Abstract
During the past decades there has been minimal improvement in prevention and treatment of hypertrophic scarring. Reasons include the lack of a validated animal model, imprecise techniques to dissect scar into the histologic components, and limited methodology for measurement of gene expression. These problems have been addressed with the Duroc/Yorkshire model of healing, laser capture microdissection, and the Affymetrix Porcine GeneChip®. Here we compared collagen gene expression in fibroproliferative healing in the Duroc breed to nonfibroproliferative healing in the Yorkshires. We made shallow and deep dorsal wounds, biopsied at 1, 2, 3, 12, and 20 weeks. We sampled the dermal cones by laser capture microdissection, extracted and amplified the RNA, and hybridized Affymetrix Porcine GeneChips®. We also obtained samples of human hypertrophic scar approximately 20 weeks postinjury. Data were normalized and statistical analysis performed with mixed linear regression using the Bioconductor R/maanova package. Genes for further analysis were also restricted with four biologic criteria, including that the 20-week deep Duroc expression match the human samples. Eleven ollagen genes and seven collagen types were differentially over expressed in deep Duroc wounds including 1a1, 1a2, 3a1, 4a1, 4a2, 5a1, 5a2, 5a3, 6a3 (transcript variant 5), 14a1 and 15a1. COL7a1 gene was differentially under expressed in deep Duroc wounds. The results suggest that collagens I, III, IV, V, VI, VII, XIV, and XV1 are involved in the process of fibroproliferative scarring. With these clues, we will attempt to construct the regulatory pathway(s) of fibroproliferative healing.
We have little understanding of the pathophysiology of hypertrophic scarring that follows deep partial thickness injury; clinical prevention and treatment are marginal at best.1-9 There are at least six reasons for this lack of understanding. The major reason is that 1) in the past, there has been no accepted and validated animal model of hypertrophic scarring.9-28 Given the complexities and uncertainties of burn injuries, human tissue research, and hypertrophic scarring, it is 2) virtually impossible to obtain samples of burn wounds that will become hypertrophic from a given patient early on and serially after an injury. Therefore, studies of scar formation have usually 3) analyzed human tissues obtained months or years after injury, long after the initial causes of the process could be examined. Furthermore, 4) studies of scar have typically considered skin to be one homogenous unit whereas it has, in fact, several distinct components. Blending a multitissue layered biopsy into one uniform sample may have hidden the important changes. Related to this, 5) there has been no technology to dissect skin into its various components. And finally; 6) the methods used in the past to evaluate gene expression (Northern blot analysis and in situ hybridization) are time consuming and detect only a few genes per assay. Further, the result is likely an incomplete assessment of the process of scarring. Fortunately, these problems are to some degree resolved as we have expertise with:
The Duroc (fibroproliferative)/Yorkshire (non-fibroproliferative) porcine model of scarring which has been validated and permits us to study the process early and serially with an appropriate control.29-40 These studies have confirmed that healing in the Duroc breed proceeds with more fibrosis, pigmentation, and contraction than in the Yorkshire breed. However, they have also confirmed that the fibroproliferative healing in the Duroc is not identical to human hypertrophic scar. Therefore, we refer to “fibroproliferative scarring” in the porcine model and “hypertrophic scarring” in humans.
The anatomy of the cones of skin has been established. It is also clear that the cones of skin are physiologically active as in burns wounds; they are grossly inflamed compared with the surrounding collagen matrix. We have also established that the dermal cones are located on the same body parts, where hypertrophic scar occurs. It is then reasonable to hypothesize that they are somehow involved in hypertrophic scarring.41-43
Laser Capture Microdissection (LCM) (Molecular Devices, Sunnyvale, CA) permits dissection of skin histoanatomy down to the level of a single cell.
Affymetrix (Santa Clara, CA) has released the Porcine GeneChip® with 23,937 probes that interrogate 20,201 genes.
Our understanding of the porcine transcriptome has expanded significantly during the past few years, as a large amount of expressed sequence data has entered the public databases. These sequence data have also permitted the development of several different transcript-profiling platforms. Using these tools, the pig transcriptome has been analyzed in a multitude of tissues including liver, brain, muscle, lymph node, adipose, and reproductive tissues.44 A new Affymetrix Porcine GeneChip® that can interrogate over 20,000 different transcripts has been recently developed, and this microarray is the most comprehensive platform with published data.45 Couture O and Tuggle CK (Personal Communication, 2007) annotated the Affymetrix assay elements so that a direct comparison to human biology could be made. Confirmatory analysis using Quantitative-Polymerase chain reaction (Q-PCR) showed that the Affymetrix platform was accurate, and with the comprehensive annotation of the Genechip®, this platform should be highly useful for groups studying a variety of tissues and biological questions.
We have evaluated differential gene expression in the dermal cones in shallow and deep wounds in Duroc and Yorkshire pigs at 1, 2, 3, 12, and 20 weeks postinjury. Because hypertrophic scar is fundamentally excessive collagen matrix, it seems logical that the collagen genes may be termed the “signature genes” and their expression is either directly or indirectly involved in the process. We hypothesize that 1) the collagen genes involved in fibroproliferative scarring are expressed in the dermal cones, 2) the expression is different between deep partial thickness and shallow partial thickness wounds, and 3) the expression is also different between breeds. This may be expressed as ΔbreedΔdepth ≠ 0, or ΔΔ ≠ 0.
METHODS
Wound and Breed Model
The experimental wounds are deep partial thickness, leaving the deep portion of the dermal matrix and the deep aspect of the cones. Shallow partial thickness wounds are the control wounds (Figure 1).
Figure 1.
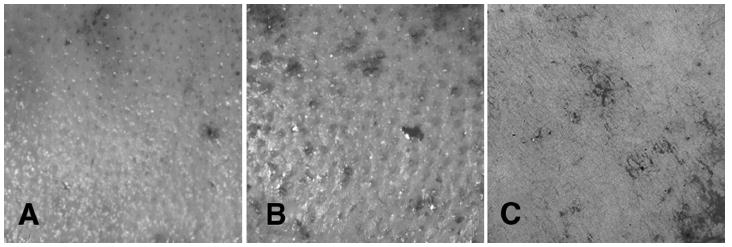
The shallow wound (A) demonstrates the fine punctate, pin cushion appearance of the cones in the upper dermis. The deep dermal wound (B) shows the cobblestone appearance of the large diameter cones in the deep dermis. The full thickness wound (C) demonstrates that the dermal matrix and cone pattern is absent.
The Duroc breed forms thick, fibroproliferative scar and is the experimental breed. The Yorkshire breed forms nonproliferative scar and is the control breed.
Animal Care and Wounding
All animal studies were performed as previously described.31-33,35 In accord with the Animal Care Committee, three female Duroc and three female Yorkshire pigs, 6 weeks old, approximately 16 to 18 kg, were purchased (Q-Bar Farm, http://www.viclink.com/~qbarfarm) and housed in the Harborview Medical Center Research and Training Vivarium with 12-hour light/dark cycles. The animals were observed for 1 week and fed lab porcine grower diet and water ad lib. At 7 weeks, anesthesia was established with oxymorphone (0.1 mg/kg 30 in) I.M. before the procedure, Telazol® reconstituted with 5 ml xylazine (100 mg/ml) at a dosage of 1 ml/18 kg body weight I.M. (Phoenix Pharmaceuticals, Inc., St. Joseph, MO), and Isoflurane. The entire anesthetic procedure was performed by the veterinarian staff of the University of Washington. The hair on the back was clipped and skin cleansed with Betadine® solution and rinsed with 70% alcohol.
