Abstract
Trimerization of the human immunodeficiency virus (HIV-1) envelope glycoproteins is mediated by the ectodomain of the gp41 transmembrane glycoprotein. Here we investigate oligomer-specific conformations of gp41 by using monoclonal antibodies (MAbs) from HIV-1-infected humans. Human MAbs directed against the cluster I region of gp41 recognized trimeric, dimeric, and monomeric forms of soluble envelope glycoproteins; thus, the integrity of the cluster I epitopes is minimally affected by the oligomeric state. In contrast, human MAbs to the cluster II region were all oligomers specific. One cluster II MAb, 126-6, recognized exclusively the trimeric form of envelope glycoproteins, whereas the others recognized both trimeric and dimeric forms. Thus, a distinct trimer-specific conformation exists in the cluster II region of gp41. Analysis of soluble envelope glycoprotein mutants revealed that gp41 sequences immediately N-terminal to isoleucine 646 contribute to the formation of both the trimer and the trimer-specific conformational epitope.
Introduction
Human immunodeficiency virus type 1 (HIV-1) entry into the host cell is mediated by the viral envelope glycoproteins, gp120 and gp41, which constitute a trimeric complex on the viral surface. The gp120 exterior envelope glycoprotein is retained on the trimer via noncovalent interactions with the ectodomain of the gp41 transmembrane envelope glycoprotein.1–3 The ectodomain of the gp41 glycoprotein contains a hydrophobic, glycine-rich amino terminus (fusion peptide), and two heptad repeat (HR) regions, designated HR1 and HR2, connected by a 25- to 30-residue region characterized by a disulfide-bonded loop and several N-linked glycosylation sites. HR1 is immediately carboxy-terminal to the fusion peptide and HR2 is close to the viral membrane-spanning region.4,5 It is thought that gp41 exists in a native, prefusogenic state prior to receptor binding, and this prefusogenic conformation may be stabilized by extensive interaction with the inner domain of gp120.6 Upon the interaction of gp120 with its cellular receptors CD4 and one of the chemokine receptors, CCR5 or CXCR4, the trimeric HIV-1 envelope glycoprotein complex undergoes extensive conformational transitions that culminate in the formation of a gp41 six-helix bundle, in which the HR2 regions pack into the well-conserved, largely hydrophobic grooves on the outer surface of the HR1 coiled coil.1–5 The formation of the six-helix bundle structure is thought to approximate the viral and the target cell membranes and eventually drive membrane fusion.7
The gp41 glycoprotein ectodomain is very immunogenic, inducing high-titer antibodies in essentially all HIV-1-infected individuals. Several distinct antigenic determinants in the gp41 ectodomain were identified and mapped by human monoclonal antibodies (MAbs),8–11 or by MAbs produced by immunization of mice with envelope glycoproteins.12 Two regions in the gp41 ectodomain appear to be immunodominant: (1) the region between HR1 and HR2 that contains the intrachain disulfide bond (denoted cluster I epitopes) and (2) the region containing HR2 (designated cluster II epitopes).10 Antibodies to cluster I recognize peptides containing amino acid residues 579–604, whereas the binding of antibodies to cluster II is generally dependent on gp41 conformation and can be disrupted by changes in the gp41 region between residues 644 and 663.8,10 Most human MAbs to cluster II epitopes do not react with the HR2 peptide (aa 624–666) designated C43, but do react with the complex that is formed by N51 (aa 540–590) and C43 peptides, which is thought to approximate the six-helix bundle core of the postfusogenic form of gp41.13 Human MAbs to both cluster I and cluster II have been shown to bind HIV-1-infected cells14–16 and intact virions,17,18 and can mediate Ab-dependent cellular cytotoxicity (ADCC)15 and complement-dependent virolysis.18 Most human MAbs to gp41 do not neutralize HIV-1 viruses.19 Exceptions to this are human MAbs 2F5 and 4E10,11,20,21 which recognize nearby but distinct epitopes on the membrane-proximal external region (MPER) of gp41 at the C-terminal end of cluster II.11,20,21 One anti-cluster II MAb 98-6 has been reported to neutralize some primary HIV-1 isolates.22 Cluster II human MAbs have been shown to block MAb 2F5 binding to gp41 epitopes to variable degrees.23 Other weakly neutralizing gp41-directed MAbs have been reported.24,25
The mature, trimeric spikes of gp120/gp41 represent the functional form of the HIV-1 envelope glycoproteins. However, it has been suggested that HIV-1 particles also bear nonfunctional gp120/gp41 monomers and gp120-depleted gp41 stumps on their surface.17,26 The formation of heterotrimeric complexes of HIV-1 gp120/gp41 presumably causes quaternary structural changes that could lead to new antigenic properties compared with the monomeric forms of these molecules.27 Indeed, recognition of trimeric HIV-1 envelope glycoproteins on cell or virion surfaces has been shown to correlate better with the neutralizing activity of antibodies than recognition of monomeric envelope glycoproteins.28–30 Thus, understanding the antigenic structure of the mature, trimeric envelope glycoproteins has implications for vaccine development and for the study of the immune response against HIV-1.
In the mature HIV-1 envelope glycoprotein trimer, the structure of gp41 is likely to be strongly influenced by quaternary interactions with gp120 and other gp41 subunits; as an example of the latter; contacts involving the three gp41 ectodomains are largely responsible for envelope glycoprotein trimerization.31 In an effort to understand the structure of the HIV-1 envelope glycoprotein complex, soluble trimers lacking the transmembrane anchor and cytoplasmic tail have been produced; the stability of these soluble trimers (sgp140) has been increased by disruption of the proteolytic cleavage site between gp120 and gp41 and, in some cases, by adding heterologous trimerization domains at the C-terminus.32 For cleavage-defective sgp140(-) glycoproteins without heterologous sequences, the relative amount of the glycoprotein in trimeric, dimeric, or monomeric forms was influenced by the extent of gp41 ectodomain sequences included in the constructs; for example, deletions affecting the carboxyl terminus of the sgp140(-) glycoprotein resulted in disruption of the trimers.31 Trimer-specific HIV-1 gp41 epitopes recognized by human monoclonal antibodies have not yet been described,13 and no previous work has carefully studied the oligomers dependence of a panel of human anti-gp41 MAbs. Here, we screened a panel of human anti-gp41 MAbs with cross-linked HIV-1 sgp140(-) envelope glycoproteins. We demonstrated that a human monoclonal antibody, MAb 126-6, exclusively recognizes trimeric forms of the HIV-1 envelope glycoproteins. We analyzed the gp41 regions critical for the integrity of the oligomer-specific epitopes by using sgp140(-) mutants, and found that the trimer-specific conformations were mainly presented within the cluster II region of the gp41 ectodomain.
