Abstract
While the recognition of stop codons by class 1 release factors (RFs) on the ribosome takes place with extremely high fidelity, the molecular mechanisms behind this remarkable process are poorly understood. Here we performed structural probing experiments with Fe(II)-derivatized RFs to compare the conformation of cognate and near-cognate ribosome termination complexes. The structural differences that we document provide an unprecedented view of signal transduction on the ribosome that depends on authentic stop codon recognition. These events initiate with very close interactions between RF and the small subunit decoding center (DC), lead to increased interactions between the switch loop of the RF and specific regions of the subunit interface and end in the precise orientation of the RF for maximal catalytic activity in the large subunit peptidyl transferase center (PTC).
Keywords: Ribosome, termination, release factor, conformational rearrangement, hydroxyl radical probing
Introduction
Translation termination takes place when one of three stop codons is recognized by a proteinaceous class 1 release factor (RFs) in the A site of the ribosome. The RFs are bifunctional molecules, like tRNA, carrying a “tripeptide anticodon” motif at one end (domain 2 and 4) that is responsible for deciphering stop codons in the small subunit decoding center (DC) and a “GGQ”motif at the other end (domain 3) that promotes the hydrolytic reaction to release the growing polypeptide chain (reviewed in ref. 1). Both tRNA and RF selection take place with extremely high fidelity through mechanisms which remain incompletely understood (see ref. 2).
More is generally known both structurally and biochemically about the high fidelity selection of tRNAs by the ribosome during the process of elongation3. High-resolution structures of tRNA anticodon stem loops (ASLs) bound to cognate-codon-programmed 30S subunit reveal striking conformational rearrangements in the decoding center4. These local structural changes, triggered by cognate codon recognition, lead to global rearrangements (domain closure) in the ribosome that result in accelerated rates of GTPase activation (by EFTu) and tRNA accommodation, and thus to accurate tRNA selection during the elongation cycle. How does the cognate recognition complex differ from the related near-cognate complex that fails to promote rapid GTPase activation and accommodation? In studies performed with isolated 30S subunits bound to cognate and near-cognate ASLs, domain closure was only observed for the cognate species5. And, again, these views yielded detailed information only on the structural state of the decoding center and for the near-cognate case only when paromomycin was simultaneously bound. What remains unresolved is how the structural differences detected in the small subunit decoding center (where a single mismatch in the codon:anticodon helix is being sensed) are transmitted to the large subunit to affect downstream events. These questions are directly relevant to those that we address here, but in experiments exploring translation termination.
RF catalyzed peptide release is a similarly highly accurate procedure. Release factors have been found to catalyze premature translation termination on sense codons with an extremely low frequency of ~10−5 in vivo6. This level of discrimination has been largely recapitulated in in vitro experiments showing that RFs can very effectively discriminate against near-stop codons (differing at a single position from authentic stop codons)7 through differential effects on binding (Km) and catalysis (kcat). The observed effects on catalysis argue that high fidelity recognition by RFs relies on strict coupling between recognition of stop codons in the decoding center and catalysis of hydrolysis (i.e. peptide release) in the peptidyl transferase center some 75 Å away. Indeed, one recent study showed that RF1 recognition of stop, but not near-stop, codons triggered conformational rearrangements in the decoding center that impact the rates of peptide release8. These observations together argue for communication between these distinct locales, and the body of the RF and the ribosome represent obvious candidates for transmitting this detailed molecular information.
Recent atomic resolution structures of RF1 and RF2 bound to the ribosome9–11 provided critical insight into the molecular details of stop codon recognition. Of particular interest for understanding signal transduction is a region connecting domains 3 and 4 (termed the “switch loop”) which is largely unstructured in isolated RF112, but which assumes a more rigid alpha-helical conformation when bound to the ribosome10. What remains missing from this list of snapshots of the terminating ribosome is an equivalent view of “near-stop” RF-ribosome recognition complexes. Here we use a structural probing approach to compare the conformation of these two closely related complexes, an approach that has been used in other systems13,14. From these data, we see a clear signature of signal transduction between the two key functional centers of the ribosome, thus providing a structural interpretation for stop codon recognition and the associated rate enhancements for RF-catalyzed peptide release.
Results
Identification of appropriate single-cysteine RF1 variants
To develop a tethered Fe(II) probing approach for comparing the interactions of RF1 with various ribosome “termination” complexes, we set out to identify single cysteine-containing RF1 variants that conformed to several different criteria. First, the chosen single cysteine variants needed to be readily derivatized with the bromoacetyl activated iron-chelating reagent, 1-(p-bromoacetamidobenzyl)-EDTA (FeBABE). Second, when the Fe(II)-derivatized RF1 variants were bound to different ribosome complexes (containing either a stop- or near-stop-codon-programmed A site), we were interested in those that exhibited differential patterns of hydroxyl-radical cleavage.
Our choices were first guided by considering the position of RF1 within the ribosome9–11, and then targeted regions of particular functional interest such as the tripeptide anticodon motif, the GGQ motif and the switch loop. Second, we considered positions evaluated in earlier efforts that used tethered Fe(II) probing to initially determine the position of class 1 RFs on the ribosome15. Starting with an RF1 variant lacking the three natural cysteines, we used site-directed mutagenesis to introduce unique cysteine residue at a number of positions including 156, 187 and 194 proximal to the tripeptide anticodon motif, 220, 226 and 229 near the GGQ motif, and finally 289 and 292 near and within the recently identified switch loop10 (Supplementary Fig. 1a). All of these chosen variants were derivatized with FeBABE16 and further characterized.
