Abstract
The ability to quantify or visualize newly synthesized proteins has important uses in cell biology. For example, a researcher may wish to quantify basal or inducible rates of translation of a specific gene of interest, or detect subcellular locations of newly synthesized copies of a protein in order to study the role of new protein synthesis in the growth of specialized cellular structures. In this unit, we describe the TimeSTAMP method for labeling of newly synthesized copies of a protein of interest. In the TimeSTAMP method, the experimenter expresses a protein of interest as a fusion with a cis-acting protease and an epitope tag, both of which are removed by protease activity by default. Addition of a specific protease inhibitor then allows preservation of the tag on subsequently synthesized proteins. Finally, the tag is detected by immunological methods.
INTRODUCTION
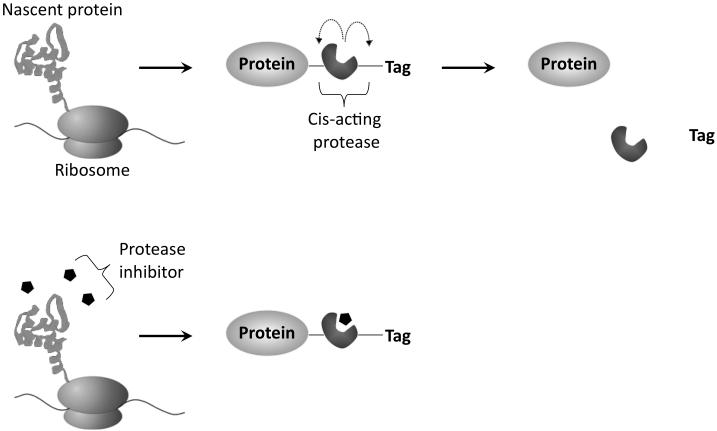
This unit describes a protocol for carrying out the “TimeSTAMP” method of tagging newly synthesized copies of a protein of interest in a temporally regulated manner (Lin et al., 2008). Here, a protein of interest is produced as a fusion to an epitope tag, wherein the epitope tag is attached via a cassette composed of the hepatitis C virus (HCV) protease flanked on both sites by cognate protease recognition sites (Figure 1). This fusion construct rapidly undergoes cis cleavage by the HCV protease by default, resulting in removal of the tag from the protein of interest. After a chosen time, tags can be preserved on newly synthesized proteins by the simple administration of the cell-permeable small-molecule HCV protease inhibitor BILN-2061. When applied at low micromolar concentrations, BILN-2061 is able to bind to and inhibit protease activity before tag removal occurs. After lysate preparation, amounts of new and old proteins can be quantified by immunoblotting. Alternatively, after fixation, locations of new proteins can be visualized by immunocytochemistry.
Figure 1.
Schematic of the TimeSTAMP strategy. A protein of interest is expressed as a fusion with a cis-acting protease and an epitope tag, both of which are removed by protease activity by default. Addition of a specific protease inhibitor then allows selective preservation of the tag on subsequently synthesized proteins.
Basic Protocol: Testing and use of TimeSTAMP tags by immunoblotting
Immunoblotting provides a straightforward way to test that the TimeSTAMP cassette is operating properly when fused to a protein of interest. It also provides a quantitative method for measuring rates of protein synthesis from a reporter construct within a defined time window under different stimulation or treatment conditions.
Materials
TimeSTAMP cassette DNA (from the authors: Michael Z. Lin, mzlin@stanford.edu, linlab.stanford.edu; Roger Y. Tsien, rtsien@ucsd.edu, tsienlab.ucsd.edu)
BILN-2061 (Boehringer Ingelheim), 10-30mM in DMSO
reagents for cloning and PCR
cell type of interest
cell culture media
transfection reagent
2x SDS lysis buffer
2-mercaptoethanol
PBS or HBSS
sonicator with microtip, or 25U/μL benzonase nuclease (Novagen)
SDS-PAGE and immunoblotting equipment and reagents
rat 2F2 (Roche) or mouse 12CA5 (Roche) or mouse HA-7 (Sigma) or chicken anti-HA (ICL) antibody
mouse anti-T7 antibody (Novagen)
secondary antibodies for detection by chemiluminescence or fluorescence chemiluminescence substrate (if performing chemiluminescence)
autoradiography film (for chemiluminescence) or immunoblot imaging system (for chemiluminescence and fluorescence)
Construct a TimeSTAMP fusion to the gene of interest
1. Identify whether the protein of interest is best tagged at its N-terminus or C-terminus.
A terminus that is unlikely to interfere with the activity or localization of the protein should be selected. (See “Critical Parameters.”)