Materials And Methods
Envelope glycoprotein constructs
The soluble HIV-1 envelope glycoprotein sgp140(-) consists of the complete gp120 and gp41 ectodomain with alterations in the gp120/gp41 cleavage site (the arginines at amino acid positions 508 and 511 changed to serines).30 The mutations resulting in C-terminal truncations introduced two stop codons at the desired position. Soluble gp140(-) constructs from the YU2, JR-FL, and HXBc2 HIV-1 strains were studied. Amino acid residue numbers correspond to those of the prototypic HXBc2 HIV-1 envelope glycoproteins, as per current convention.33
Antibodies and reagents
Human MAbs 50-69, 98-6, 240D, 246D, 126-6, 1281, and 1379 were derived from HIV-1-infected individuals from the United States.10,13,34 MAb F240 was obtained from Drs. Marshall Posner and Lisa Cavacini through the NIH AIDS Research and Reference Reagent Program.35 MAbs 2F5 and 5F3 were obtained from Dr. Hermann Katinger through the NIH AIDS Research and Reference Reagent Program.20,36,37 Two subclones were obtained from the original 126-6 heterohybridoma line: one producing an IgG2 MAb (described in Xu et al.10) and the other producing an IgG1 MAb (which has not previously been described). Both have specificity for cluster II of gp41. The MAb used here was from the subclone of 126-6 producing the IgG1 MAb. This latter MAb is now available from the AIDS Research and Reference Reagent Program as 126-7 to avoid confusion.
Expression and cross-linking of soluble envelope glycoproteins
The soluble envelope glycoproteins were transiently expressed in 293T cells by transfecting with env-expressing plasmids using Lipofectamine 2000 transfection reagent, following the manufacturer's protocol (Invitrogen). Twenty-four hours after the start of transfection, the expressed envelope glycoproteins were radiolabeled with [35S]methionine/cysteine for 36 h prior to harvesting. The harvested culture supernatants were centrifuged at low speed to clear the cell debris, and the envelope glycoproteins in the supernatant were cross-linked with glutaraldehyde (GA), as previously described.29,30 Briefly, 100 or 200 μl of supernatants was diluted to 400 μl with phosphate-buffered-saline (PBS) containing 1 mM EDTA, and incubated with GA at a final concentration of 10 mM at room temperature for 5 min, followed by the addition of glycine to a final concentration of 100 mM to quench the unreacted GA. The cross-linked soluble envelope glycoproteins in the culture supernatant were then subjected to immunoprecipitation.
Immunoprecipitation of soluble envelope glycoproteins
The radiolabeled soluble envelope glycoproteins in the culture supernatant were diluted with PBS and incubated with 1 μl of a mixture of pooled sera (PS) from HIV-1-infected individuals or with 1 μg of individual monoclonal antibodies. The antibody-glycoprotein complex was precipitated by protein A-Sepharose beads (Amersham Biosciences) and washed twice with PBS containing 0.2% Tween-20 before analysis by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The radiolabeled envelope glycoprotein bands on the SDS-PAGE gels were exposed to a PhosphorImager Screen and analyzed with a STORM system (Molecular Dynamics).
For the depletion of specific MAb-reactive envelope glycoproteins, the supernatants containing radiolabeled soluble envelope glycoproteins were incubated with the MAb and protein A-Sepharose beads. The envelope glycoproteins reactive with the MAb were precipitated, and the remaining supernatants were incubated with the same fresh MAb and protein A-Sepharose beads. After precipitation, the supernatants were subjected to one or two additional precipitations with the same MAb. The envelope glycoproteins left in the supernatants after the depletion were then either precipitated directly by PS or cross-linked and precipitated by PS. The precipitates were washed and subjected to SDS-PAGE, as described above.
Results
Oligomer-specific epitopes on soluble HIV-1 envelope glycoproteins recognized by human anti-gp41 antibodies
Previous studies showed that soluble, cleavage-defective HIV-1 sgp140(-) envelope glycoproteins form trimers, dimers, and monomers, and these three oligomeric forms of sgp140(-) can be readily distinguished by cross-linking and SDS-PAGE.30 We utilized this approach to investigate potential oligomer-specific epitopes in the soluble envelope glycoproteins, and to study oligomer-specific conformations of the gp41 ectodomain. Because the sgp140(-) glycoprotein from the primary HIV-1 strain JR-FL contains measurable amounts of trimers, dimers, and monomers, we chose this glycoprotein to screen a panel of human anti-gp41 MAbs for their interaction with different oligomeric forms of sgp140(-). The sgp140(-) glycoprotein was transiently expressed, radiolabeled in 293T cells, and then cross-linked with 10 mM GA. We showed previously that GA at this concentration fully cross-linked soluble HIV-1 envelope glycoprotein oligomers.30
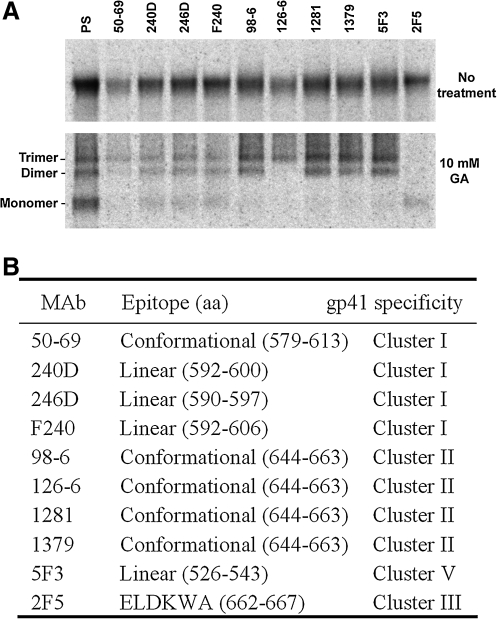
The untreated and cross-linked soluble envelope glycoproteins were immunoprecipitated by either PS from HIV-1-infected humans or a panel of human MAbs that recognizes different epitopes of the gp41 envelope glycoprotein (Fig. 1). The PS serves as control for precipitating all the trimers, dimers, and monomers of the envelope glycoproteins in the supernatant, as it is known to recognize both native and denatured forms of the HIV-1 envelope glycoproteins.38–42 The amount of the untreated sgp140(-) envelope glycoproteins precipitated by the individual MAbs was similar, except that MAbs 50-69 and 126-6 precipitated less of the envelope glycoproteins than the other MAbs. Different preferences in precipitating the cross-linked, oligomeric forms of the soluble envelope glycoproteins were observed for the MAbs tested (Fig. 1A). The MAbs 50-69, 240D, 246D, and F240 directed against cluster I epitopes recognized trimeric, dimeric, and monomeric forms of cross-linked sgp140(-) envelope glycoproteins, although only a small portion of each oligomer was precipitated by these MAbs compared to that precipitated by PS (Fig 1A). The 50-69 MAb precipitated monomeric and dimeric sgp140(-) glycoproteins less efficiently than the trimeric sgp140(-) glycoproteins, which is consistent with previous findings.43 Among the MAbs that recognize cluster II epitopes, MAbs 98-6, 1281, and 1379 efficiently precipitated trimers and dimers, whereas MAb 126-6 precipitated only the trimeric form of cross-linked sgp140(-) envelope glycoproteins (Fig. 1A). The amount of each oligomer precipitated by these cluster II monoclonal antibodies was equivalent to that precipitated by PS. That the MAb 126-6 reacted only with the trimeric form of sgp140(-) envelope glycoproteins explained the lower amounts of the untreated envelope glycoproteins precipitated by this antibody. MAb 5F3, which recognizes a linear gp41 epitope adjacent to the N-terminal fusion peptide, preferentially precipitated cross-linked trimeric and dimeric forms of the sgp140(-) glycoproteins. MAb 2F5, which is specific for the ELDKWA epitope in the membrane-proximal region of the gp41 ectodomain,9 interacted inefficiently with the cross-linked envelope glycoproteins and precipitated only a trace amount of the monomeric cross-linked envelope glycoprotein.