Ribosomes were next programmed with a short mRNA directing initiator tRNA (tRNAfMet) to the P site and either an authentic stop codon (UAA) or three different near-stop codons (CAA, UCA or UAC) poised in the A site decoding center8. A toe-printing assay confirmed that ribosomes in each sample were positioned equivalently on the mRNA with the anticipated stop or near-stop codon in the A site (Supplementary Fig. 1b).
Fe(II)-derivatized RF1 variants were then bound to the various stop and near-stop ribosome complexes, and supplied with hydrogen peroxide and ascorbic acid to generate localized hydroxyl radicals to target the surrounding RNA for cleavage. In each case, the Fe(II) derivatized RF1 was added at a concentration known to be saturating for peptide release activity on that particular codon (typically 75 µM, data not shown). Cysteine-free RF1 subjected to the same FeBABE derivatization and ribosome incubation procedures provided the required negative control. Cleavage sites within 16S and 23S rRNA were mapped using a primer extension assay17, while cleavage of the P-site tRNA was directly monitored using a tRNAfMet radioactively labeled at the 3′-end18 (probed within equivalent ribosome complexes) and analyzed by denaturing PAGE.
In an initial pass, we looked broadly at the rRNA cleavage patterns resulting from derivatization of RF1 at the selected positions, a number of which had previously been explored15. In general, our results recapitulated the earlier probing patterns, though what we were most interested in identifying were variants that exhibited different probing patterns or intensities on the stop- and near-stop-programmed ribosome complexes.
For the RF1 variants near the tripeptide anticodon motif (H156C, V187C and G194C), the sites of cleavage were very similar to those previously observed for Fe(II) probing in the decoding center15. Moreover, each of these variants showed differential intensities of cleavage on the stop and near-stop ribosome complexes and were thus potential candidates for this study. Because the Fe(II)-derivatized V187C- and G194C-RF1 exhibited some defects in peptide release (rates reduced by a factor of 4, data not shown), they were not chosen for subsequent analysis.
Of the RF1 variants within and proximal to the switch loop region (S292C and A289C, respectively), only the Fe(II)-S292C-RF1 exhibited differential cleavage intensities on the stop and near-stop ribosome complexes. The Fe(II)-A289C-RF1 exhibited equivalent patterns on the different ribosome complexes and will be included as a control as detailed below.
For the variants near the catalytic GGQ motif, the Fe(II)-S229C-RF1 showed cleavages indistinguishable from those reported previously, though there were no apparent differences on the stop and near-stop complexes; because this variant also exhibits substantial catalytic defects after modification (reduction of kcat by more than two orders of magnitude, data not shown), it was not further characterized. Two other new positions in this region were evaluated (226 and 220). No differential cleavage patterns on the stop and near-stop complexes were observed for the Fe(II)-A220C-RF1 and so it was not further characterized; the Fe(II)-T226C-RF1 exhibited interesting and diverse patterns on these same complexes and was chosen for subsequent analysis.
Based on this preliminary analysis, we settled on an in-depth characterization of four different RF1 variants (H156C, T226C, A289C and S292C), which reveal informative differences among various complexes. For these variants, the efficiency of FeBABE derivatization ranged from 80%~90% (determined using a thiol alkylation (NTCB) protection assay19) and the peptide release activity8 was unaffected for three variants (H156C, A289C and S292C), and modestly affected (reduced by a factor of about 10) for the T226C-RF1 after modification. All four variants exhibited high catalytic specificity for stop-codon-programmed ribosome complexes (relative to near-stop) making them good candidates for further analysis (Supplementary Fig. 1c,d).
Probing the decoding center with Fe(II) tethered H156C-RF1
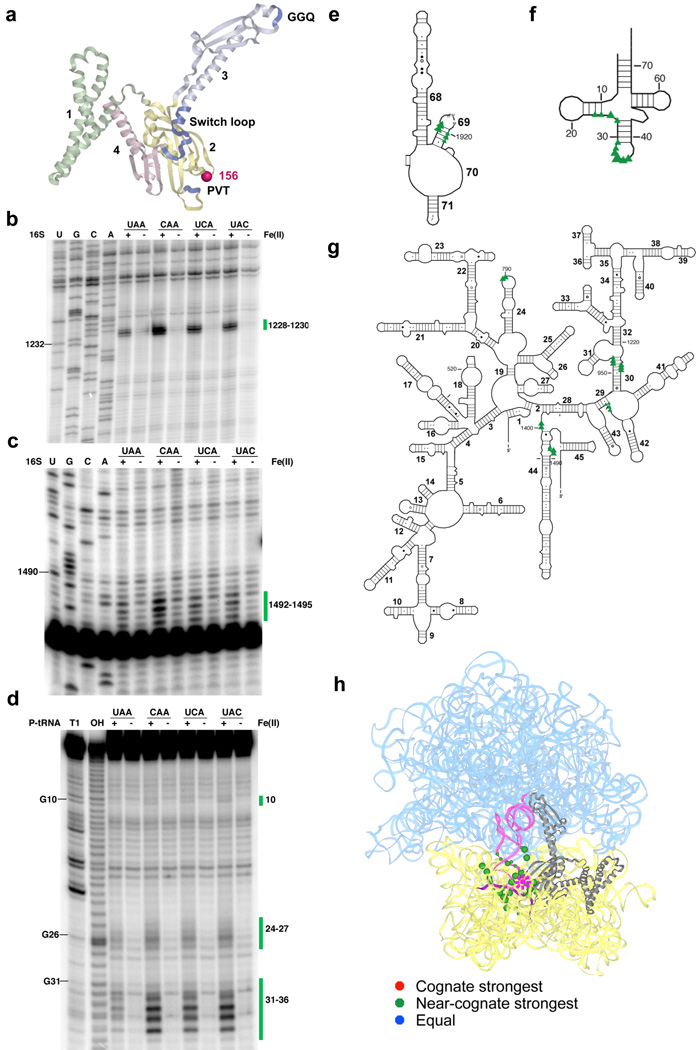
Hydroxyl radicals originating from position 156 near the tripeptide anticodon motif (Fig. 1a) cleaved predictable RNA elements in the small subunit (decoding center within h44, h30, h29 and h24 of 16S rRNA) and the large subunit (H69 of 23S rRNA), as well as the D-stem and anticodon loop of P-site tRNA (Fig. 1b–h and Supplementary Fig. 2). What is striking about the cleavage patterns is that in each case, the intensity of cleavage is considerably greater on the near-stop ribosome complexes (relative to those carrying an authentic stop codon). Indeed, the pattern of some of these target sites correlates inversely with the known release activity of the different codons (UAA>UCA=UAC>CAA) (see Supplementary Fig. 1c for relative rates of release on various complexes), with strongest cleavage for the first position mismatch CAA complex, less strong cleavage for the second and third position mismatches (UCA and UAC) and the least strong cleavage for the authentic stop codon (UAA).