2. Obtain an expression plasmid for the protein of interest.
Depending on the nature of the experiment, the promoter could be a native promoter (such as that of the gene for the protein of interest), a constitutive viral promoter (such as CMV), or an artificial promoter (such as a tet-responsive promoter). Other elements should be included depending on the nature of the experiment, e.g. introns or untranslated regions if the investigator wishes to reconstitute regulation of mRNA splicing or transport.
3. Select for fusion either the TimeSTAMPa or TimeSTAMPt cassette, for analysis of proteins undergoing low or high rates of synthesis, respectively, or test both versions.
The TimeSTAMPa cassette is typically used for low abundance proteins or proteins with slow ongoing synthesis rates whereas the TimeSTAMPt is typically used for highly expressed proteins or proteins with faster ongoing synthesis rates. For proteins whose characteristics are not well understood, both versions can be tried in parallel. (See “Critical Parameters.”)
4. Subclone each complete TimeSTAMP cassette in frame to either the N- or C-terminus of the gene.
The complete cassette consists of N-terminal T7 epitope tag, N-terminal cleavage site, NS3 protease, C-terminal cleavage site, and C-terminal HA tag. In the original TimeSTAMP cassettes, SrfI and AscI restriction sites can be used to transfer the entire cassette, if SrfI and AscI sites are first inserted into the beginning or end of the coding region of the gene of interest. The proper translation frames reading through the SrfI and AscI sites are +2 and +1, where the first nucleotide of the +1 frame is the first nucleotide of the recognition site (Figure 1). Alternative sites that do not exist in the sequence of the TimeSTAMP cassette (available from the authors) can also be used, in which case the TimeSTAMP cassette can be amplified with PCR with primers containing these sites in place of SrfI and AscI.
5. Verify that the cloning junctions and TimeSTAMP sequences are free of unwanted mutations by sequencing, and prepare plasmid DNA for transfection by anion-exchange chromatography.
Commercial anion-exchange chromatography “maxiprep” kits such as those available from Qiagen or Invitrogen are preferred. Silica adsorption chromatography as found in most “miniprep” kits and some “maxiprep” kits do not yield as consistent results in transfections.
Verify proper functioning of the TimeSTAMP cassette
6. Grow a transfectable cell type to the proper density for transfection in a 12-well plate. Prepare two wells for each construct to be tested, plus one well to serve as an untransfected control.
For instance, HEK293 or 293T cells are reliably transfectable at 80-90% confluence by commercial liposomal reagents such as Lipofectamine 2000 (Invitrogen) or Fugene (Roche), or by calcium phosphate. Two wells are required per construct, as one well will be treated with BILN-2061 and the other left untreated.
7. For each construct to be tested, transfect the two allocated wells with 0.8 μg DNA each with a liposomal reagent or with calcium phosphate.
Perform liposomal or calcium phosphate transfections according to standard protocols, except use only 0.8 μg of DNA to avoid toxicity, adjusting all other volumes in the transfection mixture accordingly.
8. For each construct being tested, prewarm two tubes each of 1 mL of standard culture media with serum. To one tube, add BILN-2061 to a final concentration of 3 μM. Two hours after addition of transfection reagent to cells, for each construct, remove the media in the two wells. To one well, add the 1mL of media without BILN-2061, while to the other well, add the media without BILN-2061. Return the plate to the incubator for 24 hours.
If HEK293 or 293T cells are being used, apply new solutions slowly to the wells in order to prevent detaching the cells from the surface.
9. For each construct, prepare 500 μL 2x reducing SDS lysis buffer by adding 2-mercaptoethanol to 2x SDS lysis buffer to a final volume of 10%, then heating to 85-95° C in a chemical safety flow hood. Prepare for each well 1 mL of wash buffer in the form of PBS or HBSS.
A hot solution of SDS and 2-mercaptoethanol, rather than a non-ionic detergent solution, is used to lyse the cells, as this solution causes immediate denaturation of cellular proteins. Destruction of the protease activity within the TimeSTAMP cassette allows the omission of BILN-2061 from the lysis buffer.
10. Remove the transfected cells from the incubator, then, working quickly, replace the media in each well with 1mL wash buffer, dispensing the wash buffer gently onto the wall of the well so as not to wash the cells off the surface. After accomplishing this for all wells, quickly aspirate away the wash buffer. Immediately move the plate to the hood, then quickly dispense into the center of each well 200 μL of the 2x reducing SDS lysis buffer. After lysis buffer has been dispensed into all the wells, agitate the plate to spread the lysis buffer around the entire surface of the wells.
