Englund for sharing unpublished data and helpful discussions on the trypanosome helicases. NS was supported by the Wenner-Gren Foundation and MB by the institution training grant NIH 2 T32 CA00952. Research on Pif1 family helicases in the Zakian lab is supported by NIH RO1 GM026938.
Abbreviations2
- Sc
Saccharomyces cerevisiae
- Sp
Schizosaccharomyces pombe
- Tb
Trypanosoma brucei
- mtDNA
mitochondrial DNA
- ssDNA
single-stranded DNA
- rDNA
ribosomal DNA
- ChIP
chromatin immunoprecipitation
- RFB
replication fork barrier
- kDNA
kinetoplast DNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- 1.Foury F, Kolodynski J. pif mutation blocks recombination between mitochondrial rho+ and rho− genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1983;80:5345–5349. doi: 10.1073/pnas.80.17.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lahaye A, Stahl H, Thines-Sempoux D, Foury F. PIF1: a DNA helicase in yeast mitochondria. EMBO J. 1991;10:997–1007. doi: 10.1002/j.1460-2075.1991.tb08034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schulz VP, Zakian VA. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 4.Ivessa AS, Zhou J-Q, Zakian VA. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100:479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- 5.Keil RL, McWilliams AD. A gene with specific and global effects on recombination of sequences from tandemly repeated genes in Saccharomyces cerevisiae. Genetics. 1993;135:711–718. doi: 10.1093/genetics/135.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt KH, Kolodner RD. Requirement of Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol Cell Biol. 2004;24:3213–3226. doi: 10.1128/MCB.24.8.3213-3226.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang DH, Zhou B, Huang Y, Xu LX, Zhou JQ. The human Pif1 helicase, a potential Escherichia coli RecD homologue, inhibits telomerase activity. Nucl Acids Res. 2006;34:1393–1404. doi: 10.1093/nar/gkl029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger JM. SnapShot: nucleic acid helicases and translocases. Cell. 2008;134:888–888. doi: 10.1016/j.cell.2008.08.027. e881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J-Q, Qi H, Schulz V, Mateyak M, Monson E, Zakian V. Schizosaccharomyces pombe pfh1 + encodes an essential 5′ to 3′ DNA helicase that is a member of the PIF1 sub-family of DNA helicases. Mol. Biol. Cell. 2002;13:2180–2191. doi: 10.1091/mbc.02-02-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J-Q, Monson EM, Teng S-C, Schulz VP, Zakian VA. The Pif1p helicase, a catalytic inhibitor of telomerase lengthening of yeast telomeres. Science. 2000;289:771–774. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- 12.Myung K, Chen C, Kolodner RD. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001;411:1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- 13.Mangahas JL, Alexander MK, Sandell LL, Zakian VA. Repair of chromosome ends after telomere loss in Saccharomyces. Mol. Biol. Cell. 2001;12:4078–4089. doi: 10.1091/mbc.12.12.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makovets S, Blackburn EH. DNA damage signalling prevents deleterious telomere addition at DNA breaks. Nat Cell Biol. 2009 doi: 10.1038/ncb1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomaska L, Nosek J, Kramara J, Griffith JD. Telomeric circles: universal players in telomere maintenance? Nat Struct Mol Biol. 2009;16:1010–1015. doi: 10.1038/nsmb.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eugster A, Lanzuolo C, Bonneton M, Luciano P, Pollice A, Pulitzer JF, Stegberg E, Berthiau AS, Forstemann K, Corda Y, Lingner J, Geli V, Gilson E. The finger subdomain of yeast telomerase cooperates with Pif1p to limit telomere elongation. Nat Struct Mol Biol. 2006;13:734–739. doi: 10.1038/nsmb1126. [DOI] [PubMed] [Google Scholar]
- 17.Boule J, Vega L, Zakian V. The Yeast Pif1p helicase removes telomerase from DNA. Nature. 2005;438:57–61. doi: 10.1038/nature04091. [DOI] [PubMed] [Google Scholar]
- 18.Vega L, Phillips J, BR T, DP T, Onigbanjo M, Zakian V. Sensitivity of yeast strains with long G-tails to levels of telomere-bound telomerase. PLoS Genet. 2007;3:1065–1075. doi: 10.1371/journal.pgen.0030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diede SJ, Gottschling DE. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- 20.Marcand S, Brevet V, Mann C, Gilson E. Cell cycle restriction of telomere elongation. Curr. Biol. 2000;10:487–490. doi: 10.1016/s0960-9822(00)00450-4. [DOI] [PubMed] [Google Scholar]
