Abstract
MicroRNAs (miRNAs), small 19–25 nucleotide RNAs that influence gene expression through posttranscriptional regulation of mRNA translation and degradation, have recently emerged as important regulators of neural development. Using conditional knock-out of Dicer, an RNase III enzyme required for miRNA maturation, previous studies have demonstrated an essential role for miRNAs in mouse cortical, inner ear, and olfactory development. However, a previous study (Damiani et al., 2008) using a Chx10cre mouse to delete Dicer in retinal progenitors reported no defects in the retina before the second postnatal week, suggesting that miRNAs are not required for mouse retinal development. In an effort to further study the role of miRNAs during retinal development and resolve this apparent conflict, we conditionally knocked out Dicer using a different (αPax6cre) line of transgenic mice. In contrast to the previous study, we demonstrate an essential role for miRNAs during mouse retinal development. In the absence of Dicer in the embryonic retina, production of early generated cell types (ganglion and horizontal cells) is increased, and markers of late progenitors are not expressed. This phenotype persists into postnatal retina, in which we find the Dicer-deficient progenitors fail to generate late-born cell types such as rods and Müller glia but continue to generate ganglion cells. We further characterize the dynamic expression of miRNAs during retinal progenitor differentiation and provide a comprehensive profile of miRNAs expressed during retinal development. We conclude that Dicer is necessary for the developmental change in competence of the retinal progenitor cells.
Introduction
A fundamental question in developmental neurobiology is how multipotent progenitors differentiate to form the myriad of cell types in the mature nervous system. With its simple structure and limited number of cell types, the retina is an ideal model for studying the regulation of neurogenesis. Like many neural progenitors, retinal progenitors demonstrate intrinsic changes in competence, generating different neurons at different developmental stages. Early cell types include ganglion and horizontal cells, and cone photoreceptors, followed by amacrine cells, rod photoreceptors, bipolar cells, and finally Müller glia. Although many transcription factors and signaling pathways underlying neuronal differentiation have been identified in the retina (Ohsawa and Kageyama, 2008), the molecular mechanisms that control retinal progenitor competence are not completely understood.
Recently, microRNAs (miRNAs), 19–25 nucleotide small RNAs have emerged as important regulators of developmental processes. miRNAs mediate their effects by guiding the RNA-induced silencing complex to the 3′ untranslated region (UTR) of target mRNAs, leading to target cleavage or translational repression (He and Hannon, 2004). Because miRNAs may target the 3′ UTR of many genes, they may play a role in global changes in gene expression or in fine-tuning developmental programs (Lim et al., 2005). miRNAs are thus good candidates for regulators of progenitor cell competence and neuronal cell fate determination, and recent studies support such a role (Johnston et al., 2005; Li and Carthew, 2005; Li et al., 2006; Makeyev et al., 2007).
Mammalian miRNA processing requires the RNase III enzyme Dicer. Thus, through conditional knock-out (CKO) of Dicer, mice can be generated that have lost all mature miRNAs in a specific set of cells. Damiani et al. (2008) used this strategy to remove miRNAs from Chx10cre-expressing mouse retinal progenitors cells (RPCs). They found that loss of Dicer caused no obvious developmental abnormalities, although retinal neurons began to undergo apoptosis beginning the second postnatal week. This lack of a developmental phenotype is in contrast to a subsequent study in which Dicer was reduced in developing Xenopus retina using morpholinos, leading to developmental changes in cell cycle, lamination, and timing of differentiation (Decembrini et al., 2008). This latter study is more consistent with results in other neural tissues, such as developing neocortex, in which Dicer CKO leads to defective layering, impairment of differentiation, and a smaller cortex (De Pietri Tonelli et al., 2008).
To better study the role of microRNAs during retinal development and better understand these species differences, we conditionally knocked out Dicer using αPax6cre transgenic mice. Unlike the mosaic Chx10cre line used by Damiani et al. (2008), Dicer is removed in large, continuous domains in αPax6cre transgenic mice. In addition, by crossing these mice onto the R26EYFP reporter line, we were able to identify Dicer-deficient cells. In contrast to the previous study in mice, our data demonstrate a requirement for Dicer during retinal development, with distinct changes in gene expression along with developmental deficits in differentiation and lamination. Our data support a model in which microRNAs regulate both retinal progenitor cell competence as well as differentiation and maturation of retinal neurons.
Materials and Methods
Generation of αPax6cre; R26EYFP; Dicerfl/fl animals.
αPax6cre mice were obtained from Ruth Ashery-Padan (Tel-Aviv University, Tel-Aviv, Israel) and have been described previously (Marquardt et al., 2001). Genotyping was done for cre using the following primers: forward, TGCCAGGATCAGGGTTAAAG; reverse, TCCTTAGCGCCGTAAATCAA. Dicerfl/fl mice were purchased from The Jackson Laboratory and genotyped as described previously (Harfe et al., 2005). R26EYFP mice have been described previously (Srinivas et al., 2001) and were obtained from Rachel Wong (University of Washington, Seattle, WA). Yellow fluorescent protein (YFP) genotyping was performed using the following primers: forward, GACTTCTTCAAGTCCGCCATGCC; reverse, GTGATCCCGGCGGCGGTCACG. Dicerfl/fl mice were crossed to αPax6cre mice to generate αPax6cre; Dicerfl/+ mice. These were backcrossed to αPax6cre mice to generate Dicerfl/+ mice carrying two copies of the αPax6cre allele. Dicerfl/fl mice were crossed to R26EYFP mice to generate R26EYFP; Dicerfl/+ mice, which were backcrossed to generate R26EYFP; Dicerfl/fl mice. These were then crossed to produce Dicerfl/fl mice carrying two copies of the R26EYFP allele. αPax6cre;Dicerfl/+ mice were crossed to either Dicerfl/fl or R26EYFP; Dicerfl/fl mice to generate αPax6cre;R26EYFP; Dicerfl/fl and αPax6cre; Dicerfl/fl mice. Dicerfl/+ [heterozygous (Het)] littermates were used as controls and showed no difference from wild-type animals. Animal housing and care was performed by the Department of Comparative Medicine at the University of Washington. All procedures were done in compliance with the standards and protocols set forth by the University of Washington Institutional Animal Care and Use Committee.
BrdU birthdating.
For bromodeoxyuridine (BrdU) (Sigma) birthdating, two subsequent injections of BrdU (100 mg/kg each) were administered subcutaneously to postnatal day 2 (P2) mice.
Histological analysis.
We used both frozen and paraffin sections for histological analysis. For frozen sections, whole embryo heads were collected at embryonic day 16 (E16), and eyes were collected at P0/P1, P4, P5, and P32 and placed in 2% or 4% paraformaldehyde (Electron Microscopy Systems) in PBS (EMD Biosciences) for 60–120 min at room temperature (RT). The samples were rinsed in PBS, followed by 10% sucrose (Mallinckrodt) in PBS for 1 h, 20% sucrose for 2 h, and 30% sucrose overnight at 4°C. Samples were then embedded in OCT (Sakura Finetek) and quickly frozen using dry ice and 85% ethanol. Samples were sectioned to 12 μm using a Leica CM 1850 cryostat and collected on Superfrost/Plus Microscope Slides (Thermo Fisher Scientific), followed by storage at −20°C. For Ascl1 staining, P0 retinas were cultured as described below for 48 h with DMSO and fixed for 20 min at RT, followed by a sucrose series and embedding for frozen sectioning as described above.
Other retinas were analyzed from paraffin sections. For this analysis, eyes were collected at P1 and P5 and placed in modified Carnoy's fix (60% ethanol, 11% formaldehyde, and 10% glacial acetic acid) overnight at 4°C. Samples were rinsed for 1 h each in 70, 80, 90, and 100% ethanol at RT, followed by an overnight wash in 100% ethanol at 4°C. Samples were then washed in xylene twice for 15 min each at RT, followed by three washes in melted Paraplast X-Tra paraffin (Thermo Fisher Scientific). Samples were sectioned to 8 μm using a Leitz 1516 rotary microtome onto StarFrost Platinum Microscope Slides (Mercedes Medical), followed by storage at RT. Retinal diameter was calculated by counting the number of retinal sections made. Before immunohistochemistry, paraffin sections were baked for 2 h at 68°C and deparaffinized as follows: two 10 min xylene washes followed by two 10 min 100% ethanol washes.
Immunohistochemistry.
Nonspecific labeling was prevented by pretreatment of the sections for 1 h in a solution containing the supernatant of a solution containing 5% dry milk, 0.5% Triton X-100 (Sigma), sodium azide, and PBS centrifuged for 15 min at 12,000 rpm. For BrdU antigen retrieval, cells were incubated in this same solution with 200 U/ml bovine pancreatic deoxyribonuclease I (Sigma) for 75 min. Slides were incubated with primary antibodies diluted in this same solution overnight at 4°C, followed by two 5 min PBS rinses. Slides were subsequently incubated with secondary antibodies in the same solution for 1–2 h at RT, followed by a 5 min PBS rinse. Counterstaining with 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma) was performed by incubating slides with a 10,000× dilution in water for 30 s, followed by two 5 min PBS rinses. Slides were mounted for microscopy in Fluormount-G (Southern Biotechnology Associates). Primary and secondary antibodies used, as well as dilutions, are listed in supplemental Methods (available at www.jneurosci.org as supplemental material).
Microscopy and statistics.
Slides were visualized by confocal microscopy using either a Carl Zeiss LSM 510 confocal microscope or a Nikon A1 confocal microscope. Images were acquired as 1024 × 1024 Tiff files and analyzed using Nikon NIS-Elements and Photoshop CS4 (Adobe Systems).
At E16, cells were counted within the neuroblastic layer of peripheral blocks of YFP-positive (YFP+) retina and normalized per millimeter YFP+ retina by measuring along the outer edge of the retina. At least two sections per animal were averaged. At P1, cells were counted in YFP+ retina and normalized to area (square micrometers) of the corresponding YFP+ neuroblastic layer of retina to account for the lack of the ganglion cell and inner plexiform layers in Dicer CKO retinas. Ectopic Brn3+ cells were defined as those above the inner plexiform layer in control or at least one-third of the retinal thickness from the vitreal surface in Dicer CKO. At least two sections per animal were averaged for all counts at P1. For proportional counts, GFP+ cells were counted in central retina and expressed as a ratio of total Pax6+ cells in the inner nuclear layer. Statistical analysis on cell counts was performed using an unpaired two-tailed t test with Prism 4 (GraphPad Software). Data are presented as mean ± SEM, control versus CKO.
Fluorescence-activated cell sorting of retinal cells.
E16 retinas were dissected in HBSS (Sigma), rinsed in calcium- and magnesium-free HBSS, and dissociated by trituration after incubation in 0.25% trypsin–EDTA (Invitrogen). Cells were collected by centrifugation at 1500 rpm and resuspended in HBSS. Cells were sorted for YFP by flow cytometry using a BD Influx flow cytometer (BD Biosciences) with 60 mW of 488 nm argon excitation and a bandpass filter of 525/30 nm. Cells were collected by centrifugation and placed in Trizol (Invitrogen) or Qiazol and processed using the miRNeasy Mini kit according to the instructions of the manufacturer (Qiagen).