Ten 7 ×7 cm tangential wounds were created on the back of each pig with a Padgett® dermatome (Integra LifeSciences Corporation, Plainsboro, NJ) with the dermatome set to 0.020 in; five were 0.020 in and five were 0.060 in. One pass of the dermatome is sufficient for the shallow wounds but two or three passes are necessary to create the deep wounds. It is known that the actual wound depth obtained with a dermatome is variable.46 Therefore we refer to total dermatome setting rather than wound depth. The deep and shallow wounds were alternated on the animals to avoid repeatedly placing one wound depth in the same anatomic location. The wounds were allowed to heal without application of topical agents or dressings. Postoperatively, a fentanyl transdermal patch (100 μg) was applied for analgesia. Buprenorphine (0.005–0.002 mg/kg) body weight (BW), was administered i.m. or i.v., SID or BID as needed for distress until the patch had time to take effect within 24 hours.
Under general anesthesia as described above, 8 to 10 mm surgical biopsies were collected at 1, 2, 3, 12, and 20 weeks postwounding from one shallow and one deep wound. The biopsies were obtained near the center of the wounds and each wound was biopsied only once. At 22 weeks, the pigs were returned to the breeder. Uninjured skin was also biopsied at each time point.
Human Tissues
Since 1994, with University of Washington Human Subjects Committee approval, we have collected human hypertrophic scar tissue in the operating room that would otherwise have been discarded. Three of these hypertrophic scar samples were obtained 6 to 10 months after injury and, therefore, are analogous to the deep Duroc samples obtained at 5 months postinjury since both involve fibroproliferative healing.
Laser Capture Microdissection
The full-thickness human and porcine wound samples were snap-frozen in chilled Tissue-Tek® Optimal Cutting Temperature embedding medium (Sakura Finetechnical U.S.A., Torrance, CA) (Figure 2). The biopsies were sectioned at 7 μm, fixed in 75% ethanol, dehydrated, and air dried. The slides were loaded into the Arcturus® AutoPix LCM system (Molecular Devices Corporation, Sunnyvale, CA). A laser spot size of 25 μm diameter was used with 85 to 95mW power and 8,500 to 10,000 ms duration. The offset and overlap was 25%. The efficiency of the microdissection was evaluated by examining the cap after capture and by examining the tissue before and after lift off from the cap. 10 to 20 cones were captured per sample or 500 to 1000 cells (Figure 2).
Figure 2.
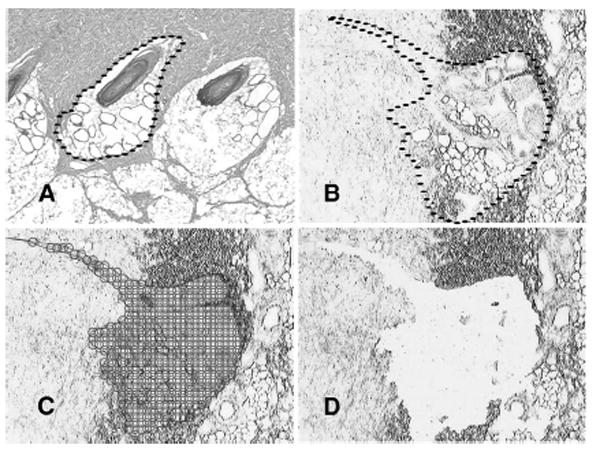
Laser capture microdissection. (A) The outline of a deep cone in uninjured skin, Hematoxylin and eosin, ×4. (B) The outline of deep cone under the laser microscope, × 4. (C) The many spots to be laser capture microdissected, ×4. (D) The residual defect, ×4.
RNA Isolation and Quality Assessment
Following tissue collection, the cap was incubated in 50 μl PicoPure™ RNA extraction buffer (Molecular Devices Corporation, Sunnyvale, CA) at 42°C for 30 minutes. After DNase (Qiagen, Valencia, CA) treatment, total RNA was eluted in 12 μl of Elution Buffer. RNA quality was monitored with Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA), the Agilent RNA 6000 Pico chip, and Eukaryote Total RNA Pico by evaluating the features of the electropherogram and the RNA integrity numbers. 28S and 18S rRNA bands were expected and the intensity of the 28S bands was expected to be higher than the intensity of the 18S. The RNA integrity numbers were expected to be greater than 6.0. Samples that failed these criteria were repeated.
Linear Amplification of RNA
The LCM RNA and 500 pg control RNA were amplified using the RiboAmp™ HS kit (Molecular Devices Corporation, Sunnyvale, CA). RNA was reverse transcribed and double-stranded cDNAs synthesized and purified. The first-round cDNA was transcribed using T7 RNA polymerase. The resulting RNA was purified and used in the second round of amplification. Transcription and labeling of complementary RNA was performed with biotinylated UTP and CTP (Affymetrix IVT labeling Kit). The target complementary RNA was fragmented, washed, and stained according to the manufacturer instructions. The quality and quantity of labeled complementary RNA was evaluated spectrophotometrically and with the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA). The required A260/280 ratio was in the range 1.8 to 2.1. Samples that failed these criteria were repeated.
Hybridization to the Affymetrix GeneChips®
The porcine tissue RNA was analyzed using the Porcine GeneChip® and the human tissue RNA using the Human GeneChip® Human Genome U133 plus 2.0. Fragmentation of cRNA and preparation of the hybridization cocktail containing fragmented, biotinylated cRNA (60 ng/μl) were performed according to the GeneChip® Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA). Spike controls were added to complementary RNA before hybridization. Labeled targets were hybridized for 16 to 18 hours at 45°C and 60 rounds per minute (Hybridization Oven 640, Affymetrix, Santa Clara, CA). Washing and staining with streptavidine-phycoerythrine was performed in an automatic fluidics station (GeneChip® Fluidics Station 400, Affymetrix, Santa Clara, CA). Scanning was performed with GeneChip® Scanner 3000 (Affymetrix, Santa Clara, CA). The quality of the hybridization and overall chip performance was evaluated by visual inspection of the raw scanned data and the quality control measures in the Affymetrix RPT report file; except that we did not monitor the Affymetrix 3′5′ ratios since there are no recommended values for amplified porcine tissues.
The human tissue was processed in a similar way using the Affymetrix Human GeneChip® Human Genome U133 plus 2.0.
Quantitative Real Time RT-Polymerase Chain Reaction Verification of Array Data
We performed Quantitative Real Time-Polymerase Chain Reaction (qRT-PCR) on Duroc data with three probe sets and Yorkshire data with one probe set to confirm the array data. Total RNA from the laser capture microdissected cells was isolated as described above. The first strand cDNA synthesis was generated using SuperScript™ III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen, Carlsbad, CA). Purified total RNA was added to RT Enzyme mix and 2 × RT Reaction Mix according to manufacturer instructions, followed by incubation at 25°C for 10 minutes, at 50°C for 30 minutes and terminated at 85°C for 5 minutes. After treatment with E. coli RNase H, cDNA was either stored at −20°C or used immediately for PCR.
Polymerase Chain Reaction Primer Design
Affymetrix has identified Probe Selection Regions in the 3′ ends of transcripts, which are frequently contained within the last exon of genes. To choose the primers from the exon–exon junctions, the regions of mRNA that include nucleotide sequences from the 5′ border of one exon and 3′ border of the neighboring exon, we blast-searched the specific areas of the gene sequence on NCBI Blast. The primers were designed by the Primer3 website (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and synthesized by Operon Biotechnologies (Huntsville, AL) (Table 1). None of the GAPDH probe sets were differentially expressed and, therefore, GAPDH was used as the internal calibrator for equal RNA loading and to normalize relative expression data.
Table 1.