FIG. 1.
Interaction of human anti-gp41 monoclonal antibodies (MAbs) with oligomeric forms of soluble envelope glycoproteins. (A) Radiolabeled sgp140(-) glycoproteins from the HIV-1 JR-FL strain were either untreated (top panel) or cross-linked with 10 mM GA (bottom panel) before being precipitated by PS or the indicated MAbs. The precipitates were subjected to SDS-PAGE and autoradiography. (B) The gp41 epitopes of the human MAbs used in (A) are indicated.
The MAbs used in this study that recognize the cluster I gp41 epitopes react with a stretch of 24 amino acids from positions 590 to 61310,13 (Fig. 1B). Three of these, MAbs 240D, 246D and F240, react with linear epitopes; by contrast, MAb 50-69 recognizes a gp41 conformation stabilized by the disulfide bond between the cysteines at positions 598 and 604.10,35 The 240D, 246D, and F240 MAbs interacted with cross-linked trimers, dimers, and monomers, indicating that the epitopes recognized by these MAbs were intact and accessible on the different oligomeric forms of soluble envelope glycoproteins. The preferential recognition of the sgp140(-) trimers by the 50-69 MAb supports previous studies showing that the conformation of the cluster I region can be influenced by the oligomeric state of the HIV-1 envelope glycoproteins.13
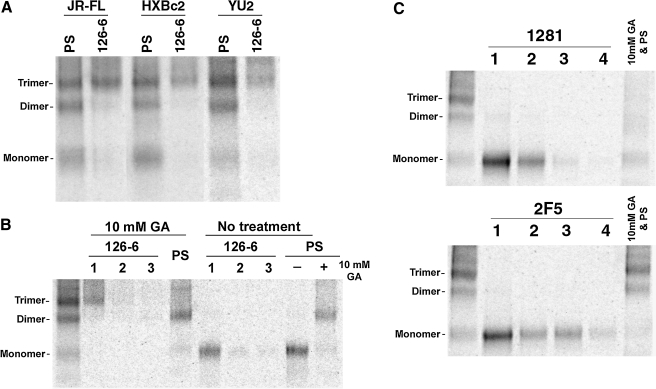
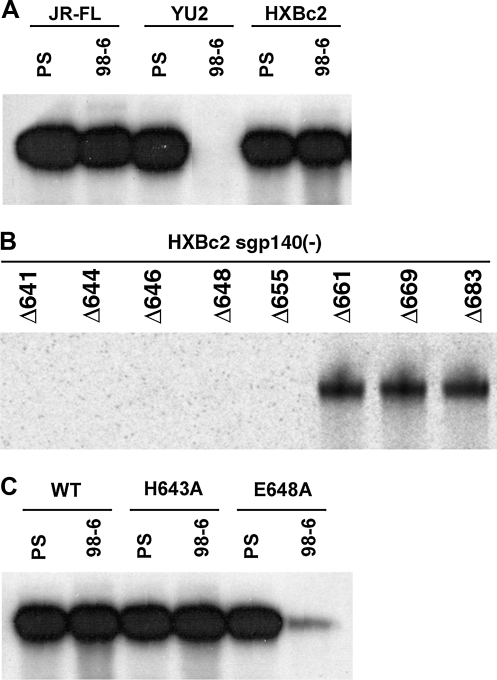
The MAbs that recognize the cluster II gp41 epitopes are all specific for conformational structures determined by amino acids between positions 644 and 66310,13 (Fig. 1B). The different specificities of these MAbs for distinct oligomeric forms of sgp140(-) indicate that the conformation of this region is strongly influenced by the oligomeric state of the HIV-1 envelope glycoprotein; for example, the epitope recognized by MAb 126-6 is present and accessible only on envelope glycoprotein trimers. As MAb 126-6 preferentially recognizes soluble envelope glycoprotein trimers from several HIV-1 strains, such as JR-FL, YU2, and HXBc2 (Fig. 2A), this trimer-specific epitope is apparently conserved on the envelope glycoproteins of clade B HIV-1 strains.
FIG. 2.
MAb depletion of soluble envelope glycoproteins. (A) The radiolabeled sgp140(-) glycoproteins from the JR-FL, HXBc2, and YU2 strains of HIV-1 were cross-linked with 10 mM GA before being precipitated by PS or MAb 126-6. (B) The sgp140(-) glycoproteins from the JR-FL strain were cross-linked with 10 mM GA (left panel) or untreated (right panel) before sequential precipitation (1–3, from left to right) by MAb 126-6. The remaining envelope glycoproteins in the supernatant were either directly precipitated by PS, or cross-linked with 10 mM GA and then precipitated by PS. (C) The JR-FL sgp140(-) glycoproteins were sequentially precipitated (1–4, from left to right) by the indicated MAb. The remaining envelope glycoproteins in the supernatant were cross-linked with 10 mM GA and then precipitated by PS (10 mM GA and PS). The precipitates were analyzed by SDS-PAGE.
As GA targets primary amines on the protein component,44–46 this may exert subtle effects on the envelope glycoprotein epitopes. To avoid any potential artifact generated by GA cross-linking, we performed MAb-depletion experiments to examine whether the oligomer-specific epitopes exist in native HIV-1 envelope glycoproteins. Untreated sgp140(-) envelope glycoproteins from the YU2 HIV-1 strain were repeatedly precipitated by MAbs 126-6, 1281, or 2F5. As shown in Fig. 2B and C, three or four successive immunoprecipitations with each of the MAbs were sufficient to remove almost all of the envelope glycoproteins precipitable by the particular MAb. The remaining envelope glycoproteins in the supernatant were then precipitated directly by PS or first cross-linked with 10 mM GA and then precipitated by PS. When the sgp140(-) envelope glycoproteins were cross-linked with 10 mM GA, MAb 126-6 only precipitated the trimeric form of the envelope glycoprotein, and the dimeric and monomeric forms of the envelope glycoproteins remained in the supernatant (Fig. 2B, left panel). In Fig. 2B (right panel), after repeated precipitation of the untreated sgp140(-) envelope glycoprotein by MAb 126-6, dimeric and monomeric forms of the sgp140(-) envelope glycoprotein remained in the supernatant (Fig. 2B, right panel). This result supports the conclusion that MAb 126-6 binds only to native envelope glycoprotein trimers. In contrast, repeated precipitation by MAb 1281 left primarily monomeric forms of the sgp140(-) envelope glycoprotein in the supernatant (Fig. 2C, upper panel). Thus, MAb 1281 reacts with native envelope glycoprotein trimers and dimers. Repeated precipitation by MAb 2F5 left trimeric and dimeric envelope glycoproteins in the supernatant (Fig. 2C, lower panel), suggesting that the epitope recognized by this MAb is present mainly on monomers. In summary, the MAb-depletion results were consistent with the results of immunoprecipitation of the cross-linked envelope glycoproteins.
