Abstract
The Bioconductor project is an “open source and open development software project for the analysis and comprehension of genomic data” Gentleman et al. (2004), primarily based on the R programming language. Infrastructure packages, such as Biobase, are maintained by Bioconductor core developers, and serve several key roles to the broader community of Bioconductor software developers and users. In particular, Biobase introduces a S4 class, the eSet, for high dimensional assay data. Encapsulating the assay data as well as meta-data on the samples, the features, and the experiment in the eSet class definition ensures propagation of the relevant sample and feature meta-data throughout an analysis. Extending the eSet class promotes code reuse through inheritance, interoperability with other R packages, and is less error prone. Recently proposed class definitions for high throughput SNP arrays extend the eSet class. This chapter highlights the advantages of adopting and extending Biobase class definitions through a working example of one implementation of classes for the analysis of high throughput SNP arrays.
Keywords: SNP array, copy number, genotype, S4 classes
Introduction
The Bioconductor project is an “open source and open development software project for the analysis and comprehension of genomic data”, primarily based on the R programming language, and provides open source software for researchers in the fields of computational biology and bioinformatics-related disciplines Gentleman et al. (2004). Infrastructure packages such as Biobase settle basic organizational issues for high throughput data and facilitates interoperability of R packages that utilize this infrastructure. Transparency and reproducibility are emphasized in Bioconductor through package vignettes.
A key element of infrastructure for high throughput genomic data is the eSet, a virtual class for organizing high-throughput genomic data defined in Biobase. An instance of an eSet-derived class contains the high throughput assay data and the corresponding meta-data on the experiment, samples, covariates, and features (e.g., probes) in a single object. While much of the development of the eSet has been in response to high-throughput gene expression experiments that measure RNA (or cDNA) abundance, the generality of the eSet class enables the user to extend the class to accommodate a variety of high-throughput technologies. Here, we focus on single nucleotide polymorphism (SNP) microarray technology, and the eSet-derived classes specific to this technology.
SNP microarrays provide estimates of genotype and copy number at hundreds of thousands of SNPs along the genome, and several recent papers describe approaches for genotype (Di et al. (2005); Rabbee and Speed (2006); Affymetrix (2006); Carvalho et al. (2007)) and copy number estimation (Nannya et al. (2005); Huang et al. (2006); Laframboise et al. (2006); Carter (2007)). In addition to probes targeting the polymorphic regions of the genome, the latest Affymetrix and Illumina platforms contain a set of non-polymorphic probes for estimating copy number.
The S4 classes and methods proposed here are organized around the multiple levels of SNP data. In particular, we refer to the raw files containing probe intensities as the features-level data and the processed data containing summaries of genotype calls and copy number as SNP-level data. Finally, there is a third level of analytic data obtained from methods that smooth the SNP-level summaries as a function of the physical position on the chromosome, such as hidden Markov models (HMMs). Algorithms at the third tier are useful for identifying genomic features such as deletions (hemizygous or homozygous), amplifications (more than 2 copies), and copy-neutral loss of heterozygosity.
This chapter is organized as follows. We begin with a brief overview of S4 classes, illustrating concepts such as inheritance using minimal class definitions for the high throughput SNP data. With these minimal definitions in place, we discuss their shortcomings and motivate the development of the current class definitions. We conclude with an example that illustrates the following workflow: (i) creating an instance of a SNP-level class from matrices of genotype calls and copy number, (ii) plotting the SNP-level data as a function of physical position along the chromosome, (iii) fitting a hidden Markov model to identify alterations in copy number or genotype, and (iv) plotting the predicted states from the hidden Markov model alongside the genomic data.
S4 Classes and Methods
In the statistical environment R, an object can be a value, a function, or a complex data structure. To perform an action on an object, we write a function. For instance, we could write a function to calculate the row means of a matrix. When the object and functions become complex, classes and methods become useful as an organizing principle. A S4 class formally defines the ingredients of an object. A method for a class tells R which function should be performed on the object. A useful property of classes and methods is inheritance. For instance, a matrix is an array with only two dimensions: rows and columns. Using the language of classes, we say that array is a parent class (or superclass) that is extended by the class matrix. Inheritance refers to the property that any methods defined for the parent class are available to the children of the parent class. In this section, we will discuss two approaches that can be used to construct classes that extend a parent class, illustrate the concept of inheritance by minimally defining S4 classes for storing estimates of genotype and copy number, provide examples of how to construct methods to access and replace elements of an instantiated class, and show how methods that check the validity of an instantiated objects can be used to reduce errors. This section provides a very brief overview of S4 classes and methods, see Chambers (1998) for a detailed description. The classes defined in this section are solely for the purpose of illustration and are not intended to be used for any analytic data.