Retinal culture and RNA extraction.
E14.5 C57BL/6 embryonic eyes were enucleated, and the retinas were dissected, cultured, and treated with the γ-secretase inhibitor DAPT (N-[N-(3,5-difluorophenacetyl-L-alanyl)]-S-phenylglycine t-butyl ester) (Sigma) for 48 h as described previously (Nelson et al., 2007). Retinas were pooled in Qiazol and processed using the miRNeasy Mini kit according to the instructions of the manufacturer.
MicroRNA microarray.
Whole RNA samples were labeled using the miRCURY LNA microRNA array labeling kit and hybridized onto miRCURY v.11.0 LNA microRNA Arrays (Exiqon) according to the instructions of the manufacturer by the Genomics Resource DNA Array Laboratory at the Fred Hutchinson Cancer Research Center (Seattle, WA). This array contains probes for 613 mouse miRNAs, 851 human miRNAs, and 349 rat miRNAs, which represent a coverage of miRBase v.13.0 of 96.2, 89, and 100%, respectively. It also contains probes for 428 “miRPlus” human miRNAs, proprietary miRNAs not included in the miRBase miRNA database. Because of the proprietary nature of these miRNAs, they have only been included in our primary array data contained in supplemental Table 1 (available at www.jneurosci.org as supplemental material) and excluded from additional analysis.
For mean signal intensity, array data were normalized by local background subtraction, and signal of the four probe spots was averaged. For intersignal comparison, array data was normalized by background subtraction, loess intraslide normalization, and scale interslide normalization using GEPAS (for Gene Expression Pattern Analysis Suite; http://gepas.bioinfo.cipf.es; Department of Bioinformatics and Genomics, Principe Felipe Centro De Investigacion, Valencia, Spain). An arbitrary cutoff value for mean fluorescence intensity of 2000 was used to determine real signal.
Quantitative reverse transcription-PCR.
mRNA quantitative PCR (qPCR) was performed as described previously (Nelson et al., 2007). For miRNA reverse transcription (RT), whole RNA was reverse transcribed in multiplex using custom designed stem-loop reverse transcription primers as described previously (Chen et al., 2005) using SuperScript II RT and associated buffers (Invitrogen). For mRNA RT, RNA was reverse transcribed using oligo-dT primers (Invitrogen) and SuperScript II RT following the protocols of the manufacturer. qPCR was performed using SYBR Green PCR Mastermix (Applied Biosystems) according to the protocols of the manufacturer on an Opticon DNA Engine (Bio-Rad). Each sample was run in triplicate, and n = 3 for number of samples. Statistical analysis on ΔCt values was performed using an unpaired two-tailed t test with Prism 4. Data are presented as mean ± SEM. Product specificity was confirmed by agarose gel electrophoresis, and all positive signals were at least five cycles higher than RT− controls.
Results
Dicer CKO retinas show developmental changes at E16
Previous studies reported that αPax6cre mediates recombination at two distinct time points during retinal development: (1) beginning at approximately E10.5, cre recombinase is active in retinal progenitors in a peripheral domain of variable extent, and (2) beginning at approximately E14.5 and extending into adulthood, cre recombinase is also active in a subset of amacrine cells (Marquardt et al., 2001; Yaron et al., 2006; Lefebvre et al., 2008). The αPax6cre transgene also drives expression of an IRES–GFP when active (αPax6:GFP), allowing for visualization of areas of active cre recombinase. Although we can use the GFP expressed from the IRES in the transgene to visualize the active cre recombinase in the progenitors, this does not reveal the extent of previous recombination. Therefore, we crossed αPax6cre; Dicerfl/fl (CKO) mice to the R26EYFP reporter line, which allowed us to visualize the region of the retina derived from Dicer-deficient progenitors. We compared αPax6cre; R26EYFP; Dicerfl/fl (CKO) and αPax6cre; R26EYFP; Dicerfl/+ (Het) retinas for YFP at E16. YFP staining showed large continuous domains of peripheral recombination, frequently extending from the periphery to cover one-third to one-half or more of the total retinal area at this stage. Thus, a large domain of retinal cells are derived from progenitors that had undergone cre-mediated recombination. In addition, there were often smaller, separate areas of YFP expression in more central retinal regions (Fig. 1A,B). Patterns and degree of expression varied from animal to animal, with some animals showing larger domains of YFP expression in nasal or temporal retina, although this was not correlated with their Dicer genotype. The extent of YFP expression was similar in both the Dicer CKO and heterozygous animals at this age.
Figure 1.
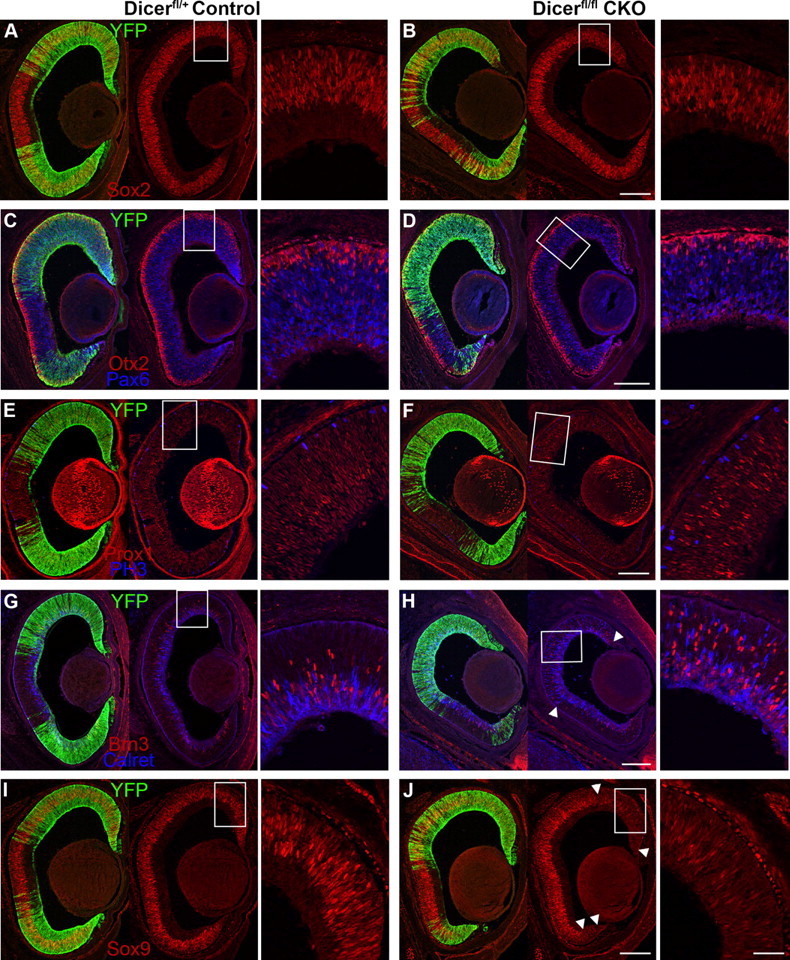
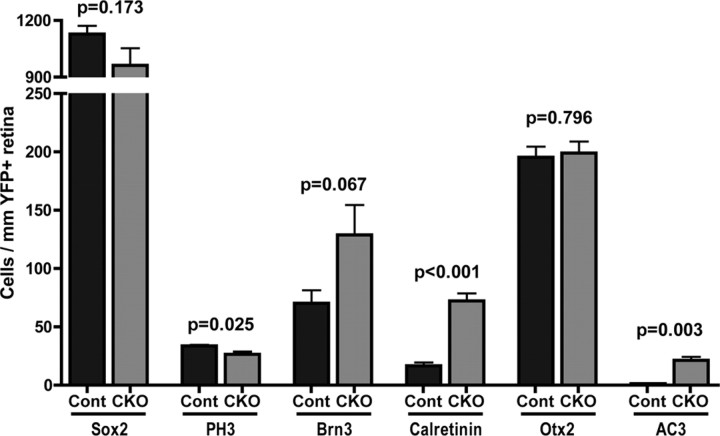
Immunofluorescence staining of E16 control and Dicer CKO retinal cryosections. A–J, YFP staining (green) indicates areas of cre-mediated recombination and Dicer CKO. A, B, Sox2 RPC staining (red) is normal in Dicer-deficient areas. C, D, Otx2 photoreceptor (red) and Pax6 RPC and amacrine cell (blue) staining is normal in Dicer-deficient areas. E, F, Bright Prox1+ horizontal cells (red) are increased in the Dicer-deficient areas, whereas faint Prox1+ RPCs are normal. Mitotic (PH3+) cells (blue) are still present. G, H, Brn3+ ganglion cells (red) and calretinin+ horizontal cells (blue) are increased in the Dicer-deficient areas (arrowheads). Note the sharp transition between normal and Dicer-deficient retina (bottom arrowhead). I, J, Sox9 RPC staining (red) is downregulated in Dicer-deficient areas (arrowheads). All images are oriented the same, with nasal retina at the top of the image. Boxes represent areas of magnification. Scale bars: 100× images, 200 μm; 400× insets, 50 μm.
To determine whether the loss of Dicer in the RPCs had any effects on the early stages of retinal development, we analyzed retinal sections using immunofluorescence with antibodies to several different markers for progenitors and developing retinal neurons. Most markers of retinal progenitors were not different in the Dicer-deficient regions of retina when compared with the retinas from Dicer heterozygous (Het) littermate controls. Staining with antibodies that recognize Sox2 (Fig. 1A,B) and Pax6 (Fig. 1C,D), two genes expressed in RPCs, revealed normal numbers and lamination of these cells in Dicer-deficient regions (Fig. 2) (1128.3 ± 43.4 vs 961.8 ± 90.8 Sox2+ cells/mm YFP+ retina; n = 3 vs n = 3; p = 0.173). The RPCs also showed near normal numbers of M-phase cells because staining and quantification of phosphohistone H3 (PH3) revealed only a small decrease in Dicer-deficient retina (Figs. 1E,F, 2) (33.6 ± 0.9 vs 26.5 ± 2.4 cells/mm YFP+ retina; n = 4 vs n = 3; p = 0.025). Staining with the proliferation marker Ki67 revealed a small, but not significant, decline (supplemental Fig. 1C,D, available at www.jneurosci.org as supplemental material) (674.8 ± 40.6 vs 560.3 ± 33.9 cells/mm YFP+ retina; n = 3 vs n = 3; p = 0.096).
Figure 2.
Quantification of immunofluorescence staining of E16 control and Dicer CKO retinas. Counts of Sox2+ retinal progenitor cells, PH3+ M-phase cells, Brn3+ ganglion cells, calretinin+ horizontal cells, Otx2+ photoreceptors, and apoptotic cells (AC3+) in the neuroblastic layer per millimeter of YFP+ retina. Error bars indicate mean ± SEM.