Mapping of the oligomer-specific regions
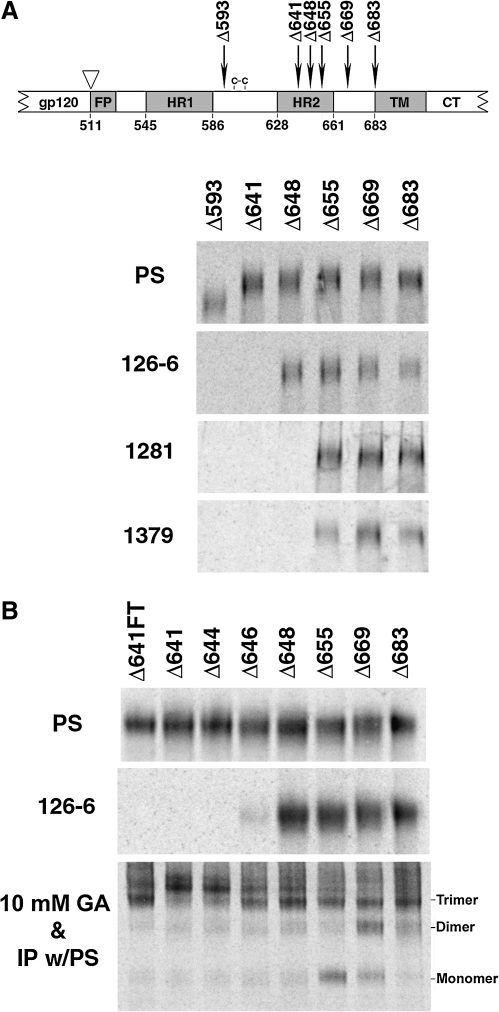
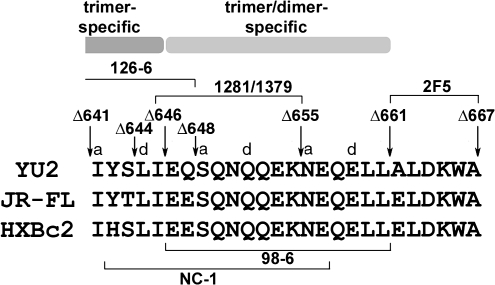
As shown above, the epitopes in the cluster II region of the HIV-1 gp41 ectodomain exhibited oligomer-specific conformations. To map the oligomer-specific regions, a series of YU2 sgp140(-) C-terminal truncation mutants, with deletions of varying portions of the gp41 ectodomain, were analyzed by immunoprecipitation (Fig. 3A). The sgp140(-) glycoprotein mutants with C-terminal deletions up to amino acid position (aa) 655 [sgp140(-)Δ655] still reacted with MAbs 126-6, 1281, and 1379. When the deletion extended further to aa 648, the truncated sgp140(-) mutant [sgp140(-)Δ648] lost reactivity to MAbs 1281 and 1379, but still bound to MAb 126-6. Only when the carboxyl terminus of sgp140(-) was deleted up to aa 641 did the glycoprotein [sgp140(-)Δ641] lose reactivity with MAb 126-6 (Fig. 3B). Therefore, amino acid residues between positions 641 and 648 in the cluster II region of the gp41 ectodomain are critical for the trimer-specific epitope recognized by MAb 126-6; the amino acid residues located between positions 648 and 655 are required for recognition by MAbs 1261 and 1379.
FIG. 3.
Mapping the oligomer-specific cluster II epitopes. (A) The gp41 glycoprotein is depicted, with the gp120-gp41 cleavage site (triangle), fusion peptide (FP), heptad repeats (HR1 and HR2), transmembrane region (TM), and cytoplasmic tail (CT) shown. The locations of the C-termini of the mutants are noted. (B) The YU2 sgp140(-) mutants truncated immediately C-terminal to the indicated amino acid residues were precipitated by either PS or the indicated MAbs. The precipitates were analyzed by SDS-PAGE. (C) The YU2 sgp140(-) mutants truncated immediately C-terminal to the indicated amino acid residues were precipitated by PS or MAb 126-6 (top two panels). The Δ641FT glycoprotein is identical to sgp140(-)Δ641 except that a fibritin trimer (FT) motif has been added to the C-terminus. In the bottom panel, the soluble envelope glycoproteins were cross-linked with 10 mM GA and then precipitated with PS prior to analysis by SDS-PAGE.
To map the region critical for the integrity of the trimer-specific epitope of MAb 126-6 further, two more sgp140(-) mutants with truncations of the gp41 ectodomain between aa 641 and 648, sgp140(-)Δ646 and sgp140(-)Δ644, were examined for interaction with MAb 126-6. As shown in Fig. 3C, very little of the sgp140(-)Δ646 glycoprotein and none of the sgp140(-)Δ644 glycoprotein were precipitated by MAb 126-6. Thus, important determinants of the MAb 126-6 epitope reside between gp41 residues 644 and 648.
As MAb 126-6 interacts only with trimeric forms of the HIV-1 envelope glycoproteins, we examined if this interaction correlated with the formation of trimers by soluble envelope glycoprotein variants. Cross-linking of sgp140(-) glycoprotein variants with 10 mM GA showed that wild-type and mutant glycoproteins containing gp41 ectodomain sequences up to aa 646 formed trimers. By contrast, the sgp140(-)Δ644 and sgp140(-)Δ641 mutant glycoproteins mainly formed higher-order oligomers (Fig. 3C). These higher-order, presumably aggregated, forms of envelope glycoproteins were apparent at low levels in the case of the sgp140(-)Δ648 mutant, and their proportion increased in the sgp140(-) mutants with further deletions of the carboxyl terminus (Fig. 3C). The sgp140(-)Δ641FT glycoprotein, which consists of the sgp140(-)Δ641 glycoprotein with a carboxy-terminal heterologous trimeric motif derived from fibritin,47 formed significant amounts of trimers (Fig. 3C). As the sgp140(-)Δ646 and sgp140(-)Δ641FT glycoproteins formed trimers but reacted inefficiently with MAb 126-6 (Fig. 3C), we conclude that trimer formation is not sufficient for recognition of soluble HIV-1 envelope glycoproteins by MAb 126-6.
