Abstract
Independent component analysis (ICA) is becoming increasingly popular for analyzing functional magnetic resonance imaging (fMRI) data. While ICA has been successfully applied to single-subject analysis, the extension of ICA to group inferences is not straightforward and remains an active topic of research. Current group ICA models, such as the GIFT (Calhoun et al., 2001) and tensor PICA (Beckmann and Smith, 2005), make different assumptions about the underlying structure of the group spatio-temporal processes and are thus estimated using algorithms tailored for the assumed structure, potentially leading to diverging results. To our knowledge, there are currently no methods for assessing the validity of different model structures in real fMRI data and selecting the most appropriate one among various choices. In this paper, we propose a unified framework for estimating and comparing group ICA models with varying spatio-temporal structures. We consider a class of group ICA models that can accommodate different group structures and include existing models, such as the GIFT and tensor PICA, as special cases. We propose a maximum likelihood (ML) approach with a modified Expectation-Maximization (EM) algorithm for the estimation of the proposed class of models. Likelihood ratio tests (LRT) are presented to compare between different group ICA models. The LRT can be used to perform model comparison and selection, to assess the goodness-of-fit of a model in a particular data set, and to test group differences in the fMRI signal time courses between subject subgroups. Simulation studies are conducted to evaluate the performance of the proposed method under varying structures of group spatio-temporal processes. We illustrate our group ICA method using data from an fMRI study that investigates changes in neural processing associated with the regular practice of Zen meditation.
Keywords: Independent component analysis, Multi-subject data, Functional magnetic resonance imaging (fMRI), Group comparison, Maximum likelihood estimation, Likelihood ratio test, EM algorithm
Introduction
Independent component analysis is becoming increasingly popular for analyzing functional neuroimaging data. Compared to the conventional analysis tools such as general linear model (GLM), a key advantage of ICA is that it is a data-driven approach and does not rely on a priori model of brain activity. Therefore, ICA is applicable to cognitive paradigms where prior knowledge of the expected brain time course is not available. ICA could also be used as an exploratory tool to identify and distinguish various types of signals.
ICA has been successfully applied to single-subject fMRI analysis (Beckmann and Smith, 2004; McKeown et al., 1998; Petersen et al., 2000). However, the extension of ICA to group inferences is not as straightforward as in the case of GLM because ICA does not have a pre-specified design matrix, and both the time courses and the spatial maps need to be estimated for each subject (Calhoun and Adali, 2006). Several methods have been proposed to perform group ICA analysis on fMRI data aggregated across multiple subjects. Calhoun et al. developed the GIFT approach (Calhoun et al., 2001), which consists in an initial data-reduction through PCA for each subject, followed by the temporal concatenation of the reduced data across subjects, and a final ICA decomposition of the concatenated data. Back-construction and statistical comparison of individual maps is performed following the ICA estimation. More recently, Beckman and Smith (2005) proposed a tensor probabilistic ICA (PICA) that factors the multi-subject data as a trilinear combination of three outer products, representing the loadings in the temporal, spatial and subject domains, respectively. Tensor PICA is derived from parallel factor analysis (PARAFAC) (Harshman and Lundy, 1984) and it is a natural extension of the two-way product factoring of the single subject ICA, which factors the data as a combination of two outer products of loadings in the temporal and spatial domains. Other group methods that have been proposed include the approach by Svensén and colleagues (Svensén et al., 2002), which concatenates multi-subject fMRI data in the spatial domain and extracts independent components with subject-specific spatial maps associated with common time courses across subjects. Schmithorst and Holland also proposed a group ICA method, which performs PCA reduction and ICA decomposition on the data averaged across subjects (Schmithorst and Holland, 2002).
Among the existing methods, the GIFT and tensor PICA are the most frequently used for performing group ICA analysis of multi-subject fMRI data. These two methods share several similarities: both methods are spatial ICA approaches, i.e., they assume statistical independence of the spatial maps of the extracted components and both methods provide estimation of group spatial maps by performing ICA on the aggregated group data. On the other hand, the GIFT and tensor PICA have important distinctions. A major difference lies in the structure of the group spatio-temporal processes that is assumed in the ICA decomposition. The tensor PICA approach decomposes the multi-subject fMRI data as a trilinear combination of three outer products representing group spatial maps, group time courses and subject loadings. That is, subjects are associated with the same set of group spatial maps and time courses but differ in the magnitude of loading on the group spatio-temporal processes. The GIFT decomposition of the data, on the other hand, implies group spatial maps and subject-specific time courses. Furthermore, the estimation and statistical inference procedure for the GIFT and tensor ICA model are also distinctive and are tailored to their specific model structure. The GIFT approach belongs to the classical noise-free ICA framework which assumes that data are completely characterized by the estimated sources and the mixing matrix. Based on the noise-free assumption, the GIFT reconstructs a subject’s spatial map from the group ICA estimation by multiplying the inverse of the block of the mixing matrix corresponding to the subject with the observed data from the subject. The statistical inference for spatial activation is then performed through a “random effects” inference on the individual maps. The tensor PICA approach is a probabilistic ICA approach which assumes the observed data is the combination of a set of statistically non-Gaussian sources aggregated through the mixing matrix and additive Gaussian noises. Voxel-wise Z-scores are calculated by dividing the estimated spatial maps by the noise standard deviation. The Z-scores are then modeled with the Gaussian/Gamma mixture where the Gaussian component represents the background noise and the Gamma distribution models brain activation. The statistical inference for spatial activation is performed through calculation of the posterior probability for activation based on the mixture model for the voxel-wise Z-scores.
Since the existing group ICA models assume different group structures in the ICA decomposition and their estimation and statistical inference procedures are based on the particular model structure, these methods may produce different results when applied to the same fMRI data. It is desirable to develop a more general framework for group ICA that could accommodate varying structures of group spatio-temporal processes. It is also important to develop a statistical method to select an appropriate group structure for a particular data set.
An important task in analyzing multi-subject fMRI data is to characterize and compare brain activity between subjects from different groups, such as subjects with/without certain psychiatric conditions or subjects assigned to various treatment arms. Current group ICA methods do not take into account subjects’ group identification in the ICA decomposition. Group comparisons are typically performed as post-ICA-estimation analysis often by comparing independent components estimated separately in each group. There are several limitations associated with the existing group comparison approaches. First, when independent components are estimated separately in each group, it is necessary to first identify matching components in different groups. Because independent components are not ordered as principle components, this is often done by identifying independent components in each group that are associated with a pre-specified spatial or temporal template, which requires some prior information on the spatial distribution or temporal dynamics of the underlying source signal (Calhoun et al., 2004). Furthermore, it is possible that spatial ICA may split a spatio-temporal structure into two or more temporally correlated components, which creates another difficulty in selection of matching components from different groups. Recently, Calhoun and colleagues proposed an approach that performs a group ICA on combined data from both subject groups and then reconstruct subject-specific maps and time courses for group comparisons (Calhoun et al., 2008). This new approach avoids the need for matching components between groups. However, the group identification is still not incorporated directly in the ICA decomposition. The second issue with the existing group ICA comparison approaches lies in the interdependence of group comparisons on the temporal and spatial domains. Preferably, group comparison in one of the domains should be performed while controlling for the group difference in the other domain. For example, GLM estimates and compares group spatial map by regressing each subject’s data against the same temporal paradigm. Such approach does not naturally apply to ICA because both the time courses and spatial maps are estimated from data. Within common group ICA comparison approaches (but see (Calhoun et al., 2008)), group independent components are estimated separately in each group and thus differ both in their spatial images and time courses. Hence, group comparison in either the temporal or the spatial domain is confounded by the group difference in the other domain. The above limitations of the existing approaches arise mainly because the group comparisons are performed indirectly as a second-stage analysis after estimating independent components separately in each group. Hence, it is desirable to develop a group ICA method that could directly incorporate subjects’ group information in the ICA decomposition and therefore provide a formal statistical method for group comparisons.
In this paper, we propose a unified framework for fitting group ICA models that are based on varying structures of group spatio-temporal processes. We consider a class of group spatial ICA models, assuming independence in the spatial domain. The proposed models decompose multi-subject fMRI data into group spatial maps and a group mixing matrix that reflects the assumed structure of group spatio-temporal processes, such as the trilinear product structure of tensor PICA. This class of models incorporates existing methods such as the GIFT and tensor PICA as special cases. Furthermore, by specifying an appropriate structure for the group mixing matrix, the proposed model could directly incorporate subjects’ group information in the ICA decomposition. For model estimation and statistical inference, we propose a maximum likelihood approach. Latent spatial source signals are modeled with Gaussian mixture distributions where the various Gaussian components model the probability density of background noise and BOLD effects respectively. We develop a modified EM algorithm to obtain the maximum likelihood estimates of the parameters in the group ICA models.
To evaluate the validity of various structures of group spatio-temporal processes for a particular data set, we present statistical tests for making model comparisons between group ICA models assuming different group structures. The statistical tests are based on the difference in the maximum log-likelihoods under two ICA model structures and are known as the likelihood ratio test (LRT). The LRT can be used to assess the validity of a group structure in an fMRI data set. Furthermore, a statistical test based on the LRT is developed to examine group differences in the spatial modes’ associated time courses between subject groups. We also develop a local LRT for comparisons of different group structures for a subset of independent components. The local LRT is motivated by the fact that ICA components extracted from fMRI data often reflect different kinds of signals including task-related, transiently task-related, physiology-related and artifact-related ones. (Calhoun and Adali, 2006; McKeown and Sejnowski, 1998). Given the varying characteristics of ICA components, it may be desirable to assume a specific group structure only upon components that are of interest or expected to have particular properties while not imposing this structure upon components whose properties are unknown or unlikely to conform to the chosen structure. Another motivation for the local LRT is that we may be interested only in a subset of the signals that are relevant to the study objectives. The local LRT provides more precise evaluation of an assumed group structure on the selected components.
The remainder of this paper is organized as follows. In the Methods section, we introduce aclass of group ICA models and show that existing group ICA models such as the GIFT andtensor PICA can be viewed as special cases within this class. We then present a maximum likelihood (ML) approach with a modified Expectation-Maximization (EM) algorithm for the estimation of the proposed class of models. To compare between group ICA models, we introduce likelihood ratio tests and show how to use the LRT to assess the goodness-of-fit of a group structure in a data set and to test group differences between subject groups. The Results section evaluates the performance of the proposed method on simulated data, and also illustrates its application to real fMRI data from a study investigating changes in neural processing associated with the regular practice of Zen meditation. Finally, a concluding section summarizes and provides further discussion about the presented method and findings.