The purpose of the wash step is to remove serum components, primarily albumin, whose presence in the lysate causes anomalous migration during electrophoresis and ghosting during immunoblotting. It is important to work quickly from the time wash buffer is applied to the cells until it is removed. The wash buffer does not contain BILN-2061, so BILN-2061 will begin to diffuse out of cells during the wash. This is not a concern as long as the wash is less than 1 minute in duration, as a negligible proportion of the complex of NS3 and BILN-2061 will dissociate in this time (Flores et al., 2009).
11. With the plate tilted, scrape down the lysate, which will be viscous, to the bottom of each well, then transfer into a 1.5mL microcentrifuge tube and place on ice.
The wide end of a 1mL pipette works well for scraping down the lysate.
12. Shear the DNA in each sample by sonication with a microtip sonicator. Alternatively, add 1 μL of 25U/μL benzonase nuclease and Mg2+ to 1 mM final concentration to each sample, mix well, and incubate at room temperature for 30 minutes.
Sonication is faster but requires the availability of a sonicator and familiarity with its use. Benzonase is a convenient substitute for laboratories without a sonicator, but requires purchase of the enzyme.
In Branson or Misonix sonicators, 5 s with the power set to the microtip limit should be sufficient. Switch the power on and off while the tip is immersed in the sample to prevent foaming.
Addition of 1 mM Mg2+ enhances benzonase activity. Check that the sample is no longer viscous by pipetting with a 200 μL pipette tip or flicking the tube. Repeat sonication or benzonase digestion if viscosity persists.
13. Heat the samples again on a heat block at 85-95° C for 2 minutes. Mix and centrifuge quickly in a microcentrifuge to collect the sample at the bottom of the tube.
At this point, the lysates can be stored at −20° C for analysis by immunoblotting at another time.
14. Perform SDS-PAGE with 2-4 μL of each sample in duplicate, followed by immunoblotting, according to standard protocols. For one set of lysates, perform immunoblotting with anti-T7 antibody. For the other set, perform immunoblotting with anti-HA antibody. Use any immunodetection method of choice.
Analyze the immunoblots to determine if the TimeSTAMP cassette is being efficiently removed in the absence of drug, and completely retained in the presence of drug, and that no unexpected cleavage occurs in the protein of interest. For fusion proteins with the TimeSTAMP cassette at the C-terminus, the T7 epitope detects protein produced both in the absence and presence of BILN-2061. Proteins produced in the presence of drug should not have undergone TimeSTAMP cassette removal, and should appear 30kD larger than proteins produced in the absence of drug when probed with anti-T7. The untransfected well serves to identify which bands identified by the anti-T7 antibody are non-specific background bands. The HA epitope, in contrast, is removed by protease activity, and therefore is present only on proteins produced in the presence of BILN-2061. Anti-HA protein should only detect the full-length uncleaved protein in the lysate exposed to BILN-2061. For proteins bearing the TimeSTAMP cassette at the N-terminus, the roles of the T7 and HA epitopes are reversed.
Quantify target protein synthesis rates over defined times with TimeSTAMP
15. To study protein production rates over a defined time window with treatments of interest, repeat steps 6-7 above, calculating how many wells are needed for all experimental conditions (which may be a combination of treatments to be tested and time windows in which protein synthesis will be observed).
For instance, if cells are to be either untreated and treated with a growth factor and protein synthesis over two different time windows analyzed, then at least seven wells are necessary: two time points with BILN-2061 multiplied by two treatment conditions, plus one well as a transfected but untreated and uninhibited control, a transfected well that is treated continuously with BILN-2061, and an untransfected well.
16. Following transfection for two hours, move the cells into media. For the control well treated continuously with BILN-2061, include the BILN-2061 at that time. For other wells, add the BILN-2061 later at the desired time points. Perform experimental treatments as desired.
If a well is to be cultured for longer than two days following BILN-2061 addition, refresh BILN-2061 every two days by removing 0.5 mL of the culture media with old BILN-2061 and replacing with 0.5 mL of culture media with fresh 3μM BILN-2061. When adding BILN-2061 at later time points, it is best to make a 30μM solution of BILN-2061 in media to be used as a 10x mix instead of adding BILN-2061 directly from the DMSO stock onto the cells. Direct application of the stock DMSO-based solution of BILN-2061 can result in DMSO-mediated toxicity in regions of high DMSO concentrations and uneven dispersion of BILN-2061.