- 21.Taggart AKP, Teng S-C, Zakian VA. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- 22.Boule JB, Zakian VA. The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res. 2007;35:5809–5818. doi: 10.1093/nar/gkm613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huppert JL. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem Soc Rev. 2008;37:1375–1384. doi: 10.1039/b702491f. [DOI] [PubMed] [Google Scholar]
- 24.Rawal P, Kummarasetti VB, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson JE, Smith JS, Kozak ML, Johnson FB. In vivo veritas: using yeast to probe the biological functions of G-quadruplexes. Biochimie. 2008;90:1250–1263. doi: 10.1016/j.biochi.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ribeyre C, Lopes J, Boulé J, Piazza P, Guédin A, Zakian V, Mergny J-L, Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fry M, Loeb LA. Human Werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 28.Sun H, Karow J, Hickson I, Maizels N. The Bloom's Syndrome Helicase Unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 29.Sun H, Bennett R, Maizels N. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucl. Acids Res. 1999;27:1978–1984. doi: 10.1093/nar/27.9.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azvolinsky A, Dunaway S, Torres J, Bessler J, Zakian VA. The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev. 2006;20:3104–3116. doi: 10.1101/gad.1478906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Budd ME, Reis CC, Smith S, Myung K, Campbell JL. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol Cell Biol. 2006;26:2490–2500. doi: 10.1128/MCB.26.7.2490-2500.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McElhinny S.A. Nick, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18:2764–2773. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bae SH, Bae KH, Kim JA, Seo YS. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. doi: 10.1038/35086609. [DOI] [PubMed] [Google Scholar]
- 35.Rossi ML, Pike JE, Wang W, Burgers PM, Campbell JL, Bambara RA. Pif1 helicase directs eukaryotic Okazaki fragments toward the two-nuclease cleavage pathway for primer removal. J Biol Chem. 2008;283:27483–27493. doi: 10.1074/jbc.M804550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pike JE, Burgers PM, Campbell JL, Bambara RA. Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. J Biol Chem. 2009;284:25170–25180. doi: 10.1074/jbc.M109.023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foury F, Dyck EV. A PIF-dependent recombinogenic signal in the mitochondrial DNA of yeast. EMBO J. 1985;4:3525–3530. doi: 10.1002/j.1460-2075.1985.tb04112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Dyck E, Foury F, Stillman B, Brill SJ. A single-stranded DNA binding protein required for mitochondrial DNA replication in S. cerevisiae is homologous to E. coli SSB. EMBO J. 1992;11:3421–3430. doi: 10.1002/j.1460-2075.1992.tb05421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Rourke TW, Doudican NA, Mackereth MD, Doetsch PW, Shadel GS. Mitochondrial dysfunction due to oxidative mitochondrial DNA damage is reduced through cooperative actions of diverse proteins. Mol Cell Biol. 2002;22:4086–4093. doi: 10.1128/MCB.22.12.4086-4093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Rourke TW, Doudican NA, Zhang H, Eaton JS, Doetsch PW, Shadel GS. Differential involvement of the related DNA helicases Pif1p and Rrm3p in mtDNA point mutagenesis and stability. Gene. 2005;354:86–92. doi: 10.1016/j.gene.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 41.Cheng X, Dunaway S, Ivessa AS. The role of Pif1p, a DNA helicase in Saccharomyces cerevisiae, in maintaining mitochondrial DNA. Mitochondrion. 2007;7:211–222. doi: 10.1016/j.mito.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 42.Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112:391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 43.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sabouri N, Viberg J, Goyal DK, Johansson E, Chabes A. Evidence for lesion bypass by yeast replicative DNA polymerases during DNA damage. Nucleic Acids Res. 2008;36:5660–5667. doi: 10.1093/nar/gkn555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng X, Qin Y, Ivessa AS. Loss of mitochondrial DNA under genotoxic stress conditions in the absence of the yeast DNA helicase Pif1p occurs independently of the DNA helicase Rrm3p. Mol Genet Genomics. 2009;281:635–645. doi: 10.1007/s00438-009-0438-6. [DOI] [PubMed] [Google Scholar]
- 46.Lahaye A, Leterme S, Foury F. PIF1 DNA helicase from Saccharomyces cerevisiae. Biochemical characterization of the enzyme. J Biol Chem. 1993;268:26155–26161. [PubMed] [Google Scholar]