Methods
In this section, we first describe the class of group ICA models that is able to subsume varying structures of the group spatio-temporal processes. We then present the maximum likelihood approach for model estimation and likelihood ratio tests for performing model comparisons.
A class of group ICA models
In the fMRI application of ICA, the data is usually decomposed as a product of spatial maps and associated time courses. Statistical independence is assumed for either the spatial maps or the time courses, leading to the terminology of spatial ICA or temporal ICA, respectively (Calhoun and Adali, 2006). For fMRI data, spatial ICA has become dominant because the spatial independence assumption is well suited to the sparse distributed nature of the spatial pattern for most cognitive activation paradigms (McKeown and Sejnowski, 1998). In this paper, we also assume spatial independence in our group ICA model. To set notation, let i = 1,…, N index subjects, s = 1,…,T index time points, and v = 1,…,V index voxels. Let Xi be the T ×V matrix representing the observed fMRI data from subject i. We consider the following group ICA model
| (1) |
where is the NT ×V group data matrix formed by concatenating N subjects’ data in the temporal domain, S is q×V matrix containing q statistically independent spatial maps in its rows, is the TN× q group mixing matrix, where Ai is the T×q submatrix corresponding to the i th subject, and is the NT×V noise matrix where Ei is the T×V noise matrix corresponding to the i th subject. Define ev as the v th column of the E matrix representing the N subjects’ noise term at voxel v. The noise term is assumed to follows a zero-mean multivariate Gaussian distribution within each subject and is independent across subjects, i.e. ev ~ MVN(0,IN ⊗ Σv) where Σv is the T×T error covariance matrix for the v th voxel. The ICA model in (1) is referred to as the noisy or probabilistic ICA (Beckmann and Smith, 2004; Hyvarinen et al., 2001) since it includes the Gaussian noise term to account for the background noise that is not represented in the source signals. In noisy ICA, the noise term is generally assumed to be additive and Gaussian distributed since structured noise usually appear in the data as structured non-Gaussian variability and hence can be modeled as one of the source signals (Beckmann and Smith, 2004; Beckmann and Smith, 2005; Hyvarinen,1998; Hyvarinen et al., 2001). Comparing to the classical noise-free ICA, the noisy ICA model can help address the issue of overfitting and provides a convenient framework for formal statistical tests of significance for source signal estimates (Beckmann and Smith, 2004).
Our group ICA model decomposes the multi-subject fMRI data as the product of the group mixing matrix M and the group spatial map matrix S. The group spatial map matrix S is estimated by aggregating information from all subjects’ data and represents the group spatial patterns of brain function. The group mixing matrix M is formed by concatenating the submatrices Ai which represent the mixing processes for the i th subject (i = 1,…, N). The relationship between the subject mixing matrices reflects the nature of the between-subject heterogeneity in the modeled neural activity. Therefore, by specifying different kinds of regularities across subjects for the group mixing matrix, we obtain group ICA models with varying structures of the group spatio-temporal processes. In the following, we consider several special cases within this class of group ICA models.
Connection with existing group ICA methods
By specifying an appropriate structure for the group mixing matrix, the class of group ICA models in (1) relates naturally to some of the existing group ICA methods. For example, the group structure assumed in the GIFT approach is equivalent to the setting of
| (2) |
where Ai is the T×q submatrix corresponding to the i th subject. Since there are no restrictions on the relationship between the subject submatrices, the group structure of the GIFT corresponds to the most general model within our class of group ICA models. Note, however, we note that the GIFT model is different from the proposed group ICA model in that the former is a noise-free classical ICA whereas the latter includes a noise term.
The tensor PICA is another special case within our class of group ICA models. The trilinear product model of the tensor PICA is equivalent to assuming the following Khatri-Rao product structure (Bro, 1998) for the group mixing matrix,
| (3) |
where C is the N×q subject loading matrix with ciℓ representing the i th subject loading on the ℓ th independent component (i = 1,…, N, ℓ = 1,…, q), and A is the T×q matrix containing the group time courses associated with the q independent components. The Khatri-Rao product C|⊗|A is a TN × q matrix formed by N copies of the A matrix stacked and column-wise scaled by the rows of the subject loading matrix C , i.e. C|⊗|A = ((Adiag(C1·))t,…,Adiag(CN·))t)t (Bro, 1998).In other words, we assume that each subject’s mixing matrix is equal to the common mixing matrix A scaled by the subject’s loading vector, i.e.Ai = Adiag(Ci·))t.
Extended group tensor model
In many imaging studies, it is desirable to describe and compare the spatio-temporal processes related to brain activity among subject groups with different characteristics or treatment assignments. Group comparisons are more challenging with ICA than with the standard GLM analysis. In GLM analysis, the subjects’ group identifying labels are incorporated into the design matrix and group-specific estimates of brain activations are readily derived. For ICA, the group information is generally not directly taken into account in the extraction of the independent components and group-specific independent components are typically estimated separately in each group.
Within the model framework of (1), we propose a group ICA model that incorporates the subjects’ group information in the structure of the group mixing matrix and thus factors in the group labels in the ICA decomposition of the multi-group data. For illustration purposes, we present the model for a two-group scenario but the model could easily be generalized for more than two groups. Suppose we collected fMRI data from two groups of subjects. (i = 1,…,N g) denote the T×V fMRI data matrix from subject i in group g (g = 1,2). be the N gT×V data matrix obtained by temporally concatenating the data from the subjects in group g (g = 1,2) and X = [X(1)t, X(2)t]t be the (N1 + N2)T×V data matrix obtained by stacking the data from both groups. We propose a group ICA model within the model framework (1) with the group mixing matrix specified as,
| (4) |
Here A(g) (g = 1,2) is the T×q matrix containing the group time courses associated with the q independent components for group g and C(g) is the Ng × q subject loading matrix for group g where represents the loading of the i th subject in group g on the ℓ th independent component (i = 1,…,Ng , ℓ = 1,…,q). With the group mixing matrix (4), we assume a trilinear product structure for the group spatio-temporal processes within each group, with representative time courses being allowed to be different between the two groups. The proposed model can be viewed as an extension of the tensor PICA model for multi-group data, allowing us to capture different temporal responses between subject groups.
To provide a quick overview of the aforementioned special cases within the proposed group ICA models, we summarize their key properties and defined structure for the group mixing matrix in Table 1 below. We will henceforth refer to these models using the given model names in Table 1.
Table 1.
Descriptions of some important cases of the proposed class of group ICA models.
| Model Name | Definition equation | Properties | Defined structure for the group mixing matrix | Connection with existing methods |
|---|---|---|---|---|
| Tensor Model | Eq. (3) | Each subject’s mixing matrix is equal to the common group time course matrix A scaled by the subject’s loadings, which means that the subjects’ time courses associated with an IC are scaled repetitions of the common group time course. | Tensor PICA (Beckman and Smith,2005) | |
| Group Tensor Model | Eq. (4) | Subjects within a group are associated with a common group time course matrix, with representative time courses being allowed to be different between the two groups. The model can be generalized to multiple groups. | Extension of Tensor PICA for multi-group data | |
| Full Model | Eq. (2) | The most general model within our class of group ICA models with no restrictions on the relationship between the subject mixing matrices. | GIFT (Calhoun et al., 2001) |
Estimation
The proposed group ICA models are estimated using maximum likelihood estimation. The maximum likelihood (ML) approach has been used for single-subject ICA analysis (Belouchrani and Cardoso, 1995; Cichocki et al., 1998; Hyvarinen, 1998; Moulines et al., 1997). Here, we extend the ML method for ICA decomposition of multi-subject fMRI data. The likelihood function is constructed based on a data augmentation scheme where unobserved spatial source signals are treated as missing data. The distribution of the source signals is modeled by a mixture of Gaussian distributions. We developed a modified expectation-maximization (EM) algorithm to obtain the maximum likelihood estimates for these sources.
The complete likelihood function
We first construct the likelihood function for group ICA models in (1). Given the spatial signal sv, the observed multi-subject fMRI data at voxel v follows a multivariate Gaussian distribution with mean Msv and variance I N ⊗ Σ v. Therefore, the complete log-likelihood for the observed data and the latent spatial signals in the group ICA model (1) is as follows
| (5) |
Where ψ(·) is the multivariate Gaussian density function, f(sv;φ) represents the probability density function (pdf) for the spatial signal sv, with φ denoting the parameters involved in the pdf, and θ = (M,{Σv},φ) represents all the parameters in the likelihood function. The complete likelihood in (5) is composed of two parts. The first part represents the likelihood function of the observed fMRI data given the parameters and the spatial signals and the second part is the likelihood for the latent spatial signals based on a prior distribution function. In the following, we discuss the choice of the distribution function for the spatial signals.
Gaussian mixtures for the latent spatial source signals
We follow two major criteria in selecting an appropriate distribution function for the latent spatial signals. First, the distribution should be able to capture the salient features of the underlying spatial source signals; for example, in fMRI most task-related signals are usually attributable to a small percentage of voxels of the brain, whereas the rest of the brain areas are thought to exhibit task-unrelated background fluctuations (Biswal and Ulmer, 1999; Suzuki et al., 2001). Secondly, due to the enormous amount of data in multi-subject fMRI studies, it is desirable to select a distribution that offers numerical efficiency in the estimation of the model parameters. Based on the above two criteria, we model the spatial source signals with a mixture of Gaussian distributions. The Gaussian mixture distribution is well suited to capture different patterns of neural activity across the brain (Xu et al., 1997). Furthermore, the Gaussian mixture offers tractable mathematic properties that facilitate the model estimation, as will be detailed below.
Denote with svℓ the spatial source signal for ℓ th (ℓ = 1,…, q) independent component at voxel v. The pdf of svℓ , based on Gaussian mixture distribution, is,
| (6) |
where m is the number of Gaussian density components in the mixture. Our experiments and previous work (Beckmann and Smith, 2004) suggest that two to three mixtures are typically appropriate to capture the distribution of the spatial signals. is the Gaussian density function with mean μℓj and variance is the proportion of the j th Gaussian density which satisfies represents the parameters associated with the Gaussian mixture distribution for the ℓ th independent spatial source signal. To facilitate our later derivation with the mixture distribution, it is helpful to assume that a latent class variable wvℓ exists, taking values in [1,…,m] with probability p(wvℓ =πℓj) (1 ≤ j ≤ m). The latent class variable wvℓ indicates the membership of voxel v to the Gaussian mixture, i.e. when wvℓ= j, the conditional distribution of svℓ corresponds to the j th Gaussian component in the mixture,
| (7) |
Due to the statistical independence between the components, the joint distribution of q independent components at voxel v equals the product of the pdf of each component,i.e.,
| (8) |
where sv = [sv1 ,… , svq]t and φ = (φ1,…,φq). We can then substitute the Gaussian mixture density function in (8) into the complete log-likelihood function (5).