In addition to characterizing the RPCs in the Dicer CKO retinas, we also used immunofluorescence to analyze the development of the various classes of differentiated neurons. The first cell types generated in the retina are ganglion cells, horizontal cells, and cone photoreceptors, and antibodies against Brn3, Prox1, and Otx2 were used to characterize their development in Dicer CKO and littermate control retinas. Strikingly, staining for Brn3 showed a substantial increase in the number of differentiating ganglion cells in Dicer-deficient regions when compared with similar regions of retina from Dicer heterozygous animals (Figs. 1G,H, 2) (70.1 ± 11.1 vs 129 ± 25.6 cells/mm YFP+ retina; n = 4 vs n = 3; p = 0.067). Likewise, Dicer-deficient areas showed a significant increase in calretinin+ cells (Figs. 1G,H, 2) (16.6 ± 2.8 vs 72.3 ± 6.4 cells/mm YFP+ retina; n = 4 vs n = 3; p = 0.0003), although these did not colocalize with Brn3. Subsequent analysis demonstrated that Prox1+ cells were also increased in Dicer-deficient areas (Fig. 1E,F) and that these cells colabeled with calretinin (supplemental Fig. 1A,B, available at www.jneurosci.org as supplemental material). The position of these Prox1/calretinin cells in the upper half of the neuroblastic layer suggested that they were differentiating horizontal cells. A boundary effect was often observed within individual Dicer CKO retinas, in which increased horizontal and ganglion cell staining appeared only in recombined (YFP+) areas (Fig. 1H, arrowheads), with normal expression in non-recombined areas. No boundary effects were ever observed in Het control retinas, however, suggesting that loss of a single copy of Dicer is insufficient to produce a developmental phenotype.
We also labeled sections for Otx2 to identify photoreceptors. Staining and quantification revealed normal numbers and lamination of developing photoreceptors (Figs. 1C,D, 2) (195 ± 9.1 vs 199 ± 9.8 cells/mm YFP+ retina; n = 4 vs n = 3; p = 0.796). However, at this age, we cannot determine whether these are cone or rod photoreceptors, because the specific opsins for these cell types are not yet expressed.
To determine whether the changes we observed in ganglion and horizontal cell numbers were attributable to a decline in their death at this stage, we used labeled sections with antibodies against activated caspase 3 (AC3). We detected an increase in apoptosis in Dicer-deficient retinas (Fig. 2) (supplemental Fig. 1E,F, available at www.jneurosci.org as supplemental material) (1.37 ± 0.45 vs 21.4 ± 2.96 cells/mm YFP+ retina; n = 3 vs n = 3; p = 0.0026), and colabeling with cell-specific markers indicated that apoptotic cells included retinal progenitor cells, ganglion cells, and photoreceptors (data not shown). These data suggest that Dicer is required for the survival of retinal neurons as well as progenitor cells. Thus, the small reduction in PH3-labeled cells we observed (above) might be attributable in part to cell death in the progenitors. However, the observed increase in ganglion cells in the Dicer-deficient regions cannot be explained by a reduction in ganglion cell death because we observe an increase in apoptosis.
The results from the above analysis indicate that Dicer-deficient retinal progenitor cells are present in numbers close to those in normal mice at E16. However, the increase in ganglion and horizontal cells at this age led us to further characterize the developmental state of the RPCs. Both Pax6 and Sox2 are expressed in RPCs throughout their development. However, Sox9 is normally not expressed in RPCs until E14.5, marking a later stage of their development (Poché et al., 2008). Therefore, we also labeled sections of Dicer CKO retinas for Sox9 expression. We found that Sox9 expression was reduced in the periphery of Dicer CKO retinas in the Dicer-deficient domain (Fig. 1I,J, arrowheads), whereas Sox2 expression remained normal, consistent with the hypothesis that Dicer-deficient RPCs remain in an early progenitor state. Together, our data indicate that, in the absence of microRNA processing, early retinal development is perturbed, with an overproduction of the earliest-born neurons and a decrease in expression of Sox9, a marker of late stage RPCs.
Neonatal Dicer CKO retinas show defects in progenitors
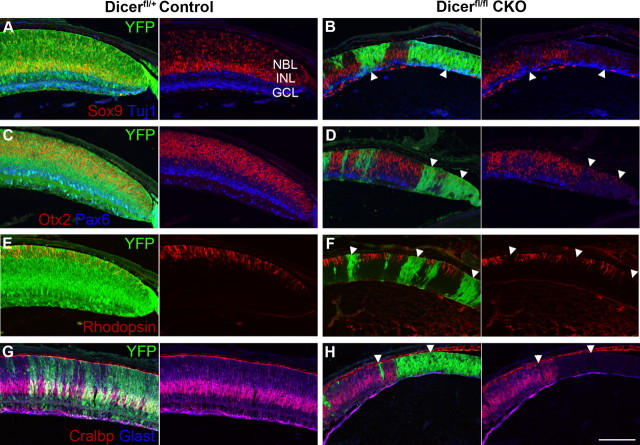
We next analyzed retinas from early postnatal pups to determine whether similar changes to those seen at E16 are observed later in development. At P1, Dicer CKO retinas were smaller in diameter than control retinas (1917 ± 28 vs 1616 ± 57 μm; n = 3 vs n = 3; p = 0.0089), and the peripheral retina often lacked both a distinct ganglion cell layer and inner plexiform layer and was markedly thinner than control retinas (Fig. 3A,B). Despite the reduced thickness of the retina at P1, we found that the progenitor cells were still present at P1 in the Dicer-deficient regions by labeling sections with progenitor and cell cycle markers. Sox2 staining revealed that retinal progenitor cells were still present in the Dicer-deficient areas, although Sox2 protein levels were higher in Dicer-deficient RPCs than in the adjacent Dicer-containing cells, often showing a sharp boundary that correlated with YFP expression (Fig. 3C,D, arrowheads). In addition, Pax6 levels appeared normal in the progenitors (Fig. 3E,F, arrowhead). However, PH3 staining was reduced in Dicer-deficient areas (Figs. 3G,H, arrowhead, 4I) (4.32 ± 0.31 vs 2.45 ± 0.32 cells/104 μm2; n = 4 vs n = 4; p = 0.0055), indicating a decrease in cells entering M-phase, and proliferating cell nuclear antigen (PCNA), a marker of cycling cells, was expressed at a reduced level (Fig. 3G,H, arrowheads). These data indicate that progenitors persist to P1 without Dicer, although cell cycle markers are reduced.
Figure 3.
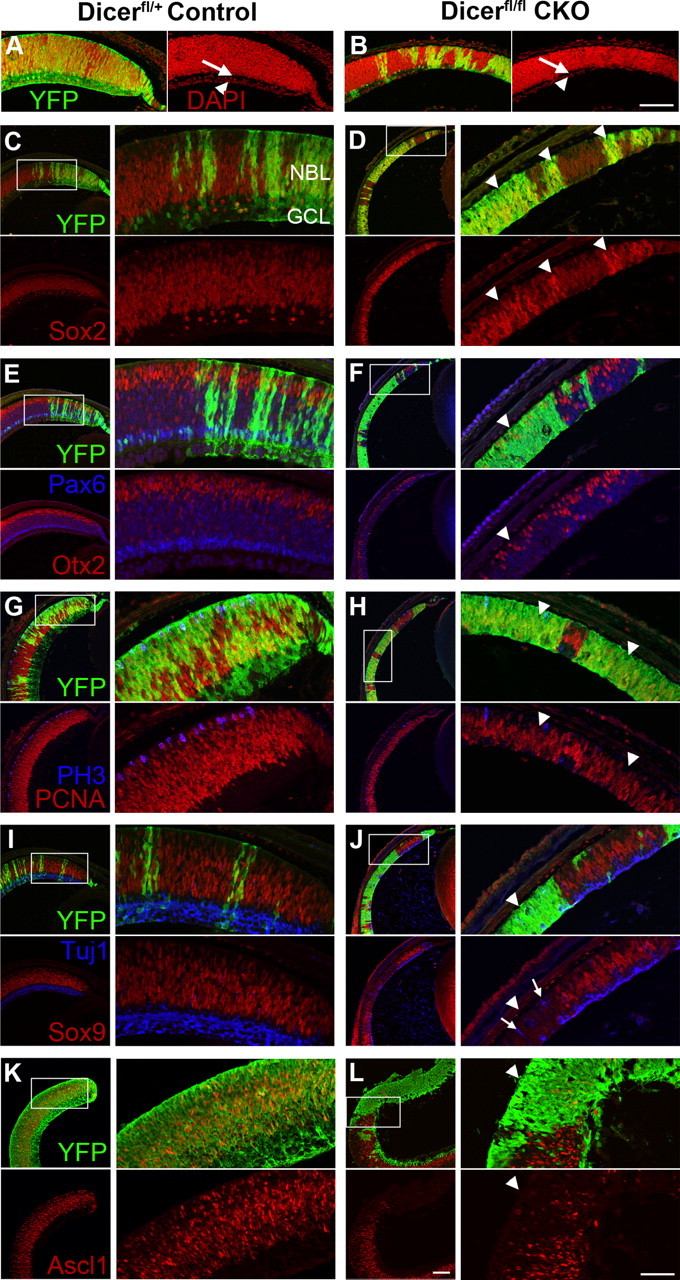
Immunofluorescence staining of early postnatal control and Dicer CKO retinas. A–J, P0/P1 retinal paraffin sections. YFP staining (green) indicates areas of cre-mediated recombination and Dicer CKO. K, L, P0 + 2 DIV retinal cryosections. YFP staining in green. A, B, Retinal lamination is affected in Dicer CKO retinas, with an absent ganglion cell layer (arrowhead) and inner plexiform layer (arrow) as revealed by nuclear DAPI (red) staining. C, D, Sox2 RPC staining (red) is increased in Dicer-deficient areas (arrowheads). E, F, Otx2+ photoreceptors (red) are decreased in Dicer-deficient areas (arrowhead). Bright Pax6+ amacrine cells (blue) are absent from Dicer-deficient areas, although fainter RPC Pax6 staining appears normal (arrowhead). G, H, Dicer-deficient areas (arrowheads) show reduced PCNA staining (red) and have fewer PH3+ cells (blue). I, J, Dicer-deficient areas (arrowhead) lack Sox9+ RPCs (red) and have ectopic Tuj1+ neurons (blue, arrows). K, L, Dicer-deficient areas (arrowhead) lack Ascl1+ RPCs (red). NBL, Neuroblastic layer; GCL, ganglion cell layer. All images are oriented the same, with the ganglion cell layer on the bottom and peripheral retina to the right. Boxes represent areas of magnification. Scale bars: 200× images, 100 μm; 400× insets, 50 μm.
Figure 4.