Figure 1.
Directed hydroxyl radical probing of the interaction between the codon recognition domain of RF1 and the ribosome in complexes programmed with various codons in the A site. (a) Ribbon diagram of RF1 (PDB entry: 3D5A10). Functionally important motifs including “PVT” tripeptide anticodon, catalytic “GGQ” and switch loop are shown in blue. Each domain is labeled by number and shown in different colors. Sphere indicates Cα of residue 156 where Fe(II) is tethered. Three-dimensional structures were prepared in Pymol (http://pymol.sourceforge.net). (b–c) Primer extension analysis of cleavage pattern of 16S rRNA from saturating levels of Fe(II)-H156C-RF1 in ribosome complexes with UAA, CAA, UCA or UAC codons in the A site. (U, G, C and A) are sequencing lanes. (+) represents the Fe(II)-H156C-RF1 sample while (−) represents the sample treated with mock labeled Cysless-RF1. Primers used begin extension at position 1391(b) and 1508 (c). (d) Denaturing sequencing gel analysis of cleavage of P-site tRNA from Fe(II)-H156C-RF1. (T1) cleavage by RNaseT1; (OH) alkaline hydrolysis ladder. Bars and numbers at right indicate cleavages corresponding to nucleotides within 16S rRNA or P-site tRNA. (e–g) Directed hydroxyl radical cleavage sites from Fe(II)-H156C-RF1 shown on the secondary structure25 of (e) part of the 3′ half of 23S rRNA, (f) P-site tRNA and (g) 16S rRNA. Triangle size reflects cleavage intensity relative to the strongest hits for that particular complex. (h) All cleavage sites modeled on tertiary structure of RF1 bound ribosome complex (PDB entry: 3D5A10 and 3D5B10). 16S rRNA from 30S subunit and 23S rRNA from 50S subunit are shown in yellow and cyan, respectively, while the ribosomal proteins are omitted. RF1 is shown in gray; P-site tRNA is in pink and mRNA is deep purple. Star represents the position to which Fe(II) is tethered, spheres indicate targeted sites (with size reflecting relative cleavage intensity). Overall cleavage patterns are indicated with various colors (stronger on cognate (red), stronger on near-cognate (orange), and equivalent on all (blue)). Unless otherwise indicated, same labeling scheme used throughout manuscript.
In a previous study, cleavages produced by RF1 derivatized with Fe(II) at nearby position 187 were compared in similar ribosome complexes (either with the stop codon (UAG) or non-stop codon (UUU))15. In this case, cleavage was only observed with the authentic stop codon complex, likely because of the relatively lower concentration (1.5 µM versus 75 µM in our case) of labeled RF1 used.