Modified E-M algorithm
When the likelihood function involves unobserved latent variables, the expectation-maximization (EM) algorithm (Dempster et al., 1977; Dempster et al., 1981) is often used to find maximum likelihood estimates of parameters. The EM algorithm is an iterative algorithm that alternates between performing an expectation step (E-step) and a maximization step (M-step). In the E-step, one computes an expectation of the log-likelihood conditioning on the posterior distribution of latent variables S given the observed data and the current parameter estimates θ̂(k). At the M-step, the updated maximum likelihood estimates of the parametersθ̂(k+1) is computed by maximizing the expected log-likelihood found on the E-step. The parameter estimates θ̂(k+1) found on the M-step are then used to begin another E-step, and the process is iterated until convergence, i.e. until the parameter estimates θ̂(k) and θ̂(k+1) in two consecutive iterations are considered sufficiently close.
Since the log-likelihood in (5) involves unobserved spatial source signals S , we consider the EM framework for model estimation. Due to the special features of the imaging data and our proposed group ICA model, we develop a modified EM algorithm.
At the E-step, the expected log-likelihood function is computed as
| (9) |
To calculate the expectation, we first need to derive the posterior distribution of the latent spatial signals. Given the assumed mixture of Gaussian distributions for sv and the multivariate Gaussian distribution of the data, we show that the posterior distribution of sv is again a mixture of Gaussian distributions,
| (10) |
rth realization of latent labels wv = (wv1,…,wvq, where r = 1,…mq
The expectation of the log-likelihood function is then obtained by integrating the log-likelihood function over the Gaussian mixture posterior distribution. However, the integration in (9) does not have a tractable form. When the E-step is intractable, typical solutions include the Monte Carlo EM (MCEM) algorithm (Wei and Tanner, 1990) or the stochastic EM algorithm (Delyon et al., 1999), which approximate the intractable expectation function by simulating random variables from the posterior distribution. Due to the large number of voxels with imaging data, implementations of these algorithms are practically infeasible. As an alternative, we propose to approximate the intractable expectation function Q(θ | θ̂(k)) using a second-order Taylor expansion approximation, which gives the following explicit form for Q(θ | θ̂(k)),
| (11) |
Our modified E-step does not require iterative numerical simulations and hence is computationally more efficient than the existing algorithms.
At the M-step of the standard EM algorithm, the updated estimates are obtained by maximizing the expected log-likelihood function computed in the E-step,
| (12) |
In our class of group ICA models, the group mixing matrix M may have a specific structure based on the chosen assumption for the group spatio-temporal processes. For example, the Tensor Model assumes that the matrix M can be written as a Khatri-Rho product of the group time course matrix and the subject loading matrix. In order to estimate the group mixing matrix with a specific structure, an additional step is performed after the M-step of the standard E-M algorithm to project the estimated group mixing matrix onto the space of the specified model structure. Continuing with our example, to estimate the group mixing matrix for the Tensor Model such that M^ = C^ |⊗|A^, a rank-1 approximation can be employed (Beckmann and Smith, 2005).
The steps for the proposed modified E-M algorithm are summarized in the following:
start with initial parameter values θ(0)= (M(0),{Σv(0)}, φ(0)). The initial values could be obtained by first analyzing the multi-subject fMRI data using existing group ICA software such as the GIFT (http://icatb.sourceforge.net/) or FSL’s MELODIC routine (http://www.fmrib.ox.ac.uk/fsl/melodic).
modified E-step: calculate Q(θ |θ^(k)) based on a Taylor expansion approximation (11)
M-step: update parameter estimates by maximizing the expectation function obtained through step 2, i.e. θ̂(k+1) arg max θ Q(θ |θ^(k))
project the estimated group mixing matrix M̂(k+1) onto the space specified by the model structure. The step is done by taking each column of M̂(k+1) that corresponds to a particular independent component and projecting it to the specified space.
iterate between steps 2–4 until convergence
After obtaining the parameter estimates θ̂ = (M̂, {Σ̂v}, φ̂) from the modified E-M algorithm, the spatial source signal and its variance can be estimated based on its posterior distribution (10),
| (13) |
| (14) |
The spatial IC maps can then be obtained by plotting the raw IC estimates ŝ v or the IC standardized by their standard deviation ŝv / s .d..(sv), to evaluate the relative activity of various brain regions in each signal.
An important goal in fMRI analysis is to identify "active" voxels, i.e. voxels that display significant changes of activity (positive or negative), typically task -related but not necessarily so (e.g., in resting state studies). We model the spatial source signals using Gaussian mixtures, with different Gaussian components modeling the probability density of background noise and BOLD effects, respectively. In this scheme, the activation of a voxel can be evaluated by calculating the probability for the voxel to belong to the Gaussian component that represents activation. Gaussian components that model the activation vs. background noise could be easily identified by examining the estimated mean associated with each Gaussian. Gaussian with a mean close to 0 represents background noise and Gaussians with mean far away from 0 represent activation corresponding to positive or negative BOLD effects. Suppose that the j1 th Gaussian component models the activation of interest in each signal, the probability of activation at voxel v in the ℓ th signal can be estimated as the posterior probability for the latent class variable wvℓ to point to the j1 th Gaussian component , i.e.,
| (15) |
A statistical inference for the distribution of the activated voxels can be obtained by plotting the thresholded posterior probability of their signal.
Statistical tests between group ICA models
Based on the maximum likelihood method, we can compare various group ICA models using the likelihood ratio test (LRT). The LRT can be used to assess the goodness-of-fit of an assumed spatio-temporal structure in a group data set. Furthermore, we derive a statistical test based on the LRT to test group differences in the fMRI signal time course between subject groups.
Likelihood ratio test
The likelihood ratio test is a statistical test between two nested models in which the simple model is a special case of the general model, in the sense that the general model differs from the simple model only by the addition of one or more parameters. In the likelihood ratio test, the two nested models are framed as a null hypothesis H0 and an alternative hypothesis H1 where the simple model is represented by the null hypothesis H0 and the general model is represented by the combination of the null and alternative hypotheses, i.e. H0 ∪H1. The likelihood ratio test statistic is the difference between the maximum log-likelihoods of the data under the two models. Under the null hypothesis H0 , the likelihood ratio test statistics (LR) approximately follows a Chi-square distribution with degrees of freedom equal to the number of additional parameters in the general model. If the LR is greater than the α upper percentile of the Chi-square distribution with the given degrees of freedom, the null hypothesis is rejected and we conclude that the general model is significantly better supported by the observed data than the simple model.
To perform the likelihood ratio test between nested group ICA models with different structures for the group mixing matrix M, we calculate the marginal likelihood of the observed data by integrating the complete likelihood function with respect to the density of the unobserved spatial source signals. The marginal likelihood for voxel v can be written as:
| (16) |
Hence, the marginal log-likelihood for our class of group ICA models is
| (17) |
where c is a constant term that does not involve the parameters. The LR test statistic for comparing two nested group ICA models is,
| (18) |
where θ̂0 and θ̂1 are the MLE of the parameters under the simple model and the general model, respectively. The LR statistic follows a Chi-square distribution, i.e. LR ~ χ2 (τ) , with the number of degrees of freedom τ given by the difference in the dimensionality of θ̂0 and θ̂1, which essentially equals the difference in the number of parameters in the group mixing matrix under the two ICA models.
Goodness-of-fit test for a group ICA model
Using the LRT, we could evaluate the goodness-of-fit for a specific structure of the group spatio-temporal processes in a multi-subject fMRI data set by comparing the model with the specific group structure against the Full Model. In the following, we present the goodness-of-fit test for the Tensor Model. Similar tests can be performed to evaluate the appropriateness of other group structures.
The Tensor Model assumes the trilinear product structure for the group ICA decomposition. When the assumption of trilinear structure is violated, the validity of the estimated group spatio-temporal processes becomes questionable. To evaluate the goodness-of-fit of the Tensor Model in a data set, we compare the group ICA model with the trilinear product structure against the Full Model using the likelihood ratio test. The hypotheses of the test are:
| (19) |
The null hypothesis Ho states that the group mixing matrix can be expressed as the Khatri-Rho product of the subject loading matrix and the group time course matrix, which is equivalent to specifying a trilinear product structure for the group spatio-temporal processes. The general model in the test, i.e. H0 ∪ H1 , is the Full Model, where the group mixing matrix is equal to the concatenated mixing matrices from the individual subjects, without any assumption of a shared structure among them. The trilinear model is a special case of the Full Model when each subject’s mixing matrix can be expressed as the group time courses matrix column-wise scaled by the subject’s loading vector. Hence, the test is equivalent to testing whether the subjects’ individual mixing matrices satisfy the relationship Ai=A diag(Ci·) for i =1,…,N. The likelihood ratio test statistic is defined in (18) with θ̂0 = {Â, Ĉ, {Σ̂v0}, φ̂0} and θ̂1 = M̂, {Σ̂v}, φ̂} representing the set of estimated parameters for the models under the hypotheses H0 and H0 ∪ H1 , respectively. The LR statistic follows a Chi-square distribution with the number of degrees of freedom equal to the difference in the number of parameters under the two hypotheses, which is τ = TNq −(T + N)q. If , the null hypothesis is rejected and it can be concluded that the data do not satisfy the trilinear product structure.