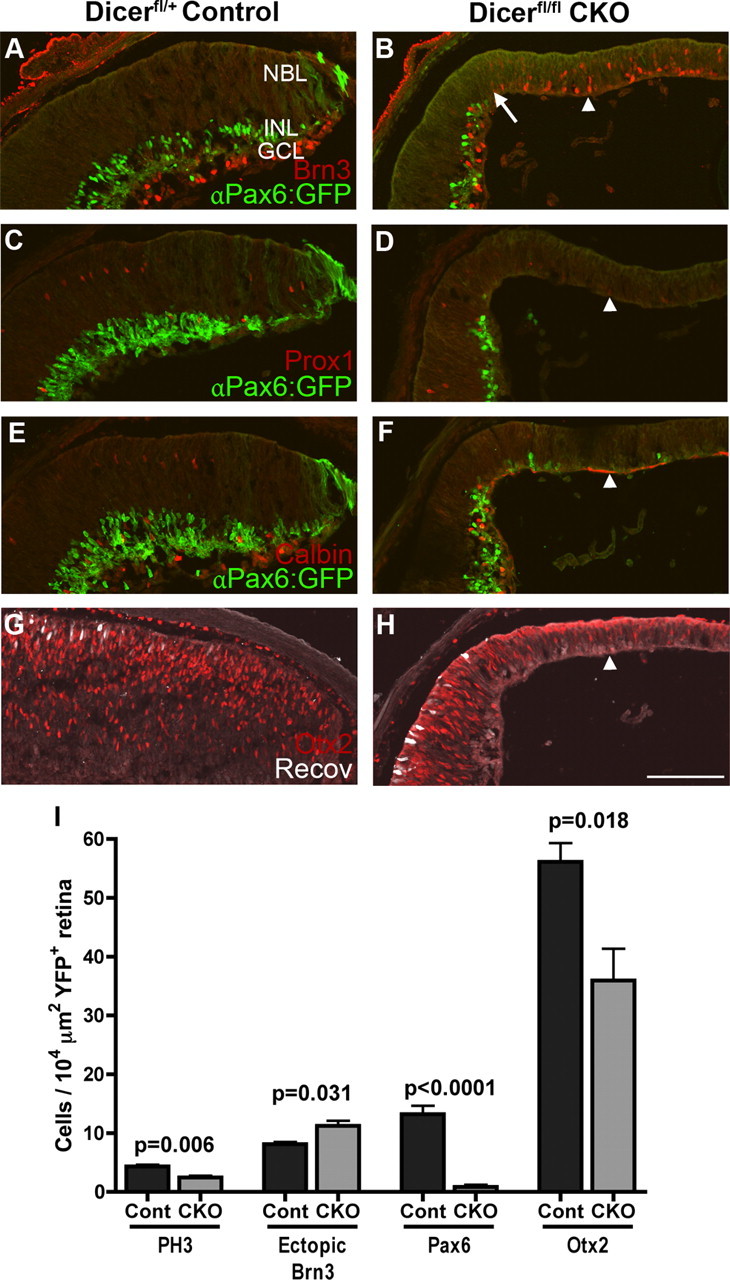
Immunofluorescence staining of P1 control and Dicer CKO retinal cryosections for neuronal markers. A–H, αPax6:GFP labeling (green, amacrine cells). A, B, Brn3+ ganglion cells (red) and GFP+ amacrine cells show a marked transition in Dicer CKO retinas (arrow). Ganglion cells are increased in Dicer-deficient areas, whereas amacrine cells are missing (arrowhead). C, D, Prox1+ amacrine and horizontal cells (red) are absent from Dicer-deficient areas (arrowhead). E, F, Calbindin+ amacrine and horizontal cells (red) are gone from Dicer-deficient areas (arrowhead). G, H, Otx2+ photoreceptors (red) are reduced in Dicer-deficient areas, whereas mature recoverin+ photoreceptors (white) are missing (arrowhead). I, Quantification of immunofluorescence staining of P1 control and Dicer CKO retinas. Counts of PH3+ M-phase cells, ectopic Brn3+ ganglion cells, Pax6+ amacrine cells, and Otx2+ photoreceptors per 104 μm of YFP+ retina. Error bars indicate mean ± SEM. NBL, Neuroblastic layer; INL, inner nuclear layer; GCL, ganglion cell layer. All images are oriented the same, with the ganglion cell layer on the bottom and peripheral retina to the right. Scale bar, 100 μm.
As noted above, the data from the analysis at E16 suggested that RPCs had not progressed from their early developmental state: there was a decline in the late progenitor marker Sox9 and an increase in the numbers of early born cell types, ganglion cells, and horizontal cells. We found a similar phenotype at P1 in the Dicer-deficient regions of retina. Sox9 staining was absent or reduced from Dicer-deficient retinal progenitor cells at P1 (Fig. 3I,J, arrowhead). In addition, we stained with Ascl1, which, like Sox9, is expressed only in later staged RPCs (Jasoni and Reh, 1996). We found that Ascl1 expression was missing from Dicer-deficient progenitors, showing a sharp boundary effect similar to that often observed for Sox9 (Fig. 3K,L, arrowhead).
Together, our data indicate that, in the absence of microRNA processing, the progenitor cells in the P1 retina show changes in cell cycle and progenitor markers. Whereas retinal progenitor cells are still present and mitotically active (albeit reduced), RPCs show reduced or absent expression of late-stage progenitor markers. These data suggest that miRNAs have a role in regulating progenitor phenotype and possibly developmental timing.
Neonatal Dicer CKO retinas have ectopic ganglion cells but lack most other mature neurons
At E16, the loss of Dicer led to an increase in the number of ganglion cells and horizontal cells and a loss in the expression of late stage progenitor markers. The analysis of RPCs at P1 suggests that this phenotype might persist into postnatal development. To test for this possibility, we labeled αPax6cre; Dicerfl/fl (CKO) retinas for Brn3. This labeling showed that ganglion cells were still present, located both in their normal position along the vitreal surface and ectopically throughout the neuroblastic layer (Fig. 4A,B, arrowhead). Quantification revealed that there were significantly more ectopic ganglion cells in Dicer-deficient regions of retina than in control retina (Fig. 4I) (8.1 ± 0.39 vs 11.2 ± 0.85; n = 3 vs n = 4; p = 0.031). To further characterize this ganglion cell phenotype, we stained for HuC/D, a marker of mature ganglion and amacrine cells, which revealed the presence of mature ganglion cells along the vitreal surface of the retina (supplemental Fig. 2A,B, arrowheads, available at www.jneurosci.org as supplemental material). Furthermore, YFP+ axons were present in the optic nerve of both control and Dicer CKO retinas (data not shown), suggesting that ganglion cell maturation is normal in Dicer-deficient areas.
At E16, we found that horizontal cells also increased in number; however, at P1, Prox1+ cells were absent from Dicer-deficient areas (Fig. 4C,D, arrowhead), as was calbindin, another marker of horizontal and amacrine cells (Fig. 4E,F, arrowhead). These data suggest that, although horizontal cells may be generated in excess in Dicer-deficient areas earlier in development, they do not survive to birth. Indeed, staining with the apoptotic marker AC3 at P1 revealed substantial apoptosis in Dicer CKO retinas (supplemental Fig. 2C,D, available at www.jneurosci.org as supplemental material).
Although these data are consistent with the possibility that the loss of Dicer leads to a persistence of the early progenitor state in the RPCs, we further tested this possibility by labeling sections for markers of later born neurons. Previous studies have shown that retinal progenitors express Pax6 at low to moderate levels, whereas amacrine cells show a much higher level of labeling. In the Dicer-deficient areas, Pax6-expressing amacrine cells were severely reduced or absent (Figs. 3E,F, 4I) (13.2 ± 1.4 vs 0.84 ± 0.38 cells/104 μm2; n = 4 vs n = 4; p = 0.0001). As noted above, at P1, the αPax6cre transgene can be used to directly visualize a subset of amacrine cells through their GFP expression (Fig. 4A–F, αPax6:GFP). Staining Dicer CKO retinas for GFP in animals lacking the R26EYFP reporter shows an abrupt absence of GFP+ amacrine cells in peripheral retina (Fig. 4B, arrow).
Most rod photoreceptors are generated in the postnatal retina, and so we also analyzed their development at this age. Although there was no significant decline in Otx2+ cells at E16 in the Dicer-deficient regions of retina, suggesting that early photoreceptors (i.e., cones) are generated in the absence of Dicer, at P1 we found a significant reduction in the number of Otx2+ cells in Dicer-deficient areas (Figs. 3E,F, arrowheads; 4G,H, arrowheads; 4I) (56.1 ± 3.2 vs 35.9 ± 5.4 cells/104 μm2; n = 4 vs n = 4; p = 0.018), indicating a reduction in photoreceptors in the postnatal retina. The Otx2+ cells we did find were less intensely labeled than in control regions of retina. In addition, another marker of photoreceptors, recoverin, was absent in the Dicer-deficient areas (Fig. 4G,H), suggesting that the later stages of photoreceptor differentiation require miRNAs.
Together, our analysis at P1 shows that the Dicer-deficient RPCs fail to express markers of the late progenitor state (Sox9 and Ascl1) and that the differentiation of those cells types normally generated at this stage (amacrine cells and rod photoreceptors) is reduced. Nevertheless, the progenitors persist and express Sox2 and Pax6, as well as cell cycle markers PH3 and PCNA (albeit at reduced numbers and levels), and the differentiation of at least one early-born cell type, ganglion cells, is maintained.
Postnatal day 5 Dicer CKO retinas show a similar phenotype as that found at P1
To determine whether the changes observed at P1 were persistent or attributable to a delay in developmental timing, we stained Dicer CKO retinas for similar markers at P5. A nearly identical phenotype was observed, with Dicer-deficient areas lacking horizontal cells and amacrine cells (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). Dicer-deficient retinal progenitor cells showed severely reduced Sox9 staining (Fig. 5A,B, arrowheads) but still expressed Sox2 and Pax6, both of which were increased in intensity compared with non-deficient areas (Fig. 5C,D, arrowheads) (supplemental Fig. 3A,B, available at www.jneurosci.org as supplemental material). Otx2+ photoreceptors were present, but reduced, in Dicer-deficient areas, whereas recoverin, a marker of mature photoreceptors, and rhodopsin, a specific marker of rod photoreceptors, were both absent (Fig. 5C–F, arrowheads) (supplemental Fig. 3I,J, available at www.jneurosci.org as supplemental material). Surprisingly, ectopic Brn3+ ganglion cells were still present in Dicer-deficient areas, well beyond the normal window of ganglion cell genesis, suggesting continued ganglion cell production at P5 (supplemental Fig. 3C,D, arrowheads, available at www.jneurosci.org as supplemental material). Staining for cellular retinaldehyde-binding protein (Cralbp) and the glutamate transporter Glast revealed an absence of Müller glia in Dicer-deficient areas (Fig. 5G,H, arrowheads), suggesting that their differentiation is also impaired in the absence of microRNA processing or possibly attributable to the lack of Sox9 expression in the progenitors.
Figure 5.