Probing the switch loop region with Fe(II) tethered RF1 variants
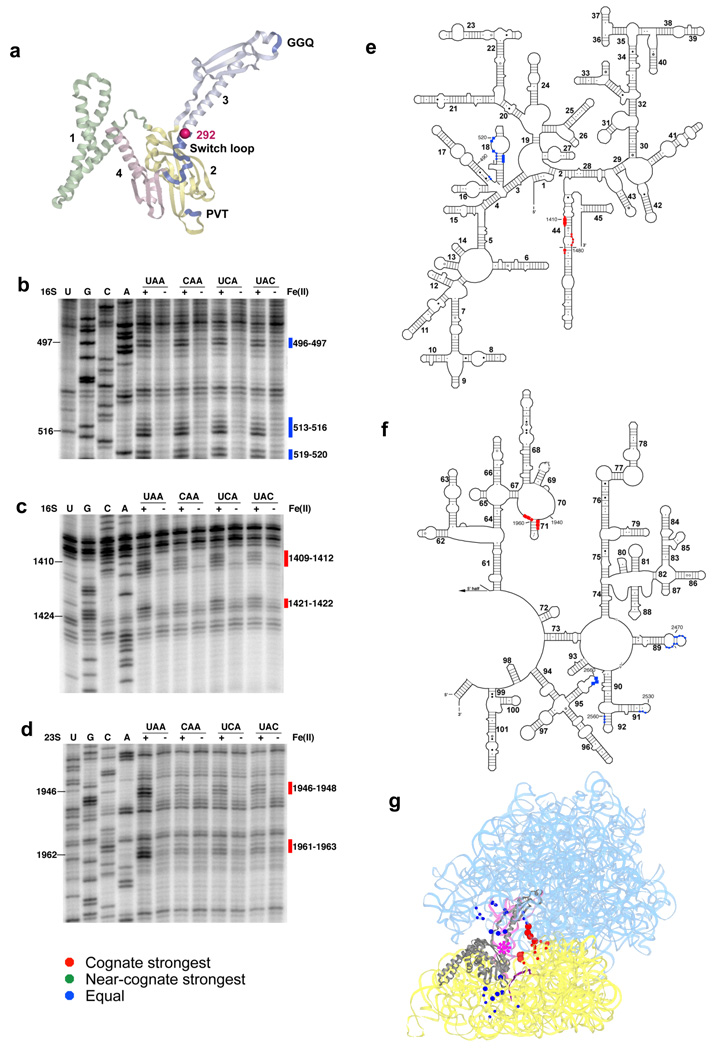
Hydroxyl radicals originating from position 292 of the switch loop led to two classes of cleavage pattern among the stop and near-stop complexes (Fig. 2a). First, we observed a set of cleavages localized within h18 of 16S rRNA, the loops of H89, H91 and H95 (the sarcin-ricin loop) and the stem of H92 of 23S rRNA, where the intensity was equal among all ribosome complexes (Fig. 2b and Supplementary Fig. 3a–c). These same cleavages were also observed with the Fe(II)-A289C-RF1, located somewhat further up the switch loop (Supplementary Fig. 4a,c–f). What was surprising about all of these cleavages is that these sites are somewhat remote from the site of radical generation based on an analysis of recent crystal structures9–11. At a minimum, the observation that the intensity of cleavage is equivalent on all structures suggests that the stop and non-stop complexes are equivalently bound (or sampled) by these Fe(II)-derivatized RF1s.
Figure 2.
Directed hydroxyl radical probing of the ribosome environment of the switch loop of RF1 in ribosome complexes programmed with various codons in the A site. (a) Ribbon diagram of RF1. Sphere indicates Cα of residue 292 where Fe(II) is tethered. (b–d) Primer extension analysis of cleavage of 16S rRNA (b–c) and 23S rRNA (d) from saturating levels of Fe(II)-S292C-RF1 incubated with the indicated ribosome complexes with primers (b) 565 (c) 1490 and (d) 2042. (e–f) Directed hydroxyl radical cleavage sites from Fe(II)-S292C-RF1 shown on secondary structure of (e) 16S rRNA and (f) the 3′ half of 23S rRNA. (g) All cleavage sites modeled on tertiary structure of RF1 bound ribosome complex (PDB entry: 3D5A10 and 3D5B10), as in Figure 1.
The second type of cleavage pattern that we observed for the Fe(II)-S292C-RF1 consisted of cleavages in RNA elements at h44 of 16S rRNA as well as H70 and H71 of 23S rRNA, entirely consistent with the relevant X-ray structures. Interestingly, these cleavages were stronger on authentic stop codon complexes (Fig. 2c–g and Supplementary Fig. 3d). These RNA elements overlap with the functionally important ribosome subunit contact site B3 through which the decoding center of the small subunit is connected to the A loop of the large subunit20. We tested the specificity of this observation by mutating amino acids that were shown to interact directly with the stop codon10 (changing two amino acids, E123 and T190 within the tripeptide anticodon motif, to P and A, respectively) within the body of S292C-RF1; when used in a tethered Fe(II) probing experiment, the pattern of strong cleavage observed on the stop codon complexes at subunit contact site B3 was lost (Supplementary Fig. 3e); control cleavages seen on stop and near-stop complexes were still visible (Supplementary Fig. 3f) indicating that the variant RF1 still bound effectively. These data argue for the importance of stop codon recognition in specifying conformational changes in the switch loop region. Moreover, we note that the stop codon-dependent cleavage pattern was specific for the actual switch loop, since the same pattern was not generated by the A289C-RF1, tethered with Fe(II) only 5 Å away (Supplementary Fig. 4b, e, f).