Mouse MAb NC-1 and human MAb 126-6 recognize different trimer-specific epitopes
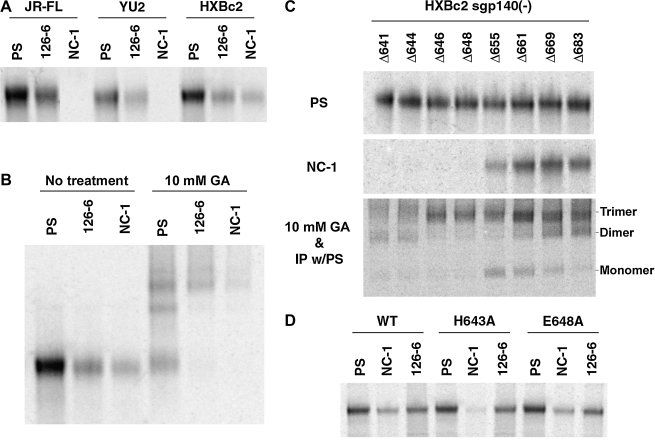
Jiang et al. previously generated a conformation-dependent monoclonal antibody, MAb NC-1, by immunizing a mouse with a polypeptide that models the HIV-1 gp41 six-helix bundle.48 MAb NC-1 binds specifically to oligomeric forms of HIV-1 gp41, a property similar to that of the human MAbs that recognize cluster II epitopes. We compared the HIV-1 envelope glycoprotein regions required for recognition by MAb 126-6 and MAb NC-1. Surprisingly, MAb NC-1 did not react with the sgp140(-) glycoproteins from the primary R5 HIV-1 strains, JR-FL or YU2; it reacted only with the sgp140(-) glycoprotein from the laboratory-adapted X4 strain, HXBc2 (Fig. 4A). This result implied that MAb NC-1 and MAb 126-6 recognize different epitopes, because MAb 126-6 recognizes the sgp140(-) glycoproteins from all three HIV-1 strains. Subsequent experiments involving MAb NC-1 were carried out with soluble envelope glycoproteins from the HXBc2 strain of HIV-1. Immunoprecipitation with cross-linked sgp140(-) glycoproteins revealed that MAb NC-1, similar to MAb 126-6, reacted preferentially with the trimeric form of the envelope glycoproteins (Fig. 4B). Immunoprecipitation with a series of C-terminally deleted sgp140(-) mutants showed that MAb NC-1 bound efficiently to sgp140(-) mutants with C-termini truncated up to aa 661 [sgp140(-)Δ661]. The recognition of sgp140(-)Δ655 by MAb NC-1 was significantly decreased, and sgp140(-)Δ648 was not precipitated by MAb NC-1 (Fig. 4C, middle panel). Thus, the HIV-1 gp41 region between aa 648 and 661 is important for recognition by MAb NC-1. The C-terminal deletion mutants of sgp140(-) from the HXBc2 strain exhibited the same pattern of trimer formation as that from the primary strain YU2; wild-type sgp140(-) and mutants with C-termini truncated up to aa 646 efficiently formed trimers, whereas the sgp140(-)Δ644 and sgp140(-)Δ641 glycoprotein mutants oligomerized primarily into nontrimeric states (Fig. 4C, lower panel).
FIG. 4.
Mapping the epitope of MAb NC-1. (A) The sgp140(-) glycoproteins from the JR-FL, YU2, and HXBc2 strains of HIV-1 were immunoprecipitated by PS, MAb 126-6, or MAb NC-1. (B) The HXBc2 sgp140(-) glycoproteins were either untreated or cross-linked with 10 mM GA before being precipitated by PS, MAb 126-6, or MAb NC-1. (C) The HXBc2 sgp140(-) truncated immediately C-terminal to the indicated amino acid residues were either precipitated by PS or MAb NC-1 (top two panels) or cross-linked with 10 mM GA before precipitation by PS (bottom panel). (D) The wild-type (WT) HXBc2 sgp140(-) glycoproteins or mutants containing the indicated changes were precipitated by PS, MAb NC-1, or MAb 126-6. The precipitates were analyzed by SDS-PAGE.
MAb NC-1 recognizes only envelope glycoproteins from the HXBc2 HIV-1 strain, not those from the YU2 or JR-FL strains, yet the amino acid residues in the region that is important for antibody recognition, aa 648 to 661, are identical among these three strains (Fig. 5). Therefore, other regions must contribute to the epitope recognized by MAb NC-1. Comparing the amino acid sequences in the nearby gp41 region of the three HIV-1 strains, the closest amino acid residue that differs between HXBc2 and the other two strains is at aa 643, which is histidine in HXBc2 and tyrosine in YU2 or JR-FL (Fig. 5). A change of histidine to alanine (H643A) in the HXBc2 sgp140(-) glycoprotein decreased the binding of MAb NC-1 (Fig. 4D). In contrast, a change of glutamic acid to alanine at aa 648 (E648A) in sgp140(-) did not affect MAb NC-1 recognition (Fig. 4D). We conclude that the epitope recognized by MAb NC-1 includes amino acid residues from aa 643 to 655. Some amino acid residues in the aa 655 to 661 region also contribute to the MAb NC-1 epitope because the deletion of this region significantly reduced antibody binding.
FIG. 5.
A map of gp41 epitopes influenced by oligomerization state. The sequences of part of the HR2 region and adjacent membrane-proximal region of gp41 from the indicated HIV-1 strains are aligned. The a and d positions of the HR2 heptad repeat are shown. The positions of the C-termini of some of the sgp140(-) mutants tested in this study are indicated. The gp41 regions implicated in the integrity of the indicated MAbs are delineated, as are the regions proposed to contribute to the formation of trimer-specific and trimer/dimeric-specific epitopes.
Characterization of the gp41 region important for human MAb 98-6 recognition
Among the human MAbs tested in this study, MAbs 98-6, 1281, and 1379 recognize epitopes in the cluster II region of gp41 and react with trimeric and dimeric forms of sgp140(-) envelope glycoproteins from the JR-FL HIV-1 strain (Fig. 1). However, unlike MAbs 1281 or 1379, MAb 98-6 did not precipitate the sgp140(-) glycoprotein from the YU2 HIV-1 strain (Fig. 6A); thus, MAb 98-6 recognizes a gp41 epitope in the cluster II region different from that recognized by the MAbs 1281 and 1379. Immunoprecipitation experiments showed that MAb 98-6 bound to the HXBc2 sgp140(-) mutants with C-termini truncated up to aa 661; further deletion of the amino acid residues at the C-terminus resulted in the disruption of antibody recognition (Fig. 6B). Thus, amino acid residues between positions 655 and 661 in the gp41 ectodomain are important for recognition by MAb 98-6.
FIG. 6.
Mapping of the MAb 98-6 epitope. (A) The sgp140(-) glycoproteins from the JR-FL, YU2, and HXBc2 strains of HIV-1 were precipitated by PS or MAb NC-1 98-6. (B) The HXBc2 sgp140(-) mutants truncated immediately C-terminal to the indicated amino acid residues were precipitated by MAb 98-6. (C) The wild-type (WT) HXBc2 sgp140(-) glycoproteins or mutants containing the indicated changes were precipitated by PS or MAb 98-6. The precipitates were analyzed by SDS-PAGE.