17. Detect newly synthesized proteins following BILN-2061 addition by performing steps 9-14 above.
If utilizing chemiluminescence or fluorescence for antibody detection in the immunoblot, the relative quantities of old and new proteins in each sample can be calculated from the emission counts collected from the faster and slower migrating bands, respectively, detected by the constitutive tag (T7 for TimeSTAMP fusions to the C-terminus of the protein of interest, HA for fusions to the N-terminus).
Support Protocol: Immunocytochemical detection of newly synthesized proteins by drug control of TimeSTAMP tags
Immunocytochemistry allows visualization of the spatial distributions of newly synthesized proteins of interest.
Materials
TimeSTAMP fusion construct (see Basic Protocol)
12mm coverslips and 24-well plate, or 35mm glass-bottom tissue-culture dishes (Mattek)
cell type of interest
cell culture media
transfection reagent
BILN-2061 (Boehringer Ingelheim), 10-30 mM in DMSO
PBS with 0.1% Triton X-100
8% paraformaldehyde in PBS, pH 7.2-7.5 (see Reagents and Solutions)
Blocking solution (see Reagents and Solutions)
rat 2F2 anti-HA (Roche)
mouse anti-T7 antibody (Novagen)
rabbit or chicken antibody to protein of interest (if available)
Alexa Fluor 488-conjugated goat anti-mouse IgG, highly cross-absorbed to rat serum proteins (Invitrogen)
Alexa Fluor 568-conjugated goat anti-rat IgG, highly cross-absorbed to mouse serum proteins (Invitrogen)
Alexa Fluor 647-conjugated goat anti-rabbit or anti-chicken IgG (Invitrogen)
Vectashield mounting solution (Vector Laboratories)
Fluorescence microscope with filters for fluorescein, rhodamine/Texas Red, and Cy5 wavelengths.
Donkey secondary antibodies can be used instead of goat secondary antibodies if the experimenter desires to use a goat primary antibody to the protein of interest.
Transfect cells with TimeSTAMP reporters
1. For cell types to be transfected in adherent monolayers by liposomal reagents or calcium phosphate, grow a transfectable cell type to the proper density for transfection on 12 mm coverslips in a 24-well plate or on 35 mm glass-bottom tissue-culture dishes, using enough wells or dishes for the combinations of time points and treatment conditions desired. One well or dish should be reserved for a no-drug control control. For cell types to be transfected by electroporation (e.g. using the Amaxa Nucleofector protocol), prepare the coverslips or plates separately and obtain the proper number of cells for each transfection condition according to the electroporation protocol for the desired cell type.
2. Transfect with the reporter plasmid expressing the protein of interest fused to a TimeSTAMP cassette, whose responsiveness to drug was previously verified by immunoblotting (Basic Protocol). Any time after transfection is complete (e.g. 2 hours for adherent cells, or after electroporated cells have adhered), but prior to performing drug administration, transfection media should be replaced with fresh media to remove transfection reagents or dead cells.
3. Perform biological treatments (e.g. growth factor treatment if investigating the effects of a growth factor on synthesis of a protein of interest) as desired. If the experimenter desired to perform a time course of treatment, e.g. 1, 3, 6, 12, and 24 hours, it is convenient to perform the treatment on different wells at those time intervals prior to a common experimental end time. Add BILN-2061 to begin the time interval in which newly synthesized proteins are to be tagged. Typically, this is immediately after the biological treatment or stimulation, but it is also possible to delay the beginning of the tag preservation window relative to the treatment time.
It is best to make a 30μM solution of BILN-2061 in media to be used as a 10x mix instead of adding BILN-2061 directly from the DMSO stock onto the cells.
4. At the end of the time window in which newly synthesized proteins are to be visualized, add 1 volume of 8% paraformaldehyde, equilibrated to room temperature. Incubate at room temperature for 15 min.
Adding paraformaldehyde to the media with BILN-2061 achieves both fixation and destruction of protease activity without allowing escape from inhibition.
5. Rinse with PBS 3 times for 3 minutes each, then permeabilize and block with blocking solution for 30-60 minutes at room temperature.
6. Prepare primary antibody solution by diluting rat 2F2, mouse anti-T7, and rabbit antibody to protein of interest (if available) to 1 μg/mL each in blocking solution. Incubate cells in primary antibody solution at room temperature for 1-2 hours or at 4° C for 12-24 hours.