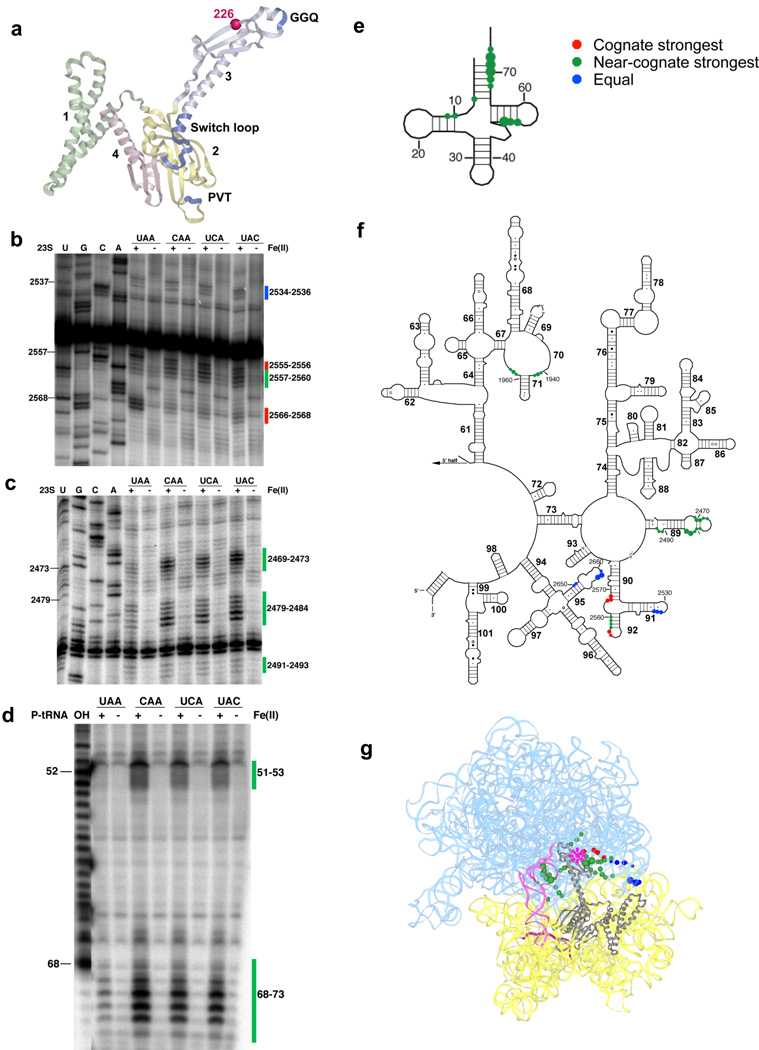
Probing the large subunit catalytic center with Fe(II) tethered T226C-RF1
Hydroxyl radicals originating from Fe(II) tethered to T226C-RF1 result in diverse cleavage patterns in several different regions of the ribosome (Fig. 3a). First, the H91 and H95 (sarcin-ricin) loops in 23S rRNA are similarly cleaved in all complexes (Fig. 3b,f,g and Supplementary Fig. 5b). These data serve as controls to indicate that binding of the derivatized T226C-RF1 was equivalent in each case. In other regions, H70, H89 and part of the stem of H92 of 23S rRNA, as well as the acceptor stem and elbow region of P-site tRNA, were preferentially cleaved in near-stop complexes, while regions of the 23S rRNA near to the A loop (2555–2556 and 2566–2568) were cleaved more efficiently in true stop complexes (Fig. 3b–g and Supplementary Fig. 5a,c). These differential patterns were of substantial interest as they provide a structural correlate for previous kinetic results showing increased rates of catalysis (kcat) on authentic stop codon complexes7,8.
Figure 3.
Directed hydroxyl radical probing of the ribosome environment of the catalytic motif “GGQ” of RF1 in ribosome complexes programmed with various codons in the A site. (a) Ribbon diagram of RF1. Sphere indicates Cαof residue 226 where Fe(II) is tethered. (b–c) Primer extension analysis of 23S rRNA from saturating levels of Fe(II)-T226C-RF1 incubated with the indicated ribosome complexes with primers (b) 2639 and (c) 2542. (d) Denaturing sequencing gel analysis of cleavage of P-site tRNA from Fe(II)-T226C-RF1. (T1) cleavage by RNaseT1; (OH) alkaline hydrolysis ladder. (e–f) Directed hydroxyl radical cleavage sites from Fe(II)-T226C-RF1 shown on secondary structure of (e) P-site tRNA and (f) the 3′ half of 23S rRNA. (g) All cleavage sites modeled on tertiary structure of RF1 bound ribosome complex (PDB entry: 3D5A10 and 3D5B10), as in Figure 1.
Discussion
Here we present a chemical probing analysis of RF1 interacting with various ribosome complexes that allows us to visualize for the first time the events of signal transduction on the ribosome following RF-mediated stop codon recognition. Analysis of ribosome (rRNA) and P-site tRNA cleavage patterns resulting from Fe(II) tethered on RF1 in functionally relevant positions revealed substantial differences (both in position and intensity) on the stop (UAA) and near-stop (CAA, UCA or UAC) programmed ribosome complexes. These data unambiguously establish that there are discrete conformational differences among these complexes, and that these reflect differences in the interactions between the class 1 RF, the codon positioned in the A site and the ribosome.
First, hydroxyl radicals generated from position 156 close to the tripeptide anticodon motif of RF1 (and targeting primarily the decoding center of the small ribosomal subunit), cleave surrounding RNA elements in near-stop complexes much more strongly than in authentic stop complexes. In contrast, hydroxyl radicals originating from position 292 (in the so called switch loop) cleave authentic stop complexes much more strongly than near-stop complexes at regions of the ribosome involved in bridging interactions (bridge B3) between the large and small subunit. Finally, hydroxyl radicals originating from position 226 proximal to the catalytic GGQ motif generated diverse cleavage patterns in the stop and near-stop complexes; certain positions were more strongly modified on stop complexes whereas others were more strongly modified on near stop complexes.
These observations can be brought together to envision how RF-mediated stop codon recognition in the small subunit decoding center is communicated to other functional centers on the ribosome. Our initial expectation in these studies was that the RNA cleavage patterns would be stronger on authentic stop codon complexes, reflecting the greater stabilization of the interaction (binding is certainly tighter for RFs on authentic stop codons, data not shown). Moreover, we knew from structural studies that the RF very closely engages the stop codon through direct interactions with amino acids (the PVT or SPF motifs found there) in the tip of domain 2 (ref. 9–11,21). As such, we were struck by the results from the Fe(II)-H156C-RF1 in the decoding center wherein stronger cleavage patterns were observed for the near-stop ribosome complexes. These results are consistent with two distinct molecular explanations: 1. RF1 engages the decoding center more snugly on near-stop complexes, and thus pulls away from the region following authentic stop codon recognition; or 2. RF1 engages the decoding center so snugly on authentic stop codons that solvent is excluded from this region and Fenton chemistry is therefore precluded. We favor the latter interpretation which is consistent with what is known from X-ray structures9–11 as well as previous reports indicating that paromomycin is directly displaced from the decoding center on recognition of true stop codons by the class 1 RFs8.