Immunofluorescence staining of P5 control and Dicer CKO retinas. A–H, Retinal paraffin sections. YFP staining (green) indicates areas of cre-mediated recombination and Dicer CKO. A, B, Retinal progenitor Sox9 staining (red) is reduced in Dicer-deficient areas (arrowheads), whereas Tuj1+ neurons (blue) are ectopically located. C, D, Otx2+ photoreceptors (red) are reduced in Dicer-deficient areas (arrowheads), whereas bright Pax6+ amacrine cells are absent (blue). Faint Pax6 staining in RPCs is increased in Dicer-deficient regions. E, F, Mature rhodopsin+ rods (red) are absent from Dicer-deficient areas (arrowheads). G, H, Müller glia, stained with Cralbp (red) and Glast (blue), are absent from Dicer-deficient areas (arrowheads). NBL, Neuroblastic layer; INL, Inner nuclear layer; GCL, ganglion cell layer. All images are oriented the same, with the ganglion cell layer on the bottom and peripheral retina to the right. Boxes represent areas of magnification. Scale bars, 100 μm.
These data suggest that, in the absence of miRNA processing, development of retinal cell types generated late in retinal histogenesis is permanently reduced and not simply delayed relative to normal development. To further test this hypothesis, we also analyzed the Dicer-deficient regions for the presence of bipolar cells, another cell type generated late in histogenesis. Using antibodies against PKC, Vsx1, and Chx10, we attempted to stain for bipolar cells at P5. Although nascent bipolar cells were present in central retina, these three markers are not expressed in the peripheral retina at this age, preventing us from making conclusions on their expression in the peripheral Dicer-deficient areas. Attempts to stain for these markers at P8, an age at which their domain of expression has extended into the periphery of the retina, were unsuccessful because nearly all Dicer-deficient cells have died by this age (data not shown). Nevertheless, overall, our analysis shows that late cell types are not generated in Dicer-deficient regions of the retina, and loss of expression of late RPC markers suggests that miRNAs play an important role in the regulation of progenitor competence.
Dicer CKO progenitors produce ganglion cells beyond their normal competence window
The presence of ectopic Brn3+ cells as late as P5 in Dicer-deficient regions of retina suggested a continuing production of ganglion cells beyond the normal window of ganglion cell genesis. To test this hypothesis directly, pups were given a single pulse of BrdU, a thymidine analog that incorporates into the DNA of cells undergoing S-phase, at P2 and killed 48 h later. Previous birthdating studies have shown that this is at the very tail end of retinal ganglion cell genesis (Young, 1985), and, as expected, BrdU+ ganglion cells were only occasionally found in control retinas. When present, these BrdU+ ganglion cells were primarily found at the very peripheral edge of the retina (Fig. 6A, arrowhead). In contrast, Dicer CKO retinas showed significantly more BrdU+ ganglion cells (Fig. 6B,C) (0.69 ± 0.26 vs 3.3 ± 0.33 cells per peripheral retinal field; n = 4 vs n = 3; p = 0.0014). Surprisingly, these BrdU+ ganglion cells were found throughout the Dicer-deficient region, as far as 600 μm from the peripheral edge of the retina (Fig. 6B, arrowheads). The persistence of ganglion cell production by the Dicer-deficient postnatal progenitors supports the hypothesis that progenitors require miRNAs to make the transition from an early to late stage in their development.
Figure 6.
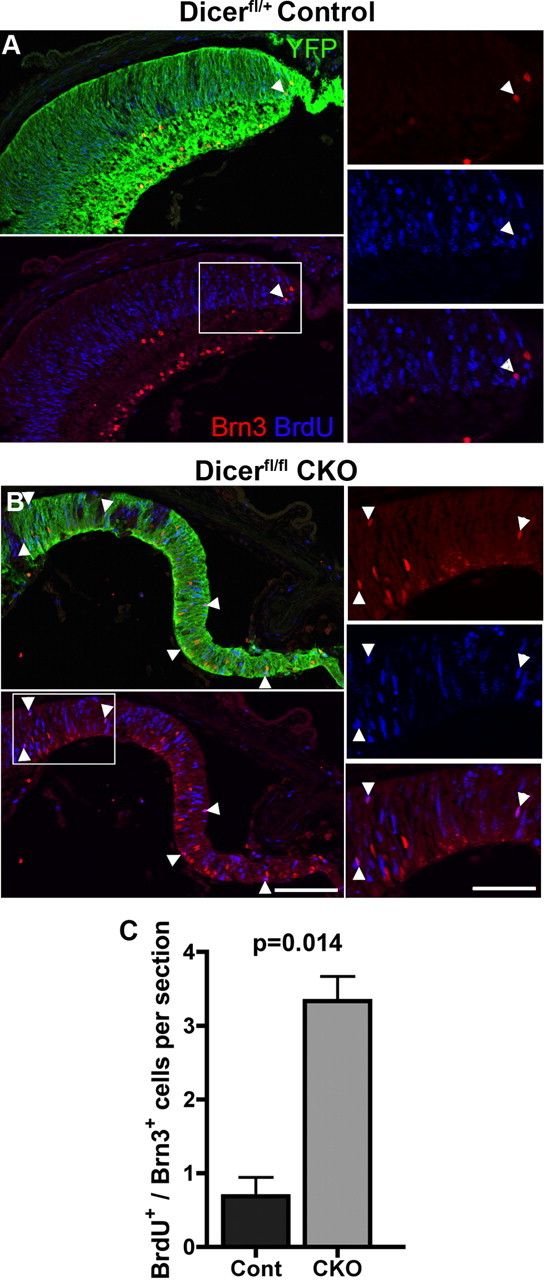
Immunofluorescence staining of BrdU and Brn3 at P4. A, B, Retinal cryosections. YFP staining (green) indicates areas of cre-mediated recombination and Dicer CKO. Labeling for BrdU (blue), injected 48 h previously, reveals colabeled Brn3+ (red) ganglion cells throughout Dicer-deficient areas (arrowheads), in contrast to their localization at the far peripheral edge (arrowhead) in control. C, Quantification of BrdU/Brn3 costaining in control and Dicer CKO retinal sections. Scale bars: 200× images, 100 μm; 400× insets, 50 μm. Error bars indicate mean ± SEM.
Most Dicer-deficient cells die by adulthood
To determine the ultimate fate of Dicer-deficient retinal cells, we looked at P32 Dicer CKO retinas. During dissection, Dicer CKO eyes were easily identifiable, with smaller optic nerves than control eyes and oblong and contracted pupils (data not shown). Sectioning revealed that Dicer CKO retinas were often detached from the overlying pigmented epithelium, and many had a thin filament extending from their peripheral margin (Fig. 7B, arrow), which connexin 43 staining revealed to be part of the ciliary epithelium (Fig. 7B, asterisk). In contrast to P5, in which large blocks of Dicer-deficient cells could be visualized by YFP staining, only scattered YFP+ cells remained at P32, suggesting that most Dicer-deficient cells had undergone apoptosis. Indeed, Dicer CKO retinas were thinner than their Het littermates (Fig. 7A,B), indicating extensive retinal remodeling.
Figure 7.
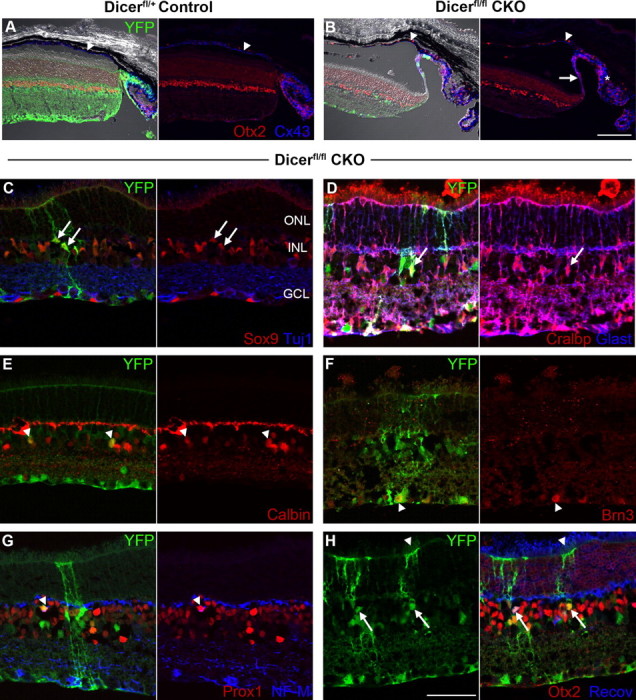
Immunofluorescence staining of P32 control and Dicer CKO retinal cryosections. A–H, YFP staining (green) indicates areas of cre-mediated recombination and Dicer CKO. A, B, Dicer CKO retinas are thinner than controls and are often detached from the overlying retinal pigmented epithelium (arrowheads). Connexin 43 (blue) staining reveals that Dicer CKO retinas have long, filamentous extensions (arrow) of the ciliary body (asterisk). C, D, Scattered YFP+ Müller glia are present in Dicer CKO retinas (arrows), as evidenced by colabeling with Sox9 (C, red), Cralbp (D, red), and Glast (D, blue). E, Scattered YFP+ amacrine cells are present in Dicer CKO retinas (arrowheads), as evidenced by colabeling with calbindin (red). F, Scattered YFP+ ganglion cells are present in Dicer CKO retinas (arrowhead), as evidenced by colabeling with Brn3 (red). G, Scattered YFP+ horizontal cells are present in Dicer CKO retinas (arrowhead), as evidenced by colabeling with Prox1 (red) and neurofilament-M (NF-M; blue). H, Scattered YFP+ photoreceptors are present in Dicer CKO retinas (arrowhead), as evidenced by colabeling with Otx2 (red) and recoverin (Recov; blue) in the outer nuclear layer. Scattered YFP+ bipolar cells are present in Dicer CKO retinas (arrows), as evidenced by colabeling with Otx2 (red) and recoverin (blue) in the inner nuclear layer. ONL, Outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. All images are oriented the same, with the ganglion cell layer on the bottom. Scale bars: 200× images, 100 μm; 400× images, 50 μm.
Dicer CKO retinas were generally morphologically normal, although occasional rosettes or regions of disrupted lamination were observed (data not shown). These areas usually did not correlate with the presence of YFP+ cells, suggesting that they were the result of retinal remodeling subsequent to the loss of Dicer-deficient cells, not a direct result of Dicer deficiency. The majority of remaining cells had the morphology of Müller glia (Fig. 7C,D, arrows) or amacrine cells (Fig. 7E, arrowheads), but costaining with cell-type-specific markers also revealed the presence of scattered ganglion cells (Fig. 7F, arrowhead), horizontal cells (Fig. 7G, arrowhead), photoreceptors (Fig. 7H, arrowhead), and bipolar cells (Fig. 7H, arrows). Interestingly, although ganglion cell survival appeared normal at P5, few YFP+ ganglion cells were still present at P32, suggesting that their ultimate survival is also dependent on microRNA processing.
Because many of these cells were not present in Dicer-deficient areas at earlier ages, the remaining cells may have escaped Dicer recombination or have been rescued in some other way. Indeed, occasional areas were observed at P1 and P5 that lacked the Dicer-deficient phenotype (supplemental Fig. 4, arrowheads, available at www.jneurosci.org as supplemental material). Whereas large blocks of Dicer-deficient cells showed tight correlation of the described phenotype with YFP expression (supplemental Fig. 4, asterisks, available at www.jneurosci.org as supplemental material), occasional groups of cells did not. These cells were most commonly found in small groups, surrounded by wild-type cells, or at the border of a YFP+ group. Although the source of this variation is not clear, it is possible that these cells represent clones that did not undergo complete recombination for Dicer or that have been rescued in a non-cell-autonomous manner (see Discussion). Thus, the YFP+ cells remaining at P32 may be the progeny of these cells.
