Abstract
Human immunodeficiency virus type 1 (HIV-1) group M is responsible for the current AIDS pandemic and exhibits exceedingly high levels of viral genetic diversity around the world, necessitating categorization of viruses into distinct lineages, or subtypes. These subtypes can differ by around 35% in the envelope (Env) glycoproteins of the virus, which are displayed on the surface of the virion and are targets for both neutralizing antibody and cell-mediated immune responses. This diversity reflects the remarkable ability of the virus to adapt to selective pressures, the bulk of which is applied by the host immune response, and represents a serious obstacle for developing an effective vaccine with broad coverage. Thus, it is important to understand the underlying biological consequences of intersubtype diversity. Recent studies have revealed that some of the HIV-1 subtypes exhibit phenotypic differences stemming from subtle changes in Env structure, particularly within the highly immunogenic V3 domain, which participates directly in viral entry. This review will therefore explore current research that describes subtype differences in Env at the genetic and phenotypic level, focusing in particular on V3, and highlighting recent discoveries about the unique features of subtype C Env, which is the most globally prevalent subtype.
Introduction
For 2007, the UNAIDS organization estimated that 33.2 million people were living with HIV worldwide, including 2.5 million new infections and 2.1 million AIDS deaths in that year alone, underscoring the profound nature of the global HIV pandemic.1 One unexpected challenge that has arisen from the HIV pandemic is the incredible amount of viral genetic diversity, which is generated through an error-prone viral-encoded polymerase,2,3 high levels of persistent virus replication,4,5 and frequent genomic recombination events6 that allow the virus to rapidly adapt to changing selective pressures. Viruses of the HIV-1 group M lineage are responsible for the current global pandemic,7,8 and the last common ancestor for group M HIV-1 was dated to the early twentieth century.9 Based on the phylogenetic characterization of HIV-1 sequences recovered from frozen specimens in west-central Africa, divergent HIV-1 subtypes were already circulating in this region by the 1960s.10,11 The cumulative genetic variability of HIV-1 is managed on paper by classifying viral sequences into one of 13 currently recognized subtypes or subsubtypes (A1–A4, B, C, D, F1–F2, G, H, J, K) or 43 circulating recombinant forms.12 As of 2004, HIV-1 subtype A, C, and D accounted for 65% of worldwide HIV-1 infections, with subtype C alone being responsible for half of all global infections.13 However, due to the prominence of subtype B HIV-1 in North America and Europe, these viruses have historically been most thoroughly characterized.12,13 Thus, much of our understanding of HIV-1 has been based on subtype B, although recent studies continue to reveal evidence that the viral subtypes have different phenotypic properties, such as coreceptor utilization,14–29 in vitro replication fitness,30,31 rate of disease progression,32–35 biology of transmission,36–38 antigenicity,39–41 genital shedding,42 and mutational patterns.43–48 For a summary of biological properties that differ between subtypes B and C, refer to Table 1.
Table 1.
Comparison of Subtype B and C Biological Properties
| |
|
Intraclade heterologous breadth of sera |
Interclade heterologous breadth of sera |
Breadth of mAb activity |
|
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subtype | Early autologous Nab potency | Early infection | Chronic infection | Pooled plasma | Early infection | Chronic infection | Pooled plasma | b12 | 2F5 | 2G12 | 4E10 | Hypervariable domain length | Coreceptor usage |
| B | Moderate–High40,61,63,115,125,126 | Low– moderate40,115 | Low– moderate87,125 | Moderate– high87,118 | Low107 | Low87,118 | Broad77,86,87 | Broad77,86,87 | Broad77,86,87 | Broad77,86,87 | Longer40,56,88 | R5, but 50% X4 in late infection83,104–106 | |
| C | High40,89,107 | Low40,89,107,119 | Moderate– high88,89 | Very high88,118 | Low107 | High88,118 | Moderate77,88,120 | None77,88,120 | None77,88,120 | Broad77,88,120 | Shorter40,56,88 | Mainly R5 even in later stage of infection16–19,27,28,107–109 | |
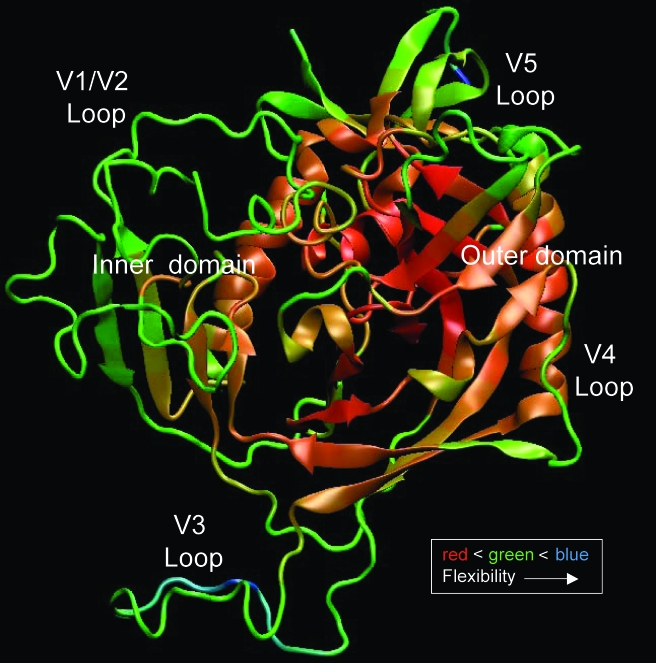
Most of these differences reflect variability in the env gene, which encodes the envelope (Env) surface glycoprotein 120 (gp120) and transmembrane glycoprotein 41 (gp41).49 Together, these Env proteins form a complex that protrudes from the virion surface as a trimer. Much of what is currently known about the conformation of gp120 is based on crystal structures of the truncated, deglycosylated, CD4-liganded subtype B protein core or the truncated, glycosylated, unliganded core of simian immunodeficiency virus (SIV).50–52 Structures of CD4-liganded, truncated gp120 with an intact, antibody bound V3 domain53 and a truncated gp120 bound to monoclonal antibody b12, which recognizes a neutralizing epitope overlapping the CD4 binding site, also have been recently deduced.54 In all of these structures, the outer domain of gp120 appears to be similar; however, the inner domain undergoes significant conformational change upon binding to CD4, as reflected by its relative flexibility as compared to the outer domain (Fig. 1). The structure and position of the V1 and V2 “hypervariable” domains contained within gp120 have been difficult to determine because of their conformational flexibility. Even when the conformations of other hypervariable loops have been determined (V3 and V4), they may have been stabilized by crystalline contacts or bound antibodies. It is, therefore, not fully understood how these variable domains might influence the overall conformation of the native Env protein in the context of the functional trimer. The Env glycoproteins can exhibit 35% amino acid diversity between subtypes and 20% within a subtype, with most of the genetic variation occurring in gp120.55 This level of diversity could lead to subtle but important structural differences in Env across subtypes.39,47,56,57 To investigate these differences, structural homology models of gp120 can be generated from the X-ray structures of subtype B gp120 using the consensus or specific sequences of other subtypes. Even though these models can be used to provide structural insights about the outer domain, the lack of N-linked glycosylation, absence of accurate information on the position of the loop domains, and the conformational fluidity of the inner domain limit their utility.
FIG. 1.
Atomic fluctuations in gp120. Backbone flexibility of the YU2 gp120 molecule was calculated from long time scale equilibrium molecular dynamics simulations. These all atom simulations were carried out with gp120 solvated in explicit solvent molecules. The calculated B-factors correspond to backbone atomic fluctuations and are graphically mapped on an arbitrary structure of a liganded gp120 with modeled loops using a color gradient. The red to blue indicates small to large atomistic fluctuations (rigid to flexible) in the backbone of the structure. The outer domain is relatively more rigid than the inner domain, while the loop regions are also more flexible than the core. Even though the starting conformation of gp120 corresponds to that of the CD4-liganded structure, the CD4 molecule was not included in the calculations. Despite the incomplete sampling of the gp120 conformational space, significant flexibility is observed in the inner domain, some of which is associated with the relief of the conformational constraints induced by binding to CD4.