The switch loop cleavage patterns fit nicely with our initial conception of what might be observed; on recognition of a true stop codon, nearby regions of the rRNA are pulled closer, as though the ribosome were folding around the factor to stabilize the interaction (i.e. “RF selection” has occurred). Finally, in the large subunit functional center, certain cleavages from Fe(II)-T226C-RF1 are stronger (in the A loop region, 2555–2556 and 2566–2568) for authentic stop codon complexes, while many others (in the P-site tRNA, H70, H89 and H92 of 23S rRNA) are stronger for near-stop codon complexes. These differential cleavage patterns are readily explained by two different docking positions of the class 1 RF on these distinct complexes. In the authentic stop codon complexes, the cleavage pattern correlates with a bound state that yields a maximal rate constant for peptide release, while in the near-stop complexes, the RF is clearly mispositioned correlating with slower catalysis7.
Using the atomic resolution structures of class 1 RFs bound to stop codon programmed ribosomes, we can predict all nucleotides that should be within probing range (~24 Å) of the tethered Fe(II) in the various positions (Supplementary Fig. 6a–d). A comparison of these predicted patterns with the observed patterns in our ribosome complexes reveals some interesting discrepancies. For instance, the observed cleavages for position 156 in domain 2 in the decoding center are entirely consistent with what is predicted from the X-ray structures (Supplementary Fig. 6a,e). These data argue that this domain of the RF is quite stably bound in the conformation observed crystallographically, consistent with the long lived single FRET state observed in recent single molecule studies using a fluorescent probe at nearby position 167 (ref. 22). By contrast, the cleavage patterns observed from the Fe(II)-derivatized S292C-, A289C- and T226C-RF1s (variants in the switch loop and GGQ motif, both found in domain 3) included a number of hits not predicted by the X-ray structures (Supplementary Fig. 6b–d,f–h). We note that these nucleotides all are found on the side of the ribosome where RF1 enters the A site. These data argue for more mobility in this region of the RF so that multiple binding states are sampled within the entry corridor (the loops of H91 and H95)23, at least in the post-termination complex that we analyze here. We anticipate that these extraneous cleavages might be affected by the addition of the class 2 release factor, RF3, thought to participate in downstream events in termination24. As a final point, we note that the predicted cleavage pattern for the Fe(II)-T226C-RF1 (Supplementary Fig. 6c) overlaps better with cleavages observed on the stop codon complex (than on the near-stop), as might be anticipated given that the X-ray structure represents an authentic stop codon complex; positioning of the GGQ motif thus depends on appropriate docking in the small subunit decoding center.
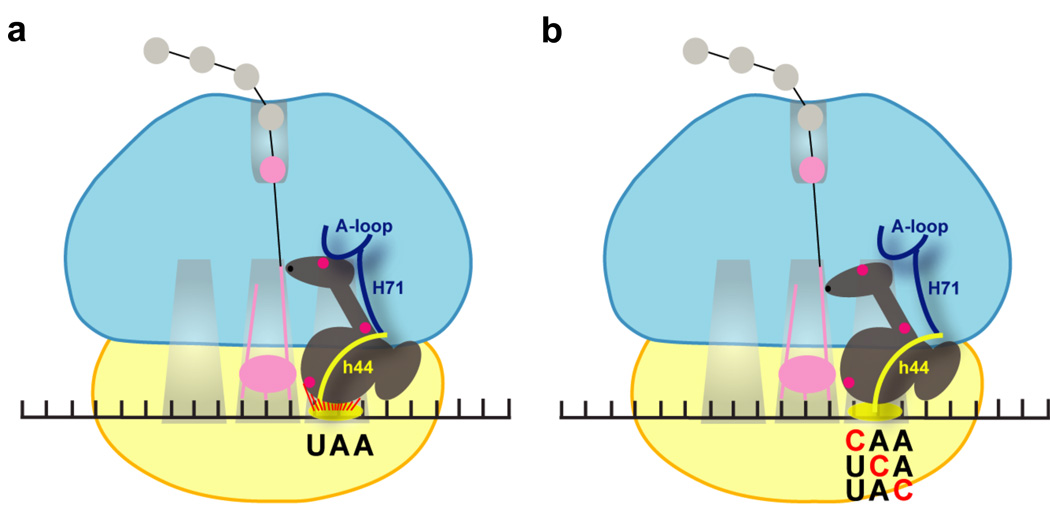
While recent atomic resolution structures have provided much insight into the conformation of class 1 RFs bound to the ribosome after the recognition of authentic stop codons, what has been difficult to understand is the extent to which the precise mode of binding depends on key recognition events in the decoding center. The data we present here provide compelling evidence for stop codon recognition being essential in determining the ultimate conformation of the recognition complex -- in the final bound state, the ribosome appears to collapse around RF1 in a fashion that strictly depends on signals originating in the decoding center (Fig. 4). The molecular features important for these rearrangements may be related to those essential for tRNA selection on cognate codons.
Figure 4.
Cartoon representation of structural rearrangements in ribosome complexes following RF1 recognition of stop codons (a) or near-stop codons (b). 30S subunit and 50S subunits are shown in yellow and cyan, respectively. RF1 is shown in the A site with Fe(II) tethering positions (156, 226 and 292) indicated in pink. Specific functionally important rRNA helices and loops that exhibited differential cleavage patterns in the cognate and near-cognate complexes are highlighted.