Testing group differences between subject groups
We hereby derive a statistical test of differences between subject groups. The proposed test is constructed as the likelihood ratio test between the Tensor Model and the Group Tensor Model. In the Tensor Model, the defining assumption is that all subjects are associated with the same set of group spatial maps and group temporal responses. As an extension, the Group Tensor Model (4) allows heterogeneous temporal responses between different subject groups. Hence, by comparing the two models using the LRT, we could examine whether there is a significant difference in the temporal responses between two groups. The hypotheses for the proposed group test are
| (20) |
The Tensor Model in the null hypothesis is a special case of the Group Tensor Model when A(1) = A(2) , i.e. when the group time courses of the two groups are assumed to be the same. Hence, the test is equivalent to testing whether A(1) = A(2). The LRT test statistic is defined as in (18) with θ̂0 = {Â, Ĉ, {Σ̂v0}, φ̂0} and θ̂1 = {Â1, Ĉ1, Â2, Ĉ2, {Σ̂v}, φ̂} representing the maximum likelihood estimates of the parameters under the Tensor Model and the Group Tensor Model, respectively. The difference in the two models is that the null hypothesis provides for only one group time course matrix for all the subjects, while the alternative hypothesis specifies two time course matrices, one for each group. Hence, the number of degrees of freedom of the Chi-square distribution equals the number of parameters in the additional group time course matrix required by the alternative hypothesis, i.e. τ = Tq. If the null hypothesis is rejected and it can be concluded that the temporal responses are significantly different between the two groups.
Local likelihood ratio test for a subset of independent components
Observed fMRI data represent the combination of various kinds of signals including task-related, transiently task-related, physiology-related and artifact-related ones. Since the signals stem from different underlying sources, they may be associated with various types of between-subject variability. For example, the temporal dynamics of task-related signals tend to be more consistent across subjects (when the experimental paradigm is fixed), since the neural activity is temporally locked to the same stimulus presentation schedule for all subjects. Other types of signals, such as those representing physiological fluctuations (e.g., cardiac or respiratory effects on the BOLD signal, or spontaneous mental activity unrelated to the task) are typically more heterogeneous among subjects. Given the varying characteristics of the signals, it may be desirable to assume different group spatio-temporal structures for independent components related to different kinds of signals. For example, one may choose to assume the trilinear product structure only for a subset of independent components (e.g., task-related ones), while not imposing this structure upon components for which it seems unreasonable (e.g., head-motion related ones). The assessment of the goodness-of-fit of the trilinear structure is therefore only relevant to the selected subset of components. Similarly, when testing group differences between subject groups, the interest of the researcher may focus on only those signals that are relevant to the study objectives.
To address the above issues, we propose the local LRT that is targeted to a subset of the independent components. The local LRT allows for comparisons between structures of group spatio-temporal processes for a subset of components. Suppose that a researcher is interested in testing the group structures for q1 independent components where 1 ≤q1 ≤ q. Without loss of generality, it can be assumed that the selected components correspond to the first q1 columns in the group mixing matrix due to the permutation invariance of ICA. Hence, the group mixing matrix can be partitioned into two sub-matrices such that M = [M1,M2] , where M1 is the TN × q1 mixing matrix corresponding to the selected q1 components and M2 is the TN × q2 mixing matrix corresponding to the other components with q2 = q−q1. The local LRT focuses on the comparisons of different group structures for M1 related to the selected components.
As an example, let us consider the local goodness-of-fit test for the trilinear product structure. The hypotheses being tested are:
| (21) |
whereC* is the N × q1 subject loading matrix representing the subjects’ loadings on the q1 selected independent components, A* is the T × q1 matrix containing the group time courses associated with the selected components, and is the T × q1 individual mixing matrix corresponding to the i th subject for the q1 selected independent components. The local LRT test statistic is then constructed based on the difference in the maximum log-likelihood under the hypotheses H0 and H0 ∪ H1 , respectively. The LR statistic follows a Chi-square distribution with the number of degrees of freedom τ = TNq1 − (T + N) q1. If , the null hypothesis is rejected and it can be concluded that the selected subset of independent components does not satisfy the trilinear product structure assumption.
Similarly, the local LRT can be set up to test group differences between subject groups restricted to a subset of selected independent components. The testing hypotheses are in this case:
| (22) |
where A(g)* (g = 1,2) is the T × q1 matrix containing the group time courses associated with the q1 selected independent components for group g and C(g)* is the Ng × q1 subject loading matrix for group g for the selected components. The LR statistic for this test follows a Chi-square distribution with the number of degrees of freedom τ = Tq1.
To perform the local LRTs on a subset of selected components, we need to fit group ICA models with a component-specific group spatio-temporal structure. This can be achieved by identifying, in step 4 of the modified E–M algorithm, the columns in the updated group mixing matrix that correspond to the selected signals and project these columns onto the space specified by the group structure assumed for the selected signals. To identify the column that corresponds to a selected signal, the estimated spatial maps for the independent components can be ranked according to their correlation with a spatial template of the signal of interest.
Data dimension reduction and whitening
ICA analysis usually incorporates several preprocessing steps including centering, dimension reduction and whitening in order to reduce the complexity for the subsequent ICA decomposition. In group ICA analysis of multi-subject fMRI data, two stages of dimension reduction are often performed to reduce the computational load and avoid overfitting (Beckmann and Smith, 2005; Calhoun et al., 2001). At the first stage, a dimension reduction in performed in the temporal domain within each subject. Following Beckman and Smith (2005), we first perform a probabilistic PCA (PPCA) of the group data matrix obtained by spatially concatenating the individual subjects’ data, i.e. YT×NV =[X1 … XN]. Each subject’s data is then projected onto the common subspace spanned by the first R eigenvectors from the PPCA analysis UR , where R is determined using Laplace approximation (Minka, 2000),
| (23) |
where X̃i is the reduced data for the i th subject with the dimensionality of R×V.
The reduced data from all the subjects are then concatenated to form the matrix and a second stage dimension reduction and whitening is performed as in a single subject PICA analysis (Beckmann and Smith, 2004),
| (24) |
where q is the number of independent components extracted from the concatenated dataset X̃ that is again estimated using Laplace approximation, Uq and Λq contain the first q eigenvectors and eigenvalues based on the singular value decomposition of X̃, and the error variance σ2 represents the variability in X̃ accounted by the q independent components and is estimated by the average of the NR−q smallest eigenvalues of X̃.
The two-stage dimension reduction and whitening is equivalent to multiplying the original group fMRI data by the following transformation matrix,
| (25) |
Hence, the group ICA model in (1) can be re-expressed on the reduced and sphered space as following:
| (26) |
where X* = HX,M* = HM, and E* = HE. The transformed noise term in model (26) still follows a multivariate Gaussian distribution, i.e. The new parameter vector for the group ICA model (26) is Note that φ, which represents the parameters related to the Gaussian mixture distribution of the spatial source signals, is not affected by the transformation since the dimension reduction is performed in the temporal domain. The parameter vector θ* can be estimated using the proposed modified EM algorithm. Note that a slight modification is needed in the projection step (Step 4) of the EM algorithm because the group mixing matrix on the reduced dimension is no longer composed of concatenated subject submatrices. Therefore, in Step 4 ,M̂*(k+1) needs first be transformed back to the original scale with is then projected onto a specified subspace and the new estimates are obtained on the reduced dimension as After obtaining the parameter estimates θ̂* from the modified EM, the parameters estimates θ̂ can be computed by back-transforming θ̂* to the original scale.
Simulation Studies
To evaluate the performance of the proposed maximum likelihood method and the likelihood ratio tests for the group ICA models, we conducted two sets of simulation studies. In the first set, we generated a multi-subject dataset for one group of subjects and considered four simulation cases with different structures for the group mixing matrices, representing various types of subject heterogeneity. We fitted the Full Model and the Tensor Model for each simulation case with the proposed maximum likelihood approach. The accuracy of the model estimation was assessed by calculating the correlations between the true and estimated spatial maps and time courses. The goodness-of-fit of the Tensor Model was evaluated through the proposed LRT for each simulation case. In the second set of simulations, we generated data for two groups of subjects. The Tensor Model and Group Tensor Model were fitted and the proposed LRT was applied for testing group differences.
Single-group simulation study
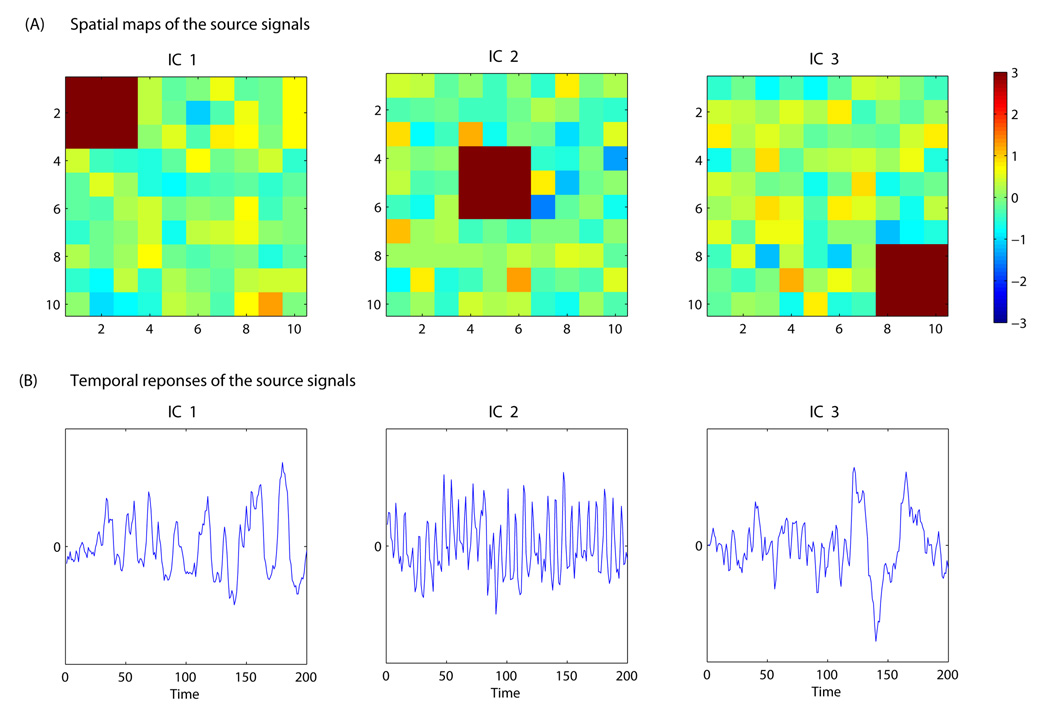
Three source signals were generated with the spatial maps and associated time courses depicted in Figure 1. The time courses in Figure 1 were estimated time sources from the ICA analysis of an fMRI dataset and hence represent realistic temporal dynamics of source signals. While the simulated spatial maps are simpler than typical 3D brain activation images from real fMRI data, they are devised to reproduce the sparse nature of activations in fMRI, i.e. the fact that the source signal is attributable to a small number of voxels while the majority of the voxels exhibit background fluctuations. Gaussian background noises were added to the generated source signals. We simulated data for a group of 9 subjects. We considered four simulation cases with different structures for the group mixing matrix, representing varying types of between-subject heterogeneity:
Figure 1.