This review will explore current research that describes subtype variation in Env at the genetic, phenotypic, and structural level, highlighting recent discoveries about the unique features of subtype C Env. Studies of the third hypervariable domain (V3) of gp120 will be emphasized because of the importance of this domain in viral entry, eliciting antibodies, and the plethora of available sequence data from this region. Understanding how and why Env regions differ between subtypes is necessary to tackle the genetic diversity of HIV-1 in vaccine design and treatment.12
Mutational patterns in Env across subtypes
Env is a target for both cell-mediated and humoral immune responses during HIV-1 infection, and much of the sequence evolution that occurs is thought to be in response to immune pressure.57–64 The pattern of adaptive evolution in the env gene was examined recently by Choisy et al.44 who performed pairwise comparisons of the most prominent HIV-1 group M subtypes using representative sequences from the Los Alamos HIV Database (www.hiv.lanl.gov). The positions in Env that reflected positive selection (an increase in the frequency of an advantageous residue, leading to sequence diversity and greater fitness) were similar between subtypes A, B, C, and D, suggesting that these viruses are exposed to analogous selective pressures in the infected host. However, the magnitude of selection at these positions was statistically different when subtype B was compared with A or C, indicating that there are discrete features of immune pressure and/or adaptive evolution between subtypes. Patterns of selective pressure were more fully explored in a study by Travers et al.43 using full-length env sequences deemed representative of the diversity present in each group M HIV-1 subtype (A, B, C, D, F1, F2, G, H, J, and K). In that study, each subtype was systematically compared to the rest of the group M subtypes. Using this approach, two residues in subtype C, two residues in subtype F1, and three residues in subtype G were identified as undergoing positive selection (defined above) in that particular subtype while undergoing purifying selection (decrease in the frequency of a deleterious residue, leading to sequence conservation and maintenance of fitness) in all other subtypes. Conversely, six residues in subtype A and 45 in subtype K were found to have undergone purifying selection while being under positive selection in all other subtypes.43 It is interesting to note that these residues occurred throughout the entire gp160 coding region, which includes gp120 and gp41. These findings suggest that the adaptive pressures that have shaped Env in each lineage are distinct, and this may have formed the basis for conformational differences in Env between subtypes.
Mutational patterns in gp41 across subtypes
According to HXB2 numbering, gp41 encompasses residues 512–856 in gp160. It contains heptad repeat regions 1 and 2 (HR1 and HR2), which reside in the ectodomain portion of gp41 (external to the viral membrane), and form a six helix bundle that facilitates entry of the virus into the target cell after gp120 binding to receptor molecules and insertion of the fusion peptide into the target cell membrane.49 The FDA-approved fusion inhibitor enfuvirtide targets the interaction between HR1 and HR2. Resistance to this drug has been shown to involve mainly mutations within a specific subregion of HR1 (amino acids 36 to 45),65 but can be influenced by residues in HR2,66 leading to increased interest in sequence variation across subtypes in this region. HR2 is typically more variable than HR1 across subtypes,67,68 but subtype-specific patterns of sequence polymorphism have been demonstrated in both regions,67,69 suggesting that selection pressures could differ between subtypes. Expanding upon this finding, Razzolini et al. studied amino acid sequence polymorphisms in gp41 from 102 subtype B and 95 non-subtype B enfuvirtide-naive HIV-1-infected Italian patients.70 Examination of the degree of amino acid conservation between the four best-represented subtypes (B, C, F1 and CRF02_AG), revealed differences in gp41 conservation levels, with the majority of polymorphisms occurring within HR2. These data are further supported by the work of Eshleman et al. who sequenced the HR1 and HR2 regions of 126 HIV-1-infected patients from around the world representing at least nine different subtypes and CRFs (A/A2, B, C, D, F, G, CRF01_AE, CRF02_AG, and other recombinant forms).71 In the HR1 region, 19 polymorphisms were found to occur infrequently and generally involved the same amino acid substitution. In HR2, however, 8 out the 15 polymorphisms detected occurred in most of the nine subtypes examined, but the amino acids accounting for these polymorphisms varied between subtypes. These data indicate that HR2 is more variable than HR1 across subtypes, and that there are subtype-specific patterns of mutation in these regions. However, the majority of these polymorphisms are not predicted to engender primary enfuvirtide resistance. Indeed, viruses from most subtypes are susceptible to enfuvirtide in vitro.68,72–74 Thus, substantial differences in enfuvirtide susceptibility in the clinic would not be expected based on viral subtype.
A region in gp41 with relevance to vaccine design is located where the ectodomain meets the viral membrane, known as the membrane proximal external region (MPER). This region contains epitopes that are recognized by some patient sera.75,76,76a The epitopes of two monoclonal antibodies that target this region and have broad neutralization activity against subtype B viruses have been characterized.78,79 2F5 recognizes the motif DKW; however many non-B subtypes, including subtypes C and D, contain a substitution in this region and are therefore not susceptible to neutralization.77 Interestingly, the presence of DKW was not always sufficient for neutralization by 2F5. Monoclonal antibody 4E10 requires the epitope WFXI, and unlike the 2F5 epitope, this sequence is well conserved across subtypes and recombinant forms.77 4E10-resistant virus was recovered from a subtype C-infected patient that had neutralization activity against the MPER, and this was attributed to a substitution in the epitope (F to L) as well as changes in the gp41 cytoplasmic tail.75 Thus, subtype-specific differences in the MPER could limit the utility of this region for vaccine design.
Mutational patterns in V3 across subtypes
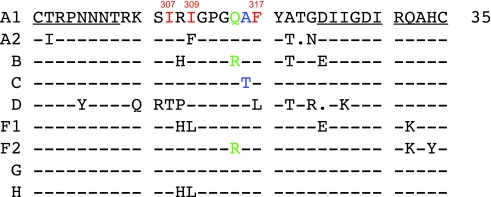
The third hypervariable domain (V3) of HIV-1 gp120 is a cysteine-bounded loop structure usually composed of 35 amino acids (Fig. 2), traditionally categorized as the base (residues 1–8 and 25–35; Fig. 2, underlined residues), stem (residues 9–14 and 18–24), and turn (residues 15–17) regions. Of the five gp120 hypervariable domains, V3 is relatively conserved and does not exhibit the dramatic insertions, deletions, and shifts in glycosylation that are characteristic of other domains, perhaps because V3 participates directly in coreceptor binding.80–82 V3 has long been a target of interest for entry-based inhibitors because of its critical role in defining the specificity of Env interaction with cellular coreceptor molecules, usually CCR5 or CXCR4, to facilitate entry into target cells. Coreceptor specificity may also be important for viral transmission, since CCR5-utilizing viruses are frequently (but not always) present during acute/early HIV-1 infection.29,83–88 V3 is also highly immunogenic for eliciting antibodies in infected patients89,90 and following immunization of animals.91–93
FIG. 2.
V3 consensus sequences for HIV-1 group M subtypes. Consensus amino acid sequences were obtained from the Los Alamos HIV Database and aligned using Seqpublish. Dashes indicate conserved residues relative to the A1 consensus; dots indicate a deleted residue relative to A1; and amino acid differences from A1 are indicated. The base regions of V3 are underlined. Red residues indicate those participating in a potential “hydrophobic cluster”; green indicates the R/Q substitution that distinguishes B from many non-B subtypes; blue indicates a single difference between the subtype A1 and C consensus.