Initializing classes
To construct classes for SNP-level summaries of genotype calls and copy number estimates after pre-processing, we can use the following classes as minimal definitions:
> setClass(“MinimalCallSet”, representation(calls = “matrix”)) [1] “MinimalCallSet” > setClass(“MinimalCopyNumberSet”, representation(copyNumber = “matrix”)) [1] “MinimalCopyNumberSet”
An instance of MinimalCallSet contains a slot for the matrix of genotype calls, and an instance of MinimalCopyNumberSet contains a slot for the matrix of copy number estimates.
Extending classes
A parent class of MinimalCallSet and MinimalCopyNumberSet, called SuperSet, is created by the function setClassUnion:
> setClassUnion(“SuperSet”, c(“MinimalCallSet”, “MinimalCopyNumberSet”)) [1] “SuperSet” > showClass(“SuperSet”) Virtual Class “SuperSet” No Slots, prototype of class “NULL” Known Subclasses: “MinimalCallSet”, “MinimalCopyNumberSet” > extends(“MinimalCallSet”, “SuperSet”) [1] TRUE
MinimalCallSet and MinimalCopyNumberSet extend SuperSet. Note that SuperSet is a virtual class, and therefore we cannot instantiate an object of class SuperSet. However, instantiating one of the derived classes requires only a matrix of the SNP-level summaries. Using a recent version of R (≥ 2.7), one may obtain an example dataset from the VanillaICE R package.
> source(“http://www.bioconductor.org/biocLite.R”) > biocLite(“VanillaICE”, type = “source”) > library(VanillaICE) > data(sample.snpset) > gt <- calls(sample.snpset)[1:3, 1:3] > gt[gt == 1] <- “AA” > gt[gt == 2] <- “AB” > gt[gt == 3] <- “BB” > cn <- copyNumber(sample.snpset)[1:3, 1:3] > colnames(cn) <- colnames(gt) <- sapply(colnames(gt), function(x) strsplit(x, “_”)[[1]][1]) > callset <- new(“MinimalCallSet”, calls = gt) > cnset <- new(“MinimalCopyNumberSet”, copyNumber = cn) > attributes(callset) $calls NA17101 NA17102 NA17103 SNP_A-1507972 “AB” “BB” “AB” SNP_A-1641761 “AB” “AB” “AB” SNP_A-1641781 “AB” “AA” “AA” $class [1] “MinimalCallSet” attr(,“package”) [1] “.GlobalEnv” > attributes(cnset) $copyNumber NA17101 NA17102 NA17103 SNP_A-1507972 3.176972 2.775924 3.051108 SNP_A-1641761 1.705276 1.793427 1.647903 SNP_A-1641781 2.269756 1.741290 1.806562 $class [1] “MinimalCopyNumberSet” attr(,“package”) [1] “.GlobalEnv”
As MinimalCallSet and MinimalCopyNumberSet extend SuperSet, methods defined at the level of the parent class are inherited. For instance, we define show, and call this function on the instantiated objects of MinimalCallSet and MinimalCopyNumberSet.
> setMethod(“show”, “SuperSet”, function(object) attributes(object)) [1] “show” > show(callset) $calls NA17101 NA17102 NA17103 SNP_A-1507972 “AB” “BB” “AB” SNP_A-1641761 “AB” “AB” “AB” SNP_A-1641781 “AB” “AA” “AA” $class [1] “MinimalCallSet” attr(,“package”) [1] “.GlobalEnv” > show(cnset) $copyNumber NA17101 NA17102 NA17103 SNP_A-1507972 3.176972 2.775924 3.051108 SNP_A-1641761 1.705276 1.793427 1.647903 SNP_A-1641781 2.269756 1.741290 1.806562 $class [1] “MinimalCopyNumberSet” attr(,“package”) [1] “.GlobalEnv”
The contains argument in the function setClass can be used to extend an existing parent class. For instance,
> setClass(“MinimalSnpSet”, contains = “SuperSet”, representation(calls = “matrix”, + copyNumber = “matrix” )) [1] “MinimalSnpSet”
By defining methods that access specific elements of a class at the level of the parent class, it is not necessary to define these methods for any of the derived classes.