The remaining amacrine cells may also belong to the αPax6:GFP-expressing population previously described, having undergone recombination as mature neurons, not RPCs. To determine whether Dicer is required for the survival of mature amacrine cells, we quantified the proportion of Pax6-expressing cells that also express GFP at P0 and P32 in the inner nuclear layer of central retina. Although no significant difference was observed at P0 (0.45 ± 0.05 vs 0.37 ± 0.03; n = 5 vs n = 5; p = 0.18), GFP+ amacrines represented a significantly smaller proportion of Pax6+ cells when comparing CKO with control at P32 (0.19 ± 0.01 vs 0.12 ± 0.01; n = 4 vs n = 7; p = 0.0045). These data suggest a postnatal requirement for Dicer in the maintenance of mature amacrine cells, which is similar to the postnatal degenerative phenotype seen by Damiani et al. (2008).
Specific miRNAs are upregulated and downregulated during differentiation
Because Dicer conditional knock-out leads to a deficiency in the processing of all microRNAs, determination of the mechanisms underlying the observed developmental changes requires a better description of the miRNAs present in retinal progenitors and their differentiated progeny. Indeed, our data suggest a role for microRNAs in both the survival and differentiation of retinal neurons and the definition of retinal progenitor cell phenotype. Although a recent study by Hackler et al. (2009) has provided a profile of miRNA expression across retinal development, very little is known about the role and expression of miRNAs in retinal progenitors and their differentiated progeny.
Numerous examples suggest that miRNAs play a role in determination and definition of cell fate, and thus we hypothesized that microRNA profiles would change early in differentiation. Because Dicer-deficient retinal progenitor cells would be unable to make these early differentiation miRNAs, these miRNAs might be potential mediators of the observed phenotype. We have shown previously that inhibition of the Notch signaling pathway using the γ-secretase inhibitor DAPT leads to coordinated differentiation of retinal progenitor cells appropriate for their developmental stage (Nelson et al., 2007). Microarray and qPCR profiling of cultured retinas demonstrated that DAPT treatment leads to the rapid downregulation of progenitor genes concomitant with an upregulation of genes involved in neuronal differentiation, such as the proneural basic helix–loop–helix transcription factors. By extension, we hypothesized that DAPT treatment would lead to a downregulation of progenitor microRNAs and an upregulation of proneural microRNAs.
Microarray analysis was performed on E14.5 C57BL/6 retinas cultured for 48 h in DMSO or DAPT. Using our criteria, 110 mouse miRNAs (18% of those represented on the array) were expressed in either condition, as well as 2 rat and 48 human miRNAs without mouse homologs (supplemental Table 1, available at www.jneurosci.org as supplemental material). Highly expressed miRNAs included the neuron-specific miR-124, several members of the let-7 family, and miR-9, as well as several human miRNAs not described previously in mouse (Table 1). Additional analysis revealed a number of microRNAs that were upregulated >1.5-fold during coordinated differentiation, suggesting an important role in neuronal differentiation (Table 1). These include miR-7a and miR-7b, conserved miRNAs that also function in Drosophila photoreceptor differentiation (Li and Carthew, 2005), the photoreceptor-expressed miR-96/182/183 cluster (Xu et al., 2007), and miR-125b, which has been shown to promote neuronal differentiation (Eda et al., 2009; Le et al., 2009). Also upregulated were numerous members of the miR-17 family and miR-26a, which has been shown recently to regulate calcium channel expression in cones (Shi et al., 2009). Numerous miRNAs were also downregulated during DAPT treatment (Table 1), including miR-9 and miR-9*, suggesting that they are expressed in retinal progenitors. Indeed, previous in situ hybridization studies have shown that both miR-9 and miR-9* are expressed in mouse cortical neural progenitors (Deo et al., 2006) and in the proliferative ciliary marginal zone of the zebrafish retina (Kapsimali et al., 2007).
Table 1.
Selected miRNAs expressed in E14.5 retina
| MicroRNA | Fold change | DMSO mean signal | DAPT mean signal | Previous retinal description |
|---|---|---|---|---|
| mmu-miR-7b | 12 | 256 | 3210 | 1, 4 |
| mmu-miR-96 | 8.7 | 903 | 7813 | 5, 7, 8, 9, 10, 11, 12 |
| mmu-miR-183 | 6.9 | 1410 | 9513 | 3, 5, 7, 8, 9, 10, 11, 12 |
| mmu-miR-182 | 6.7 | 2585 | 16660 | 3, 5, 6, 7, 8, 9, 10, 11, 12 |
| mmu-miR-99b | 2.9 | 1130 | 3325 | 8, 11 |
| mmu-miR-125b | 2.4 | 2747 | 6389 | 2, 3, 5, 7, 8, 10, 11 |
| mmu-miR-191 | 2.2 | 1296 | 2865 | 7, 8, 10, 11 |
| mmu-miR-101a | 2.1 | 2360 | 4845 | 10 |
| mmu-miR-30a | 2 | 1363 | 2784 | 8, 11 |
| mmu-miR-93 | 1.9 | 1924 | 3763 | 11 |
| mmu-miR-136 | 1.9 | 1091 | 2133 | 7, 11 |
| mmu-miR-125a-5p | 1.7 | 5779 | 9667 | 7, 8, 10, 11 |
| mmu-miR-26a | 1.7 | 3856 | 6304 | 3, 8, 10, 11, 13 |
| hsa-miR-18a | 1.7 | 1298 | 2212 | 11 |
| mmu-miR-7a | 1.6 | 3713 | 5965 | 1, 4, 8, 11 |
| mmu-miR-183* | 1.6 | 1814 | 2905 | 11 |
| mmu-miR-16 | 1.5 | 2813 | 4238 | 8, 10, 11 |
| mmu-miR-17 | 1.5 | 2710 | 4026 | 10, 11 |
| mmu-miR-124 | 1.5 | 46541 | 59667 | 2, 3, 4, 5, 6, 7, 8, 10, 11 |
| mmu-miR-26b | 1.4 | 6116 | 8484 | 7, 8, 10 |
| mmu-miR-691 | 1.2 | 26100 | 28373 | 7, 11 |
| hsa-miR-1827 | 1.2 | 21945 | 24384 | |
| hsa-miR-939 | 1.2 | 18256 | 20599 | |
| mmu-let-7e | 1.2 | 7790 | 8865 | 7, 8, 10, 11 |
| mmu-miR-709 | 1.1 | 61041 | 59742 | 11 |
| hsa-miR-665 | 1.1 | 16351 | 16962 | |
| hsa-let-7a | 1.1 | 11428 | 11448 | 8, 10, 11 |
| mmu-let-7f | 1.1 | 13281 | 14137 | 8, 10, 11 |
| mmu-miR-200b* | 1.1 | 8369 | 8868 | 11 |
| mmu-miR-106b | 1.1 | 7470 | 7781 | 7, 8, 10 |
| mmu-let-7a | 1 | 15218 | 14691 | 8, 10, 11 |
| mmu-miR-875-3p | 0.94 | 14148 | 12400 | |
| hsa-miR-550 | 0.91 | 15240 | 12914 | |
| mmu-miR-503* | 0.89 | 7079 | 6026 | |
| mmu-let-7c | 0.89 | 7032 | 6543 | 8, 10, 11 |
| mmu-miR-690 | 0.84 | 52868 | 40030 | 11 |
| mmu-miR-720 | 0.71 | 7477 | 5086 | |
| hsa-miR-1826 | 0.62 | 63450 | 35396 | |
| hsa-miR-1308 | 0.57 | 5902 | 3270 | |
| mmu-miR-674 | 0.57 | 4118 | 2322 | |
| mmu-miR-9* | 0.53 | 2457 | 1301 | 7, 8, 10, 11 |
| mmu-miR-805 | 0.51 | 4266 | 2176 | |
| mmu-miR-9 | 0.49 | 8014 | 3755 | 5, 6, 8, 10, 11 |
| mmu-miR-668 | 0.49 | 6285 | 3010 | |
| mmu-miR-2142 | 0.41 | 45906 | 17305 |
Included are the top 20 most highly expressed miRNAs in both DAPT- and DMSO-treated E14.5 retina, as well as all miRNAs upregulated >1.5-fold or downregulated >0.67-fold after 48 h DAPT treatment. Reference numbers are as follows: 1, Li and Carthew, 2005; 2, Makarev et al., 2006; 3, Ryan et al., 2006; 4, Arora et al., 2007; 5, Kapsimali et al., 2007; 6, Karali et al., 2007; 7, Loscher et al., 2007; 8, Xu et al., 2007; 9, Loscher et al., 2008; 10, Shen et al., 2008; 11, Hackler et al., 2009; 12, Jin et al., 2009; 13, Shi et al., 2009. For review, see Huang et al, 2008; Rapicavoli and Blackshaw, 2009; Xu, 2009.
Select miRNAs are lost from Dicer CKO areas at E16
To determine whether the molecular changes observed with immunofluorescent analysis at E16 could also be studied using qPCR, we collected Dicer-deficient retinal cells using flow sorting for YFP. As expected, qPCR showed a drastic decrease in Dicer mRNA levels in Dicer-deficient cells compared with Het cells (Fig. 8A). Furthermore, as predicted, Sox9 mRNA levels were decreased in Dicer-deficient cells, whereas Sox2 mRNA levels were not changed (Fig. 8A). In addition, Brn3b mRNA levels trended upward in Dicer-deficient cells but were not statistically significant (Fig. 8A).
Figure 8.