7. Prepare secondary antibody solution by diluting Alexa Fluor 488-conjugated anti-mouse IgG, Alexa Fluor 568-conjugated anti-rat IgG, and Alexa Fluor 647-conjugated anti-rabbit IgG to 0.5 μg/mL each. Remove primary antibody solution from cells, rinse cells with PBS with 0.1% Triton X-100 3 times for 3 minutes each, then incubate cells in secondary antibody solution at room temperature for 30-60 minutes.
Secondary antibody solution can be prepared in PBS with 0.1% Triton X-100 without normal goat serum as well.
8. Wash with PBS with 0.1% Triton X-100 3 times for 10-15 minutes each. Remove final wash by aspiration. For coverslips, invert onto a 3 μL drop of Vectashield on a glass side, aspirate away excess Vectashield from the coverslip edges, and seal the edges with fingernail polish. For glass-bottom dishes, cover the glass area with Vectashield, replace the lid on the dish, and seal with wax film to prevent evaporation. Samples can now be stored at 4° C for up to a week or imaged on a microscope.
Here, Alexa Fluor 488 is used to detect the N-terminal T7 tag while Alexa Fluor 568 detects the C-terminal HA tag. If the TimeSTAMP cassette is at the N-terminus of the protein of interest, Alexa Fluor 488 fluorescence would reveal newly synthesized protein copies while Alexa Fluor 568 fluorescence would reveal all protein copies. If the TimeSTAMP cassette is at the C-terminus, the opposite relationship holds. Signal from the Alexa Fluor 647-conjugated anti-rabbit or anti-chicken secondary antibody (invisible to the human eye) can be used to compare distributions and levels of endogenous and transfected protein of interest, if a specific antibody is available. These are suggested wavelengths; researchers may rearrange the wavelengths used for their convenience. Alexa Fluor 555 may be used in place of Alexa Fluor 568, but in our experience is dimmer and slightly more prone to aggregation. Note anti-T7 antibodies typically show some cross-reactivity to cell nuclei.
REAGENTS AND SOLUTIONS
8% paraformaldehyde in PBS pH 7.2-7.5
For 10 ml of a 8% paraformaldehyde stock solution, weigh out 0.8 g of paraformaldehyde in a chemical safety hood, or carefully while wearing an aerosol mask. In a 15 ml polypropylene conical tube in a chemical safety hood, mix the powder, 8 ml of distilled or MilliQ water, and 5 μl of 1 M NaOH. Heat to 70° C and mix frequently until the solution clears, then allow the solution to cool down to room temperature. Add water to 9 ml, followed by 1 ml of PBS 10X, then mix. Use fresh or store at 4° C for up to 1 week or −20° C for up to 1 month. Check pH with indicator paper and adjust to pH 7.2-7.5 with HCl or NaOH if necessary.
Blocking solution
PBS
0.1% Triton X-100
5% normal goat serum
Commentary
Background Information
Spatially regulated protein synthesis and delivery occur during the establishment and regulation of polarized cellular structures, e.g. cell migration, epithelial cell maintenance, or activity-dependent modification of neuronal connections (Mili and Macara, 2009). Researchers investigating these processes may desire to specifically detect proteins synthesized after a defined time. This requires a means to label newer protein molecules separately from older molecules in the cell, preferably in a time-controlled manner. The TimeSTAMP method allows tagging and visualization of newly synthesized proteins, with onset of tagging triggered by drug administration (Lin et al., 2008).
Along with TimeSTAMP, several methods exist for age-specific protein labelling. The detection of specific protein species synthesized within specific time windows has traditionally been performed by metabolic incorporation of radioactively labeled amino acids. More recently, the incorporation of amino acid analogues with reactive azide or alkyne chemical groups has been introduced as non-radioactive chemical method for time-specific protein labeling (Dieterich et al., 2007). Both the above methods are well suited for labeling the entire set of proteins undergoing synthesis, but allow for the specific detection of a particular protein of interest only by further biochemical purification steps. While these methods can be used in conjunction with gel electrophoresis for quantification of synthesis rates or detection of post-translational modifications, they generally are not compatible with visualizing the locations of new protein copies in an intact cell.