Spatial maps and time courses used for the simulation study. Panel (A) presents the spatial maps of the three independent components where the areas in dark red represents activated locations in each component. The spatial signals at other locations represent Gaussian background noise. Panel (B) plots the time courses associated with each of the independent components. The time courses are the estimated temporal responses of independent components extracted from real fMRI data.
Case 0
In this case, we generated data according to the Tensor Model. Hence, the group mixing matrix is the Khatri-Rao product of the subject loading matrix C and a group time series matrixA. The group time series matrix consists of the three time courses as depicted in Figure 1, i.e. A = (t1,t2,t3), which are associated with the three spatial maps. The subjects’ loadings on the three sources are (1, 0.5, 0.3), (0.3, 1, 0.5), (0.5, 0.3, 1), (0.9, 0.4, 0.3), (0.3, 0.9, 0.4),(0.4, 0.3, 0.9), (0.8, 0.4, 0.2), (0.2, 0.8, 0.4), and (0.4, 0.2, 0.8) times the noise standard deviation for the 9 subjects.
Case 1
Each subject’s mixing time course Ai (i = 1,…,9) is the composition of the group time courseA = (t1,t2,t3) in Figure 1 and individualized Gaussian noise. Hence, a subject’s time course associated with each source signal is no longer proportional to the group time course and the group model thus deviates from the Tensor Model.
Case 2
We transformed the group time courses in Figure 1 into the frequency domain where we retained the frequency but randomized the phase for each subject, and then transformed it back to the temporal domain to obtain each subject’s time course. This case is closer to the resting-state fMRI BOLD signals where the temporal responses of different subjects may have similar frequency features but different phase patterns.
Case 3
Each subject contains the three spatial maps in Figure 1 but modulated by individual time courses Ai (i = 1,…,9) that were generated from a Gaussian process and hence do not reflect common underlying temporal dynamics.
Multi-group simulation study
In the second set of simulation study, we generated data from two groups of subjects each with 3 subjects. Here, we considered a relatively small group size so that we can evaluate the performance of the proposed methods in small sample studies. In typical multi-group fMRI studies, the number of subjects in each group is usually larger than 3, resulting in generally better performance in model estimation and statistical tests. The generated data represented the combination of three independent components. The spatial maps of the components are portrayed in Figure 1. Gaussian background noises were added to the spatial source signals. We considered two simulation cases, exemplifying different types of between-group heterogeneity:
Case 1
We generated data according to the Tensor Model. Hence, the group mixing matrix of all subjects is the Khatri-Rao product of the subject loading matrix C and a group time series matrix A. The time series matrix consists of three time courses as depicted in Figure.1, i.e. A = t1, t2, t3, which are associated with the three spatial maps. The subject loading on the three sources are (1, 0.5, 0.3), (0.3, 1, 0.5), (0.5, 0.3, 1), (1, 0.6, 0.8), (0.6, 1, 0.2), and (0.3, 0.4, 1) times the noise standard deviation for the six subjects in the two groups. Note that the same group time course matrix A is shared by subjects of both groups, indicating fMRI signals with the same temporal structure in the two groups.
Case 2
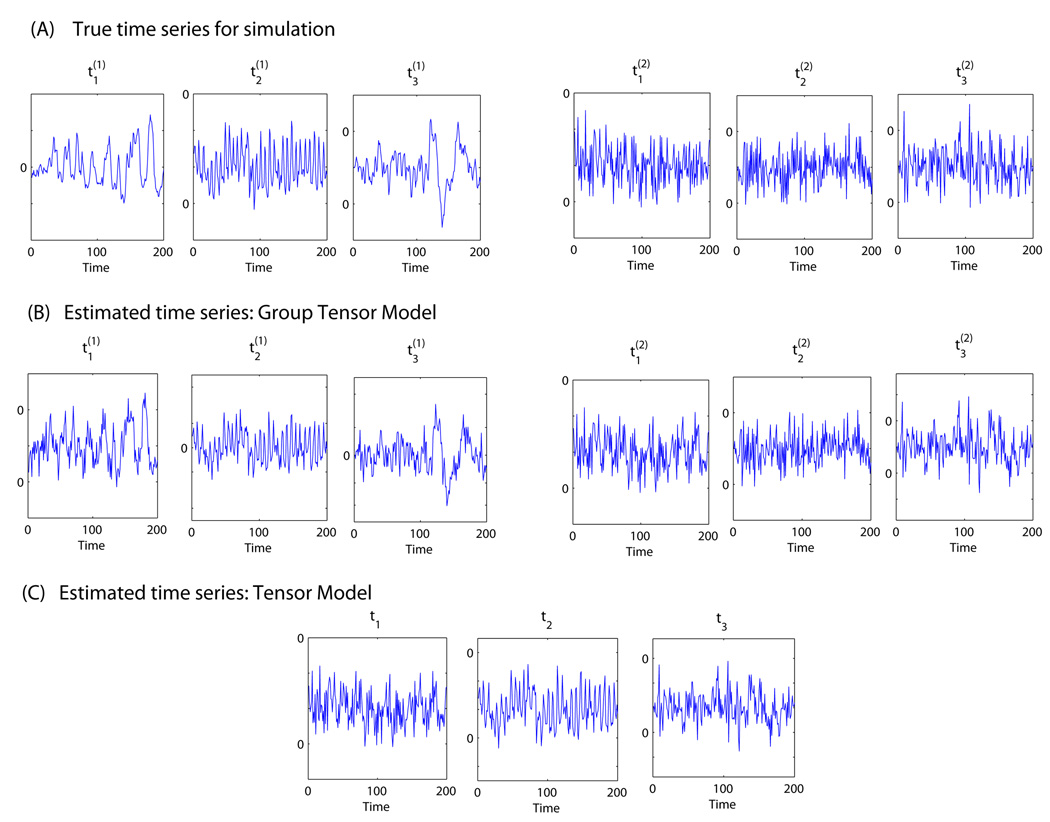
We generated data from the Group Tensor Model. Within group g (g = 1,2), the group mixing matrix of the 3 subjects is the Khatri-Rao product the subject loading matrix C(g) and the group time series matrix A(g). The group time series matrices A(g) =(t1(g) ,t2(g) ,t3(g)) (g = 1,2)contain two different sets of time courses (Figure 3) associated with the three spatial source signals. Therefore, the simulated fMRI time courses are similar within each group but different between the two groups.
Figure 3.
Accuracy of estimation of the two groups’ time courses for Case 2 in the multi-group simulation study based on the Tensor Model and the Group Tensor Model. Panel (A) presents the true time series for the three independent components for group 1 and group 2 ; Panel (B) depicts the estimated time series for the two groups based on Group Tensor Model; Panel (C) presents the estimated group time series based on the Tensor Model. The Group Tensor Model accurately estimates the group time courses from both groups while the Tensor Model only captures the time course from one group due to the restriction in its model assumption.
Results
Simulation studies
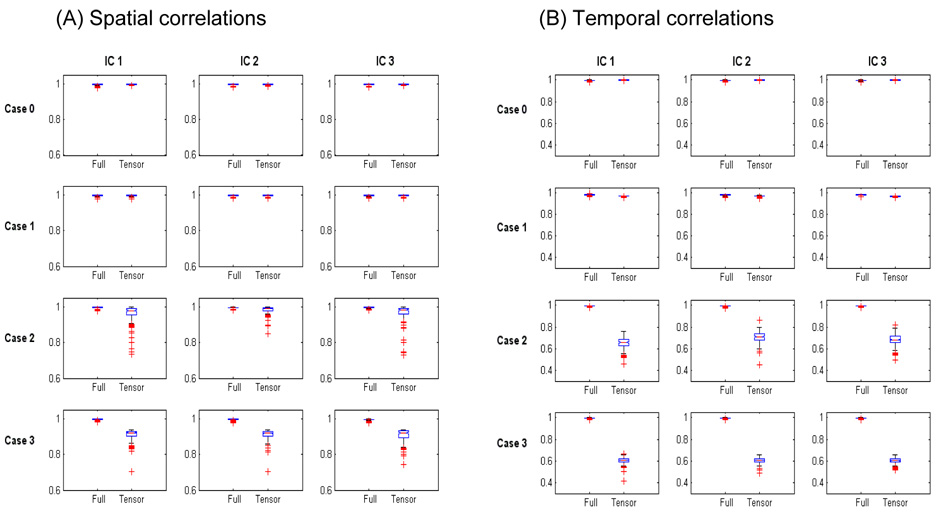
In the following, we compare the performance of different group ICA models under the various types of spatio-temporal structure embedded in the simulated datasets. We first consider the results from the single-group simulation study and compare the results from the Tensor Model and Full Model. The accuracy of the model estimates is measured by the spatial correlations between the estimated and true spatial maps (Figure 2A) and the temporal correlations between the estimated and true time courses (Figure 2B). We then consider the multi-group simulation study where subjects were from two groups and compare the results from the Tensor Model and Group Tensor Model. In the simulation study, the proposed likelihood ratio tests were performed between nested group ICA models. The performance of the LRT was evaluated by the empirical type I error and power of the test. More specifically, we recorded the rejection rate of the LRT, that is, the proportions of simulated data where the LRT rejected the null hypothesis, which is the simple model, in favor of the alternative hypothesis, which is the more general model. The rejection rate is an empirical estimate of the type I error when the null hypothesis is true and the data were generated from the simple model, and an empirical estimate of the power of the LRT when the null hypothesis is false and the data were generated from the general model.
Figure 2.
Accuracy of estimation of the spatial maps and time courses of the three independent components for the single-group simulation study using the maximum likelihood method. Boxplots of correlations between the true and estimated spatial signals (A) and temporal signals (B) for the Full Model (Full) and the Tensor Model (Tensor) within the class of group ICA models. Both the Full and Tensor Models have high accuracy for Case 0 where the true model is the Tensor Model and Case 1 where the true model deviates slightly from the Tensor Model. For Case 2 and 3 where the true model is not related to the Tensor Model, the accuracy of Tensor Model decreases because the structure of the group spatio-temporal processes differ significantly from the assumed trilinear product structure of the Tensor Model. The Full Model still maintains high accuracy in Case 2 and 3 due to its general model structure.