Primer sequences for qRT-PCR
| Gene and Probe Set | Sequences | Base Pairs | GenBank Accession Number |
|---|---|---|---|
| COL1a1 | (F) acctcaagatgtgccactcc | 106 | AF201723 |
| Ssc.1091.1.s1_at | (R) cctgtctccatgttgcagaa | ||
| COL7a1 | (F) gtgacttgttccgtgggtct | 119 | CO945178 |
| Ssc.29731.1.A1_at | (R) acgcagtcacatttgctcac | ||
| COL14a1 | (F) ggtctggcataagaccttgg | 78 | BE693212 |
| Ssc.6778.1.s1_at | (R) ggtctggcataagaccttgg | ||
| GAPDH | (F) ctcaacgaccacttcgtcaa | 113 | AF017079 |
| (R) tccaggggctcttactcctt |
qRT-PCR, quantitative real-time polymerase chain reaction.
Quantitative Real Time RT-Polymerase Chain Reaction
Q-PCR was performed using SYBR®Greener™ qRT-PCR SuperMix (Invitrogen, Carlsbad, CA) according to manufacturer instructions in an ABI Prism 7900H sequence detection system (Applied Biosystems, Scottsdale, AZ), using 384-well microtiter plates. All samples were run in triplicate. The amplification conditions were (stage 1) 95°C for 10 minutes and (stage 2) 50 cycles at 95°C for 15 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 30 seconds. The final cycle (stage 3), a melting-curve analysis verified the specificity of the PCR reaction. The copy ratio of each analyzed cDNA was determined as the mean of three experiments. The qRT-PCR data were quantified using relative quantification (2-ΔCT) method as described.47 Negative controls comprised of primers without sample did not produce amplicons.
Annotation of the Porcine GeneChip®
Affymetrix annotated the pig sequence data used to create the probes included in their porcine Gene-Chip® and updates the annotation quarterly. This annotation is available at the Affymetrix NetAffx website (http://www.affymetrix.com/analysis/index.affx). However, this annotation is rather incomplete. Global and current annotations of porcine gene sequences are available at three main public websites including: 1) National Center for Biotechnology Unigene (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene), 2) Dana-Farber Porcine Gene Index (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=pig), and 3) the new Sino-Danish Pig Genome Sequencing Project (http://pigest.ku.dk). The first two sites have analyzed data for about 600,000 pig sequences, whereas the SinoDanish site summarizes data on one million sequences. We have used the available Affymetrix consensus sequences for each GeneChip® probe in sequence matching using BLAST against the updated Genbank Reference Sequence database and increased the annotation to 17,598 probes (unpublished data) (Couture O, Tuggle CK. Personal Communication. 2007) using a conservative BLAST score cut-off of e−10.
Statistical Analysis and Biologic Requirements
Management of the raw data was accomplished with Filemaker Pro 8.5 (http://www.filemaker.com), ChartMaker Pro 7v3 (http://www.briandunning.com/chartmaker), and Microsoft Excel (http://www.microsoft.com).
We normalized the 60 chips with the Bioconductor “affy” package48 and the gcrma algorithm. This data set includes paired measures (shallow and deep wounds on each pig) and repeated measures (using the same pigs at each time point). To deal with these experimental conditions, we calculated the log ratio of the deep wound signal over the shallow for each probe and each sample. We performed mixed linear regression on the log ratios using the Bioconductor R/maanova package,48 the F-test, and 1000 permutations. We used P < .1 to cast a very wide net in this early screening process.49 We did not include a False Discovery Rate correction accepting more false positives in favor of minimizing false negatives as did Johnston.50 Statistical analysis of the PCR data was accomplished with the Student’s t-test comparing deep to shallow wounds, using P < .05.
Statistical analysis is necessary but not sufficient to deal with all of the biologic confounding variables; we added four additional biologic requirements for probe selection for further study: 1) The Affymetrix software reports each probe as Present (P) or Absent (A). We concatenated these strings into Pig1ShallowDeep_Pig2ShallowDeep_Pig3ShallowDeep for each breed and each time point, which results in strings such as “PP_PP_PP” and “AA_AA_AA.” A “logical” string wound be, for example, PA_PA_PA meaning present in all three shallow wounds and absent in all three deep wounds for the breed and the time point. An illogical string might be AP_PA_AA wherein no pigs matched. We require these strings to be “logical.” 2) For each breed at each time point we obtained three log ratios of deep or shallow. We also required that the numerical direction of the three log ratios be the same. For example, a set of three log ratios −2, −1.5, and −3 would be accepted whereas three log ratios −2, +1, and −1.5 would not. 3) In addition, we required that the absolute value of the mean of the three log ratios be greater than 0.5 indicating a fold change of at least 1.4. 4) Finally, we required that the 5-month deep Duroc data match the early human hypertrophic scar data as described below.
Verification in Human Hypertrophic Scar
To completely verify the porcine findings with human hypertrophic scar, we would need human biopsies at 1, 2, 3, 12, and 20 weeks. This is impossible to accomplish. However, as explained above, we do have samples of early human hypertrophic scar and compared these with the deep Duroc tissues at 20 weeks postinjury since both involve fibroproliferative healing. The choice of the 20-week time point for comparison is discussed further in Discussion. The comparison cannot be statistical, given that the two sets of data are obtained from different GeneChips®. Instead, as directed by Affymetrix tech support, we required the Present or Absent calls to match. We, therefore, included probe sets for which at least two of the three deep Duroc, 5 month samples matched the three human samples. This is explained further in Discussion.
RESULTS
The analysis with mixed linear regression returned 2043 probe sets. The application of the four biologic criteria reduced the number to 1593; 1019 of these have been annotated. Nine hundred fifty-three remain after removal of duplicates. For our initial analysis we have focused on the signature genes of hypertrophic scarring, the collagen genes.
Differential Expression of Collagen Genes
The Affymetrix Porcine GeneChip® includes 64 probe sets representing 28 collagen genes and 19 collagen types with the annotation and criteria described above. Sixteen collagen probe sets passed the statistical and biologic selection criteria representing 12 collagen genes and 8 collagen types (Table 2). The 12 collagen genes differentially expressed include 1a1, 1a2, 3a1, 4a1, 4a2, 5a1, 5a2, 5a3, 6a3 transcript variant 5, 7a1, 14a1, and 15a1. All differentially expressed probe sets demonstrated over expression in deep Duroc wounds except COL7a1, which was under expressed in deep Duroc wounds. Of equal interest, we did not find differential expression of the probe sets for collagen genes 2a1, 8a1, 9a1, 9a2, 9a3, 10a1, 11a1, 12a1, 16a1, 17a1, 18a1, 21a1, and 24a1.
Table 2.