V3 has traditionally been considered a hypervariable domain, based mostly on examination of subtype B sequences. However, the entropy exhibited by the V3 loop of CCR5 utilizing subtype B viruses is more similar to the conserved regions of gp120 than to the other hypervariable domains V1V2, V4, and V5.53 An even lower level of sequence variation has been reported for subtype C V3 in studies using sequences deposited in the Los Alamos HIV Database. When V3 and its flanking regions were analyzed for mutational trends, the subtype D V3 domain was more divergent than the other subtypes analyzed, while the subtype C V3 domain was relatively well conserved.45 Examination of nonsynonymous to synonymous substitution ratios (dN/dS; a measure of positive selection) in V3 revealed much higher diversifying selection in subtype B than in subtype C, which was particularly conserved within the turn region.46 The predominant sequence of this turn region also varies between subtypes (Fig 3; residues in green). Subtypes A and C usually contain a highly conserved GPGQ amino acid motif, while GPGR is predominant in subtype B Envs.45,94 Subtype D Envs, on the other hand, carry a mixture of residues at the R/Q position (www.hiv.lanl.gov).
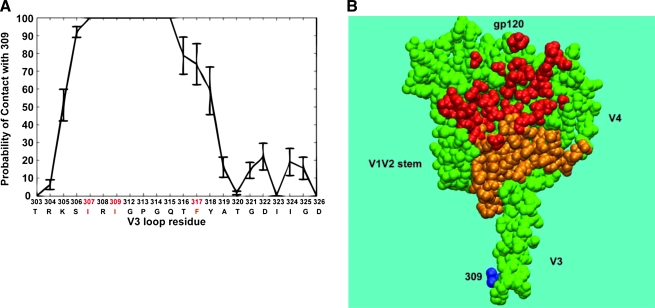
FIG. 3.
Structure-based analyses of local and global V3 interactions. (A) Contact profile of Ile 309 within the V3 loop. The graph shows the probability of contact plotted on the vertical axis between Ile 309 and the individual residues within V3. The HXB2 amino acid position and the subtype C consensus sequence for V3 are shown on the horizontal axis. The contact profile was obtained from an all-atom molecular dynamics simulation of subtype C consensus gp120 in aqueous solution. The error bars show SEM obtained from 1 ns block analysis. (B) Regions in core gp120 that could potentially interact with Ile 309. A coarse-grained model was used, and residues that showed any contact probability with Ile 309 are mapped onto the gp120 structure (2B4C;53 in orange). Residues that have been previously shown to participate in CD4 binding are red. The position of Ile 309 at the V3 crown is highlighted in blue.
Different mutational patterns in V3 across subtypes may have clinical significance by influencing the effectiveness of CCR5 inhibitors such as maraviroc,95 which was recently approved by the FDA for HIV treatment. Escape from small molecule CCR5 inhibitors is usually associated with changes in the V3 domain.96–99 One mechanism of escape is adaptation to use the inhibitor-bound form of CCR5,99–101 while another is to utilize an alternate coreceptor for entry.102,103 Certainly the presence of viruses that utilize CXCR4 will influence the clinical utility of CCR5-targeted inhibitors, and this property appears to differ between subtypes. For instance, the subtype B V3 domain facilitates a switch in tropism, from CCR5 to CXCR4 usage in about 50% of patients,83,104–106 whereas CXCR4 usage among subtype C viruses is less frequent17–19,27,107,108 even in later stage patients.16,28,109 Subtype D has a higher prevalence of X4 tropism than subtype A, which is mostly R5-tropic.20,110–112 CRF02_AG-like isolates from Ghana were also found to be predominantly R5-tropic.112 CRF14_BG isolates and BG URFs from Spain were frequently X4-tropic, while CRF02_AG isolates were mostly R5-tropic.113 Thus, unlike the universal susceptibility to enfuvirtide discussed above, the efficacy of R5 inhibitors such as maraviroc will depend on the phenotype of the circulating virus within the treated patient population. Once treated, inhibitor escape could also be influenced by the ability of different subtypes to tolerate certain sequence changes in V3.
It is important to note, however, that among these studies cited above, a variety of methods have been used to assess coreceptor usage. The earlier studies used induction of syncytia in the MT-2 cell line and/or replication in macrophages as indicators of tropism, while more recent studies have included infection of cell lines stably expressing CCR5 or CXCR4, or the use of coreceptor-specific inhibitors, to indicate tropism. In addition, sequence characteristics of the V3 domain, such as an increased net positive charge or substitutions at specific residues, have been used as a surrogate for directly evaluating coreceptor usage. Recent studies based on sequence alone have demonstrated that the subtype C V3 domain exhibits less variation compared to subtype B.47,94,114 Comparisons of the V3 region of subtype B and C viruses have also demonstrated a greater number of covarying residues in subtype B sequences as compared to C.47,114
The different mutational patterns between subtypes B and C could simply be due to more frequent CXCR4 usage in subtype B, because expanded coreceptor tropism is linked with sequence variation in the V3 domain.53 To control for this factor, Patel et al. analyzed 391 B and 351 C sequences for differences in mutational patterns after excluding V3 sequences predicted to utilize CXCR4. Using this subset of V3 sequences, the base region (closest to the core) exhibited almost identical entropy between subtypes B and C. However, significant differences in entropy, amino acid composition, and patterns of covariation were apparent in the stem and turn regions of subtype B and C V3. Interestingly, the authors also demonstrated that some subtype B derived anti-V3 monoclonal antibodies were able to bind to representative subtype B and C V3 peptides, but could bind only the subtype B gp120 molecule, indicating that the subtype C V3 within its cognate protein adopts a distinct conformation. In the X-ray structure of the CD4-liganded subtype B gp120 molecular with an intact V3, this loop projects away from the core, suggesting that it could act as a “molecular hook” that engages coreceptor after CD4 binding.53 However, in the unliganded gp120 trimer that is recognized by neutralizing antibody, the V3 loop could be more flexible, adopting multiple conformations that are influenced by interactions with the gp120 core or the other variable loops, such as V1V2. Thus, while lineage-specific genetic differences in the V3 domain have been established, their structural consequences are less clearly understood.
Mutational patterns in the α2 helix in subtypes B and C
Interestingly, in subtype C, the structural domain encoded immediately downstream from V3 in the C3 region (the α2 helix) not only exhibits higher dN/dS ratios than B,46 but also higher entropy at variable positions, as shown in a comparison using 582 C-Envs and 634 B-Envs from the HIV database.56 The amino acid composition of the 18 residue α2 helix also differed between these two subtypes. In subtype C, the α2 helix is amphipathic (it maintains distinct polar and nonpolar faces), and variable positions on the surface can accommodate a positively or negatively charged residue. In contrast, sequence variation in the subtype B helix does not strictly preserve the amphipathicity of the α2 helix and variable positions maintain a similar charge.56 The interior positions of the helix are well conserved in both subtypes, indicating critical contacts with the gp120 core. In a separate study, using a mutual information analysis of 73 subtype C Envs from Zambian donor–recipient transmission pairs, sequence variation at five residues within the α2 helix tracked with neutralization resistance against linked donor plasma in a pseudoviral assay; however, domain exchange studies showed that the α2 helix was not sufficient to confer neutralization resistance.57 These studies suggest that this structure has a prominent but unidentified role in escape from immune pressure during subtype C infection.