Methods
Reagents and RF1 modification
Tight-couple 70S ribosomes were purified from exponentially grown MRE600 cells as previously described26 and stored in HiFi buffer (50 mM Tris-HCl pH 7.5, 70 mM NH4Cl, 30 mM KCl, 3.5 mM MgCl2, 0.5 mM spermidine, 8 mM putrescine, and 2 mM DTT). tRNAs were purchased from Sigma-Aldrich. mRNAs were generated by PCR and T7 transcription to produce the sequence 5′-GGGUUAACUUUAGAAGGAGGUAAAAAAA AUG NNN UUU UUC UUU-3′ with the indicated codon (NNN) as specified. To construct single cysteine RF1 variants, a cysteine-less version of RF115 was prepared and single cysteine residue was introduced by site-directed mutagenesis using the Quickchange Kit (Stratagene). All the variants were purified as previously reported8 and stored in RF1 storage buffer (30 mM HEPES-KOH pH7.6, 70 mM NH4Cl, 30 mM KCl, 10 mM MgCl2, 3.5 mM DTT and 50% (v/v) glycerol) at −20° C. RF1 variants were derivatized with FeBABE according to previously published procedures16. Typically, RF1 variants at 30 µM were incubated with 1 mM FeBABE in modification buffer (80 mM HEPES-KOH pH 7.6, 1 M KCl and 0.01% (v/v) Triton) at 37°C for 1 hr. Excess FeBABE was removed by dialysis overnight into storage buffer (80 mM HEPES-KOH, pH 7.6, 70 mM NH4Cl, 30 mM KCl, and 10 mM MgCl2). Modified protein was divided into aliquots and stored at −80°C.
Termination complex formation
Ribosome complexes were formed in buffer A (50 mM Tris-HCl pH 7.5, 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2, and 1 mM DTT) by incubating 1 µM 70S ribosomes, 1.5 µM tRNAfMet, and 3 µM mRNA with various codons in the A site for 30 min at 37°C. Mock-derivatized RF1 (Cys-) or FeBABE-derivatized single cysteine RF1 variants (75 µM) were bound to ribosome complexes (37°C for 10 min, 0°C for 10 min). To identify hydroxyl radical cleavages on the P-site tRNA for Fe(II)-H156C-RF1 and Fe(II)-T226C-RF1 variants, another batch of complexes was formed using tRNAfMet labeled with [32P] at the 3′ end18.
Directed hydroxyl radical probing and cleavage analysis
Localized hydroxyl radicals were generated from Fe(II) tethered to RF1 by the addition of H202 (0.05% (v/v)) and ascorbic acid (5 mM) to RF1-bound ribosome complexes (0°C, 10 min). To identify hydroxyl radical cleavages on the rRNA, RNAqueous lysis buffer was added to the reaction and RNA was extracted as per instructions (Ambion). rRNA was then scanned for cleavage by primer extension using reverse transcriptase as previously described17. To examine the sites of hydroxyl radical cleavage on P-site tRNA, an equal volume of loading buffer (85% (v/v) formamide, 10 mM EDTA, 0.1% (w/v) bromophenol blue and 0.1% (w/v) xylene cyanol) was added to the reactions and the sample was loaded on a 10% (w/v) denaturing PAGE sequencing gel. Cleavage intensities were visually assigned as weak, medium or strong according to the intensity of each band relative to the intensity of control sequencing lanes.
Toe-printing assay
Toe-printing analysis was carried out essentially as described27. Ribosome complexes formation was carried out as described above except that mRNAs used had extra sequences at the 3′-end to allow for oligonucleotide priming, and were provided in an equimolar ratio to the ribosomes. Ribosome complexes were first annealed to a trace amount of cDNA primer labeled with [32P] at its 5′ end. Primer extension reactions were then carried out in primer extension mix (50 mM Tris-HCl pH 7.5, 70 mM NH4Cl, 30 mM KCl2, 7 mM MgCl2, and 1 mM DTT, 600 µM dNTPs and 1 unit µ−1 of AMV-RT) at 37°C for 15 mins, followed by the addition of 100 mM NaOH and incubation at 90 °C for 10 min to digest the RNA. The reaction was ethanol precipitated before analysis on long format 6% (w/v) denaturing PAGE.
NTCB cleavage assay
The cleavage assay was loosely based on a previous study with some modifications19. Either derivatized or underivatized single cysteine RF1 variants (15 µg) were reacted with 50 mM NTCB (2-nitro-5-thiocyanobenzoic acid, Sigma-Aldrich) in reaction buffer (6 M Guanidine-HCl and 300 mM Bicine-NaOH, pH 8.5) at 45°C for 10 min. The cyanylated proteins were trichloroacetic acid-precipitated and resuspended in 5 µl of cleavage buffer (800 mM NH4OH and 8 M Urea). After 1 hr incubation at 45°C, 15 µl of loading buffer (300 mM Tris pH 6.8, 2% (w/v) SDS, 8% (v/v) Glycerol and Brilliant blue G250) was added to quench the cleavage, and the samples were examined by SDS-PAGE.