Time-specific labeling of a defined protein species, on the other hand, can be achieved through fluorescence recovery after photobleaching (FRAP) of fluorescent protein tags (Kourtis and Tavernarakis, 2009). Here, fluorescent signal from preexisting protein copies is first removed by applying sufficient excitation light to result in complete bleaching throughout the entire cell. Subsequent fluorescence signal is then inferred to arise from newly synthesized proteins. A similar procedure can be carried out using photoconvertible fluorescence proteins which change the color of their fluorescence upon illumination (in effect losing fluorescence at one wavelength and acquiring it at another). However, FRAP for the purpose of visualizing new proteins requires complete removal of all pre-existing fluorescence from all points of a cell, and the light intensities and durations required to achieve this may cause toxicity (Kourtis and Tavernarakis 2009). Furthermore, the fluorescence that develops on newly synthesized proteins after FRAP is subject to a delay due to the maturation kinetics of fluorescent proteins.
Compared to these existing methods, TimeSTAMP shares similarities in usage to both isotopic and chemical pulse-chase methods and FRAP. Like pulse-chase, TimeSTAMP uses cell-permeable small molecules to control the labeling window and is compatible with electrophoretic analysis. Like FRAP, TimeSTAMP allows visualization of new copies of a genetically tagged protein of interest in the spatial context of the cell.
Critical Parameters
The protease used in the TimeSTAMP cassette is the NS3 protease of HCV, chosen for its high specificity and the availability of cell-permeable inhibitors including BILN-2061. NS3 protease has an unusually strict substrate preference, with the 5 amino acids proceeding and the 2 amino acids following the cut site contributing to substrate recognition (Ingallinella et al., 1998; Ingallinella et al., 2000), so cut sites in endogenous proteins are rare. To further prevent cleavage of endogenous proteins, the TimeSTAMP cassette includes only NS3 and not its NS4A cofactor, which increases the catalytic rate of the protease and is necessary for trans-cleavage of substrates (Bartenschlager et al., 1995; Landro et al., 1997). Supporting the lack of transprotease activity by NS3 alone, permanent localization of NS3 to the neuronal postsynaptic density, a structure composed of thousands of proteins held together by protein-protein interactions, by fusion to PSD-95, the most abundant protein in the postsynaptic density, did not result in noticeable defects (Lin et al., 2008). In the TimeSTAMP cassette, the existence of protease sites on both sides of the protease assures that the protease is released from the protein of interest and any associated proteins once it is active.
The experimenter may choose among two TimeSTAMP cassettes, TimeSTAMPt and TimeSTAMPa, to optimize the signal-to-background ratio. TimeSTAMPt contains the wild-type NS3 domain, whereas TimeSTAMPa contains the NS3 domain with a T54A mutation which reduces the catalytic rate constant (Tong et al., 2006). TimeSTAMPa exhibits higher signals after prolonged incubation with BILN-2061, presumably due to less cleavage during transient escape from inhibition, but also exhibits small amounts of signal on overexpressed proteins in the absence of BILN-2061. For proteins that are rapidly turning over (half-life ≤ 24 hours), synthesis rates may be high enough that TimeSTAMPa would produce some background in the absence of BILN-2061, and prolonged incubation with BILN-2061 for more than a few hours is usually not necessary to detect a pulse of protein synthesis. Therefore TimeSTAMPt is typically preferred in order to minimize background signal. For proteins with slow turnover (≥ 24 hours), synthesis rates are slow compared to even the reduced rate of cleavage by the T54A mutant of NS3, and prolonged incubation in BILN-2061 may be necessary, so the increased sensitivity of TimeSTAMPa may be beneficial. Whether a particular protein in the context of an expression system is suitable for tagging with TimeSTAMPa can be determined by examining whether cleavage is complete in the absence of BILN-2061 by immunoblotting (Basic Protocol). If cleavage is incomplete, the protein is expressed at rates that exceed the processing capacity of the T54A mutant of NS3, and TimeSTAMPt would be more appropriate.
The TimeSTAMP cassette is 287 amino acids in length with a molecular weight of 30 kD. This is the same size as a fluorescent protein [such as GFP?], and, as with a fluorescent protein, fusion of a TimeSTAMP cassette may interfere with a protein’s function or localization. In cases where a protein has not been successfully fused before to a fluorescent protein or similarly sized tag, the experimenter will want to verify that fusion to TimeSTAMP does not alter a protein’s distribution in the cell or biological activity. On the other hand, if it had been fused previously at one terminus to a fluorescence protein while maintaining protein function or localization, that terminus can be selected for fusion. Then the localization of TimeSTAMP fusions should still be compared to the fluorescent protein fusion to confirm the absence of unexpected effects.
During immunocytochemistry (Support Protocol), epitope tags on uncleaved TimeSTAMP fusions are retained via fixation of the fusion protein. Cleaved epitope tags are not fixed and detected, most likely due to their rapid degradation in the cytosol by thimet oligopeptidase and aminopeptidases (Rock et al., 2004). Any epitope tags that avoid degradation may also escape fixation and be lost after permeabilization.