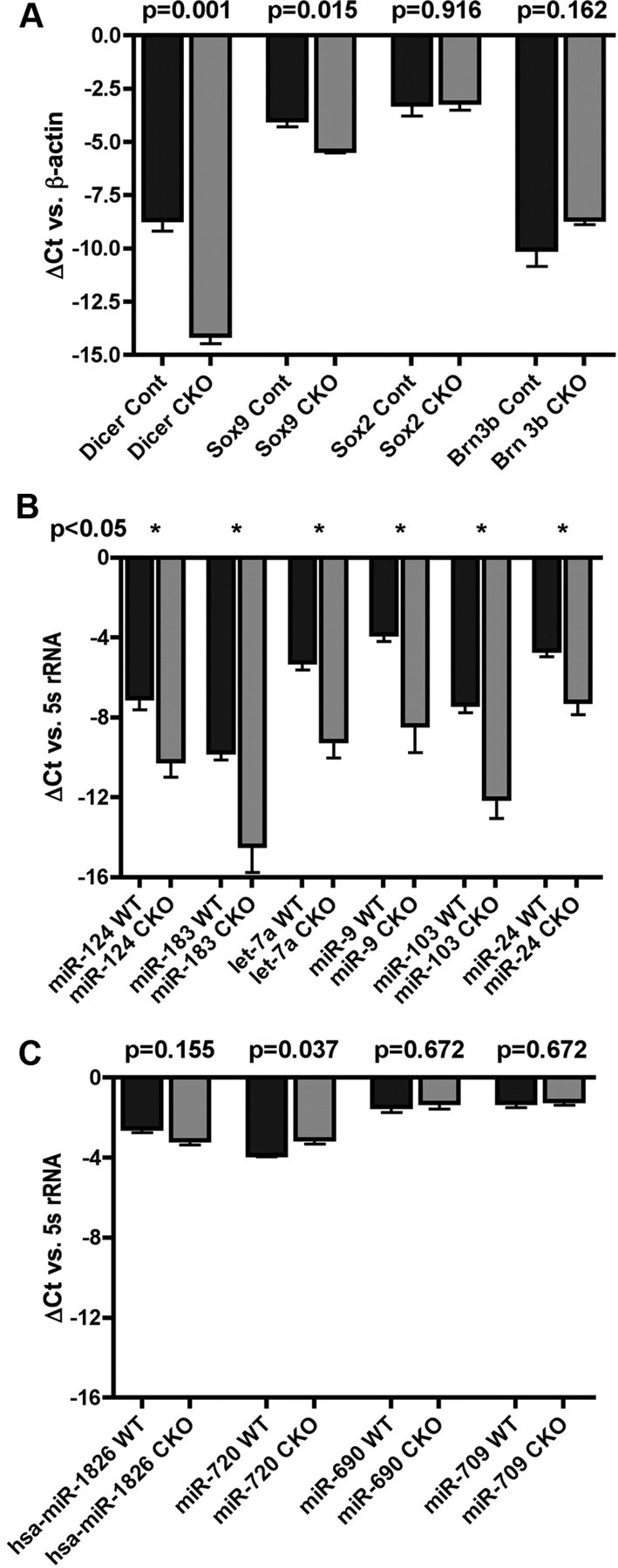
Quantitative RT-PCR of fluorescence-activated cell sorted E16 control and Dicer CKO retinas. A, ΔCt values for mRNAs. B, C, ΔCt values for miRNAs. Error bars indicate mean ± SEM.
To confirm that the loss of Dicer led to a reduction in miRNA processing, we used qPCR on several miRNAs that had been identified through our microarray analysis or had been shown previously in other studies to be expressed in the developing retina (Fig. 8B). Several miRNAs that we found to be upregulated during DAPT treatment, such as miR-124 and miR-183, showed significant decreases in Dicer-deficient cells, indicating a failure to generate them during normal differentiation. Other miRNAs were also reduced, including miR-9 and miR-24, which has been shown to repress apoptosis during retinal development in Xenopus (Walker and Harland, 2009).
Surprisingly, not all microRNAs showed a decrease in Dicer-deficient cells (Fig. 8C). Notably, hsa-miR-1826, miR-690, and miR-720, all among the most highly expressed miRNAs on the array, showed no change in Dicer-deficient cells. Because these miRNAs are among the most highly expressed at E14.5, it is likely that they are expressed in retinal progenitors, which make up the majority of cells at this age. In support of this, all three show a decrease upon DAPT-mediated differentiation. The fact that these three miRNAs do not change in Dicer-deficient cells suggests that some progenitor miRNAs may be stable with very little turnover and minimal requirement for continual Dicer processing to maintain their cellular levels. Alternatively, a small amount of Dicer may still be present in the cells, and these persistent miRNAs may represent a population whose precursors have higher affinity for the Dicer catalytic domain.
Discussion
Several conditional knock-out studies have established an essential role for Dicer in mammalian neural development, including the cortex (De Pietri Tonelli et al., 2008; Kawase-Koga et al., 2009), inner ear (Friedman et al., 2009; Soukup et al., 2009), and olfactory epithelium (Choi et al., 2008). However, a previous study using Chx10cre to remove Dicer from retinal progenitors concluded that Dicer was expendable for cell fate specification and migration because Dicer-deficient retinas displayed all cell types and normal lamination (Damiani et al., 2008). In contrast to the conclusions of that paper, our data demonstrate an essential role for Dicer during mouse retinal development. In the absence of Dicer, we see significant developmental defects in gene expression and neurogenesis as early as E16, which become more pronounced in neonatal animals.
The defects we observe in the Dicer-deficient regions of retina span a range of developmental processes. At E16, there is an upregulation of early neuronal types and a downregulation of late progenitor cell markers, together with decreased M-phase entry and increased apoptosis. By birth, we see significant changes in retinal morphology and lamination and a complete absence of all mature neurons except ganglion cells. We also demonstrate a differential requirement for Dicer in the generation and maturation of different cell types. By adulthood, most Dicer-deficient cells have degenerated, suggesting an essential role for Dicer in retinal cell survival, consistent with the study by Damiani et al. (2008). Our data are also consistent with a previous study showing that morpholinos targeting Dicer lead to developmental changes in cell cycle, lamination, and timing of differentiation in Xenopus retina (Decembrini et al., 2008).
Several differences exist between our study and that of Damiani et al. (2008) that may explain the differences in our data and conclusions. Whereas Damiani et al. (2008) used the Chx10cre line to mediate Dicer conditional knock-out in retinal progenitor cells, we used the αPax6cre line. Although both cre lines mediate recombination at similar times in development, the αPax6cre line shows recombination primarily in peripheral retina, whereas the Chx10cre leads to recombination throughout the retina (Rowan and Cepko, 2004). Because retina matures in a central-to-peripheral gradient, this may mean that αPax6cre deletes Dicer in a less mature population of RPCs than Chx10cre. Additionally, whereas αPax6cre is active in large continuous blocks of RPCs, Chx10cre leads to mosaic recombination. Thus, whereas Dicer-deficient cells are primarily surrounded by other Dicer-deficient cells in the αPax6cre line, the mosaic nature of the Chx10cre line leads to interspersing of Dicer-deficient cells among wild-type cells. Because Damiani et al. (2008) did not use a lineage marker to identify Dicer-deficient cells, it was difficult to determine whether the neurons were born from Dicer-deficient or normal retinal progenitor cells.
An additional explanation for the differences between our study and that of Damiani et al. (2008) is that the mosaic nature of recombination in Chx10cre animals might allow for non-cell-autonomous rescue of the observed phenotype. Indeed, any role for miRNAs in the regulation of cell–cell signaling could potentially be occluded by the mosaicism of Chx10cre animals but would be revealed in our study. Furthermore, studies suggest the possibility of non-cell-autonomous transfer of miRNAs through microvesicles (Yuan et al., 2009) or gap junctions (Valiunas et al., 2005; Kizana et al., 2009). Indeed, RPCs and many mature retinal cell types are coupled through gap junctions (Bloomfield and Völgyi, 2009; Cook and Becker, 2009). In support of a non-cell-autonomous rescue of the Dicer CKO phenotype, we occasionally observed small areas of recombined (YFP+) cells that appeared wild type, expressing normal neuronal and RPC markers, which were absent from most Dicer-deficient areas. We cannot currently rule out the possibility that these cells simply did not undergo complete recombination for Dicer, however, and we observed other examples of isolated or small groups of YFP+ cells that still displayed the Dicer-deficient phenotype, as well as sharp phenotypic borders between YFP+ and YFP-negative areas, suggesting that any non-cell-autonomous rescue may be quite limited in our system.
A surprising finding of our study is a significant effect of Dicer loss on the retinal progenitor cell phenotype, suggesting a role for miRNAs in regulating the competence state of RPCs. Dicer-deficient RPCs lost Sox9 and Ascl1 expression and showed increased Sox2 expression compared with wild-type RPCs. Because both Sox9 and Ascl1 are absent from the earliest, Sox2-expressing RPCs, this suggests that Dicer-deficient RPCs may have been stuck in an immature state. Normal RPCs go through a progressive shift in competence, first generating ganglion cells, horizontal cells, and cone photoreceptors, followed by other mature neuronal types. Dicer-deficient areas had increased production of ganglion and horizontal cells at E16, with normal production of photoreceptors. At P1, Dicer-deficient areas lacked all neuronal markers, except those for ganglion cells and immature photoreceptors. Interestingly, ganglion cells were ectopically located throughout the progenitor cell layer at late as P5 and showed a continued genesis beyond their normal competence window, strongly suggesting a role for miRNAs in regulating RPC competence. One transcription factor that controls the developmental competence of retinal progenitors, Ikaros, is a potential candidate for regulation by miRNAs in this regard, because misexpression of this gene imparts competence to generate early neurons (Elliott et al., 2008).
It is not currently clear which microRNAs underlie the observed Dicer-deficient phenotype. Because Dicer CKO leads to impaired maturation of all miRNAs, numerous cellular pathways and genes may be affected. Further complicating this analysis is the fact that some miRNAs show stability beyond loss of Dicer, suggesting that additional studies will be needed to determine the time course of individual miRNA loss after Dicer CKO. A recent study showed that miR-24a inhibits apoptosis during Xenopus retinal development (Walker and Harland, 2009), and our data indicate that the mouse homolog is significantly reduced in Dicer-deficient cells. Thus, part of the increased apoptosis observed in Dicer-deficient areas may be attributable to a loss of miR-24.
Recently, Decembrini et al. (2009) showed a role for four miRNAs in inhibiting Otx2 and Vsx1 during Xenopus retinal development. Inhibition of these miRNAs led to increased expression of Otx2 and Vsx1 and an increased generation of bipolar cells. In contrast, our study shows a decrease in the number and levels of Otx2 staining and no ectopic Vsx1 staining (data not shown) in Dicer-deficient retina. These differences may reflect species-specific mechanisms of regulation. Indeed, although Otx2 and Vsx1 show temporal differences in their mRNA and protein expression attributable to translational inhibition in Xenopus, such differences have never been reported in mouse. Furthermore, of these four miRNAs, only miR-214 was expressed in E14.5 mouse retina (supplemental Table 1, available at www.jneurosci.org as supplemental material).
Several other microRNAs identified in our screen have been established as key regulators of neurogenesis elsewhere in the nervous system. Shibata et al. (2008) found that miR-9 promotes an early cortical cell fate, the Cajal-Retzius cells, by targeting Foxg1, a transcription factor that maintains the early progenitor state. miR-9 also participates in a feedback loop in neural development, targeting Tlx, a regulator of progenitor cell self-renewal (Zhao et al., 2009), whereas miR-9* targets Baf53a, a progenitor-specific component of the SWI/SNF chromatin remodeling complex (Yoo et al., 2009). Both of these mechanisms drive neuronal differentiation and inhibit the progenitor state. These miRNAs may also play a role in the phenotypes we observe in Dicer-deficient RPCs, because both are expressed in the developing retina and are downregulated with DAPT.
let-7a also showed significant declines in Dicer-deficient retinal cells. let-7 was first identified as a regulator of developmental timing in Caenorhabditis elegans, and homologs of let-7 have been identified in vertebrates. Rybak et al. (2008) recently showed that let-7a is expressed in mouse neural progenitors, as are other components of this pathway, including homologs to lin-28 and lin-4 (miR-125). A recent knock-out study of the let-7 target lin-41 (TRIM71) showed nervous system defects (Maller Schulman et al., 2008), whereas Schwamborn et al. (2009) showed that overexpression of let-7a promotes neuronal differentiation of neural progenitors. It is tempting to speculate that a conserved heterochronic pathway in C. elegans may also function to regulate the timing of retinal cell type generation.