Single-group simulation study
We present the results for the four simulation scenarios in the single-group simulation study. Among the four cases, Case 0 is generated from the Tensor Model and Case 1 to Case 3 represent group ICA models that deviate from the Tensor Model with the degree of deviation increasing from Case 1 to Case 3. Both the Tensor Model and the Full Model were fitted in all cases and their results compared. The proposed LRT were performed to compare the Tensor Model and the Full Model. The rejection rate of the LRT for rejecting the Tensor Model in favor of the Full Model was recorded for each simulation scenario. In Case 0 where the null hypothesis (Tensor Model) is true, the rejection rate of LRT is an empirical estimate of the type I error of the LRT. In Case 1 to Case 3, the rejection rate reflects the power of the LRT for detecting the deviations from the Tensor Model.
Case 0
In this case, where the data were generated from the Tensor Model structure, both the Tensor Model and the Full Model provided accurate estimates of the spatial maps (Figure 2A) and the time courses of the simulated signals (Figure 2B). The box plots show that there were high correlations between the estimated and true signals within the spatial and temporal domains. Across the three simulated independent components, the mean (SD) of the spatial correlations was 0.995 (0.003) and 0.996 (0.003) for the Full Model and Tensor Model, respectively. The mean (SD) of the temporal correlations was 0.996 (0.004) and 0.998 (0.001) for the Full Model and Tensor Model, respectively. The LRT was performed between the two models. Since the data were generated from Tensor Model, the rejection rate of the LRT for rejecting the null hypothesis (Tensor Model) in favor of the alternative hypothesis (Full Model) is an estimate of the type I error. At the significance level of 0.05, the empirical type I error was 0.045, which is close to the nominal level.
Case 1
This model represents a slight deviation from Tensor Model in that each individual’s temporal responses are combinations of the group time courses and individual Gaussian noises. The box plots show that both the Full Model and Tensor Model were still fairly accurate in estimating the spatial maps and the temporal responses associated with each source. Across the three simulated independent components, the mean (SD) of the spatial correlations was 0.995 (0.003) and 0.996 (0.003) for the Full Model and Tensor Model, respectively. The mean (SD) of the temporal correlations was 0.980 (0.005) and 0.970 (0.003) for the Full Model and Tensor Model, respectively. The power of the LRT for detecting the deviation from the Tensor Model in Case 1 was 0.34 at the significance level of 0.05.
Case 2
This case mimics the characteristics of the temporal dynamics observed in resting state fMRI studies. The frequency information of the time courses of each independent component was similar across subjects but the phase of the time courses was randomized across subjects. The box plots show that the Full Model still provided highly accurate estimates of the signals high within both the spatial and temporal domains. In comparison, the accuracy of the Tensor Model was decreased, especially in the temporal domain because the subjects’ temporal responses were quite different from the group time courses associated with each independent component. Across the three simulated independent components, the mean (SD) of the spatial correlations was 0.995 (0.003) and 0.972 (0.034) for the Full Model and the Tensor Model, respectively. The mean (SD) of the temporal correlations was 0.994 (0.004) and 0.680 (0.05) for the Full Model and the Tensor Model, respectively. The power of the LRT for detecting the deviation from the Tensor Model in Case 2 was 0.77 at the significance level of 0.05.
Case 3
In this case, the time courses were generated from Gaussian processes for each subject and hence vary significantly across subjects. The box plots show that the Full Model provided highly accurate estimates of the signals within both the spatial and temporal domains, while the accuracy of the Tensor Model was significantly decreased in both domains because of the considerable deviation from the Tensor Model assumption. Across the three simulated independent components, the mean (SD) of the spatial correlations was 0.995 (0.003) and 0.970 (0.030) for the Full Model and Tensor Model, respectively. The mean (SD) of the temporal correlations was 0.995 (0.004) and 0.602 (0.03) for the Full Model and Tensor Model, respectively. The power of the LRT for detecting the deviation from the Tensor Model in Case 3 was 0.96 at the significance level of 0.05.
Multi-group simulation study
In the following, we describe the results from simulation studies where subjects are from two groups. We fitted both the Tensor Model as well as the Group Tensor Model and compared their results. The proposed LRT were performed between the two models. The LRT rejection rates for the Tensor Model in favor of the Group Tensor Model were assessed for each simulation scenario.
Case 1
The multi-subject data from the two subgroups were generated from the Tensor Model, that is, subjects from both group have similar signal time courses. Since the null hypothesis (Tensor Model) was true in this case, the rejection rate of LRT is an empirical estimate of the type I error of the LRT. At the significance level of 0.05, the empirical type I error was 0.08, which is close to the nominal level. Both the Tensor Model and Group Tensor Model provided accurate estimates of the source signals in the spatial and temporal domains. Across the three simulated independent components, the mean (SD) of the spatial correlations was 0.995 (0.004) and 0.996 (0.002) for the Group Tensor Model and Tensor Model, respectively. The mean (SD) of the temporal correlations was 0.998 (0.001) and 0.998 (0.001) for the Group Tensor Model and Tensor Model, respectively.
Case 2
The data were generated from the Group Tensor Model, with the two groups being characterized by different time courses. Because the null hypothesis (Tensor Model) was not true in this case, the rejection rate of LRT reflects the power of the test for detecting the deviation from the Tensor Model. The empirical power of LRT in Case 2 was 0.97 at the significance level of 0.05. Across the three simulated independent components, the mean (SD) of the spatial correlations was 0.996 (0.002) and 0.988 (0.005) for the Group Tensor Model and Tensor Model, respectively. The mean (SD) of the temporal correlations was 0.998 (0.001) and 0.732 (0.002) for the Group Tensor Model and Tensor Model, respectively. In Figure 3, we plot the true time courses associated with the two subgroups and the estimated time courses from the two models. The results show that the Group Tensor Model captured the difference in the two groups and accurately estimated the temporal responses associated with the three independent components in each group. In comparison, the Tensor Model only captured the temporal response from one group due to the restriction in its model assumption.
Application to real fMRI data
Subjects
We applied our proposed group ICA approach to real fMRI data from a study on the neurobiological correlates of meditation. Twelve Zen meditators with more than 3 years of daily practice (MEDT) were recruited from the local community and meditation centers, along with twelve control subjects (CTRL) who never practiced meditation. The groups were matched for gender (MEDT=10 M, CTRL=9 M), age (mean±SD: MEDT, 37.3±7.2 years; CTRL, 35.3±5.9 years; 2-tailed, 2-sample t-test: p=0.45), and education level (mean±SD: MEDT, 17.8±2.5 years; CTRL, 17.6±1.6 years; p=0.85). All participants were native speakers of English and right-handed, except one meditator who was ambidextrous. Subjects gave written informed consent for a protocol approved by the Emory University Institutional Review Board.
Rationale and design of the experimental task
The practice of Zen meditation is traditionally considered as conducive to a mental state of reduced conceptual processing while full awareness is retained. This state is pragmatically elicited by heedfulness to distraction from spontaneous thoughts, usually by adopting a reference attentional object such as one’s own breathing. We hypothesized that habitual meditators would exhibit an enhanced capacity to voluntarily moderate intensity and duration of the automatic conceptual processing kindled by the presentation of semantic stimuli, compared to control subjects. Under this hypothesis, we expected differential patterns of neural responses to semantic stimuli during meditation in control subject and habitual meditators.
To test this hypothesis, we adapted a simple lexical decision paradigm from Binder and colleagues (Binder et al., 2003) employing semantic and nonsemantic stimuli: 50 word and 50 phonologically and orthographically matched nonword items, were presented visually on a screen in pseudo-random temporal order and subjects were asked to respond whether the displayed item was "a real English word" via an MRI-compatible button-box with their left hand (index finger = "yes", middle finger = "no"). Subjects were instructed to use the awareness of their breathing throughout the session as a reference point to monitor and counteract attentional lapses. Whenever they would catch themselves mind-wandering and having lost their attentional focus, they were to immediately return their attention to their breathing. They were also told that when a stimulus (word/nonword) appeared on the screen, they should perform the lexical decision task as accurately as possible and return their attention to breathing without any further lingering. The experimental task can be thus conceived as having a dual-layer structure: an ongoing meditative baseline condition and a phasic perturbation of this baseline by semantic and nonsemantic stimuli. Importantly, given the length of the imaging run (~ 20 min), the schedule of stimulus presentation was sparse enough to allow a reasonable establishment of the “meditative” attentional refocusing in between the stimuli (inter-stimulus interval: range 1.4–72.9 s, median=8.2 s, IQR=12.9 s).
MRI acquisition and preprocessing
A T1-weighted high-resolution anatomical image (MPRAGE, 176 sagittal slices, voxel size: 1×1×1 mm) and a single series of functional images (echo-planar, 520 scans, 35 axial slices, voxel size: 3×3×3 mm, TR=2.35 s, TE=30), were acquired with a 3.0 Tesla Siemens Magnetom Trio scanner. The functional volumes were corrected for slice acquisition timing differences and subject movement. The anatomical image was registered to the mean of the corrected functional images and subsequently spatially normalized to the MNI standard brain space by using the segmentation routine of SPM5 (http://fil.ion.ucl.ac.uk/spm/software/spm5). The computed MNI-normalization parameters were then applied to the functional images, which were finally smoothed with an 8mm isotropic Gaussian kernel. Multi-subject fMRI data were concatenated spatially and a probabilistic PCA (PPCA) was performed on the mean covariance matrix. Based on the Laplace approximation, each subject’s data was projected onto the common subspace spanned by the first 40 eigenvectors from the PPCA.