Expression of the collagen gene probe sets
| Probe ID | Gene Symbol | P (—Means Not Expressed) | Passed Biologic Selection Criteria | qRT-PCR Confirmed |
|---|---|---|---|---|
| Ssc.1091.1.s1_at | COL1a1 | .01 | Yes | Duroc and Yorkshire data at 3 weeks |
| Ssc.1091.2.a1_at | COL1a1 | — | ||
| Ssc.1091.2.s1_at | COL1a1 | — | ||
| Ssc.1091.3.a1_at | COL1a1 | .01 | Yes | |
| Ssc.21011.1.s1_at | COL1a2 | .04 | Yes | |
| Ssc.24975.1.s1_at | COL1a2 | .27 | No | |
| Ssc.9362.1.a1_at | COL1a2 | — | ||
| Ssc.16016.1.a1_at | COL2a1 | — | ||
| Ssc.16016.1.s1_at | COL2a1 | — | ||
| Ssc.25009.1.a1_at | COL2a1 | — | ||
| Ssc.11302.1.s1_at | COL3a1 | .33 | No | |
| Ssc.11302.1.s2_at | COL3a1 | .03 | Yes | |
| Ssc.4345.1.s1_at | COL4a1 | .00 | No | |
| Ssc.4345.1.s2_at | COL4a1 | .00 | Yes | |
| Ssc.6435.1.s1_at | COL4a1 | .14 | No | |
| Ssc.3467.1.s1_at | COL4a2 | .05 | Yes | |
| Ssc.9747.1.a1_at | COL4a2 | — | ||
| Ssc.16254.1.a1_at | COL4a3 | — | ||
| Ssc.16254.1.s1_at | COL4a3 | — | ||
| Ssc.26705.1.a1_at | COL4a3 | — | ||
| Ssc.19643.1.a1_at | COL4a4 | — | ||
| Ssc.16329.1.a1_at | COL5a1 | — | ||
| Ssc.16329.1.s1_at | COL5a1 | — | ||
| Ssc.18545.1.a1_at | COL5a1 | — | ||
| Ssc.31199.1.s1_at | COL5a1 | — | ||
| Ssc.4993.1.a1_at | COL5a1 | .03 | Yes | |
| Ssc.9002.1.a1_at | COL5a1 | .02 | Yes | |
| Ssc.16328.1.a1_at | COL5a2 | — | ||
| Ssc.16328.1.s1_at | COL5a2 | — | ||
| Ssc.17300.1.s1_at | COL5a2 | .01 | Yes | |
| Ssc.12617.1.s1_at | COL5a3 | .01 | Yes | |
| Ssc.16327.1.a1_at | COL5a3 | — | ||
| Ssc.16327.1.s1_at | COL5a3 | — | ||
| Ssc.21754.1.a1_at | COL6a1 | — | ||
| Ssc.21754.2.s1_at | COL6a1 | — | ||
| Ssc.5895.1.a1_at | COL6a1 | — | ||
| Ssc.5895.1.a2_at | COL6a1 | .38 | No | |
| Ssc.5895.2.a1_at | COL6a1 | — | ||
| Ssc.12068.1.a1_at | COL6a3 transcript variant 5 | .09 | Yes | |
| Ssc.16589.1.s1_at | COL6a3 transcript variant 5 | .04 | Yes | |
| Ssc.29731.1.a1_at | COL7a1 | .06 | Yes | Duroc data at 12 wks |
| Ssc.16132.1.a1_at | COL8a1 | — | ||
| Ssc.16132.1.s1_at | COL8a1 | — | ||
| Ssc.16132.2.a1_at | COL8a1 | — | ||
| Ssc.16132.2.s1_at | COL8a1 | — | ||
| Ssc.24708.1.s1_at | COL9a1 | — | ||
| Ssc.24708.2.s1_at | COL9a1 | — | ||
| Ssc.19233.1.s1_at | COL9a2 | — | ||
| Ssc.22252.2.s1_at | COL9a3 | — | ||
| SscAffx.19.1.s1_at | COL10a1 | — | ||
| Ssc.15786.1.a1_at | COL11a1 | — | ||
| Ssc.15786.1.s1_at | COL11a1 | — | ||
| Ssc.26569.1.s1_at | COL11a1 | — | ||
| Ssc.1049.1.s1_at | COL12a1 | .57 | No | |
| Ssc.15374.1.s1_at | COL14a1 | — | ||
| Ssc.31124.1.s1_at | COL14a1 | .01 | Yes | |
| Ssc.6778.1.s1_at | COL14a1 | .00 | Yes | Duroc data at 3 wks |
| Ssc.19883.1.s1_at | COL15a1 | .00 | Yes | |
| Ssc.25168.1.s1_a_at | COL16a1 | .37 | No | |
| Ssc.27604.1.a1_at | COL17a1 | — | ||
| Ssc.27604.1.s1_at | COL17a1 | — | ||
| Ssc.4892.1.s1_at | COL18a1 | .75 | No | |
| Ssc.24270.1.s1_at | COL21a1 | .88 | No | |
| Ssc.29300.1.a1_at | COL24a1 | — |
qRT-PCR, quantitative real-time polymerase chain reaction.
Quantitative Real Time RT-Polymerase Chain Reaction Verification of Three Probe Sets
To verify the general validity of the array data, we performed quantitative real time RT-PCR on three probe sets.
Ssc.1091.1.s1_at (COL1a1)
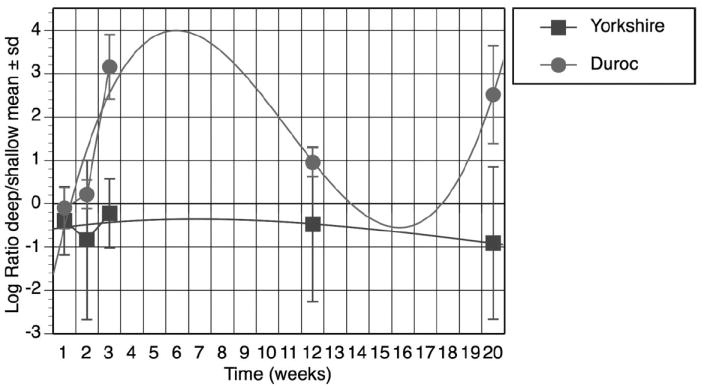
Ssc.1091.1.s1_at (COL1a1) was chosen for verification as collagen type I is fundamental to skin structure and physiology. With mixed linear regression, there was differential expression of Ssc.1091.1.s1_at (COL1a1) over time (P = .01) (Figure 3). The biphasic response was previously described by Gallant.34,38
Figure 3.
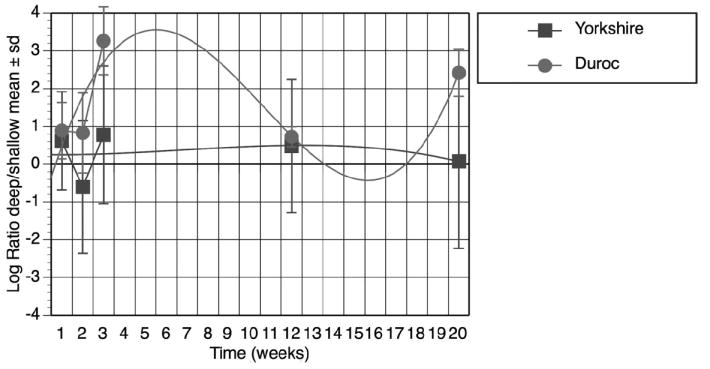
Expression of COL1a1 (Affymetrix Probe Set Ssc.1091.1.s1_at) over time. COL1a1 gene expression was calculated as the log ratio of deep or shallow signals. Data from Durocs and Yorkshires were plotted over the time course of 20 weeks. Each data point represents mean and standard deviation from three biologic replicates. COL1a1 was differentially over expressed in deep duroc wounds (P = .01). The curves are 3rd degree exponential.
To confirm the 3-week array data for Ssc.1091.1.s1_ at, we performed quantitative real time RT-PCR (Figure 4). The probe was significantly more expressed in Duroc deep wounds than in Duroc shallow wounds (P = .026). There was no difference between Yorkshire deep and shallow wounds.
Figure 4.