Autologous Nab responses during infection with subtypes B and C
In HIV-1 infection, neutralizing antibodies are directed against Env. Given the subtype differences in these proteins outlined above, it would not be surprising to find variation in the serology of infection with diverse subtypes. During natural infection, subtype B HIV-1 elicits neutralizing antibody activity against the autologous virus that is usually detectable in patient plasma within the first few months of infection.61,63,115 Subtype C HIV-1 elicits a Nab response with similar kinetics.40,89 However, when the autologous Nab response in 6 subtype B-infected seroconvertors was compared directly against 11 subtype C-infected seroconvertors from Zambia, a 3.5-fold higher 50% inhibitory titer of Nab was found in the C subjects. In terms of the breadth of the Nab response, plasma from these subtype C subjects had less cross-reactive activity against heterologous Envs of the same subtype, compared to plasma from the subtype B patients,40 suggesting that the initial Nab response in subtype C infection is directed against strain-specific epitopes. This lack of cross-reactivity in early subtype C infection was corroborated in an independent study of 14 South African patients.89 Intriguingly, in studies of South African patients, antibodies directed against the V3 domain were present in the plasma of all subjects during early infection, and were capable of binding to autologous and heterologous V3 peptides, yet these antibodies did not contribute to neutralization of autologous virus in most of the patients.107,116 The ubiquitous presence of anti-V3 antibodies could suggest recognition of “decoy” V3 epitopes exposed on defective Env forms (i.e., monomeric Env), but sequestration of V3 on the native, trimeric Env, thereby preventing neutralizing activity.117 Furthermore, Moore et al. demonstrated that Nab activity in the South African patients seems to be frequently directed toward epitopes within the C3–V4 region.116
Examination of Nab breadth during infection with different subtypes
The inability to induce broadly cross-neutralizing antibodies against HIV during natural infection, much less via immunization, has hampered attempts to generate an effective vaccine. Recent studies of the cross-neutralization properties of individual and pooled subtype-specific plasma using both pseudovirus and PBMC-based assays have determined that in general, subtype-specific relationships do exist between neutralizing antibody and virus sensitivity.77,88,107,118,119 However, one study found that for individual subtype C plasma samples, genetic relatedness between the autologous and heterologous Env within subtype C was not a determinant of cross-neutralizing activity.119 In other words, patient plasma was not more likely to cross-neutralize a heterologous virus that shared genetic similarity with the autologous virus than a heterologous virus that was more distantly related. This could reflect the poor concordance between neutralization epitopes, which are often conformational, and the linear, gap-stripped Env sequence that is analyzed with phylogenetic methods. The authors of this study did find that autologous viruses with shorter hypervariable domains (the V1V2 domain and the V1V4 region) were better able to elicit antibodies that could cross-neutralize heterologous C Envs, but there was no association between shorter loop length in the autologous virus and the ability to cross-neutralize B Envs, suggesting that the targets of neutralization differ between the two subtypes.119
A separate study that found higher in vitro autologous Nab titers in early subtype C vs. B infection (discussed above) also demonstrated that the shorter hypervariable domains in the subtype C Envs were correlated with Nab potency.40 In fact, subtype C Envs tend to have shorter hypervariable domains than B Envs in general40,56,88 and this propensity could contribute to the observed subtype differences in Env immunogenicity and susceptibility to neutralization. Intriguingly, in one study, subtype C pooled plasma was highly cross-neutralizing against viral Envs of almost all subtypes measured (A, B, C, D, AE, and AG) using a sensitive pseudovirus assay.118 As previously discussed, individual subtype C plasma samples usually do not possess high levels of intraclade or interclade neutralization activity within the first few years of infection.40,89,107 The autologous Nab response against subtype C Env is typically potent yet focused, and data from our laboratory indicate that distinct epitopes may be targeted across patients in the early stages of infection (Rong et al., unpublished observations). One explanation for this apparent contradiction is that when these plasma samples are pooled, the breadth of targets recognized is increased substantially. In contrast, if autologous Nab across subtype B infected patients recognized similar targets, as suggested,61 pooling the plasma would not be expected to dramatically increase the breadth.118 A few “broadly reactive” monoclonal antibodies have been derived from subtype B-infected patients, but most lack neutralizing activity against non-B viruses.77,88,107,110,120 For example, antibodies 2F5 and 2G12 have limited activity against subtypes A, C, and D viruses, and for 2G12, simply reconstituting the epitope in subtype C Env does not necessarily result in neutralization, suggesting that conformational constraints prevent formation or exposure of this epitope.39 Together these findings indicate that Env of different subtypes has distinct antigenic properties.
Nab responses directed against V3
As discussed above, the R/Q substitution found at the V3 tip of non-B subtypes constitutes a major antigenic distinction for neutralization by some anti-V3 monoclonal antibodies.41,121 X-ray and NMR structures of subtype B V3 bound to monoclonal antibody 447-52D have proven useful in deducing the role of the GPGR motif on obtaining antibody specificity, which was linked specifically to different patterns of surface charge at the tip of the loop in one study.122 However, cross-subtype reactive anti-V3 antibodies can be elicited in patients as well, indicating that there are also conserved features of V3. Subtype A infections appear to elicit antibodies that recognize features of V3 that are conserved across subtypes. For example, plasma from Cameroonian patients infected with subtype A or CRF02_AG more frequently harbored anti-V3 antibodies that were cross-reactive than did North American patients infected with subtype B.90 This finding was confirmed and expanded in a study examining the neutralization capabilities of anti-V3 monoclonal antibodies derived from patients infected with subtype A or B Env.41 Interestingly, when these anti-V3A or anti-V3B MAbs were evaluated against viruses containing a V3 consensus sequence from multiple subtypes within a neutralization-sensitive Env background (SF162), the B consensus V3 was preferentially neutralized by all of the anti-V3 MAbs, even those elicited against subtype A V3. When the subtype C V3 consensus was placed into the same background, it was neutralized, but much less efficiently than the subtype A and B V3, even though the A and C V3 sequences differed by only one residue (Fig. 2, blue residues immediately C terminal to the GPGQ turn). Thus it appears that antibodies that are directed against subtype A and B V3 can neutralize the native trimer conformation of subtype B V3 more potently than the other subtypes, but display weak activity against the subtype C V3 in particular. Furthermore, this study illustrates how a single substitution within the turn region can produce a dramatic phenotypic effect.
Conservation of hydrophobic residues in V3 in subtypes B and C
Thus, studies from our laboratory and others have demonstrated that the subtype C V3 domain is less variable in sequence, and is under less selective pressure, than subtype B. One explanation for this phenomenon could be that subtype C V3 is less exposed on the native Env trimer. Evidence for this hypothesis comes from findings that subtype B viruses are susceptible to neutralization by anti-V3 monoclonal antibodies;41,77 however, this activity can be limited by conformational masking of V3 on the virion-associated Env trimer41,63,123 and sequence variation.121 In contrast, subtype C appears to be generally less susceptible to anti-V3-mediated neutralization.41,47,89,107 Thus, V3 could exist in multiple conformations on the unliganded Env trimer, some of which are more accessible to antibody than others.
One possible factor influencing V3 exposure could be the arrangement of hydrophobic residues within the V3 stem, particularly residues I307, I309, and F317 (Fig. 2, residues in red). These three hydrophobic residues are conserved in both subtype B and C sequences in the database, but to varying degrees.47 I307 is more highly conserved in B than C (97 vs. 54%), while the reverse is true for I309 (68 vs. 99%) and F317 (75 vs. 97%). Variation in these positions, however, is restricted to hydrophobic residues.47 Thus, while the framework for a “hydrophobic cluster” could potentially exist in both B and C V3 domains, it appears that subtype C tends to preserve specific hydrophobic residues at two out of three positions. All atom molecular dynamics simulations in our laboratory (Fig. 3A)56 and crystallographic studies in the context of V3 peptide bound to antibody by others94 have provided evidence for the proximity of these residues to one another. In the Stanfield et al. study, a naturally occurring Leu at position 309 (instead of the more common Ile at 309) of a subtype A V3 domain altered the orientation of the F317 side chain on the opposite strand, suggesting that 309 and 317 were in close contact.94 Further evidence for the proximity of I307, I309, and F317 stems from recent crystallographic studies of V3 peptide bound to MAb 3074.124 This study found that these three residues comprise part of the 3074 epitope. Moreover, coarse-grained calculations that we performed demonstrate that the subtype C V3 (and perhaps the hydrophobic cluster) has the potential to interact with multiple residues in the gp120 core, several of which are proximal to the CD4 binding site and may impact CD4 binding (Fig. 3B). A simple assessment of the gp120 backbone atomic fluctuation profile (B-factors) revealed that variable loops such as V3 show much greater flexibility compared to the core regions (Fig. 1). Properties of V3 should therefore be considered in the context of a dynamic structure.