Signatures
The signature of a generic function is a named list of classes that determines the method that will be dispatched. Consider the generic function foo in the following code chunk. The method that is dispatched when foo(object) is called depends on the class of object.
> setGeneric(“foo”, function(object) standardGeneric(“foo”)) [1] “foo” > setMethod(“foo”, signature(object = “ANY”), function(object) message(“message 1”)) [1] “foo” > setMethod(“foo”, signature(object = “matrix”), function(object) message(“message 2”)) [1] “foo” > foo(1) > foo(as.matrix(1))
More precisely, the dispatched method depends on the ‘distance’ of the class of the argument to the generic function and the signature of the method. For example, if we define a new class A that extends class matrix, message 2 will be printed as the distance between the object and class matrix is 1, whereas the distance between A and ANY is greater than 1.
> setClass(“A”, contains = “matrix”) [1] “A” > x <- as(matrix(1), “A”) > foo(x) > setMethod(“foo”, signature(object = “A”), function(object) message(“message 3”)) [1] “foo” > foo(x) [1] “genotypeCalls” [1] “genotypeCalls” NA17101 NA17102 NA17103 SNP_A-1507972 “AB” “BB” “AB” SNP_A-1641761 “AB” “AB” “AB” SNP_A-1641781 “AB” “AA” “AA”
In addition to defining methods that access information from an object, one may define a method that replaces information in an object. An example of such a method follows:
> setGeneric(“genotypeCalls<-”, function(object, value) standardGeneric(“genotypeCalls<- ”))
[1] “genotypeCalls<-”
> setReplaceMethod(“genotypeCalls”, c(“SuperSet”, “matrix”), function(object, value) {
+ object@calls <- value
+ return(object)
+ })
[1] “genotypeCalls<-”
Validity methods
Validity methods can be useful to avoid committing errors when instantiating a class that can have unfortunate consequences on downstream analyses. For instance, for objects of class MinimalSnpSet it is useful to require that the row names and column names of the copy number and genotype matrices are identical. Therefore, we can define a validity method for the class MinimalSnpSet that checks whether the names are identical and, if not, throws an error.
> setValidity(“MinimalSnpSet”, function(object) {
+ valid <- identical(rownames(object@calls), rownames(object@copyNumber))
+ if (!valid)
+ stop(“rownames are not identical”)
+ valid <- identical(colnames(object@calls), colnames(object@copyNumber))
+ if (!valid)
+ stop(“colnames are not identical”)
+ return(msg)
+ })
Class “MinimalSnpSet”
Slots:
Name: calls copyNumber
Class: matrix matrix
Extends: “SuperSet”
> colnames(gt) <- letters[20:22]
> tryCatch(new(“MinimalSnpSet”, calls = gt, copyNumber = cn), error = function(e) print(e))
<simpleError in validityMethod(object): colnames are not identical>
SNP-level Classes and Methods
When constructing S4 classes for the purpose of analysing high throughput SNP data, the following considerations are useful:
Develop as little new code as possible, reusing code that has been extensively tested and documented in other packages.
The SNP-level summaries that are available as assay data may depend on the pre-processing algorithm or the particular SNP microarray technology.
Attaching meta-data on the samples, features, and experiment to the object storing the assay data (as is commonly done with eSet derived classes) is useful for ensuring that the meta-data is attached to the assay data throughout an analysis.
Adopting standard data structures defined in widely used packages such as Biobase promotes interoperability of R packages that perform complementary tasks.
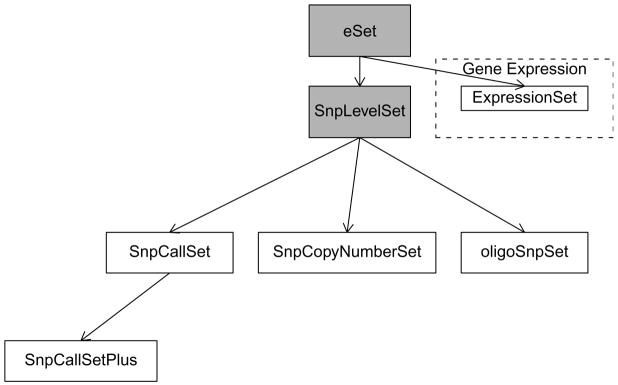
The schematic in Figure 1 illustrates the relationships of our implementation of SNP-level classes in the package oligoClasses. We briefly discuss each of these classes below.