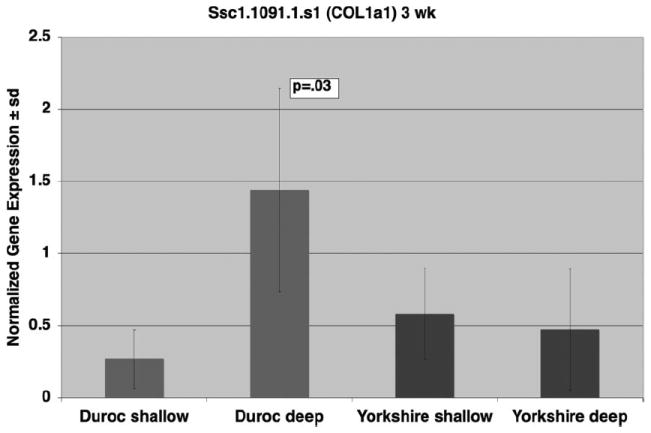
Quantitative Real Time RT-Polymerase Chain Reaction at 3 weeks for Ssc.1091.1.s1_at (COL1a1). mRNA expression levels of COL1a1 in deep wounds were compared with shallow wound in Durocs and Yorkshires at 3 weeks. Data are expressed relative to the housekeeping gene GAPDH. Bars represent data from three biologic replicates, with means and standard deviations shown. The Polymerase Chain Reaction data confirmed the array data and indicates greater expression is deep Duroc wounds compared to shallow and no difference in Yorkshire wounds.
Ssc.6778.1.s1_at (COL14a1)
Ssc.6778.1.s1_at (COL14a1) was chosen for verification, as the P value from mixed linear regression was <.001, indicating a profound difference. The gene expression was increased in Duroc deep wounds compared with Duroc shallow wounds and the difference was greatest at 3 weeks (Figure 5). There was no difference in Yorkshire wounds. Again, the biphasic response is similar to that described by Gallant.34,38
Figure 5.
Expression of COL14a1 (Affymetrix Probe Set Ssc.6778.1.s1_at) over time. COL14a1 gene expression was calculated as the log ratio of deep or shallow signals. Data from Durocs and Yorkshires were plotted over the time course of 20 weeks. Each data point represents mean and standard deviation from three biologic replicates. COL14a1 was differentially over expressed in deep duroc wounds (P < .001). The curves are 3rd degree exponential.
To confirm the 3-week array data for Ssc.6778.1.s1_ at, we performed quantitative real time RT-PCR (Figure 6). The probe was significantly more expressed in Duroc deep wounds than in Duroc shallow wounds (P = .046). There was no difference between Yorkshire deep and shallow wounds.
Figure 6.
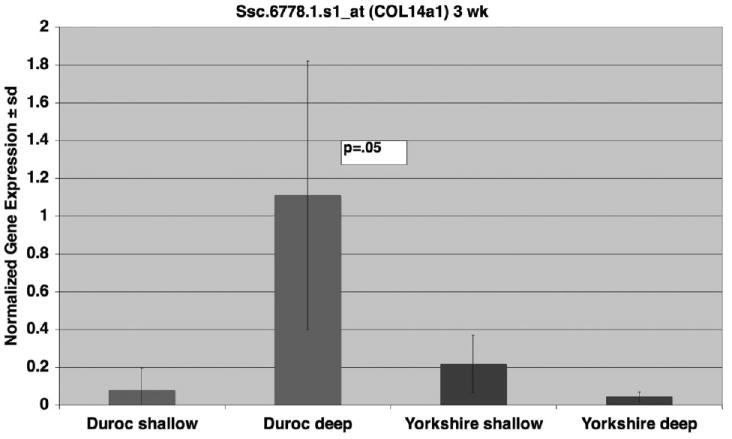
Quantitative Real Time RT-Polymerase Chain Reaction at 3 weeks for Ssc.6778.1.s1_at (COL14a1). mRNA expression levels of COL14a1 in deep wounds were compared with shallow wound in Durocs and Yorkshires at 3 weeks. Data are expressed relative to the housekeeping gene GAPDH. Bars represent data from three biologic replicates, with means and standard deviations shown. The Polymerase Chain Reaction data confirmed the array data and indicates greater expression is deep Duroc wounds compared to shallow and no difference in Yorkshire wounds.
Ssc.29731.1.a1_at (COL7a1)
Ssc.29731.1.a1_at (COL7a1) was chosen for verification because, in contrast to the other collagen genes differentially expressed, the expression in Duroc deep wounds was depressed (Figure 7). With mixed linear regression P = .06. Again, the biphasic response was previously described by Gallant.34,38
Figure 7.
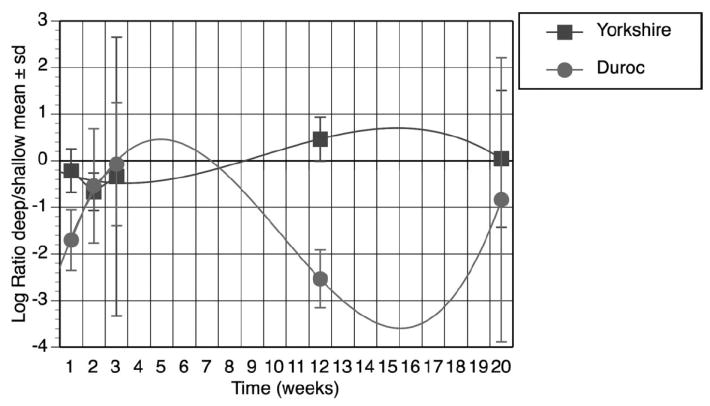
Expression of COL7a1(Affymetrix Probe Set Ssc.29731.1.a1_at) over time. COL7a1 gene expression was calculated as the log ratio of deep/ shallow signals. Data from Durocs and Yorkshires were plotted over the time course of 20 weeks. Each data point represents mean and standard deviation from three biologic replicates. COL7a1 was differentially over expressed in shallow duroc wounds (P = .06). The curves are 3rd degree exponential.
To confirm the 12-week array data for Ssc.29731.1.a1_at, we performed quantitative real time RT-PCR (Figure 8). The probe was significantly more expressed in Duroc shallow wounds than in Duroc deep wounds (P = .013). There was no difference between Yorkshire deep and shallow wounds.
Figure 8.
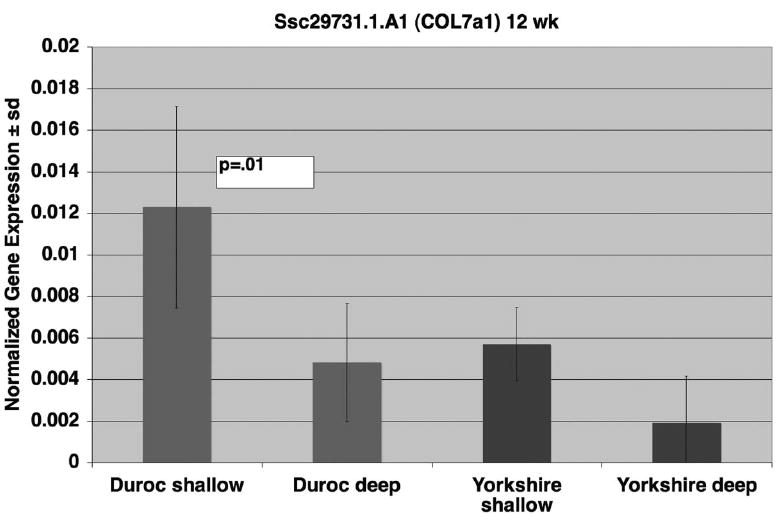
Quantitative Real Time RT-Polymerase Chain Reaction at 12 weeks for Ssc.29731.1.a1_at (COL7a1). MRNA expression levels of COL7a1 in deep wounds were compared with shallow wound in Durocs and Yorkshires at 12 weeks. Data are expressed relative to the housekeeping gene GAPDH. Bars represent data from three biologic replicates, with means and standard deviations shown. The Polymerase Chain Reaction data confirmed the array data and indicates greater expression is shallow Duroc wounds compared to deep and no difference in Yorkshire wounds.