Troubleshooting
| Problem | Possible Cause | Solution |
|---|---|---|
| Signal from the inducible TimeSTAMPa tag in the absence of inhibitor |
Protein production rate is faster than protease cleavage rate |
Switch to TimeSTAMPt. |
| Signal from the inducible TimeSTAMPt tag in the absence of inhibitor |
Protein is being expressed far over endogenous levels |
Reduce expression by substituting empty expression vector for some of the reporter plasmid. Expression levels in transient transfections vs endogenous levels can be compared by immunoblot analysis of extracts from transfected cells alongside extracts from the same mass of cells endogenously expressing the protein, using an antibody to endogenous protein. In immunocytochemistry experiments, staining with an antibody to endogenous protein can be performed to compare levels in transfected (which can be identified with the constitutive epitope tag, i.e. T7 for TimeSTAMP fusions at the C-terminus of the protein of interest or HA for fusions at the N-terminus of the protein) versus nearly untransfected cells. |
| No signal from the inducible tag in the presence of inhibitor |
Poor expression of reporter protein, actual slow turnover of protein of interest |
Confirm expression using an antibody to the tag that is always retained or an antibody to the protein of interest (which may be more sensitive if the constitutively retained tag is the T7 tag). Perform a control experiment in which BILN-2061 is applied continuously immediately after the completion of transfection. |
| Multiple bands on immunoblot probed with anti- T7 antibody |
T7 epitope is mildly crossreactive with endogenous proteins |
Perform an untransfected lysate control to determine which bands are cross-reactive endogenous proteins, or replace the T7 tag with another epitope tag of choice. |
| HEK293 or HEK293T cells lift off plate during washing |
These cell types can be loosely adherent |
Coat plates with poly-D-lysine before plating cells. To coat plates, apply 25 μg/mL poly- D-lysine (Sigma 0899 or 7886) in water for 1-24 h at 37°, then wash 3 times with water. If cells lose adherence during the final wash before lysis, they can be quickly spun down in a microfuge (200 g, 30 s), the wash buffer pipetted out, and the cells lysed by adding 2x SDS lysis buffer and immediately pipetting up and down. |
Anticipated Results
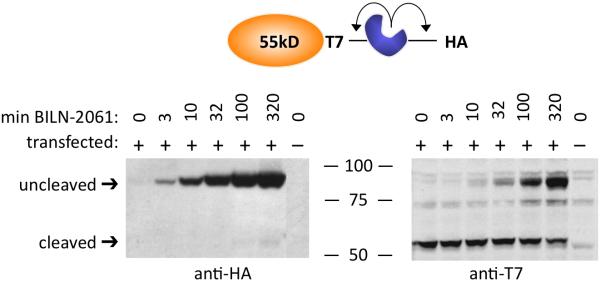
In immunoblotting of TimeSTAMP fusions (Basic Protocol), sizes of the fusion proteins in the presence or absence of the protease inhibitor BILN-2061 are first assessed in order to determine if the TimeSTAMP cassette (TimeSTAMPa or TimeSTAMPt) is being properly regulated by the drug. The expected sizes detected depend on which terminus of the protein of tinerest the TImeSTAMP cassette is fused to. For fusion proteins with the TimeSTAMP cassette at the C-terminus, the T7 epitope detects protein produced both in the absence and presence of BILN-2061. Proteins produced in the presence of drug should not have undergone TimeSTAMP cassette removal, and should appear 30kD larger than proteins produced in the absence of drug when probed with anti-T7 (Figure 2A). The HA epitope, in contrast, is removed by protease activity, and therefore is present only on proteins produced in the presence of BILN-2061. Anti-HA protein should only detect the full-length uncleaved protein in the lysate exposed to BILN-2061 (Figure 2B). For proteins bearing the TimeSTAMP cassette at the N-terminus, the roles of the T7 and HA epitopes are reversed.
The same considerations apply during the performance of immunoblotting experiments to follow production or modifications of new copies of the protein of interest during biological stimulations or treatments of cells. The primary difference in anticipated results from those obtained during testing is that treatment and BILN-2061 addition usually are performed for only a fraction of the time that cells are expressing the TimeSTAMP fusion protein, so that during immunoblotting, both lower-mobility cleaved protein produced prior to BILN-2061 addition and higher-mobility uncleaved protein produced afterwards are present in the same lysate of treated cells.