- 47.Boule JB, Zakian VA. Helicases Methods and Protocols, Methods in Molecular Biology Series. Humana Press; 2009. Characterization of the Helicase Activity and Anti-Telomerase Properties of Yeast Pif1p in vitro; pp. 359–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scholes DT, Banerjee M, Bowen B, Curcio MJ. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics. 2001;159:1449–1465. doi: 10.1093/genetics/159.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 2005;19:1905–1919. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torres JZ, Bessler JB, Zakian VA. Local chromatin structure at the ribosomal DNA causes replication fork pausing and genome instability in the absence of the S. cerevisiae DNA helicase Rrm3p. Genes Dev. 2004;18:498–503. doi: 10.1101/gad.1154704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsuda K, Makise M, Sueyasu Y, Takehara M, Asano T, Mizushima T. Yeast two-hybrid analysis of the origin recognition complex of Saccharomyces cerevisiae: interaction between subunits and identification of binding proteins. FEMS Yeast Res. 2007;7:1263–1269. doi: 10.1111/j.1567-1364.2007.00298.x. [DOI] [PubMed] [Google Scholar]
- 52.Bessler JB, Zakian VA. The amino terminus of the Saccharomyces cerevisiae DNA helicase Rrm3p modulates protein function altering replication and checkpoint activity. Genetics. 2004;168:1205–1218. doi: 10.1534/genetics.104.028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt KH, Derry KL, Kolodner RD. Saccharomyces cerevisiae RRM3, a 5′ to 3′ DNA helicase, physically interacts with proliferating cell nuclear antigen. J. Biol. Chem. 2002;277:45331–45337. doi: 10.1074/jbc.M207263200. [DOI] [PubMed] [Google Scholar]
- 54.Naryzhny SN. Proliferating cell nuclear antigen: a proteomics view. Cell Mol Life Sci. 2008;65:3789–3808. doi: 10.1007/s00018-008-8305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ivessa AS, Zhou JQ, Schulz VP, Monson EK, Zakian VA. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Azvolinsky A, Giresi PG, Lieb JD, Zakian VA. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell. 2009;34:722–734. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ivessa AS, Zhou J-Q, Schulz VP, Monson EM, Zakian VA. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and sub-telomeric DNA. Genes Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 59.Torres JZ, Bessler JB, Zakian VA. Local chromatin structure at the ribosomal DNA causes replication fork pausing and genome instability in the absence of the S. cerevisiae DNA helicase Rrm3p. Genes Dev. 2004a;18:498–503. doi: 10.1101/gad.1154704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Azvolinsky A, Giresi P, Lieb J, Zakian V. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell. 2009;34:722–374. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bochman ML, Schwacha A. The Mcm complex: unwinding the mechanism of a replicativehelicase. Microbiol Mol Biol Rev accepted. 2009 doi: 10.1128/MMBR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guy CP, Atkinson J, Gupta MK, Mahdi AA, Gwynn EJ, Rudolph CJ, Moon PB, van Knippenberg IC, Cadman CJ, Dillingham MS, Lloyd RG, McGlynn P. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol Cell. 2009;36:654–666. doi: 10.1016/j.molcel.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 64.Makovets S, Herskowitz I, Blackburn EH. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol Cell Biol. 2004;24:4019–4031. doi: 10.1128/MCB.24.9.4019-4031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- 66.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prokisch H, Scharfe C, Camp DG, 2nd, Xiao W, David L, Andreoli C, Monroe ME, Moore RJ, Gritsenko MA, Kozany C, Hixson KK, Mottaz HM, Zischka H, Ueffing M, Herman ZS, Davis RW, Meitinger T, Oefner PJ, Smith RD, Steinmetz LM. Integrative analysis of the mitochondrial proteome in yeast. PLoS Biol. 2004;2:e160. doi: 10.1371/journal.pbio.0020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taylor S, Zhang H, Eaton J, Rodeheffer M, Lebedeva M, O'rourke T, Siede W, Shadel G. The conserved Mec1/Rad53 nuclear checkpoint pathway regulates mitochondrial DNA copy number in Saccharomyces cerevisiae. Mol Biol Cell. 2005 doi: 10.1091/mbc.E05-01-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004;20:2138–2139. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 70.Heckman DS, Geiser DM, Eidell BR, Stauffer RL, Kardos NL, Hedges SB. Molecular evidence for the early colonization of land by fungi and plants. Science. 2001;293:1129–1133. doi: 10.1126/science.1061457. [DOI] [PubMed] [Google Scholar]