As mentioned above, the amino acid residues between aa 655 and 661 are identical among the YU2, JR-FL, and HXBc2 strains (Fig. 5). Examination of nearby residues revealed that aa 648 differed among these HIV-1 strains, being glutamine in YU2 and glutamic acid in JR-FL or HXBc2. Alteration of glutamine 648 to alanine (E648A) in the HXBc2 sgp140(-) glycoprotein greatly diminished the binding of MAb 98-6 (Fig. 6C). By contrast, the alteration of histidine 643 (H643A) did not affect the recognition of sgp140(-) by MAb 98-6 (Fig. 6C). Thus, the epitope recognized by the human MAb 98-6 involves amino acid residues 648 to 661, slightly carboxy-terminal to the epitope recognized by the mouse MAb NC-1.
Discussion
The functional HIV-1 envelope glycoproteins consist of trimers of heterodimeric subunits, so understanding the quaternary structure of the HIV-1 envelope glycoproteins is essential for an appreciation of the structure/function of virus entry and for rational attempts to design inhibitors and vaccines. The formation of HIV-1 envelope glycoprotein complexes presumably results in new structural moieties in gp120 and gp41 that are not present in the monomeric forms of these molecules.27 The existence of oligomer-specific conformations in the gp41 ectodomain is supported by the recognition patterns exhibited by several MAbs that selectively interact with oligomeric forms of HIV-1 envelope glycoproteins.13,49 However, due to the difficulty of separating the different oligomeric forms of envelope glycoproteins, the regions that form oligomer-specific conformations in the gp41 ectodomain have not been well defined. In this study, we used GA to cross-link the trimeric and dimeric forms of soluble HIV-1 envelope glycoproteins. These cross-linked trimeric, dimeric, and monomeric forms of envelope glycoproteins can easily be separated from one another by SDS-PAGE.30 The cross-linking approach allowed us to examine the oligomer-specific recognition of the envelope glycoproteins by monoclonal antibodies, and hence to study the oligomer-specific conformations of the gp41 ectodomain.
Although several distinct antigenic regions in the gp41 ectodomain can be recognized by MAbs produced by immunization of mice with envelope glycoproteins,12 the anti-gp41 antibodies generated in most HIV-1-infected humans mainly recognize epitopes in two regions, cluster I and cluster II; the cluster I region appears to be approximately 100-fold more immunogenic than the cluster II region.10 The data presented here indicate that most human MAbs to the cluster I region bind indistinguishably to trimeric, dimeric, and monomeric forms of the HIV-1 envelope glycoproteins; thus, the epitopes in the cluster I region are not significantly affected by the formation of the oligomeric complex.
In contrast, MAbs to cluster II regions preferentially interacted with either trimeric, or trimeric and dimeric, forms of the soluble envelope glycoproteins; thus, the conformation of the cluster II region is distinct in the different oligomeric forms. Among the MAbs examined here that recognize epitopes in the cluster II region, three (MAbs 1281, 1379, and 98-6) recognize trimeric and dimeric forms of envelope glycoprotein, and two (MAbs 126-6 and NC-1) recognize exclusively trimeric forms. The gp41 regions important for recognition by these MAbs are summarized in Fig. 5. MAb 98-6 binding to trimeric and dimeric forms of soluble envelope glycoproteins involves residues 646 to 661, indicating that at least some conformational determinants in this region are similar between the envelope glycoprotein trimers and dimers. MAb NC-1 binds only to trimeric forms of the envelope glycoproteins; the gp41 region that includes the NC-1 epitope extends from amino acid residues amino-terminal to aa 642 to a few residues carboxy-terminal to aa 655. Thus, distinct conformations of this region are achieved only in the context of envelope glycoprotein trimers. Our results suggest that the gp41 region amino-terminal to aa 648 is critical for recognition by the trimer-specific MAb 126-6, whereas the region between aa 648 and 655 is critical for the recognition by MAbs 1281 and 1379, which recognize both dimeric and trimeric soluble envelope glycoproteins. These results support a model in which the HIV-1 gp41 region amino-terminal to glutamic acid 647 forms trimer-specific structures, whereas more carboxy-terminal epitopes involving residues 646–661 can be represented on either dimeric or trimeric oligomers (Fig. 5). Compared with the epitopes for other cluster II human MAbs, the epitope for MAb 98-6 locates closer to the MAb 2F5 epitope; this explains the observation that MAb 98-6 blocks MAb 2F5 binding to a gp41 peptide more efficiently than other cluster II human Mabs.23
The human MAb 126-6 and the mouse MAb NC-1 exclusively recognize envelope glycoprotein trimers. The MAb NC-1 was generated by immunizing a mouse with a model gp41 polypeptide from the HIV-1 HXB2 strain, and was shown to bind to the gp41 six-helical bundle of several CXCR4-tropic HIV-1 strains.48,50 This MAb may be less useful for studying CCR5-tropic HIV-1 envelope glycoproteins, as we found that MAb NC-1 did not detectably interact with the CCR5-using envelope glycoproteins examined in this study.
MAb 126-6, along with other human MAbs that recognize cluster II epitopes, also binds to a gp41 six-helix bundle formed by mixing two peptides from the HR1 and HR2 regions.13 Although these data imply that these MAbs can recognize the postfusogenic form of gp41, two lines of evidence suggest that these MAbs bind as well to prefusogenic forms of gp41. First, the soluble gp140(-) envelope glycoprotein used in this study has been shown to assume a conformation resembling the prefusogenic state.31 Second, the observation that MAbs 1281, 1379, and 98-6 precipitate dimeric as well as trimeric forms of sgp140(-) also argues against these MAbs recognizing only the postfusogenic form of the gp41 envelope glycoprotein, which is a stable trimer.4,5,7 Hence, the MAbs to the epitopes in the cluster II region likely recognize both prefusogenic and postfusogenic forms of gp41, implying that the conformation of the cluster II region in gp41 may be preserved during the conformational transitions induced by receptor binding. As the cluster II region is part of the HR2 helix in the postfusogenic six-helix bundle, this region may also be helical in prefusogenic conformations of gp41. The hydrophobic face of this helix, which in the postfusogenic conformation contacts the outer groove on the HR1 coiled coil, may in prefusogenic conformations form intersubunit contacts that contribute to trimer stability. Such a model is consistent with our observation that truncated versions of sgp140(-) lacking this region do not efficiently form trimers.