Testing group differences between meditators and controls
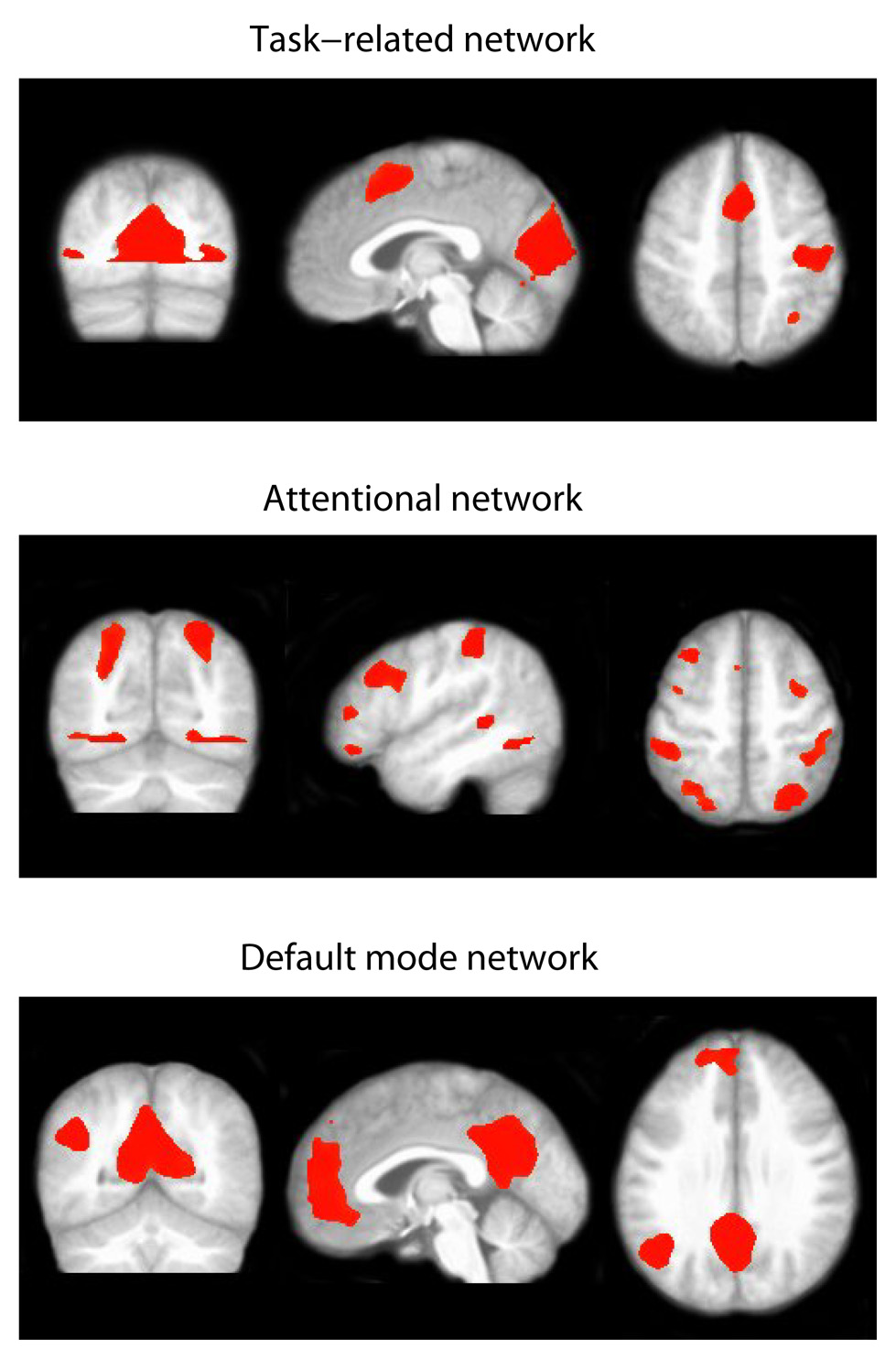
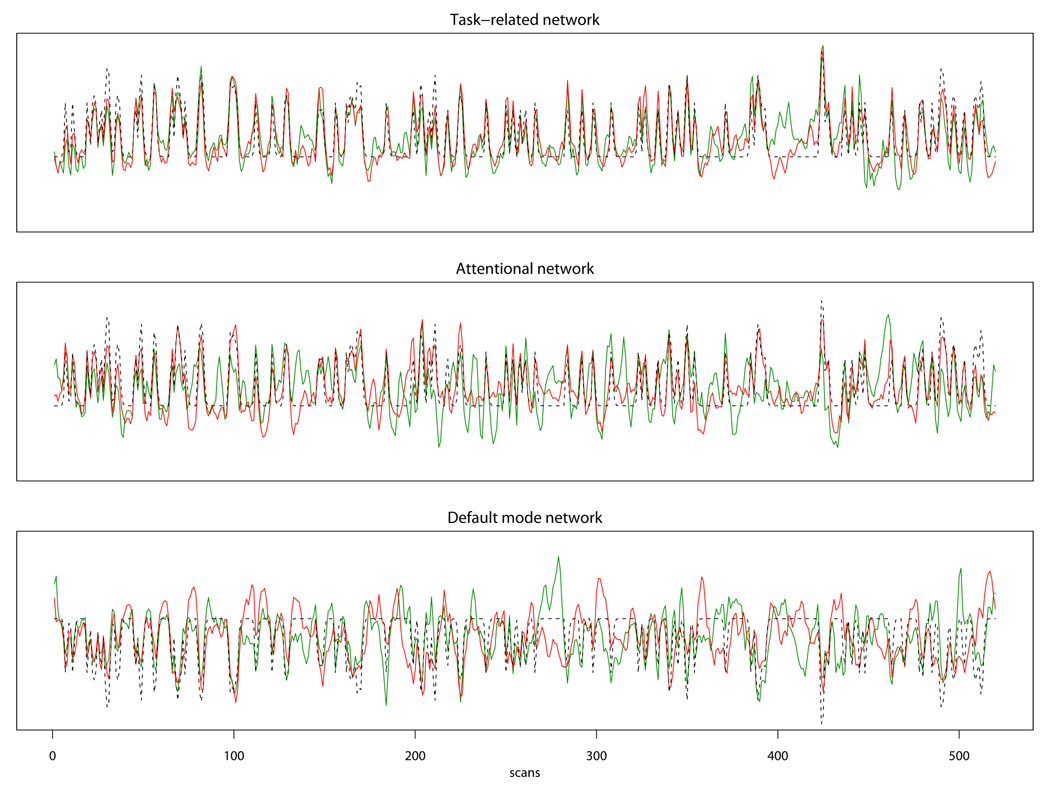
To test our hypothesis that meditators and control subjects demonstrate differential temporal patterns of neural activity related to the processing of the stimuli, we considered the Tensor Model and the Group Tensor Model, and performed the LRT to compare the two models. The Tensor Model assumes that subjects from both the meditation and control groups share the same sets of group activity time courses while the Group Tensor Model allows different time courses between the two groups. The LRT test between the two models showed that the Group Tensor Model provided a significantly better fit to the data than the Tensor Model (p=0.039), indicating a significant difference in the temporal profiles of neural activity of meditators and control subjects. The Group Tensor Model was therefore chosen for this data set. Based on the Laplace approximation, fourteen independent components were extracted. By examining the estimated spatial maps and temporal responses of the extracted fourteen components, we selected three spatio-temporal modes of interest on the basis of the putative neural systems involved in meditation and in the execution of the experimental task. The first network includes the supplementary motor area (SMA), the hand region of the right sensorimotor cortex contralateral to the (left) hand pressing the button box, and the visual cortex, and is clearly functionally related to the sensorimotor response to the lexical decision task. The second network is a fronto-parietal system including the bilateral intraparietal sulcus and the supplementary eye fields, which is consistent with the general architecture of attentional function (Corbetta and Shulman, 2002); this network was selected for the central role of attentional control in the meditative exercise. The third network represents what has come to be known as the “default mode network” (Raichle et al., 2001), a set of regions including the posterior cingulate, the medial prefrontal cortex, the lateral parietal cortex, and the hippocampi, which is characterized by high metabolism at rest and decreased activity during a variety of demanding tasks (Gusnard and Raichle, 2001); of particular interest, the default mode network has been recently implicated in the spontaneous generation of unconstrained thoughts during mind-wandering (Binder et al., 1999; Christoff et al., 2004; Mason et al., 2007), and was selected here for the purported capacity of meditative training to moderate the distracting influence of spontaneous thoughts. Spatial maps showing activated brain regions are presented in Figure 4, where the active voxels are those with an estimated posterior probability of activation exceeding 0.90.
Figure 4.
Thresholded activation maps for three selected independent components based on the Group Tensor Model for the Zen meditation fMRI data. Voxels with a posterior probability of activation exceeding 0.9 are labeled active. The three components correspond to the task-related network, attention network and default mode network.
Testing group differences for selected components: local LRT
After determining the appropriate group ICA model for analyzing an fMRI data set, we can perform further analyses by examining a selected set of independent components that are particularly relevant to the study aims. In the present case, we focused on three independent components whose spatial modes are displayed in Figure 4 and whose associated time courses for the control and the meditation groups are shown in Figure 5 (for the estimated stimulus-locked response in each network, see Figure 1 in Supplementary Material). The temporal correlations between the estimated time courses of the two groups were 0.84 for the task-related network, 0.51 for the attention network, and 0.36 for the default mode network. To examine whether there was a significant group difference in the time courses of the selected networks, we performed the local LRT test for each of the selected independent components and used Bonferroni correction to adjust for multiple testing. The results showed that while the time courses associated with the task-related network in the two groups were not statistically significantly different (p=0.054), meditators and controls were characterized by distinct temporal dynamics for the attention network (p=0.005) and the default mode network (p<0.001). We also evaluated the correlations between the IC time courses and a time series representing the expected neural response to the task stimuli (words and nonwords), i.e., the vector of delta functions corresponding to the occurrence of the task stimuli (words and nonwords) convolved with a gamma-function model of the hemodynamic response (Cohen, 1997). The correlations between the time course of the task-related network and the task time series were 0.78 and 0.86 for the control group and the meditation group, respectively; the correlations between the time course of the attention network and the task time series were 0.48 (CTRL) and 0.85 (MEDT), indicating that the activity profile of the attention network was more strongly associated with the timing of the experimental stimuli in meditators than in controls; the correlations between the time course of the default mode network and the task time series were −0.66 (CTRL) and −0.62 (MEDT).
Figure 5.
Estimated time courses for three selected independent components based on the Group Tensor Model for the Zen meditation fMRI data. The red lines correspond to the estimated time courses for the meditation group; the green lines correspond to the control group and the dotted lines are the task time series convolved with the HRF function (Note: since the default mode network is deactivated during tasks, we reverse the sign of the task time series in the third panel for depiction purpose). Local likelihood ratio tests show that the time courses associated with the task-related network are not significantly different between the meditation and control groups (p=0.054), meditators and controls are characterized by distinct temporal dynamics for the attention network (p=0.005) and the default mode network (p<0.001).
Discussion
We have presented a unified framework for group ICA analysis of multi-subject fMRI data, by introducing a class of group ICA models that is able to (1) accommodate varying types of structures of group spatio-temporal processes, including those assumed by existing group ICA methods such as the GIFT and tensor PICA; (2) provide a formal statistical framework for group comparisons by incorporating group identification in the ICA decomposition; and (3) allow component-specific group structures to provide more a flexible fit for fMRI signals related to different sources. A general maximum likelihood approach is employed for estimating group ICA models with varying model structures. The proposed group ICA method provides likelihood ratio tests to compare between group ICA models. Using the LRTs, we can perform model comparison and selection, assess the goodness-of-fit of an assumed group structure, and test group differences in fMRI signal time courses between subject subpopulations.