Stabilizing forces could drive hydrophobic residues to avoid solvent exposure by burying themselves within the V3 loop or into the gp120 core. Conservation of these and other residues in subtype C V3 could therefore constrain exposure of this domain, and could also restrict adaptability for the sequence changes that facilitate CXCR4 utilization. A higher level of positive selection, CXCR4 utilization, and susceptibility to anti-V3-mediated neutralization is observed in subtype B V3, arguing that perhaps this region is more frequently exposed on the native B Env trimer. A higher level and different pattern of sequence variation in subtype B V3 could therefore function to prevent anti-V3-mediated neutralization and to facilitate expanded tropism.
Summary
The extreme genetic diversity of HIV-1 poses a significant challenge for global vaccination approaches, and strategies to overcome this are extremely limited at present. In an effort to understand the biological consequences of intersubtype diversity, recent research has linked genetic differences in Env to both phenotypic and antigenic properties. A particular focus has been on subtypes B and C, where differences have been associated with distinct autologous humoral responses that vary in gp120 targets as well as in cross-reactive breadth, especially in the V3 domain. It is important to note that differences between subtypes that circulate in distinct geographic regions, such as B and C, could also reflect dissimilarity in the host population from which the viruses were derived, epidemic patterns, the route of infection, etc. Nevertheless, as studies continue to uncover subtype-specific differences in Env function, structure, and antigenicity, these will be important to incorporate into global vaccine design.
Acknowledgments
We would like to acknowledge Dr. Abraham Pinter for critical comments; Drs. Susan Allen and Joseph Mulenga and the project management group, staff, and participants of the Zambia Emory HIV Research Project for collaboration; and NIH Grant R01-AI-58706 for funding.
Disclosure Statement
No competing financial interests exist.
References
- 1.UNAIDS-WHO. UNAIDS Annual Report. Joint United Nations Programme on HIV/AIDS and World Health Organization; Geneva, Switzerland: 2007. [Google Scholar]
- 2.Coffin JM. Genetic diversity and evolution of retroviruses. Curr Top Microbiol Immunol. 1992;176:143–164. doi: 10.1007/978-3-642-77011-1_10. [DOI] [PubMed] [Google Scholar]
- 3.Pathak VK. Temin HM. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: Substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci USA. 1990;87(16):6019–6023. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho DD. Neumann AU. Perelson AS. Chen W. Leonard JM. Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373(6510):123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 5.Wei X. Ghosh SK. Taylor ME, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373(6510):117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 6.Ramirez BC. Simon-Loriere E. Galetto R. Negroni M. Implications of recombination for HIV diversity. Virus Res. 2008;134(1–2):64–73. doi: 10.1016/j.virusres.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Peeters M. Toure-Kane C. Nkengasong JN. Genetic diversity of HIV in Africa: Impact on diagnosis, treatment, vaccine development and trials. AIDS. 2003;17(18):2547–2560. doi: 10.1097/01.aids.0000096895.73209.89. [DOI] [PubMed] [Google Scholar]
- 8.McCutchan F. Global epidemiology of HIV. J Med Virol. 2006;78(S1):S7–S12. doi: 10.1002/jmv.20599. [DOI] [PubMed] [Google Scholar]
- 9.Korber B. Muldoon M. Theiler J, et al. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288(5472):1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 10.Worobey M. Gemmel M. Teuwen DE, et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature. 2008;455(7213):661–664. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu T. Korber BT. Nahmias AJ. Hooper E. Sharp PM. Ho DD. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature. 1998;391(6667):594–597. doi: 10.1038/35400. [DOI] [PubMed] [Google Scholar]
- 12.Taylor BS. Sobieszczyk ME. McCutchan FE. Hammer SM. The challenge of HIV-1 subtype diversity. N Engl J Med. 2008;358(15):1590–1602. doi: 10.1056/NEJMra0706737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemelaar J. Gouws E. Ghys PD. Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20(16):W13–W23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 14.Bjorndal A. Sonnerborg A. Tscherning C. Albert J. Fenyo EM. Phenotypic characteristics of human immunodeficiency virus type 1 subtype C isolates of Ethiopian AIDS patients. AIDS Res Hum Retroviruses. 1999;15(7):647–653. doi: 10.1089/088922299310944. [DOI] [PubMed] [Google Scholar]
- 15.De Wolf F. Hogervorst E. Goudsmit J, et al. Syncytium-inducing and non-syncytium-inducing capacity of human immunodeficiency virus type 1 subtypes other than B: Phenotypic and genotypic characteristics. WHO Network for HIV Isolation and Characterization. AIDS Res Hum Retroviruses. 1994;10(11):1387–1400. doi: 10.1089/aid.1994.10.1387. [DOI] [PubMed] [Google Scholar]
- 16.Morris L. Cilliers T. Bredell H. Phoswa M. Martin DJ. CCR5 is the major coreceptor used by HIV-1 subtype C isolates from patients with active tuberculosis. AIDS Res Hum Retroviruses. 2001;17(8):697–701. doi: 10.1089/088922201750236979. [DOI] [PubMed] [Google Scholar]
- 17.Cilliers T. Nhlapo J. Coetzer M, et al. The CCR5 and CXCR4 coreceptors are both used by human immunodeficiency virus type 1 primary isolates from subtype C. J Virol. 2003;77(7):4449–4456. doi: 10.1128/JVI.77.7.4449-4456.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Batra M. Tien PC. Shafer RW. Contag CH. Katzenstein DA. HIV type 1 envelope subtype C sequences from recent seroconverters in Zimbabwe. AIDS Res Hum Retroviruses. 2000;16:973–979. doi: 10.1089/08892220050058399. [DOI] [PubMed] [Google Scholar]
- 18.Coetzer M. Cilliers T. Ping LH. Swanstrom R. Morris L. Genetic characteristics of the V3 region associated with CXCR4 usage in HIV-1 subtype C isolates. Virology. 2006;356(1–2):95–105. doi: 10.1016/j.virol.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 19.Choge I. Cilliers T. Walker P, et al. Genotypic and phenotypic characterization of viral isolates from HIV-1 subtype C-infected children with slow and rapid disease progression. AIDS Res Hum Retroviruses. 2006;22(5):458–465. doi: 10.1089/aid.2006.22.458. [DOI] [PubMed] [Google Scholar]
- 20.Huang W. Eshleman SH. Toma J, et al. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: High prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J Virol. 2007;81(15):7885–7893. doi: 10.1128/JVI.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abebe A. Demissie D. Goudsmit J, et al. HIV-1 subtype C syncytium- and non-syncytium-inducing phenotypes and coreceptor usage among Ethiopian patients with AIDS. AIDS. 1999;13(11):1305–1311. doi: 10.1097/00002030-199907300-00006. [DOI] [PubMed] [Google Scholar]
- 22.Peeters M. Vincent R. Perret JL, et al. Evidence for differences in MT2 cell tropism according to genetic subtypes of HIV-1: Syncytium-inducing variants seem rare among subtype C HIV-1 viruses. J Acquir Immune Defic Syndr Hum Retrovirol. 1999;20(2):115–121. doi: 10.1097/00042560-199902010-00002. [DOI] [PubMed] [Google Scholar]
- 23.Treurnicht FK. Smith TL. Engelbrecht S, et al. Genotypic and phenotypic analysis of the env gene from South African HIV-1 subtype B and C isolates. J Med Virol. 2002;68(2):141–146. doi: 10.1002/jmv.10199. [DOI] [PubMed] [Google Scholar]
- 24.Tscherning C. Alaeus A. Fredriksson R, et al. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology. 1998;241(2):181–188. doi: 10.1006/viro.1997.8980. [DOI] [PubMed] [Google Scholar]
- 25.Utaipat U. Duerr A. Rudolph DL, et al. Coreceptor utilization of HIV type 1 subtype E viral isolates from Thai men with HIV type 1-infected and uninfected wives. AIDS Res Hum Retroviruses. 2002;18(1):1–11. doi: 10.1089/088922202753394664. [DOI] [PubMed] [Google Scholar]