DISCUSSION
It has been previously established that Duroc fibroproliferative scar is similar to human hypertrophic scar and that the Yorkshire serves as the control.29-40 The present objective was to obtain and compare microarray data on the signature collagen genes from wounds in Durocs and Yorkshires to each other and to human hypertrophic scar.
We have now obtained the microarray data from three Durocs and three Yorkshires, and confirmed the 20-week deep Duroc expression in human hypertrophic scar of approximately the same age. The data suggest that collagen genes 1a1, 1a2, 3a1, 4a1, 4a2, 5a1, 5a2, 5a3, 6a3 transcript variant 5, 7a1, 14a1, and 15a1 are directly or indirectly involved in the process of fibroproliferative scarring, and that 2a1, 8a1, 9a1, 9a2, 9a3, 10a1, 11a1, 12a1, 16a1, 17a1, 18a1, 21a1, and 24a1 are not.
Because a hallmark feature of hypertrophic scar is excessive and disorganized collagen, it is reasonable to assume that collagen synthesis, binding and/or degradation in fibroproliferative scarring is different than in nonfibroproliferative healing. It is possible that this deranged process(es) begins with deranged gene expression. But even if that is not the case, it is reasonable to assume that eventually the biological distortions will alter gene expression; and therefore microarray data may detect the problem and cast new light on the subject. But before searching the array data for alterations in the regulatory pathways, it is necessary to establish that the array data detects the fundamental problem with the signature genes. We have now established that this system will detect differences in the signature genes and will now move on to examining the regulation of these signature genes.
It is, of course, necessary to verify array data with PCR data; but it is prohibitively expensive to perform PCR on every gene discussed. The compromise is to obtain PCR data on a sufficient number of genes to verify the system and then on any gene chosen for further study. We chose 1a1 since it is fundamental to skin, 14a1 since the P value was extremely low indicating a profound difference, and 7a1 since it was the only gene differentially under expressed in deep Duroc wounds. We think three “verifies” the system. We did not do PCR on the others, so have none at this time for which PCR did not verify the array data.
We required that the porcine data to be used for further analysis matched the human data; how and why this was performed requires further explanation. There are five issues in making the comparison: 1) The pig is not human, and so undoubtedly has gene expression not present in the human. An example, perhaps trivial, is the skin gene expression necessary to make a curly tail. Any such expression must be eliminated from further study. 2) Comparing expression on human chips to expression on porcine chips, ie, between chips for different species, is very difficult. After discussions with Affymetrix, we decided the only way is to use the Present Absent calls to compare expression between species as the actual signal levels cannot be compared. 3) A chip may contain several probe sets for the same gene, so it is difficult to determine which probe set to compare between species. 4) It would be best to make the comparison at all time points. However, it is impossible to obtain human scarring and human nonscarring tissue at 1, 2, 3, 12, and 20 weeks for comparison. And finally, 5) should the pig or human comparison be made between Duroc/human or Yorkshire/human?
After considering these issues, and recalling that we did three Durocs and three human scars, we decided that two of the three Present Absent calls on any matching gene must be the same for the gene to be included. We chose two, since one seemed too loose and three seemed too tight for preliminary studies. So for gene abc, if the Duroc Present Absent calls were PPP and the human PPP, the gene was included. If the Duroc calls were PPA and the human PPA, the gene was included. If the Duroc calls were PAA and the human PPA, the gene was excluded, etc.
Then there is the question of at which time point should the comparison be made and with which breed. Because we cannot get human tissue at 1, 2, 3, and 12 weeks; but we can obtain human scar tissue from approximately 5 months postinjury, it was required to use 20-week human tissue for the comparison. And since deep Duroc wounds result in thick scar and shallow Duroc wounds and Yorkshire wounds of either depth do not, we elected to compare deep Duroc tissues to the human tissues. The next question is which time point is most appropriate for the comparison. The life span of pigs is perhaps 15 years and so it is difficult to compare the biologic and physiologic clock of the pig to the human. For example, puberty in pigs is achieved at 5 to 6 months. On the other hand, clotting time in pigs is rather comparable to humans. Consequently, it might be that the 12-week porcine samples are most comparable to 5-month human samples. However, since there is no data on this question, we simply chose to compare data from matching times.
There are many potential sources of error and bias in this study and many confounding variables including:
Species dissimilarities—although the Duroc or Yorkshire model is similar to human hypertrophic scarring, it is not identical. It is therefore, possible that the porcine array data does not match the human condition. We have, however, limited the study to those genes that have similar expression in Duroc 5-month tissue and human hypertrophic scar since both involved fibroproliferative healing.
Genes of importance are not on the GeneChip®—the GeneChip® examines approximately 30 to 50% of genes in the porcine genome, so the significant gene(s) may not be on the chip.
Depth of wound—as described above, wound depth cannot be controlled precisely. We can only create wounds that have shallow and deep appearance as shown in Figure 1.
Site of biopsy within the wound—Because wound depth cannot be precisely controlled, the location of the biopsy site within the wound is important. We attempted to place the biopsy in the deepest area, usually the center of the wound.
Time required for LCM—it is difficult and time consuming to perform LCM and during this time, RNA may become degraded. With experience we were able to complete each sample in about 40 minutes.
Relatively small numbers of cells—the deep dermis is far less cellular than many tissues and therefore the quantity of RNA is “small” and amplification is required. We obtained approximately 5 ng total RNA per sample and performed two rounds of amplification, which yielded a minimum of 15 μg. This source of variability, like the time required for laser capture microdissection, applies to both Duroc and Yorkshire samples.
However, these sources of variability apply to both deep and shallow wounds and to both Duroc and Yorkshire breeds so the comparison seems valid.
Recently Cuttle et al51 in Australia described thick, contracted, purple scars in Large White pigs after controlled, deep dermal, thermal injuries. The healed areas were 2.2 times thicker than control skin at 99 days postinjury. For decades, it has generally been held that pigs do not produce thick scar. With fibroproliferative scar in porcine wounds now reported in the United State, Canada, China, and Australia, it would seem that that idea has been laid to rest. The authors reported wounds, which like ours, are not the same as classic human hypertrophic scar, being thinner and with few nodules in the wounds up to 5 months postinjury. But the healed wounds were thicker than control and contracted and purple. Because the Yorkshire and the Large White breeds are closely related, the manuscript does raise the following question; is our using the Yorkshire as the control and their producing thick scars in Large Whites a conflict? The answer is most likely no since even though the two breeds have similar phenotypes, they are distinct genetically. However, since these two breeds are most similar than either is to the Duroc breed, it would therefore be interesting to compare Large White and Yorkshire at the gene expression level as performed here for Yorkshire and Duroc.
Future Directions
We now have the differential gene expression data comparing shallow and deep wounds and Duroc and Yorkshire pigs and it appears that the model is valid for the “signature” collagen genes. We will next attempt to “follow the money” backwards, ie trace the gene expression back to the early events of fibroproliferative healing. It is true that the “first” event might not involve gene expression. But it is reasonable to assume that as the process proceeds, eventually gene expression will be altered from that associated with normotrophic healing.