The epitope for human Mab 2F5 (ELDKWA) contains a critical lysine residue.20,21 GA could potentially interact with the primary amine on the lysine residue in our cross-linking experiments, resulting in the inefficient recognition of the cross-linked envelope glycoproteins by MAb 2F5 (Fig. 1). The MAb-depletion experiment avoids this concern (Fig. 2C). Our results showed that human MAb 2F5 reacts poorly with trimeric or dimeric forms of soluble HIV-1 envelope glycoproteins. This suggests that the epitope recognized by MAb 2F5 is occluded or disrupted as the soluble oligomers form. This result is consistent with the finding that the 2F5 and 4E10 epitopes are exposed only on a soluble gp41 construct designed to mimic a prehairpin intermediate conformation51; the prehairpin intermediate is thought to be presented only after the envelope glycoproteins bind to the CD4 receptor and before the formation of the six-helix bundle.51 However, previous studies also show that 2F5 recognizes cell-surface HIV-1 envelope glycoproteins prior to receptor binding.14,52,53 Because soluble gp41 constructs are intrinsically artificial and because nonfunctional envelope glycoproteins may exist on the surface of infected cells, neither of these disparate observations allows definitive conclusions to be reached about the state of MPER epitopes on the functional envelope glycoprotein trimers.
The gp41 subunit is far more conserved than gp120, and the membrane fusion machinery is common to all HIV-1 strains. These aspects make gp41 an attractive target for intervention. Understanding the specific conformations of the native, trimeric gp41 ectodomain will expedite progress toward this goal.
Acknowledgments
We thank Ms. Yvette McLaughlin and Ms. Elizabeth Carpelan for manuscript preparation. This work was supported by NIH grants [AI24755, AI39420, and AI40895 (J.S.) and HL59725 and AI36085 (S.Z.P.)], by a Center for HIV/AIDS Vaccine Immunology grant (AI67854), by Center for AIDS Research grants [AI42848 (DFCI) and AI27742 (NYU)], by an unrestricted research grant from the Bristol-Myers Squibb Foundation, by a gift from the late William F. McCarty-Cooper, by funds from the International AIDS Vaccine Initiative and the Department of Veterans Affairs, and by funds from the Department of Medicine, University of Virginia.
Disclosure Statement
No competing financial interests exist.
References
- 1.Wyatt R. Sodroski J. The HIV-1 envelope glycoproteins: Fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 2.Prabakaran P. Dimitrov AS. Fouts TR. Dimitrov DS. Structure and function of the HIV envelope glycoprotein as entry mediator, vaccine immunogen, and target for inhibitors. Adv Pharmacol. 2007;55:33–97. doi: 10.1016/S1054-3589(07)55002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roux KH. Taylor KA. AIDS virus envelope spike structure. Curr Opin Struct Biol. 2007;17:244–252. doi: 10.1016/j.sbi.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Root MJ. Steger HK. HIV-1 gp41 as a target for viral entry inhibition. Curr Pharm Des. 2004;10:1805–1825. doi: 10.2174/1381612043384448. [DOI] [PubMed] [Google Scholar]
- 5.Hunter E. gp41, A multifunctional protein involved in HIV entry and pathogenesis. In: Korber B, editor; Hahn B, editor; Foley B, et al., editors. Human Retroviruses and AIDS 1997. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; Los Alamos, NM: 1997. pp. III-55–III-73. [Google Scholar]
- 6.Yang X. Mahony E. Holm GH. Kassa A. Sodroski J. Role of the gp120 inner domain beta-sandwich in the interaction between the human immunodeficiency virus envelope glycoprotein subunits. Virology. 2003;313:117–125. doi: 10.1016/s0042-6822(03)00273-3. [DOI] [PubMed] [Google Scholar]
- 7.Chan DC. Kim PS. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 8.Binley JM. Ditzel HJ. Barbas CF, 3rd, et al. Human antibody responses to HIV type 1 glycoprotein 41 cloned in phage display libraries suggest three major epitopes are recognized and give evidence for conserved antibody motifs in antigen binding. AIDS Res Hum Retroviruses. 1996;12:911–924. doi: 10.1089/aid.1996.12.911. [DOI] [PubMed] [Google Scholar]
- 9.Muster T. Steindl F. Purtscher M, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu JY. Gorny MK. Palker T. Karwowska S. Zolla-Pazner S. Epitope mapping of two immunodominant domains of gp41, the transmembrane protein of human immunodeficiency virus type 1, using ten human monoclonal antibodies. J Virol. 1991;65:4832–4838. doi: 10.1128/jvi.65.9.4832-4838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zwick MB. Labrijn AF. Wang M, et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Earl PL. Broder CC. Doms RW. Moss B. Epitope map of human immunodeficiency virus type 1 gp41 derived from 47 monoclonal antibodies produced by immunization with oligomeric envelope protein. J Virol. 1997;71:2674–2684. doi: 10.1128/jvi.71.4.2674-2684.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorny MK. Zolla-Pazner S. Recognition by human monoclonal antibodies of free and complexed peptides representing the prefusogenic and fusogenic forms of human immunodeficiency virus type 1 gp41. J Virol. 2000;74:6186–6192. doi: 10.1128/jvi.74.13.6186-6192.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sattentau QJ. Zolla-Pazner S. Poignard P. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology. 1995;206:713–717. doi: 10.1016/s0042-6822(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 15.Tyler DS. Stanley SD. Zolla-Pazner S, et al. Identification of sites within gp41 that serve as targets for antibody-dependent cellular cytotoxicity by using human monoclonal antibodies. J Immunol. 1990;145:3276–3282. [PubMed] [Google Scholar]
- 16.Zolla-Pazner S. O'Leary J. Burda S, et al. Serotyping of primary human immunodeficiency virus type 1 isolates from diverse geographic locations by flow cytometry. J Virol. 1995;69:3807–3815. doi: 10.1128/jvi.69.6.3807-3815.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore PL. Crooks ET. Porter L, et al. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J Virol. 2006;80:2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spear GT. Takefman DM. Sullivan BL. Landay AL. Zolla-Pazner S. Complement activation by human monoclonal antibodies to human immunodeficiency virus. J Virol. 1993;67:53–59. doi: 10.1128/jvi.67.1.53-59.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorny MK. Zolla-Pazner S. Human monoclonal antibodies that neutralize HIV-1. In: Korber BT, editor; Brander C, editor; Haynes BF, et al., editors. HIV Immunology and HIV/SIV Vaccine Databases 2003. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; Los Alamos, NM: 2004. pp. 37–51. [Google Scholar]
- 20.Purtscher M. Trkola A. Gruber G, et al. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1994;10:1651–1658. doi: 10.1089/aid.1994.10.1651. [DOI] [PubMed] [Google Scholar]
- 21.Trkola A. Pomales AB. Yuan H, et al. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J Virol. 1995;69:6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hioe CE. Xu S. Chigurupati P, et al. Neutralization of HIV-1 primary isolates by polyclonal and monoclonal human antibodies. Int Immunol. 1997;9:1281–1290. doi: 10.1093/intimm/9.9.1281. [DOI] [PubMed] [Google Scholar]