Immunocytochemistry of TimeSTAMP fusions is performed in order to determine the subcellular locations of proteins synthesized after drug addition. As with immunoblotting, which antibody constitutively detects the fusion protein, and which one is drug-induced, depends on the orientation of the fusion. For instance, fusions with the TimeSTAMP cassette at the C-terminus are always detected with anti-T7 antibody, while anti-HA antibody specifically detects proteins synthesized after drug addition. The vice versa is true in the case of fusions with the TimeSTAMP cassette at the N-terminus. A no-drug control on transfected cells is critical to determine background levels of TimeSTAMP signal. Also a non-transfected control is important to determine any pattern of cross-reactivity of anti-T7 and anti-HA antibodies to endogenous proteins.
Figure 2.
Example of detection of total and new protein subpopulations by immunoblotting. A plasmid expressing a 55kD protein fused to the TimeSTAMPt cassette at its C-terminus was transfected into HEK293 cells. Lysates were prepared at various times after BILN-2061 addition, and analyzed by immunoblotting with anti-HA (left) or anti-T7 (right) antibodies. Anti-HA detects only the fraction of molecules synthesized after BILN-2061 addition, which appears at 85kD, 30kD larger than the size of the protein alone due to the retention of the protease and HA tag on the protein. Anti-T7 detects both the previously synthesized subpopulation (at 55kD) and newly synthesized subpopulation (at 85kD).
Literature Cited
- Bartenschlager R, Lohmann V, Wilkinson T, Koch JO. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J Virol. 1995;69:7519–7528. doi: 10.1128/jvi.69.12.7519-7528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Lee JJ, Link AJ, Graumann J, Tirrell DA, Schuman EM. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc. 2007;2:532–540. doi: 10.1038/nprot.2007.52. [DOI] [PubMed] [Google Scholar]
- Flores MV, Strawbridge J, Ciaramella G, Corbau R. HCV-NS3 inhibitors: Determination of their kinetic parameters and mechanism. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbapap.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Ingallinella P, Altamura S, Bianchi E, Taliani M, Ingenito R, Cortese R, De Francesco R, Steinkuhler C, Pessi A. Potent peptide inhibitors of human hepatitis C virus NS3 protease are obtained by optimizing the cleavage products. Biochemistry. 1998;37:8906–8914. doi: 10.1021/bi980314n. [DOI] [PubMed] [Google Scholar]
- Ingallinella P, Bianchi E, Ingenito R, Koch U, Steinkuhler C, Altamura S, Pessi A. Optimization of the P’-region of peptide inhibitors of hepatitis C virus NS3/4A protease. Biochemistry. 2000;39:12898–12906. doi: 10.1021/bi001590g. [DOI] [PubMed] [Google Scholar]
- Kourtis N, Tavernarakis N. Cell-specific monitoring of protein synthesis in vivo. PLoS One. 2009;4:e4547. doi: 10.1371/journal.pone.0004547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landro JA, Raybuck SA, Luong YP, O’Malley ET, Harbeson SL, Morgenstern KA, Rao G, Livingston DJ. Mechanistic role of an NS4A peptide cofactor with the truncated NS3 protease of hepatitis C virus: elucidation of the NS4A stimulatory effect via kinetic analysis and inhibitor mapping. Biochemistry. 1997;36:9340–9348. doi: 10.1021/bi963054n. [DOI] [PubMed] [Google Scholar]
- Lin MZ, Glenn JS, Tsien RY. A drug-controllable tag for visualizing newly synthesized proteins in cells and whole animals. Proc Natl Acad Sci U S A. 2008;105:7744–7749. doi: 10.1073/pnas.0803060105. Describes design and use of TimeSTAMP to track newly synthesized proteins of interest by immunoblotting and immunocytochemistry.
- Mili S, Macara IG. RNA localization and polarity: from A(PC) to Z(BP) Trends Cell Biol. 2009;19:156–164. doi: 10.1016/j.tcb.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock KL, York IA, Goldberg AL. Post-proteasomal antigen processing for major histocompatibility complex class I presentation. Nat Immunol. 2004;5:670–677. doi: 10.1038/ni1089. [DOI] [PubMed] [Google Scholar]
- Tong X, Chase R, Skelton A, Chen T, Wright-Minogue J, Malcolm BA. Identification and analysis of fitness of resistance mutations against the HCV protease inhibitor SCH 503034. Antiviral Res. 2006;70:28–38. doi: 10.1016/j.antiviral.2005.12.003. [DOI] [PubMed] [Google Scholar]