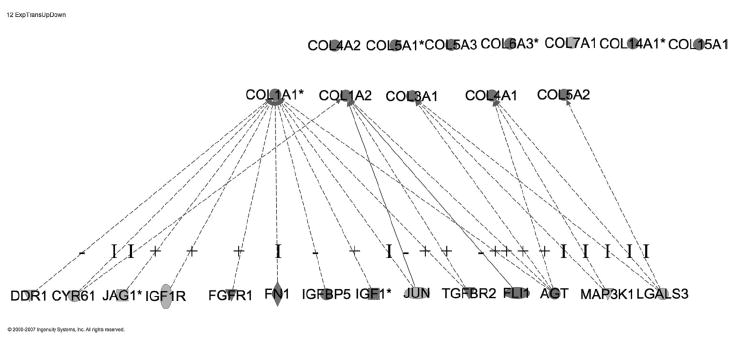
To accomplish this, we will use several techniques and several software packages including Ingenuity PathwaysAnalysis(Ingenuity®Systems,http://www.ingenuity.com). As an example of this process, we uploaded the 953 annotated, differentially expressed probe sets to Ingenuity and built the network which includes those genes that have an impact on the expression or transcription of the 12 differentially expressed collagen genes (Figure 9).
Figure 9.
Differentially expressed genes known to have an impact on the expression of the 12 differentially expressed collagen genes. Such known relationships exist for COL1A1, COL1A2, COL3A1, COL4A1 and COL5A2. No such relationships are known for the other collagen genes. Black represents differential over expression in deep Duroc wounds and gray indicates differential under expression in deep Duroc wounds. “−” refers to decreased expression of the collagen gene, “I” to involvement in expression of the collagen gene, and “+” to increased expression of the collagen gene.
We will follow the expression into deeper levels and with other relationships and attempt to develop hypotheses as to what gene expression patterns result in differential expression of the 12 collagen genes during the process of fibroproliferative healing.
Will this process contribute to our understanding of the pathogenesis of fibroproliferative healing? Maybe not. But then, maybe it will. Time will tell.
Acknowledgments
This study was supported by grants from National Institute on Disability and Rehabilitation Research/Office of Special Education and Rehabilitation Services/US Department of Education, National Institutes of Health, Washington State Council of Firefighters Burn Foundation, Northwest Burn Foundation.
Footnotes
Presented at Wound Healing Society, Tampa, Florida 2007.
References
- 1.Linares HA. From wound to scar. Burns. 1996;22:339–52. doi: 10.1016/0305-4179(95)00164-6. [DOI] [PubMed] [Google Scholar]
- 2.Rockwell WB, Cohen IK, Ehrlich HP. Keloids and hypertrophic scars: a comprehensive review. Plast Reconstr Surg. 1989;84:827–37. doi: 10.1097/00006534-198911000-00021. [DOI] [PubMed] [Google Scholar]
- 3.Murray JC. Keloids and hypertrophic scars. Clin Dermatol. 1994;12:27–37. doi: 10.1016/0738-081x(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 4.Su CW, Alizadeh K, Boddie A, Lee RC. The problem scar. Clin Plast Surg. 1998;25:451–65. [PubMed] [Google Scholar]
- 5.Dasu MR, Hawkins HK, Barrow RE, Xue H, Herndon DN. Gene expression profiles from hypertrophic scar fibroblasts before and after IL-6 stimulation. J Pathol. 2004;202:476–85. doi: 10.1002/path.1539. [DOI] [PubMed] [Google Scholar]
- 6.Rahban SR, Garner WL. Fibroproliferative scars. Clin Plast Surg. 2003;30:77–89. doi: 10.1016/s0094-1298(02)00069-x. [DOI] [PubMed] [Google Scholar]
- 7.Robson MC, Steed DL, Franz MG. Wound healing: biologic features and approaches to maximize healing trajectories. Curr Probl Surg. 2001;38:72–140. doi: 10.1067/msg.2001.111167. [DOI] [PubMed] [Google Scholar]
- 8.Robson MC. Proliferative scarring. Surg Clin North Am. 2003;83:557–69. doi: 10.1016/S0039-6109(02)00197-4. [DOI] [PubMed] [Google Scholar]
- 9.Aksoy MH, Vargel I, Canter IH, et al. A new experimental hypertrophic scar model in guinea pigs. Aesthetic Plast Surg. 2002;26:388–96. doi: 10.1007/s00266-002-1121-z. [DOI] [PubMed] [Google Scholar]
- 10.Greenhalgh DG. Models of wound healing. J Burn Care Rehabil. 2005;26:293–305. doi: 10.1097/01.bcr.0000169885.66639.b5. [DOI] [PubMed] [Google Scholar]
- 11.Mast BA. The skin. In: Cohen IK, Diegelmann RF, Lindblad WJ, editors. Wound healing: biochemical & clinical aspects. Philadelphia, PA: W.B. Saunders Co; 1992. p. 353. [Google Scholar]
- 12.Morris DE, Wu L, Zhao LL, et al. Acute and chronic animal models for excessive dermal scarring: quantitative studies. Plast Reconstr Surg. 1997;100:674–81. doi: 10.1097/00006534-199709000-00021. [DOI] [PubMed] [Google Scholar]
- 13.Polo M, Kim YJ, Kucukcelebi A, Hayward PG, Ko F, Robson MC. An in vivo model of human proliferative scar. J Surg Res. 1998;74:187–95. doi: 10.1006/jsre.1997.5251. [DOI] [PubMed] [Google Scholar]
- 14.Ha X, Li Y, Lao M, Yuan B, Wu CT. Effect of human hepatocyte growth factor on promoting wound healing and preventing scar formation by adenovirus-mediated gene transfer. Chin Med J (Engl) 2003;116:1029–33. [PubMed] [Google Scholar]
- 15.Xiang J, Wang ZY, Jia SX, Jin SW, Lu SL, Liao ZJ. Establishment of an animal model with hypertrophic scar. Zhonghua Shao Shang Za Zhi. 2004;20:281–3. [PubMed] [Google Scholar]
- 16.Kischer CW, Pindur J, Shetlar MR, Shetlar CL. Implants of hypertrophic scars and keloids into the nude (athymic) mouse: viability and morphology. J Trauma. 1989;29:672–7. doi: 10.1097/00005373-198905000-00023. [DOI] [PubMed] [Google Scholar]
- 17.Kischer CW, Sheridan D, Pindur J. Use of nude (athymic) mice for the study of hypertrophic scars and keloids: vascular continuity between mouse and implants. Anat Rec. 1989;225:189–96. doi: 10.1002/ar.1092250303. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Smith P, Pu LL, Kim YJ, Ko F, Robson MC. Exogenous transforming growth factor beta(2) modulates collagen I and collagen III synthesis in proliferative scar xenografts in nude rats. J Surg Res. 1999;87:194–200. doi: 10.1006/jsre.1999.5757. [DOI] [PubMed] [Google Scholar]
- 19.Polo M, Smith PD, Kim YJ, Wang X, Ko F, Robson MC. Effect of TGF-beta2 on proliferative scar fibroblast cell kinetics. Ann Plast Surg. 1999;43:185–90. [PubMed] [Google Scholar]
- 20.Shetlar MR, Shetlar CL, Kischer CW, Pindur J. Implants of keloid and hypertrophic scars into the athymic nude mouse: changes in the glycosaminoglycans of the implants. Connect Tissue Res. 1991;26:23–36. doi: 10.3109/03008209109152161. [DOI] [PubMed] [Google Scholar]
- 21.Shetlar MR, Shetlar CL, Hendricks L, Kischer CW. The use of athymic nude mice for the study of human keloids. Proc Soc Exp Biol Med. 1985;179:549–52. doi: 10.3181/00379727-179-rc3. [DOI] [PubMed] [Google Scholar]
- 22.Robb EC, Waymack JP, Warden GD, Nathan P, Alexander JW. A new model for studying the development of human hypertrophic burn scar formation. J Burn Care Rehabil. 1987;8:371–5. doi: 10.1097/00004630-198709000-00006. [DOI] [PubMed] [Google Scholar]