- 26.Zhong P. Bu S. Konings F, et al. Genetic and biological properties of HIV type 1 isolates prevalent in villagers of the Cameroon equatorial rain forests and grass fields: Further evidence of broad HIV type 1 genetic diversity. AIDS Res Hum Retroviruses. 2003;19(12):1167–1178. doi: 10.1089/088922203771881284. [DOI] [PubMed] [Google Scholar]
- 27.Ping LH. Nelson JA. Hoffman IF, et al. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: Underrepresentation of X4 variants. J Virol. 1999;73(8):6271–6281. doi: 10.1128/jvi.73.8.6271-6281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cecilia D. Kulkarni SS. Tripathy SP. Gangakhedkar RR. Paranjape RS. Gadkari DA. Absence of coreceptor switch with disease progression in human immunodeficiency virus infections in India. Virology. 2000;271(2):253–258. doi: 10.1006/viro.2000.0297. [DOI] [PubMed] [Google Scholar]
- 29.Williamson C. Morris L. Maughan MF, et al. Characterization and selection of HIV-1 subtype C isolates for use in vaccine development. AIDS Res Hum Retroviruses. 2003;19(2):133–144. doi: 10.1089/088922203762688649. [DOI] [PubMed] [Google Scholar]
- 30.Ball SC. Abraha A. Collins KR, et al. Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C. J Virol. 2003;77(2):1021–1038. doi: 10.1128/JVI.77.2.1021-1038.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marozsan AJ. Moore DM. Lobritz MA, et al. Differences in the fitness of two diverse wild-type human immunodeficiency virus type 1 isolates are related to the efficiency of cell binding and entry. J Virol. 2005;79(11):7121–7134. doi: 10.1128/JVI.79.11.7121-7134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasan A. Renjifo B. Hertzmark E, et al. Different rates of disease progression of HIV type 1 infection in Tanzania based on infecting subtype. Clin Infect Dis. 2006;42(6):843–852. doi: 10.1086/499952. [DOI] [PubMed] [Google Scholar]
- 33.Baeten JM. Chohan B. Lavreys L, et al. HIV-1 subtype D infection is associated with faster disease progression than subtype A in spite of similar plasma HIV-1 loads. J Infect Dis. 2007;195(8):1177–1180. doi: 10.1086/512682. [DOI] [PubMed] [Google Scholar]
- 34.Rangsin R. Piyaraj P. Sirisanthana T. Sirisopana N. Short O. Nelson KE. The natural history of HIV-1 subtype E infection in young men in Thailand with up to 14 years of follow-up. AIDS. 2007;21(Suppl. 6):S39–S46. doi: 10.1097/01.aids.0000299409.29528.23. [DOI] [PubMed] [Google Scholar]
- 35.Kiwanuka N. Laeyendecker O. Robb M, et al. Effect of human immunodeficiency virus Type 1 (HIV-1) subtype on disease progression in persons from Rakai, Uganda, with incident HIV-1 infection. J Infect Dis. 2008;197(5):707–713. doi: 10.1086/527416. [DOI] [PubMed] [Google Scholar]
- 36.Chohan B. Lang D. Sagar M, et al. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1–V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J Virol. 2005;79(10):6528–6531. doi: 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frost SD. Liu Y. Pond SL, et al. Characterization of human immunodeficiency virus type 1 (HIV-1) envelope variation and neutralizing antibody responses during transmission of HIV-1 subtype B. J Virol. 2005;79(10):6523–6527. doi: 10.1128/JVI.79.10.6523-6527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derdeyn CA. Decker JM. Bibollet-Ruche F, et al. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303(5666):2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 39.Gray ES. Moore PL. Pantophlet RA. Morris L. N-linked glycan modifications in gp120 of human immunodeficiency virus type 1 subtype C render partial sensitivity to 2G12 antibody neutralization. J Virol. 2007;81(19):10769–10776. doi: 10.1128/JVI.01106-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li B. Decker JM. Johnson RW, et al. Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J Virol. 2006;80(11):5211–5218. doi: 10.1128/JVI.00201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krachmarov CP. Honnen WJ. Kayman SC. Gorny MK. Zolla-Pazner S. Pinter A. Factors determining the breadth and potency of neutralization by v3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J Virol. 2006;80(14):7127–7135. doi: 10.1128/JVI.02619-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.John-Stewart GC. Nduati RW. Rousseau CM, et al. Subtype C Is associated with increased vaginal shedding of HIV-1. J Infect Dis. 2005;192(3):492–496. doi: 10.1086/431514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Travers SA. O'Connell MJ. McCormack GP. McInerney JO. Evidence for heterogeneous selective pressures in the evolution of the env gene in different human immunodeficiency virus type 1 subtypes. J Virol. 2005;79(3):1836–1841. doi: 10.1128/JVI.79.3.1836-1841.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choisy M. Woelk CH. Guegan JF. Robertson DL. Comparative study of adaptive molecular evolution in different human immunodeficiency virus groups and subtypes. J Virol. 2004;78(4):1962–1970. doi: 10.1128/JVI.78.4.1962-1970.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korber BT. MacInnes K. Smith RF. Myers G. Mutational trends in V3 loop protein sequences observed in different genetic lineages of human immunodeficiency virus type 1. J Virol. 1994;68(10):6730–6744. doi: 10.1128/jvi.68.10.6730-6744.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaschen B. Taylor J. Yusim K, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296(5577):2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 47.Patel MB. Hoffman NG. Swanstrom R. Subtype-specific conformational differences within the V3 region of subtype B and subtype C human immunodeficiency virus type 1 Env proteins. J Virol. 2007;82(2):903–916. doi: 10.1128/JVI.01444-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Felsovalyi K. Nadas A. Zolla-Pazner S. Cardozo T. Distinct sequence patterns characterize the V3 region of HIV type 1 gp120 from subtypes A and C. AIDS Res Hum Retroviruses. 2006;22(7):703–708. doi: 10.1089/aid.2006.22.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hunter E. Viral entry and receptors. In: Goff S, editor; Coffin JM, editor; Hughes SH, editor; Varmus HE, editor. Retroviruses. Cold Spring Harbor Laboratory Press; Plainview, NY: 1997. pp. 71–121. [PubMed] [Google Scholar]
- 50.Kwong PD. Wyatt R. Majeed S, et al. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure Fold Des. 2000;8(12):1329–1339. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- 51.Kwong PD. Wyatt R. Robinson J. Sweet RW. Sodroski J. Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen B. Vogan EM. Gong H. Skehel JJ. Wiley DC. Harrison SC. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433(7028):834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 53.Huang CC. Tang M. Zhang MY, et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310(5750):1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou T. Xu L. Dey B, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445(7129):732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Korber B. Gaschen B. Yusim K. Thakallapally R. Kesmir C. Detours V. Evolutionary and immunological implications of contemporary HIV-1 variation. Br Med Bull. 2001;58:19–42. doi: 10.1093/bmb/58.1.19. [DOI] [PubMed] [Google Scholar]