Figure 1.
Classes for SNP-level data, as defined in the Bioconductor package oligoClasses. Note that eSet and SnpLevelSet are virtual classes.
eSet: eSet is a virtual class defined in the R package Biobase Gentleman et al. (2004) and provides a basic container for high-throughput genomic data. Slots in eSet are defined for assay data ( assayData: e.g., genotype calls), characteristics of the samples (slot phenoData: e.g., phenotype), characteristics of the features (slot featureData: e.g., the name of the feature) and experimental data (slot experimentData: e.g., details of the laboratory and experimental methods). Via inheritance, each of the SNP-derived classes contain these components; accessors and replacement methods defined for the eSet can be readily applied to the eSet-derived classes.
SnpLevelSet. SnpLevelSet is a virtual class that extends eSet directly. Note that all SNP-level classes in Figure 1 extend SnpLevelSet directly. To understand why we define a virtual superclass for SNP-level data (when eSet is already available), consider that many methods are likely to be applicable to all SNP-derived classes, but perhaps not eSet-derived classes such as the ExpressionSet. For instance, the plotting methods in SNPchip and the hidden Markov model in VanillaICE rely on the chromosome and physical position of the SNP. While this information is critical for statistical methods such as a HMM that smooths SNP-levels summaries as a function of physical position on the chromosome, it may be less useful or of no use for gene expression microarrays. Furthermore, because accessors for chromosome and physical position are useful for all of the SNP-derived classes, defining these accessors at the level of SnpLevelSet eliminates the need to define accessors for each of the derived classes. Of course, the flexibility to define methods specific to each of the derived classes remains.
SnpLevelSet progeny: Progeny of SnpLevelSet, including SnpCallSet, SnpCopyNumberSet, and oligoSnpSet, are defined according to the elements in the assayData slot. Elements of the assayData in SnpCallSet include calls (genotype calls) and callsConfidence (confidence scores for the genotype calls), whereas assayData elements in SnpCopyNumberSet are copyNumber and cnConfidence (confidence scores for copy number estimates). The assay data of an oligoSnpSet is the union of the assayData elements in SnpCallSet and SnpCopyNumberSet.
Example
We suggest the Bioconductor package oligo for pre-processing high-throughput SNP array data for the various Affymetrix platforms (100k, 500k, 5.0, and 6.0). In addition to genotype calls, the crlmm function in oligo provides confidence scores of the genotype calls that can be propagated to higher level analyses, such as the hidden Markov models discussed in the following section. A method for estimating copy number in oligo is currently under development. In this section, we assume that the user has obtained SNP-level summaries of genotype and copy number by some means. We show how to create an instance of oligoSnpSet from matrices of genotype calls and copy number estimates, plot the SNP-level summaries versus physical position on the genome, and fit a HMM to identify alterations in copy number or genotype.
Instantiating an oligoSnpSet object
To create an instance of oligoSnpSet, we take advantage of an example provided with the Bioconductor package VanillaICE, using only the matrices of copy number estimates, cn, and genotype calls, gt. The data we extract from the VanillaICE package is simulated data for a chromosome 1 on the Affymetrix 100k platform. Note that the matrices are organized such that the columns are samples and the rows are SNPs. While the elements of cn can be any positive number, the elements of gt are the integers 1, 2, 3, and 4 corresponding to the genotypes AA, AB, BB, and NA (not available), respectively. The row names (here, Affymetrix identifiers for the SNP) and column names (sample identifiers) of cn and gt must be identical. Confidence scores for the copy number estimates and genotype calls, when available, are stored similarly.
> library(VanillaICE) > data(chromosome1) > copynumber <- copyNumber(chromosome1) > calls <- calls(chromosome1) > cnConf <- callsConf <- matrix(NA, nrow = nrow(copynumber), ncol = ncol(copynumber), + dimnames = list(rownames(copynumber), colnames(copynumber))) > snpset <- new(“oligoSnpSet”, copyNumber = copynumber, calls = calls, cnConfidence = cnConf, + callsConfidence = callsConf, annotation = “pd.mapping50k.hind240,pd.mapping50k.xba240” ) > validObject(snpset) [1] TRUE
The annotation slot is important for accessing the appropriate annotation package (available at Bioconductor). In this example, the SNPs originate from two Affymetrix–platforms{ the 50k Xba and 50k Hind chips. The annotation packages can be installed from Bioconductor with the following command:
> source(“http://www.bioconductor.org/biocLite.R”) > biocLite(c(“pd.mapping50k.hind240”, “pd.mapping50k.xba240”))
Because the plotting methods and the HMM both frequently access the chromosome and physical position of the SNPs in the object, it is generally more convenient to store this information in the featureData slot. The position and chromosome methods first check the variable labels in the featureData and, if not present, retrieve this information from the annotation packages.