- 23.Reiken SR, Wolfort SF, Berthiaume F, Compton C, Tompkins RG, Yarmush ML. Control of hypertrophic scar growth using selective photothermolysis. Lasers Surg Med. 1997;21:7–12. doi: 10.1002/(sici)1096-9101(1997)21:1<7::aid-lsm2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 24.Wolfort SF, Reiken SR, Berthiaume F, Tompkins RG, Yarmush ML. Control of hypertrophic scar growth using antibody-targeted photolysis. J Surg Res. 1996;62:17–22. doi: 10.1006/jsre.1996.0166. [DOI] [PubMed] [Google Scholar]
- 25.Yang DY, Li SR, Li G, et al. Establishment of an animal model of human hyperplastic scar in nude mice. Zhonghua Shao Shang Za Zhi. 2004;20:82–4. [PubMed] [Google Scholar]
- 26.Yang DY, Li SR, Wu JL, et al. Establishment of a hypertrophic sar model by transplanting full-thickness human skin grafts onto the backs of nude mice. Plast Reconst Surg. 2007;119:104–9. doi: 10.1097/01.prs.0000244828.80490.62. [DOI] [PubMed] [Google Scholar]
- 27.Hochman B, Vilas Boas FC, Mariano M, Ferreiras LM. Keloid heterograft in the hamster (Mesocricetus auratus) cheek pouch, Brazil. Acta Cir Bras. 2005;20:200–12. doi: 10.1590/s0102-86502005000300003. [DOI] [PubMed] [Google Scholar]
- 28.Lee JP, Jalili RB, Tredget EE, Demare JR, Ghahary A. Anti-fibrogenic effects of liposome-encapsulated IFN-alpha2b cream on skin wounds in a fibrotic rabbit ear model. J Interferon Cytokine Res. 2005;25:627–31. doi: 10.1089/jir.2005.25.627. [DOI] [PubMed] [Google Scholar]
- 29.Silverstein P, Goodwin MN, Raulston GL, Pruitt B. Hypertrophic scar in the experimental animal. In: Longacre JJ, editor. The ultrastructure of collagen; its relation to the healing of wounds and to the management of hypertrophic scar. Springfield IL: Thomas; 1976. pp. 213–36. [Google Scholar]
- 30.Silverstein P, Goodwin MN, Jr, Raulston GL. Hypertrophic scarring, etiology and control of a disabling complication in burned soldiers. Ann Res Progress Report of the US Army Institute of Surgical Research Section. 1972 June 30;37:1–5. [Google Scholar]
- 31.Zhu KQ, Engrav LH, Gibran NS, et al. The female, red Duroc pig as an animal model of hypertrophic scarring and the potential role of the cones of skin. Burns. 2003;29:649–64. doi: 10.1016/s0305-4179(03)00205-5. [DOI] [PubMed] [Google Scholar]
- 32.Zhu KQ, Engrav LH, Tamura RN, et al. Further similarities between cutaneous scarring in the female, red Duroc pig and human hypertrophic scarring. Burns. 2004;30:518–30. doi: 10.1016/j.burns.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 33.Liang Z, Engrav LH, Muangman P, et al. Nerve quantification in female red Duroc pig (FRDP) scar compared to human hypertrophic scar. Burns. 2004;30:57–64. doi: 10.1016/j.burns.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Gallant CL, Olson ME, Hart DA. Molecular, histologic, and gross phenotype of skin wound healing in red Duroc pigs reveals an abnormal healing phenotype of hypercontracted, hyperpigmented scarring. Wound Repair Regen. 2004;12:305–19. doi: 10.1111/j.1067-1927.2004.012311.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhu KQ, Engrav LH, Armendariz RT, et al. Changes in VEGF and nitric oxide after deep dermal injury in the female, red Duroc pig-further similarities between female, Duroc scar and human hypertrophic scar. Burns. 2005;31:5–10. doi: 10.1016/j.burns.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 36.Harunari N, Zhu KQ, Armendariz RT, et al. Histology of the thick scar on the female, red Duroc pig: final similarities to human hypertrophic scar. Burns. 2006;32:669–77. doi: 10.1016/j.burns.2006.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallant-Behm CL, Hart DA. Genetic analysis of skin wound healing and scarring in a porcine model. Wound Repair Regen. 2006;14:46–54. doi: 10.1111/j.1743-6109.2005.00087.x. [DOI] [PubMed] [Google Scholar]
- 38.Gallant-Behm CL, Olson ME, Hart DA. Cytokine and growth factor mRNA expression patterns associated with the hypercontracted, hyperpigmented healing phenotype of red Duroc pigs: a model of abnormal human scar development? J Cutan Med Surg. 2006;9:165–77. doi: 10.1007/s10227-005-0105-4. [DOI] [PubMed] [Google Scholar]
- 39.Gallant-Behm CL, Tsao H, Reno C, Olson ME, Hart DA. Skin wound healing in the first generation (F1) offspring of Yorkshire and red Duroc pigs: evidence for genetic inheritance of wound phenotype. Burns. 2006;32:180–93. doi: 10.1016/j.burns.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 40.Liang Z, Xie CY, Lin HB, Guo ZD, Yang WG. Pathomorphological observation of the hypertrophic scar induced by injury to conical structure in female red Duroc pig. Zhonghua Shao Shang Za Zhi. 2006;22:29–32. [PubMed] [Google Scholar]
- 41.Shoemaker JV. A practical treatise on diseases of the skin. New York and London: D Appleton and Company; 1905. [Google Scholar]
- 42.Jackson DM. The diagnosis of the depth of burning. Br J Surg. 1953;40:588–96. doi: 10.1002/bjs.18004016413. [DOI] [PubMed] [Google Scholar]
- 43.Matsumura H, Engrav LH, Gibran NS, et al. Cones of skin occur where hypertrophic scar occurs. Wound Repair Regen. 2001;9:269–77. doi: 10.1046/j.1524-475x.2001.00269.x. [DOI] [PubMed] [Google Scholar]
- 44.Tuggle CK, Wang Y, Couture O. Advances in swine transcriptomics. Int J Biol Sci. 2007;3:132–52. doi: 10.7150/ijbs.3.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang YF, Qu L, Uthe JJ, et al. Global transcriptional response of porcine mesenteric lymph nodes to Salmonella enterica serovar typhimurium. Genomics. 2007;90:72–84. doi: 10.1016/j.ygeno.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 46.Fang P, Engrav LH, Gibran NS, et al. Dermatome setting for autografts to cover INTEGRA. J Burn Care Rehabil. 2002;23:327–32. doi: 10.1097/00004630-200209000-00005. [DOI] [PubMed] [Google Scholar]
- 47.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitfield ML, Finlay DR, Murray JI, et al. Systemic and cell type-specific gene expression patterns in scleroderma skin. Proc Natl Acad Sci U S A. 2003;100:12319–24. doi: 10.1073/pnas.1635114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnston P, Chojnowski AJ, Davidson RK, Riley GP, Donell ST, Clark IM. A complete expression profile of matrix-degrading metalloproteinases in Dupuytren’s disease. J Hand Surg [Am] 2007;32:343–51. doi: 10.1016/j.jhsa.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 51.Cuttle L, Kempf M, Phillips GE, et al. A porcine deep dermal partial thickness burn model with hypertrophic scarring. Burns. 2006;32:806–20. doi: 10.1016/j.burns.2006.02.023. [DOI] [PubMed] [Google Scholar]