- 23.Alam SM. Scearce RM. Parks RJ, et al. Human immunodeficiency virus type 1 gp41 antibodies that mask membrane proximal region epitopes: Antibody binding kinetics, induction, and potential for regulation in acute infection. J Virol. 2008;82:115–125. doi: 10.1128/JVI.00927-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson JD. Kinkead H. Brunel FM, et al. Antibody elicited against the gp41 N-heptad repeat (NHR) coiled-coil can neutralize HIV-1 with modest potency but non-neutralizing antibodies also bind to NHR mimetics. Virology. 2008;377:107–183. doi: 10.1016/j.virol.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gustchina E. Louis JM. Lam SN, et al. A monoclonal Fab derived from a human nonimmune phage library reveals a new epitope on gp41 and neutralizes diverse human immunodeficiency virus type 1 strains. J Virol. 2007;81:12946–12953. doi: 10.1128/JVI.01260-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poignard P. Moulard M. Golez E, et al. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J Virol. 2003;77:353–365. doi: 10.1128/JVI.77.1.353-365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hurtley SM. Helenius A. Protein oligomerization in the endoplasmic reticulum. Annu Rev Cell Biol. 1989;5:277–307. doi: 10.1146/annurev.cb.05.110189.001425. [DOI] [PubMed] [Google Scholar]
- 28.Fouts TR. Binley JM. Trkola A. Robinson JE. Moore JP. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J Virol. 1997;71:2779–2785. doi: 10.1128/jvi.71.4.2779-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan W. Bazick J. Sodroski J. Characterization of the multiple conformational States of free monomeric and trimeric human immunodeficiency virus envelope glycoproteins after fixation by cross-linker. J Virol. 2006;80:6725–6737. doi: 10.1128/JVI.00118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan W. Craig S. Yang X. Sodroski J. Inter-subunit disulfide bonds in soluble HIV-1 envelope glycoprotein trimers. Virology. 2005;332:369–383. doi: 10.1016/j.virol.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 31.Mische CC. Yuan W. Strack B. Craig S. Farzan M. Sodroski J. An alternative conformation of the gp41 heptad repeat 1 region coiled coil exists in the human immunodeficiency virus (HIV-1) envelope glycoprotein precursor. Virology. 2005;338:133–143. doi: 10.1016/j.virol.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Cho MW. Subunit protein vaccines: Theoretical and practical considerations for HIV-1. Curr Mol Med. 2003;3:243–263. doi: 10.2174/1566524033479861. [DOI] [PubMed] [Google Scholar]
- 33.Kuiken CL. Foley B. Freed E, et al. HIV Sequence Compendium 2002. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; Los Alamos, NM: 2002. LA-UR 03-3564. [Google Scholar]
- 34.Gorny MK. Gianakakos V. Sharpe S. Zolla-Pazner S. Generation of human monoclonal antibodies to human immunodeficiency virus. Proc Natl Acad Sci USA. 1989;86:1624–1628. doi: 10.1073/pnas.86.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cavacini LA. Emes CL. Wisnewski AV, et al. Functional and molecular characterization of human monoclonal antibody reactive with the immunodominant region of HIV type 1 glycoprotein 41. AIDS Res Hum Retroviruses. 1998;14:1271–1280. doi: 10.1089/aid.1998.14.1271. [DOI] [PubMed] [Google Scholar]
- 36.Buchacher A. Predl R. Strutzenberger K, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses. 1994;10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 37.Purtscher M. Trkola A. Grassauer A, et al. Restricted antigenic variability of the epitope recognized by the neutralizing gp41 antibody 2F5. AIDS. 1996;10:587–593. doi: 10.1097/00002030-199606000-00003. [DOI] [PubMed] [Google Scholar]
- 38.Allan JS. Coligan JE. Barin F, et al. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science. 1985;228:1091–1094. doi: 10.1126/science.2986290. [DOI] [PubMed] [Google Scholar]
- 39.Moore JP. The reactivities of HIV-1 + human sera with solid-phase V3 loop peptides can be poor predictors of their reactivities with V3 loops on native gp120 molecules. AIDS Res Hum Retroviruses. 1993;9:209–219. doi: 10.1089/aid.1993.9.209. [DOI] [PubMed] [Google Scholar]
- 40.Moore JP. Ho DD. Antibodies to discontinuous or conformationally sensitive epitopes on the gp120 glycoprotein of human immunodeficiency virus type 1 are highly prevalent in sera of infected humans. J Virol. 1993;67:863–875. doi: 10.1128/jvi.67.2.863-875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robey WG. Safai B. Oroszlan S, et al. Characterization of envelope and core structural gene products of HTLV-III with sera from AIDS patients. Science. 1985;228:593–595. doi: 10.1126/science.2984774. [DOI] [PubMed] [Google Scholar]
- 42.Wyatt R. Sullivan N. Thali M, et al. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J Virol. 1993;67:4557–4565. doi: 10.1128/jvi.67.8.4557-4565.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pinter A. Honnen WJ. Tilley SA, et al. Oligomeric structure of gp41, the transmembrane protein of human immunodeficiency virus type 1. J Virol. 1989;63:2674–2679. doi: 10.1128/jvi.63.6.2674-2679.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Das M. Fox CF. Chemical cross-linking in biology. Annu Rev Biophys Bioeng. 1979;8:165–193. doi: 10.1146/annurev.bb.08.060179.001121. [DOI] [PubMed] [Google Scholar]
- 45.Kyte J. Structure in Protein Chemistry. Garland Publishing, Inc.; New York: 1995. [Google Scholar]
- 46.Wold F. Bifunctional reagents. Methods Enzymol. 1972;25:623–651. doi: 10.1016/S0076-6879(72)25061-3. [DOI] [PubMed] [Google Scholar]
- 47.Yang X. Lee J. Mahony EM. Kwong PD. Wyatt R. Sodroski J. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol. 2002;76:4634–4642. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang S. Lin K. Lu M. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1998;72:10213–10217. doi: 10.1128/jvi.72.12.10213-10217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Broder CC. Earl PL. Long D. Abedon ST. Moss B. Doms RW. Antigenic implications of human immunodeficiency virus type 1 envelope quaternary structure: Oligomer-specific and -sensitive monoclonal antibodies. Proc Natl Acad Sci USA. 1994;91:11699–11703. doi: 10.1073/pnas.91.24.11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Rosny E. Vassell R. Jiang S. Kunert R. Weiss CD. Binding of the 2F5 monoclonal antibody to native and fusion-intermediate forms of human immunodeficiency virus type 1 gp41: Implications for fusion-inducing conformational changes. J Virol. 2004;78:2627–2631. doi: 10.1128/JVI.78.5.2627-2631.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frey G. Peng H. Rits-Volloch S. Morelli M. Cheng Y. Chen B. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc Natl Acad Sci USA. 2008;105:3739–3744. doi: 10.1073/pnas.0800255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ofek G. Tang M. Sambor A, et al. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol. 2004;78:10724–10737. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Si Z. Phan N. Kiprilov E. Sodroski J. Effects of HIV type 1 envelope glycoprotein proteolytic processing on antigenicity. AIDS Res Hum Retroviruses. 2003;19:217–226. doi: 10.1089/088922203763315722. [DOI] [PubMed] [Google Scholar]