> featureData(snpset)$position <- position(snpset) > featureData(snpset)$chromosome <- chromosome(snpset)
Visualizing the data
The Bioconductor package SNPchip provides several useful methods for visualizing objects instantiated from one of the derived classes of SnpLevelSet (Scharpf et al., 2007). Similar to the R package lattice, the plotting method does not plot the data, rather it returns an object of class ParSnpSet that contains all of the default graphical parameters used to plot an instance of oligoSnpSet. The show method called on an object returned by plotSnp produces a plot. The following command plots the snpset object using the default graphical parameters.
> show(plotSnp(snpset))
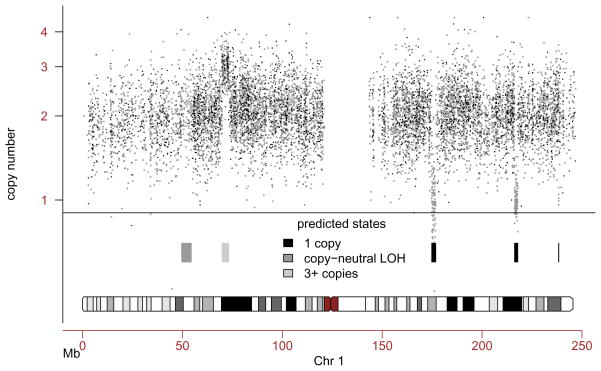
The resulting plot, together with the assessment of DNA copy number alterations, is shown in Figure 2.
Figure 2.
Simulated data for a chromosome 1 on the Affymetrix 100k platform. The x-axis denotes the loci along the chromosome, the y-axis denotes the copy number estimates. Homozygous genotype calls are plotted in light grey, heterozygous genotype calls are plotted in dark grey. Also shown is the inference for DNA copy numbers and alterations, using a hidden Markov model. This HMM captured the DNA alterations we simulated, namely (from left to right) a region of copy-neutral loss of heterozygosity, an amplification, and three deletions of various sizes on the q-arm.
Identifying chromosomal alterations
The simulated data used in this example contains 5 alterations that we utilize as benchmarks when testing the HMM model in the VanillaICE package. Details on the simulation and on the HMM model are described elsewhere (Scharpf et al., 2008). In order to fit the HMM, we must specify the hidden states and compute the emission and transition probabilities. We assume that the copy number estimates are Gaussian on the log 2 scale. To calculate the emission probabilities for copy number, we require specifying the location parameter of the Gaussian distribution (on the copy number scale) for each of the hidden states. If confidence scores for the copy number estimates are not available, the scale parameter is computed using a robust estimate of the log 2 copy number distribution and is assumed to be the same for each state. For genotype calls, one must specify the probability of a homozygous genotype call (AA or BB) for each of the hidden states. The transition probabilities, using an estimate of genomic distance, are SNP-specific.
> options <- new(“HmmOptions”, states = c(“D”, “N”, “L”, “A”), snpset = snpset, + copyNumber.location = c(1, 2, 2, 3), probHomCall = c(0.99, 0.7, 0.99, 0.7)) > params <- new(“HmmParameter”, states = states(options), initialStateProbability = 0.99) > cn.emission <- copyNumber.emission(options) > gt.emission <- calls.emission(options) > emission(params) <- cn.emission + gt.emission > genomicDistance(params) <- exp(-2*physicalDistance(options)/(100*1e+06)) > transitionScale(params) <- scaleTransitionProbability(options) > fit <- hmm(options, params) > class(fit)
The object returned by the hmm method is an instance of the class HmmPredict. HmmPredict extends SnpLevelSet directly. The following code can be used to plot the SNP-level summaries of genotype and copy number alongside the predicted states from the HMM.