- 56.Gnanakaran S. Lang D. Daniels M. Bhattacharya T. Derdeyn CA. Korber B. Clade specific differences in HIV-1: Diversity and correlations in C3–V4 regions of gp120. J Virol. 2007;81(9):4886–4891. doi: 10.1128/JVI.01954-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rong R. Gnanakaran S. Decker JM, et al. Unique mutational patterns in the envelope {alpha}2 amphipathic helix and acquisition of length in gp120 hyper-variable domains are associated with resistance to autologous neutralization of subtype C human immunodeficiency virus type 1. J Virol. 2007;81(11):5658–5668. doi: 10.1128/JVI.00257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borrow P. Lewicki H. Wei X, et al. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3(2):205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 59.Frost SD. Wrin T. Smith DM, et al. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc Natl Acad Sci USA. 2005;102(51):18514–18519. doi: 10.1073/pnas.0504658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Streeck H. Li B. Poon AF, et al. Immune-driven recombination and loss of control after HIV superinfection. J Exp Med. 2008;205(8):1789–1796. doi: 10.1084/jem.20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bunnik EM. Pisas L. van Nuenen AC. Schuitemaker H. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J Virol. 2008;82(16):7932–7941. doi: 10.1128/JVI.00757-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Navis M. Matas DE. Rachinger A, et al. Molecular evolution of human immunodeficiency virus type 1 upon transmission between human leukocyte antigen disparate donor-recipient pairs. PLoS ONE. 2008;3(6):e2422. doi: 10.1371/journal.pone.0002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei X. Decker JM. Wang S. Hui H. Kappes JC. Wu X. Salazar JF. Salazar MG. Kilby JM. Saag MS. Komarova NL. Nowak MA. Hahn BH. Kwong PD. Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 64.Rong R. Bibollet-Ruche F. Mulenga J. Allen S. Blackwell JL. Derdeyn CA. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J Virol. 2007;81(3):1350–1359. doi: 10.1128/JVI.01839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mink M. Mosier SM. Janumpalli S, et al. Impact of human immunodeficiency virus type 1 gp41 amino acid substitutions selected during enfuvirtide treatment on gp41 binding and antiviral potency of enfuvirtide in vitro. J Virol. 2005;79(19):12447–12454. doi: 10.1128/JVI.79.19.12447-12454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heil ML. Decker JM. Sfakianos JN. Shaw GM. Hunter E. Derdeyn CA. Determinants of human immunodeficiency virus type 1 baseline susceptibility to the fusion inhibitors enfuvirtide and T-649 reside outside the peptide interaction site. J Virol. 2004;78(14):7582–7589. doi: 10.1128/JVI.78.14.7582-7589.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Holguin A. De Arellano ER. Soriano V. Amino acid conservation in the gp41 transmembrane protein and natural polymorphisms associated with enfuvirtide resistance across HIV-1 variants. AIDS Res Hum Retroviruses. 2007;23(9):1067–1074. doi: 10.1089/aid.2006.0256. [DOI] [PubMed] [Google Scholar]
- 68.Holguin A. Faudon JL. Labernardiere JL. Soriano V. Susceptibility of HIV-1 non-B subtypes and recombinant variants to enfuvirtide. J Clin Virol. 2007;38(2):176–180. doi: 10.1016/j.jcv.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 69.Sanders RW. Korber B. Lu M. Berkhout B. Moore JP. Mutational analyses and natural variability of the gp41 ectodomain. In: Kuiken C, editor; Foley B, editor; Freed E, et al., editors. HIV Sequence Compendium. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; Los Alamos, NM: 2002. pp. I23–I47. [Google Scholar]
- 70.Razzolini F. Vicenti I. Saladini F, et al. Natural variability in the HR-1 and HR-2 domains of HIV type 1 gp41 from different clades circulating in Italy. AIDS Res Hum Retroviruses. 2007;23(4):558–563. doi: 10.1089/aid.2006.0273. [DOI] [PubMed] [Google Scholar]
- 71.Eshleman SH. Hudelson SE. Bruce R, et al. Analysis of HIV type 1 gp41 sequences in diverse HIV type 1 strains. AIDS Res Hum Retroviruses. 2007;23(12):1593–1598. doi: 10.1089/aid.2007.0130. [DOI] [PubMed] [Google Scholar]
- 72.Cilliers T. Patience T. Pillay C. Papathanasopoulos M. Morris L. Sensitivity of HIV type 1 subtype C isolates to the entry inhibitor T-20. AIDS Res Hum Retroviruses. 2004;20(5):477–482. doi: 10.1089/088922204323087714. [DOI] [PubMed] [Google Scholar]
- 73.Chinnadurai R. Munch J. Dittmar MT. Kirchhoff F. Inhibition of HIV-1 group M and O isolates by fusion inhibitors. AIDS. 2005;19(16):1919–1922. doi: 10.1097/01.aids.0000188425.79914.e4. [DOI] [PubMed] [Google Scholar]
- 74.Fleury HJ. Toni T. Lan NT, et al. Susceptibility to antiretroviral drugs of CRF01_AE, CRF02_AG, and subtype C viruses from untreated patients of Africa and Asia: Comparative genotypic and phenotypic data. AIDS Res Hum Retroviruses. 2006;22(4):357–366. doi: 10.1089/aid.2006.22.357. [DOI] [PubMed] [Google Scholar]
- 75.Gray ES. Moore PL. Bibollet-Ruche F, et al. 4E10-resistant variants in a human immunodeficiency virus type 1 subtype C-infected individual with an anti-membrane-proximal external region-neutralizing antibody response. J Virol. 2008;82(5):2367–2375. doi: 10.1128/JVI.02161-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yuste E. Sanford HB. Carmody J, et al. Simian immunodeficiency virus engrafted with human immunodeficiency virus type 1 (HIV-1)-specific epitopes: Replication, neutralization, and survey of HIV-1-positive plasma. J Virol. 2006;80(6):3030–3041. doi: 10.1128/JVI.80.6.3030-3041.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76a.Binley JM. Lybarger EA. Crooks ET, et al. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol. 2008;82:11651–11668. doi: 10.1128/JVI.01762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Binley JM. Wrin T. Korber B, et al. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78(23):13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zwick MB. Jensen R. Church S, et al. Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J Virol. 2005;79(2):1252–1261. doi: 10.1128/JVI.79.2.1252-1261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brunel FM. Zwick MB. Cardoso RM, et al. Structure-function analysis of the epitope for 4E10, a broadly neutralizing human immunodeficiency virus type 1 antibody. J Virol. 2006;80(4):1680–1687. doi: 10.1128/JVI.80.4.1680-1687.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cardozo T. Kimura T. Philpott S. Weiser B. Burger H. Zolla-Pazner S. Structural basis for coreceptor selectivity by the HIV type 1 V3 loop. AIDS Res Hum Retroviruses. 2007;23(3):415–426. doi: 10.1089/aid.2006.0130. [DOI] [PubMed] [Google Scholar]
- 81.Cormier EG. Dragic T. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J Virol. 2002;76(17):8953–8957. doi: 10.1128/JVI.76.17.8953-8957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Trkola A. Dragic T. Arthos J, et al. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co- receptor CCR-5. Nature. 1996;384(6605):184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 83.Connor RI. Sheridan KE. Ceradini D. Choe S. Landau NR. Change in coreceptor use coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang LQ. MacKenzie P. Cleland A. Holmes EC. Brown AJ. Simmonds P. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol. 1993;67(6):3345–3356. doi: 10.1128/jvi.67.6.3345-3356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu T. Mo H. Wang N, et al. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261(5125):1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 86.Keele BF. Giorgi EE. Salazar-Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105(21):7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li M. Gao F. Mascola JR, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79(16):10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li M. Salazar-Gonzalez JF. Derdeyn CA, et al. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol. 2006;80(23):11776–11790. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gray ES. Moore PL. Choge IA, et al. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J Virol. 2007;81(12):6187–6196. doi: 10.1128/JVI.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Krachmarov C. Pinter A. Honnen WJ, et al. Antibodies that are cross-reactive for human immunodeficiency virus type 1 clade a and clade B v3 domains are common in patient sera from Cameroon, but their neutralization activity is usually restricted by epitope masking. J Virol. 2005;79(2):780–790. doi: 10.1128/JVI.79.2.780-790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kraft Z. Strouss K. Sutton WF, et al. Characterization of neutralizing antibody responses elicited by clade A envelope immunogens derived from early transmitted viruses. J Virol. 2008;82(12):5912–5921. doi: 10.1128/JVI.00389-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ching LK. Vlachogiannis G. Bosch KA. Stamatatos L. The first hypervariable region of the gp120 Env glycoprotein defines the neutralizing susceptibility of heterologous human immunodeficiency virus type 1 isolates to neutralizing antibodies elicited by the SF162gp140 immunogen. J Virol. 