Peptide release assay
RF1 catalyzed fMet-tRNAfMet hydrolysis was perfomed as previously described8. Native IFs 1 and 3 and His-tagged IF2 were prepared as described28. Ribosome complexes were formed by incubating 2 µM 70S ribosomes, 2.6 µM f-[35S]-Met-tRNAfMet, 6 µM mRNA, 3 µM each IFs 1, 2, and 3, and 1 mM GTP in buffer A at 37°C for 30 min. Complexes were pelleted through a 1 ml sucrose cushion (1.1 M sucrose in buffer A) in a Beckman TLA 100.3 rotor at 200,000 g for 2 hr at 4°C. Pelleted complexes were resuspended in HiFi buffer and stored in aliquots at −80°C. Concentration was determined by scintillation counting of [35S]-Met. Release assays were performed in HiFi buffer at 37°C by mixing equal volumes of ribosome complexes and WT-RF1 or FeBABE modified single cysteine RF1 variants. At appropriate time points, reactions were quenched with 25% (v/v) formic acid and released f-[35S]-Met was resolved from unreacted f-[35S]-Met-tRNAfMet by electrophoretic TLC. Saturating concentrations (75 µM and 200 µM) of RF1 was used to measure the kcat (maximal kobs) of peptide release under all conditions.
Supplementary Material
Acknowledgements
We thank E. Youngman, H. Zaher, S. Djuranovic, and N. Guydosh for comments on the manuscript; NIH for funding of the project; and HHMI for salary support to R.G.
Footnotes
Author contributions
S.L.H. and R.G. designed the experiments; S.L.H. performed the experiments. Both authors discussed results and contributed to the manuscript.
References
- 1.Youngman EM, McDonald ME, Green R. Peptide release on the ribosome: mechanism and implications for translational control. Annu Rev Microbiol. 2008;62:353–373. doi: 10.1146/annurev.micro.61.080706.093323. [DOI] [PubMed] [Google Scholar]
- 2.Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 4.Ogle JM, et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 5.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 6.Jorgensen F, Adamski FM, Tate WP, Kurland CG. Release factor-dependent false stops are infrequent in Escherichia coli. J Mol Biol. 1993;230:41–50. doi: 10.1006/jmbi.1993.1124. [DOI] [PubMed] [Google Scholar]
- 7.Freistroffer DV, Kwiatkowski M, Buckingham RH, Ehrenberg M. The accuracy of codon recognition by polypeptide release factors. Proc Natl Acad Sci U S A. 2000;97:2046–2051. doi: 10.1073/pnas.030541097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Youngman EM, He SL, Nikstad LJ, Green R. Stop codon recognition by release factors induces structural rearrangement of the ribosomal decoding center that is productive for peptide release. Mol Cell. 2007;28:533–543. doi: 10.1016/j.molcel.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Weixlbaumer A, et al. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laurberg M, et al. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 11.Korostelev A, et al. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci U S A. 2008;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin DH, et al. Structural analyses of peptide release factor 1 from Thermotoga maritima reveal domain flexibility required for its interaction with the ribosome. J Mol Biol. 2004;341:227–239. doi: 10.1016/j.jmb.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 13.Fraser CS, Hershey JW, Doudna JA. The pathway of hepatitis C virus mRNA recruitment to the human ribosome. Nat Struct Mol Biol. 2009;16:397–404. doi: 10.1038/nsmb.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spanggord RJ, Siu F, Ke A, Doudna JA. RNA-mediated interaction between the peptide-binding and GTPase domains of the signal recognition particle. Nat Struct Mol Biol. 2005;12:1116–1122. doi: 10.1038/nsmb1025. [DOI] [PubMed] [Google Scholar]
- 15.Wilson KS, Ito K, Noller HF, Nakamura Y. Functional sites of interaction between release factor RF1 and the ribosome. Nat Struct Biol. 2000;7:866–870. doi: 10.1038/82818. [DOI] [PubMed] [Google Scholar]
- 16.Culver GM, Noller HF. Directed hydroxyl radical probing of RNA from iron(II) tethered to proteins in ribonucleoprotein complexes. Methods Enzymol. 2000;318:461–475. doi: 10.1016/s0076-6879(00)18070-x. [DOI] [PubMed] [Google Scholar]
- 17.Moazed D, Stern S, Noller HF. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- 18.Ledoux S, Uhlenbeck OC. [3'-32P]-labeling tRNA with nucleotidyltransferase for assaying aminoacylation and peptide bond formation. Methods. 2008;44:74–80. doi: 10.1016/j.ymeth.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverman JA, Harbury PB. Rapid mapping of protein structure, interactions, and ligand binding by misincorporation proton-alkyl exchange. J Biol Chem. 2002;277:30968–30975. doi: 10.1074/jbc.M203172200. [DOI] [PubMed] [Google Scholar]
- 20.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 21.Ito K, Uno M, Nakamura Y. A tripeptide 'anticodon' deciphers stop codons in messenger RNA. Nature. 2000;403:680–684. doi: 10.1038/35001115. [DOI] [PubMed] [Google Scholar]
- 22.Sternberg SH, Fei J, Prywes N, McGrath KA, Gonzalez RL., Jr. Translation factors direct intrinsic ribosome dynamics during translation termination and ribosome recycling. Nat Struct Mol Biol. 2009;16:861–868. doi: 10.1038/nsmb.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanbonmatsu KY, Joseph S, Tung CS. Simulating movement of tRNA into the ribosome during decoding. Proc Natl Acad Sci U S A. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freistroffer DV, Pavlov MY, MacDougall J, Buckingham RH, Ehrenberg M. Release factor RF3 in E.coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. Embo J. 1997;16:4126–4133. doi: 10.1093/emboj/16.13.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cannone JJ, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moazed D, Noller HF. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986;47:985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- 27.Dorner S, Brunelle JL, Sharma D, Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunelle JL, Youngman EM, Sharma D, Green R. The interaction between C75 of tRNA and the A loop of the ribosome stimulates peptidyl transferase activity. Rna. 2006;12:33–39. doi: 10.1261/rna.2256706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.