In the proposed method, the distribution of the spatial signals is modeled by a Gaussian mixture, on the rationale that most signals of interest are reasonably confined to a limited subset of the entire anatomical brain space. A Gaussian mixture employs different Gaussian components to capture the distribution of the small proportion of the activated voxels and the distribution of the majority of the brain areas that are not strongly related to the signal, and is therefore appropriate for modeling fMRI data. Furthermore, a mixture of Gaussians is a general distribution that can accommodate various types of source densities. For example, previous work (Xu et al., 1997) and our experiments have shown that Gaussian mixtures can model and separate sub- and super-Gaussian sources. Compared to alternative distributions for the latent source signals (Hong et al., 2005), Gaussian mixtures also have the major advantage of tractable mathematical properties. For example, the posterior distribution of the source signals given the observed fMRI data conforms to a Gaussian mixture which facilitates statistical estimation and inference.
In order to obtain the maximum likelihood estimates of the group ICA models, we have devloped a modified EM algorithm. Compared to standard EM, the modified EM algorithm includes an additional step to project the estimated components into the subspaces spanned by an assumed group structure, hence providing a general method for estimating group ICA models with varying model structures. The Likelihood ratio test statistics can also be obtained as a by-product from the modified EM algorithm to provide statistical tests between different group ICA models. The tradeoff with the modified EM is that it requires a longer computation time than some of the existing methods, due to the slow convergence of the EM algorithm. Typically, the modified EM algorithm takes approximately 10–20 times of total estimation time compared to the GIFT and tensor PICA.
An important aspect of the imaging analysis is the inference about the statistical significance of detected brain networks, which typically requires a secondary analysis of the estimated spatial maps after the ICA decomposition. For example, the GIFT identifies significantly activated brain areas by performing a “random effects” inference on the individual maps that are reconstructed from the group ICA estimation. Tensor PICA makes statistical inference on spatial activation by fitting Gamma-Gaussian mixture distributions to the estimated spatial Z-scores maps and thresholding the posterior probability of activation for each voxel. Using our proposed Gaussian mixture likelihood methods, we are able to simultaneously estimate the spatial source signals, the variability of the signals, and also the posterior probability of activation at each voxel which can be readily thresholded to identify significantly activated brain regions in source signals.
Finally, we have proposed likelihood ratio tests to compare group ICA models with different model structures. Depending on the investigators’ objectives, the proposed LRT can be performed either at global scale for all identified independent components, or at local scale for only a subset of the components. The global LRT provides an overall evaluation of the validity of the assumed group structure for source signals underlying the observed fMRI data. On the other hand, the local LRT allows testing the validity of the assumed structure on just a subset of independent components. The local LRT thus provides a more precise statistical tool when researchers are particularly interested in a specific selection of the components.
ACKNOWLEDGMENTS
The authors thank the referees and editor for helpful comments. Y.G. was partially supported by the National Institute of Mental Health (NIH grant R01-MH079251). The imaging data were collected as part of a pilot study awarded to G.P. by the Emory Center for Research on Complementary and Alternative Medicine in Neurodegenerative Diseases (NIH grant P30-AT00609)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beckmann CF, Smith SM. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging. 2004;23:137–152. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, Smith SM. Tensorial extensions of independent component analysis for multisubject FMRI analysis. Neuroimage. 2005;25:294–311. doi: 10.1016/j.neuroimage.2004.10.043. [DOI] [PubMed] [Google Scholar]
- Belouchrani A, Cardoso J-F. Maximum likelihood source separation by the expectation-maximum technique: deterministic and stochastic implementation; Proc. Int. Symp. on Nonlinear Theory and its Applications; Las Vegas: Nevada; 1995. [Google Scholar]
- Binder JR, Frost JA, Hammeke TA, Bellgowan PS, Rao SM, Cox RW A functional MRI study. Conceptual processing during the conscious resting state. J Cogn Neurosci. 1999;11:80–95. doi: 10.1162/089892999563265. [DOI] [PubMed] [Google Scholar]
- Binder JR, McKiernan KA, Parsons ME, Westbury CF, Possing ET, Kaufman JN, Buchanan L. Neural correlates of lexical access during visual word recognition. J Cogn Neurosci. 2003;15:372–393. doi: 10.1162/089892903321593108. [DOI] [PubMed] [Google Scholar]
- Biswal BB, Ulmer JL. Blind source separation of multiple signal sources of fMRI data sets using independent component analysis. J Comput Assist Tomogr. 1999;23:265–271. doi: 10.1097/00004728-199903000-00016. [DOI] [PubMed] [Google Scholar]
- Bro R. Models, Algorithms, and Applications. University of Amsterdam; 1998. Multi-Way Analysis in the Food Industry. [Google Scholar]
- Calhoun VD, Adali T. Unmixing fMRI with independent component analysis. IEEE Eng Med Biol Mag. 2006;25:79–90. doi: 10.1109/memb.2006.1607672. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Human Brain Mapping. 2001;14:140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pekar JJ. A method for comparing group fMRI data using independent component analysis: application to visual, motor and visuomotor tasks. Magn Reson Imaging. 2004;22:1181–1191. doi: 10.1016/j.mri.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Pearlson GD, Maciejewski P, Kiehl aKA. Temporal Lobe and 'Default' Hemodynamic Brain Modes Discriminate Between Schizophrenia and Bipolar Disorder. Human Brain Mapping. 2008 doi: 10.1002/hbm.20463. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoff K, Ream JM, Gabrieli JD. Neural basis of spontaneous thought processes. Cortex. 2004;40:623–630. doi: 10.1016/s0010-9452(08)70158-8. [DOI] [PubMed] [Google Scholar]
- Cichocki A, Douglas SC, Amari SI. Robust techniques for independent component analysis with noisy data. Neurocomputing. 1998;22:113–129. [Google Scholar]
- Cohen MS. Parametric analysis of fMRI data using linear systems methods. Neuroimage. 1997;6:93–103. doi: 10.1006/nimg.1997.0278. [DOI] [PubMed] [Google Scholar]
- Corbetta M, Shulman GL. Control of goal-directed and stimulus-driven attention in the brain. Nat Rev Neurosci. 2002;3:201–215. doi: 10.1038/nrn755. [DOI] [PubMed] [Google Scholar]
- Delyon B, Lavielle V, Moulines E. Convergence of a stochastic approximation version of the EM algorithm. Annals of Statistics. 1999;27:94–128. [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. Maximum Likelihood from Incomplete Data Via Em Algorithm. Journal of the Royal Statistical Society Series B-Methodological. 1977;39:1–38. [Google Scholar]
- Dempster AP, Rubin DB, Tsutakawa RK. Estimation in Covariance Components Models. Journal of the American Statistical Association. 1981;76:341–353. [Google Scholar]
- Gusnard DA, Raichle ME. Searching for a baseline: functional imaging and the resting human brain. Nat Rev Neurosci. 2001;2:685–694. doi: 10.1038/35094500. [DOI] [PubMed] [Google Scholar]
- Harshman R, Lundy M. Research methods for multimode data analysis. New York: Praeger; 1984. The PARAFAC model for three-way factor analysis and multidimensional scaling; pp. 122–215. [Google Scholar]
- Hong B, Pearlson GD, Calhoun VD. Source density-driven independent component analysis approach for fMRI data. Hum Brain Mapp. 2005;25:297–307. doi: 10.1002/hbm.20100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvarinen A. Independent component analysis in the presence of gaussian noise by maximizing joing likelihood. Neurocomputing. 1998;22:49–67. [Google Scholar]
- Hyvarinen A, Karhunen J, Oja E. Independent Component Analysis. New York: Wiley; 2001. [Google Scholar]
- Mason MF, Norton MI, Horn JDV, Wegner DM, Grafton ST, Macrae CN. Wandering minds: the default network and stimulus-independent thought. Science. 2007;315:393–395. doi: 10.1126/science.1131295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown MJ, Makeig S, Brown GG, Jung TP, Kindermann SS, Bell AJ, Sejnowski TJ. Analysis of fMRI data by blind separation into independent spatial components. Hum Brain Mapp. 1998;6:160–188. doi: 10.1002/(SICI)1097-0193(1998)6:3<160::AID-HBM5>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown MJ, Sejnowski TJ. Independent component analysis of fMRI data: examining the assumptions. Hum Brain Mapp. 1998;6:368–372. doi: 10.1002/(SICI)1097-0193(1998)6:5/6<368::AID-HBM7>3.0.CO;2-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minka T. Technical Report. Vol. 514. MIT Media Lab.; 2000. Automatic choice of dimensionality for PCA. [Google Scholar]
- Moulines É, Cardoso J-F, Gassiat E. Maximum Likelihood For Blind Separation And Deconvolution Of Noisy Signals Using Mixture Models. Proc. ICASSP, Munich. 1997:3617–3620. [Google Scholar]
- Petersen KS, Hansen LK, Kolenda T, Rostrup E, Strother SC. On the independent components of functional neuroimage; Proc. Int. Conf. on Independent Component Analysis and Blind Signal Separation; 2000. pp. 615–620. [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc Natl Acad Sci USA. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmithorst V, Holland S. Multiple networks recruited during a story processing task found using group inferences across subjects from independent component analysis the 10th Annual Meeting of ISMRM; Honolulu, HI. 2002. p. 754. [Google Scholar]
- Suzuki K, Kiryu T, Nakada T. Fast and precise independent component analysis for high field fMRI time series tailored using prior information on spatiotemporal structure. Human Brain Mapping. 2001;15:54–66. doi: 10.1002/hbm.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensén M, Kruggel F, Benali H. ICA of fMRI group study data. Neuroimage. 2002;16:551–563. doi: 10.1006/nimg.2002.1122. [DOI] [PubMed] [Google Scholar]
- Wei GCG, Tanner MA. A Monte-Carlo Implementation of the Em Algorithm and the Poor Mans Data Augmentation Algorithms. Journal of the American Statistical Association. 1990;85:699–704. [Google Scholar]
- Xu L, Cheung C, Yang H, Amari S. Maximum equalization by entropy maximization and mixture of cumulative distribution functions. Proc. Of ICNN’97, Houston. 1997:1821–1826. [Google Scholar]