2008;82(2):949–956. doi: 10.1128/JVI.02143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Derby NR. Kraft Z. Kan E, et al. Antibody responses elicited in macaques immunized with human immunodeficiency virus type 1 (HIV-1) SF162-derived gp140 envelope immunogens: Comparison with those elicited during homologous simian/human immunodeficiency virus SHIVSF162P4 and heterologous HIV-1 infection. J Virol. 2006;80(17):8745–8762. doi: 10.1128/JVI.00956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stanfield RL. Gorny MK. Zolla-Pazner S. Wilson IA. Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity. J Virol. 2006;80(12):6093–6105. doi: 10.1128/JVI.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dorr P. Westby M. Dobbs S, et al. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother. 2005;49(11):4721–4732. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kuhmann SE. Pugach P. Kunstman KJ, et al. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J Virol. 2004;78(6):2790–2807. doi: 10.1128/JVI.78.6.2790-2807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Anastassopoulou CG. Marozsan AJ. Matet A, et al. Escape of HIV-1 from a small molecule CCR5 inhibitor is not associated with a fitness loss. PLoS Pathog. 2007;3(6):e79. doi: 10.1371/journal.ppat.0030079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Marozsan AJ. Kuhmann SE. Morgan T, et al. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D) Virology. 2005;338(1):182–199. doi: 10.1016/j.virol.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 99.Tsibris AM. Sagar M. Gulick RM, et al. In vivo emergence of vicriviroc resistance in a human immunodeficiency virus type 1 subtype C-infected subject. J Virol. 2008;82(16):8210–8214. doi: 10.1128/JVI.00444-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pugach P. Marozsan AJ. Ketas TJ. Landes EL. Moore JP. Kuhmann SE. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology. 2007;361(1):212–228. doi: 10.1016/j.virol.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Westby M. Smith-Burchnell C. Mori J, et al. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J Virol. 2007;81(5):2359–2371. doi: 10.1128/JVI.02006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Daar ES. Emerging resistance profiles of newly approved antiretroviral drugs. Top HIV Med. 2008;16(4):110–116. [PubMed] [Google Scholar]
- 103.Westby M. Lewis M. Whitcomb J, et al. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J Virol. 2006;80(10):4909–4920. doi: 10.1128/JVI.80.10.4909-4920.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tersmette M. Lange JM. de Goede RE, et al. Association between biological properties of human immunodeficiency virus variants and risk for AIDS and AIDS mortality. Lancet. 1989;1:983–985. doi: 10.1016/s0140-6736(89)92628-7. [DOI] [PubMed] [Google Scholar]
- 105.Schuitemaker H. Koot M. Kootstra NA, et al. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: Progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66(3):1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Richman DD. Bozzette SA. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J Infect Dis. 1994;169:968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- 107.Bures R. Morris L. Williamson C, et al. Regional clustering of shared neutralization determinants on primary isolates of clade C human immunodeficiency virus type 1 from South Africa. J Virol. 2002;76(5):2233–2244. doi: 10.1128/jvi.76.5.2233-2244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ndung'u T. Sepako E. McLane MF, et al. HIV-1 subtype C in vitro growth and coreceptor utilization. Virology. 2006;347(2):247–260. doi: 10.1016/j.virol.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 109.Sullivan P. Decker WD. Mulenga J, et al. Coreceptor usage in seroconverting and transmitted partners in HIV transmissions in Lusaka, Zambia. AIDS Vaccine. 2008;24:70. [Google Scholar]
- 110.Blish CA. Nedellec R. Mandaliya K. Mosier DE. Overbaugh J. HIV-1 subtype A envelope variants from early in infection have variable sensitivity to neutralization and to inhibitors of viral entry. AIDS. 2007;21(6):693–702. doi: 10.1097/QAD.0b013e32805e8727. [DOI] [PubMed] [Google Scholar]
- 111.Rainwater SM. Wu X. Nduati R, et al. Cloning and characterization of functional subtype A HIV-1 envelope variants transmitted through breastfeeding. Curr HIV Res. 2007;5(2):189–197. doi: 10.2174/157016207780076986. [DOI] [PubMed] [Google Scholar]
- 112.Brandful JA. Coetzer ME. Cilliers T, et al. Phenotypic characterization of HIV type 1 isolates from Ghana. AIDS Res Hum Retroviruses. 2007;23(1):144–152. doi: 10.1089/aid.2007.23.144. [DOI] [PubMed] [Google Scholar]
- 113.Perez-Alvarez L. Munoz M. Delgado E, et al. Isolation and biological characterization of HIV-1 BG intersubtype recombinants and other genetic forms circulating in Galicia, Spain. J Med Virol. 2006;78(12):1520–1528. doi: 10.1002/jmv.20734. [DOI] [PubMed] [Google Scholar]
- 114.Gilbert PB. Novitsky V. Essex M. Covariability of selected amino acid positions for HIV type 1 subtypes C and B. AIDS Res Hum Retroviruses. 2005;21(12):1016–1030. doi: 10.1089/aid.2005.21.1016. [DOI] [PubMed] [Google Scholar]
- 115.Richman DD. Wrin T. Little SJ. Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100(7):4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Moore PL. Gray ES. Choge IA, et al. The C3–V4 region is a major target of autologous neutralizing antibodies in Hiv-1 subtype C infection. J Virol. 2007;82(4):1860–1869. doi: 10.1128/JVI.02187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Moore PL. Crooks ET. Porter L, et al. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J Virol. 2006;80(5):2515–2528. doi: 10.1128/JVI.80.5.2515-2528.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brown BK. Wieczorek L. Sanders-Buell E, et al. Cross-clade neutralization patterns among HIV-1 strains from the six major clades of the pandemic evaluated and compared in two different models. Virology. 2008;375(2):529–538. doi: 10.1016/j.virol.2008.02.022. [DOI] [PubMed] [Google Scholar]
- 119.Rademeyer C. Moore PL. Taylor N, et al. Genetic characteristics of HIV-1 subtype C envelopes inducing cross-neutralizing antibodies. Virology. 2007;368(1):172–181. doi: 10.1016/j.virol.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 120.Gray ES. Meyers T. Gray G. Montefiori DC. Morris L. Insensitivity of paediatric HIV-1 subtype C viruses to broadly neutralising monoclonal antibodies raised against subtype B. PLoS Med. 2006;3(7):e255. doi: 10.1371/journal.pmed.0030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zolla-Pazner S. Zhong P. Revesz K, et al. The cross-clade neutralizing activity of a human monoclonal antibody is determined by the GPGR V3 motif of HIV type 1. AIDS Res Hum Retroviruses. 2004;20(11):1254–1258. doi: 10.1089/aid.2004.20.1254. [DOI] [PubMed] [Google Scholar]
- 122.Gorny MK. Williams C. Volsky B, et al. Cross-clade neutralizing activity of human anti-V3 monoclonal antibodies derived from the cells of individuals infected with non-B clades of human immunodeficiency virus type 1. J Virol. 2006;80(14):6865–6872. doi: 10.1128/JVI.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pinter A. Honnen WJ. He Y. Gorny MK. Zolla-Pazner S. Kayman SC. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J Virol. 2004;78(10):5205–5215. doi: 10.1128/JVI.78.10.5205-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Burke VJ. Kim S. Williams C. Gorny MK. Zolla-Pazner S. Kong X. Structural characterization of neutralizing human anti-V3 monoclonal antibodies 3074 and 268-D. AIDS Vaccine. 2008:24. [Google Scholar]
- 125.Aasa-Chapman MM. Hayman A. Newton P, et al. Development of the antibody response in acute HIV-1 infection. AIDS. 2004;18(3):371–381. doi: 10.1097/00002030-200402200-00002. [DOI] [PubMed] [Google Scholar]
- 126.Arendrup M. Nielsen C. Hansen JE. Pedersen C. Mathiesen L. Nielsen JO. Autologous HIV-1 neutralizingantibodies: Emergence of neutralization-resistant escape virus and subsequent development of escape virus neutralizing antibodies. J Acquir Immune Defic Syndr. 1992;5(3):303–307. [PubMed] [Google Scholar]