Although the mechanisms by which Dicer CKO leads to the observed changes in retinal development remain to be fully elucidated, we have provided a comprehensive profiling of miRNAs present during retinal development and have shown that many are dynamically upregulated and downregulated during neuronal differentiation. Similar profiling was done previously for mRNA expression (Nelson et al., 2007), allowing for future comparison of miRNA changes with their predicted targets, as well as transcription factors predicted to regulate miRNA transcription. Our study presents the first evidence that microRNAs are required for mouse retinal development, with important roles in regulating retinal progenitor cell competence, as well as in the differentiation and survival of mature retinal neurons. Our data provide an important basis for future studies into the role that individual microRNAs play in the regulation of retinal neurogenesis and establish a model in which the role of microRNAs in nervous system development can be studied in greater detail.
Footnotes
This research was supported by National Institutes of Health Grant RO1 EY013475 (T.A.R.) and a National Science Foundation Fellowship (S.A.G.). We thank Dr. Ruth Ashery-Padan and Dr. Rachel Wong for their generous gift of mice used in this study, Dr. Jack Saari for Cralbp antibody, and Dr. Robert Molday for rhodopsin antibody. We also thank Donna Prunkard for assistance with cell sorting and members of the Reh Laboratory, including Dr. Joe Brzezinski IV and Dr. Deepak Lamba, for feedback on this manuscript.
References
- Arora A, McKay GJ, Simpson DA. Prediction and verification of miRNA expression in human and rat retinas. Invest Ophthalmol Vis Sci. 2007;48:3962–3967. doi: 10.1167/iovs.06-1221. [DOI] [PubMed] [Google Scholar]
- Bloomfield SA, Völgyi B. The diverse functional roles and regulation of neuronal gap junctions in the retina. Nat Rev Neurosci. 2009;10:495–506. doi: 10.1038/nrn2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi PS, Zakhary L, Choi WY, Caron S, Alvarez-Saavedra E, Miska EA, McManus M, Harfe B, Giraldez AJ, Horvitz HR, Schier AF, Dulac C. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57:41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook JE, Becker DL. Gap-junction proteins in retinal development: new roles for the “nexus.”. Physiology. 2009;24:219–230. doi: 10.1152/physiol.00007.2009. [DOI] [PubMed] [Google Scholar]
- Damiani D, Alexander JJ, O'Rourke JR, McManus M, Jadhav AP, Cepko CL, Hauswirth WW, Harfe BD, Strettoi E. Dicer inactivation leads to progressive functional and structural degeneration in the mouse retina. J Neurosci. 2008;28:4878–4887. doi: 10.1523/JNEUROSCI.0828-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decembrini S, Andreazzoli M, Barsacchi G, Cremisi F. Dicer inactivation causes heterochronic retinogenesis in Xenopus laevis. Int J Dev Biol. 2008;52:1099–1103. doi: 10.1387/ijdb.082646sd. [DOI] [PubMed] [Google Scholar]
- Decembrini S, Bressan D, Vignali R, Pitto L, Mariotti S, Rainaldi G, Wang X, Evangelista M, Barsacchi G, Cremisi F. MicroRNAs couple cell fate and developmental timing in retina. Proc Natl Acad Sci U S A. 2009;106:21179–21184. doi: 10.1073/pnas.0909167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo M, Yu JY, Chung KH, Tippens M, Turner DL. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev Dyn. 2006;235:2538–2548. doi: 10.1002/dvdy.20847. [DOI] [PubMed] [Google Scholar]
- De Pietri Tonelli D, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for the expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eda A, Tamura Y, Yoshida M, Hohjoh H. Systematic gene regulation involving miRNAs during neuronal differentiation of mouse P19 embryonic carcinoma cell. Biochem Biophys Res Commun. 2009;388:648–653. doi: 10.1016/j.bbrc.2009.08.040. [DOI] [PubMed] [Google Scholar]
- Elliott J, Jolicoeur C, Ramamurthy V, Cayouette M. Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron. 2008;60:26–39. doi: 10.1016/j.neuron.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Friedman LM, Dror AA, Mor E, Tenne T, Toren G, Satoh T, Biesemeier DJ, Shomron N, Fekete DM, Hornstein E, Avraham KB. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc Natl Acad Sci U S A. 2009;106:7915–7920. doi: 10.1073/pnas.0812446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackler L, Wan J, Swaroop A, Qian J, Zack DJ. MicroRNA profile of the developing mouse retina. Invest Ophthalmol Vis Sci. 2009 doi: 10.1167/iovs.09-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is require for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Huang KM, Dentchev T, Stambolian D. MiRNA expression in the eye. Mamm Genome. 2008;19:510–516. doi: 10.1007/s00335-008-9127-8. [DOI] [PubMed] [Google Scholar]
- Jasoni CL, Reh TA. Temporal and spatial pattern of MASH-1 in the developing rat retina demonstrates progenitor cell heterogeneity. J Comp Neurol. 1996;369:319–327. doi: 10.1002/(SICI)1096-9861(19960527)369:2<319::AID-CNE11>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Jr, Chang S, Etchberger JF, Ortiz CO, Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal fate decision. Proc Natl Acad Sci U S A. 2005;102:12449–12454. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapsimali M, Kloosterman WP, de Bruijn E, Rosa F, Plasterk RH, Wilson SW. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8:R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karali M, Peluso I, Marigo V, Banfi S. Identification and characterization of microRNAs expressed in the mouse eye. Invest Ophthalmol Vis Sci. 2007;48:509–515. doi: 10.1167/iovs.06-0866. [DOI] [PubMed] [Google Scholar]
- Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev Dyn. 2009;238:2800–2812. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizana E, Cingolani E, Marbán E. Non cell-autonomous effects of vector-expressed regulatory RNAs in mammalian heart cells. Gene Ther. 2009;16:1163–1168. doi: 10.1038/gt.2009.64. [DOI] [PubMed] [Google Scholar]
- Le MT, Xie H, Zhou B, Chia PH, Rizk P, Um M, Udolph G, Yang H, Lim B, Lodish HF. MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol Cell Biol. 2009;29:5290–5305. doi: 10.1128/MCB.01694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre JL, Zhang Y, Meister M, Wang X, Sanes JR. gamma-Protocadherins regulate neuronal survival but are dispensable for circuit formation in retina. Development. 2008;135:4141–4151. doi: 10.1242/dev.027912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123:1267–1277. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Loscher CJ, Hokamp K, Kenna PF, Ivens AC, Humphries P, Palfi A, Farrar GJ. Altered retinal microRNA expression profile in a mouse model of retinitis pigmentosa. Genome Biol. 2007;8:R248. doi: 10.1186/gb-2007-8-11-r248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscher CJ, Hokamp K, Wilson JH, Li T, Humphries P, Farrar GJ, Palfi A. A common microRNA signature in mouse models of retinal degeneration. Exp Eye Res. 2008;87:529–534. doi: 10.1016/j.exer.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarev E, Spence JR, Del Rio-Tsonis K, Tsonis PA. Identification of microRNAs and other small RNAs from the adult newt eye. Mol Vis. 2006;12:1386–1391. [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller Schulman BR, Liang X, Stahlhut C, DelConte C, Stefani G, Slack FJ. The let-y microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle. 2008;7:3935–3942. doi: 10.4161/cc.7.24.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Hartman BH, Georgi SA, Lan MS, Reh TA. Transient inactivation of Notch signaling synchronizes differentiation of neural progenitor cells. Dev Biol. 2007;304:479–498. doi: 10.1016/j.ydbio.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa R, Kageyama R. Regulation of retinal cell fate specification by multiple transcription factors. Brain Res. 2008;1192:90–98. doi: 10.1016/j.brainres.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Poché RA, Furuta Y, Chaboissier MC, Schedl A, Behringer RR. Sox9 is expressed in mouse multipotent retinal progenitor cells and functions in Müller glial cell development. J Comp Neurol. 2008;510:237–250. doi: 10.1002/cne.21746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Blackshaw S. New meaning in the message: noncoding RNAs and their role in retinal development. Dev Dyn. 2009;238:2103–2114. doi: 10.1002/dvdy.21844. [DOI] [PubMed] [Google Scholar]
- Rowan S, Cepko CL. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev Biol. 2004;271:388–402. doi: 10.1016/j.ydbio.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vis. 2006;12:1175–1184. [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates MicroRNAs and prevents self-renewal in mouse neural progenitors. Cell. 2009;136:913–925. doi: 10.1016/j.cell.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Yang X, Xie B, Chen Y, Swaim M, Hackett SF, Campochiaro PA. MicroRNAs regulate ocular neovascularization. Mol Ther. 2008;16:1208–1216. doi: 10.1038/mt.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Kurokawa D, Nakao H, Ohmura T, Aizawa S. MicroRNA-9 modulates Cajal-Retzius cell differentiation by suppressing Foxg1 expression in the mouse medial pallium. J Neurosci. 2008;28:10415–10421. doi: 10.1523/JNEUROSCI.3219-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Ko ML, Ko GY. Rhythmic expression of microRNA-26a regulates the L-type voltage-gated calcium channel α1C subunit in chicken cone photoreceptors. J Biol Chem. 2009;284:25791–25803. doi: 10.1074/jbc.M109.033993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA, Fritzsch B, Pierce ML, Weston MD, Jahan I, McManus MT, Harfe BD. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev Biol. 2009;328:328–341. doi: 10.1016/j.ydbio.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiunas V, Polosina YY, Miller H, Potapova IA, Valiuniene L, Doronin S, Mathias RT, Robinson RB, Rosen MR, Cohen IS, Brink PR. Connexin-specific cell-to-cell transfer of short interfering RNA by gap junctions. J Physiol. 2005;568:459–468. doi: 10.1113/jphysiol.2005.090985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JC, Harland RM. microRNA-24a is required to repress apoptosis in the developing neural retina. Genes Dev. 2009;23:1046–1051. doi: 10.1101/gad.1777709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S. microRNA expression in the eyes and their significance in relation to functions. Prog Retin Eye Res. 2009;28:87–116. doi: 10.1016/j.preteyeres.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- Yaron O, Farhy C, Marquardt T, Applebury M, Ashery-Padan R. Notch 1 functions to suppress cone-photoreceptor fate specification in the developing mouse retina. Development. 2006;133:1367–1378. doi: 10.1242/dev.02311. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes during neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212:199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
- Yuan A, Farber EL, Rapoport AL, Tejada D, Deniskin R, Akhmedov NB, Farber DB. Transfer of microRNAs by embryonic stem cell microvesicles. PLoS One. 2009;4:e4722. doi: 10.1371/journal.pone.0004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]