- 71.Tanaka H, Ryu GH, Seo YS, Tanaka K, Okayama H, MacNeill SA, Yuasa Y. The fission yeast pfh1+ gene encodes an essential 5′ to 3′ DNA helicase required for the completion of S-phase. Nucleic Acids Res. 2002;30:4728–4739. doi: 10.1093/nar/gkf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pinter SF, Aubert SD, Zakian VA. The Schizosaccharomyces pombe Pfh1p DNA helicase is essential for the maintenance of nuclear and mitochondrial DNA. Mol Cell Biol. 2008;28:6594–6608. doi: 10.1128/MCB.00191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gould K, Burns C, Feoktistova A, Hu C, Pasion S, Forsburg S. Fission yeast cdc24(+) encodes a novel replication factor required for chromosome integrity. Genetics. 1998;149:1221–1233. doi: 10.1093/genetics/149.3.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tanaka H, Tanaka K, Murakami H, Okayama H. Fission yeast cdc24 is a replication factor C- and proliferating cell nuclear antigen-interacting factor essential for S-phase completion. Mol Cell Biol. 1999;19:1038–1048. doi: 10.1128/mcb.19.2.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ryu GH, Tanaka H, Kim DH, Kim JH, Bae SH, Kwon YN, Rhee JS, MacNeill SA, Seo YS. Genetic and biochemical analyses of Pfh1 DNA helicase function in fission yeast. Nucl Acids Res. 2004;32:4205–4216. doi: 10.1093/nar/gkh720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu B, Liu Y, Motyka SA, Agbo EE, Englund PT. Fellowship of the rings: the replication of kinetoplast DNA. Trends Parasitol. 2005;21:363–369. doi: 10.1016/j.pt.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 77.Saxowsky TT, Choudhary G, Klingbeil MM, Englund PT. Trypanosoma brucei has two distinct mitochondrial DNA polymerase beta enzymes. J Biol Chem. 2003;278:49095–49101. doi: 10.1074/jbc.M308565200. [DOI] [PubMed] [Google Scholar]
- 78.Klingbeil MM, Motyka SA, Englund PT. Multiple mitochondrial DNA polymerases in Trypanosoma brucei. Mol Cell. 2002;10:175–186. doi: 10.1016/s1097-2765(02)00571-3. [DOI] [PubMed] [Google Scholar]
- 79.Liu B, Wang J, Yaffe N, Lindsay ME, Zhao Z, Zick A, Shlomai J, Englund PT. Trypanosomes have six mitochondrial DNA helicases with one controlling kinetoplast maxicircle replication. Mol Cell. 2009;35:490–501. doi: 10.1016/j.molcel.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li Z, Lindsay ME, Motyka SA, Englund PT, Wang CC. Identification of a bacterial-like HslVU protease in the mitochondria of Trypanosoma brucei and its role in mitochondrial DNA replication. PLoS Pathog. 2008;4:e1000048. doi: 10.1371/journal.ppat.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stith CM, Sterling J, Resnick MA, Gordenin DA, Burgers PM. Flexibility of eukaryotic Okazaki fragment maturation through regulated strand displacement synthesis. J Biol Chem. 2008;283:34129–34140. doi: 10.1074/jbc.M806668200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu B, Wang J, Yildirir G, Englund PT. TbPIF5 is a Trypanosoma brucei mitochondrial DNA helicase involved in processing of minicircle Okazaki fragments. PLoS Pathog. 2009;5:e1000589. doi: 10.1371/journal.ppat.1000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mateyak M, Zakian V. Human PIF helicase is cell cycle regulated and associates with telomerase. Cell Cycle. 2006;23:2796–2804. doi: 10.4161/cc.5.23.3524. [DOI] [PubMed] [Google Scholar]
- 84.Gu Y, Masuda Y, Kamiya K. Biochemical analysis of human PIF1 helicase and functions of its N-terminal domain. Nucleic Acids Res. 2008;36:6295–6308. doi: 10.1093/nar/gkn609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.George T, Wen Q, Griffiths R, Ganesh A, Meuth M, Sanders CM. Human Pif1 helicase unwinds synthetic DNA structures resembling stalled DNA replication forks. Nucleic Acids Res. 2009;37:6491–6502. doi: 10.1093/nar/gkp671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Futami K, Shimamoto A, Furuichi Y. Mitochondrial and nuclear localization of human Pif1 helicase. Biol Pharm Bull. 2007;30:1685–1692. doi: 10.1248/bpb.30.1685. [DOI] [PubMed] [Google Scholar]
- 87.Snow B, Mateyak M, Paderova J, Wakeham A, Iorio C, Zakian V, Squire J, Harrington L. Murine pif1 interacts with telomerase and is dispensable for telomere function in vivo. Mol Cell Biol. 2007;27:1017–1026. doi: 10.1128/MCB.01866-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 89.Hall MC, Matson SW. Helicase motifs: the engine that powers DNA unwinding. Mol Microbiol. 1999;34:867–877. doi: 10.1046/j.1365-2958.1999.01659.x. [DOI] [PubMed] [Google Scholar]
- 90.Boule JB, Zakian VA. Roles of Pif1-like helicases in the maintenance of genomic stability. Nucl Acids Res. 2006;34:4147–4153. doi: 10.1093/nar/gkl561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shapiro TA, Klein VA, Englund PT. Isolation of kinetoplast DNA. Methods Mol Biol. 1999;94:61–67. doi: 10.1385/1-59259-259-7:61. [DOI] [PubMed] [Google Scholar]