> gp <- plotSnp(snpset(options), fit) > gp$col <- c(“grey60”, “black”, “grey60”) > gp$cex <- c(2, 1.5, 2) > gp$hmm.ycoords <- c(0.6, 0.7) > gp$ylim <- c(0.4, 4.5) > gp$xlim[1] <- -10000 > gp$abline <- TRUE > gp$abline.h <- 0.9 > gp$abline.col <- “black” > gp$cytoband.ycoords <- c(0.4, 0.45) > gp$col.predict <- c(“black”, “white”, “grey60”, “grey80”) > print(gp) > legend(95*1e+06, 0.9, fill = gp$col.predict[-2], legend = c(“1 copy”, “copy-neutral LOH”,+ “3+ copies”), bty = “n”, title = “predicted states”)
Closing Remarks
The Bioconductor project has several infrastructure packages that are useful for organizing and annotating genomic data. In particular, the Biobase package introduces the virtual class eSet that provides an organization for high throughput assay data set and the corresponding meta-data on the samples, features, and experiment. Extensions of the eSet class to a variety of different platforms and architectures are feasible. As our focus is on S4classes and methods for high throughput SNP data, we discuss the classes that are currently in place and the considerations that motivated these definitions. We emphasize the importance of using standardized data structures and the ease by which code can be reused through inheritance, both of which are facilitated by utilizing S4 classes and methods. The visualization methods in the SNPchip package and the HMM in the VanillaICE package serve as useful illustrations of how one can build on these definitions.
Acknowledgments
This work was supported by NSF grant DMS034211 and training grant 5T32HL007024 from the National Heart, Lung, and Blood Institute (RBS), and NIH grant R01 GM083084 (IR).
References
- Affymetrix. BRLMM: an improved genotype calling method for the genechip human mapping 500k array set. Affymetrix, Inc. White Paper; 2006. Tech. rep. [Google Scholar]
- Carter NP. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat Genet. 2007;39(7 Suppl):S16–S21. doi: 10.1038/ng2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho B, Bengtsson H, Speed TP, Irizarry RA. Exploration, normalization, and genotype calls of high-density oligonucleotide SNP array data. Biostatistics. 2007;8(2):485–499. doi: 10.1093/biostatistics/kxl042. [DOI] [PubMed] [Google Scholar]
- Chambers JM. Programming with Data: a guide to the S language. Springer-Verlag; New York: 1998. [Google Scholar]
- Di X, Matsuzaki H, Webster TA, Hubbell E, Liu G, Dong S, Bartell D, Huang J, Chiles R, Yang G, mei Shen M, Kulp D, Kennedy GC, Mei R, Jones KW, Cawley S. Dynamic model based algorithms for screening and genotyping over 100 K SNPs on oligonucleotide microarrays. Bioinformatics. 2005;21(9):1958–1963. doi: 10.1093/bioinformatics/bti275. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JYH, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wei W, Chen J, Zhang J, Liu G, Di X, Mei R, Ishikawa S, Aburatani H, Jones KW, Shapero MH. CARAT: a novel method for allelic detection of DNA copy number changes using high density oligonucleotide arrays. BMC Bioinformatics. 2006;7:83. doi: 10.1186/1471-2105-7-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laframboise T, Harrington D, Weir BA. PLASQ: A Generalized Linear Model-Based Procedure to Determine Allelic Dosage in Cancer Cells from SNP Array Data. Biostatistics. 2006 doi: 10.1093/biostatistics/kxl012. [DOI] [PubMed] [Google Scholar]
- Nannya Y, Sanada M, Nakazaki K, Hosoya N, Wang L, Hangaishi A, Kurokawa M, Chiba S, Bailey DK, Kennedy GC, Ogawa S. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65(14):6071–6079. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- Rabbee N, Speed TP. A genotype calling algorithm for affymetrix SNP arrays. Bioinformatics. 2006;22(1):7–12. doi: 10.1093/bioinformatics/bti741. [DOI] [PubMed] [Google Scholar]
- Scharpf RB, Parmigiani G, Pevsner J, Ruczinski I. Hidden Markov models for the assessment of chromosomal alterations using high-throughput SNP arrays. Annals of Applied Statistics. 2008 doi: 10.1214/07-AOAS155. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharpf RB, Ting JC, Pevsner J, Ruczinski I. SNPchip: R classes and methods for SNP array data. Bioinformatics. 2007;23(5):627–628. doi: 10.1093/bioinformatics/btl638. [DOI] [PubMed] [Google Scholar]