Abstract
Most clustering techniques do not incorporate phenotypic data. Limited biological interpretation is garnered from the informal process of clustering biological samples and then labeling groups with the phenotypes of the samples. A more formal approach of clustering samples is presented. The method utilizes simulated annealing of the Modk-prototypes objective function. Separate weighting terms are used for microarray, clinical chemistry and histopathology measurements to control the influence of each data domain on the clustering of the samples. The weights are adapted during the clustering process. A cluster’s prototype is representative of the phenotype of the cluster members. Genes are extracted from phenotypic prototypes obtained from the livers of rats exposed to acetaminophen (an analgesic and antipyretic agent) that differed in the extent of centrilobular necrosis. Map kinase signaling and linoleic acid metabolism were significant biological processes influenced by the exposures of acetaminophen that manifested centrilobular necrosis.
Keywords: Clustering, Simulated Annealing, Gene Expression, Microarray, Phenotype
1. Introduction
Clustering of microarray gene expression data is a common practice to find groups of genes that may be under coordinate regulation. 1 Various types of clustering strategies have been employed to group the data. Hierarchical clustering uses an agglomerative, bottom-up approach for grouping the data. k-means clustering partitions the data into clusters from the top-down. Self organizing maps (SOM) uses an unsupervised neural network framework for clustering the data into patterns that have an order associated with them. Most clustering methods simply use the gene expression data alone to group the samples or genes. In other words, they do not utilize the biological information about the samples or genes along with the gene expression data for performing the clustering. Including phenotypic information into the clustering of gene expression data may lead to a more informed analysis of the biology of the samples.
An additional limitation with clustering gene expression data is that the number of clusters in the data needs to be known. For instance, in k-means clustering, the number of k-clusters in the samples needs to be defined upfront. Likewise, with SOM, the topology (n rows by m columns) of the nodes for the neural networks has to be decided upon prior to running the algorithm. Furthermore, in hierarchical clustering one has to decide at what value to cut the dendrogram in order to generate meaningful clusters. More importantly, clustering methods like k-means are not assured to find the global minimum of the objective function and hence, cannot guarantee the optimal clustering solution. These limitations can potentially jeopardize the biological interpretation garnered from the clustering of the data.
Recently, simultaneous clustering of microarray gene expression data with clinical chemistry measurements and histopathology observations was shown to be advantageous for identifying “phenotypic prototypes” that are descriptive of, and phenotypically anchored to end-points of toxicity. 2 The method called Modk-prototypes involves constructing an objective function for microarray and clinical chemistry numeric data and simple matching for histopathology categorical values. Recursion was used to update the prototypes in order to find the configuration of the initial k-prototypes which ultimately results in the reduction of the objective function closest to the global minimum. The approach works well generally but is not designed to find the global optimum of the clustering solution. Hence, the phenotypic prototypes of the clusters of samples may not be optimized.
Clustering by simulated annealing has been applied recently to optimize the grouping of genes based on gene expression data. Although simulated annealing does not guarantee the global optimum of a clustering solution, it performs quite well at avoiding local minima. Simulated annealing has been used to cluster expression data based on mutual information and fuzzy membership of the genes. 3 More sophisticated applications of clustering gene expression data was proposed where a Genetic Algorithm was employed with simulated annealing to group multiple classes of samples.4 The approach improved on the clustering of the data by maximizing internal cluster cohesion and external cluster isolation. Despite these successful applications of simulated annealing to cluster gene expression data, the use of it to improve the clustering of biological samples based on microarray data and associated phenotypic measurements has not been investigated.
A clustering approach called simulated annealing of Modk-prototypes (SA-Modk-prototypes) to group biological samples based on microarray gene expression data and classes of known phenotypic variables in an optimized fashion is proposed in this paper. The method utilizes simulated annealing for optimization of an objective function comprised of the sum of the squared Euclidean distances for microarray and clinical chemistry numeric data and simple matching for histopathology categorical values. Separate weighting terms are used for microarray, clinical chemistry and histopathology measurements to control the influence of each data domain on the clustering of the samples. The dynamic validity index and category utility measure for numeric and categorical data respectively, are used to validate the clustering of the biological samples. A cluster’s prototype, formed from the mean of the values for numeric features and the mode of the categorical values of all the samples in the group, is representative of the phenotype of the cluster members. For SA-Modk-prototypes clustering, we use a gene expression data set and phenotypic measurements (clinical chemistry and historpathology) from the livers of rats treated with acetaminophen; an analgesic and antipyretic agent that causes centrilobular necrosis. Genes extracted from the prototypes of the clusters are shown to be anchored to the phenotypes of the samples.
2. Methods and Materials
2.1 Acetaminophen microarray gene expression data and analysis
Microarray gene expression data was derived from left liver lobe mRNA samples collected from 4 male Fischer F344/N rats per dose group exposed to either 50mg/kg, 150mg/kg (low doses), 1500mg/kg or 2000mg/kg (high doses) body weight acetaminophen during a light period (between 12 noon and 1 pm) as well as liver mRNA collected from control (vehicle-treated) male rats. 5 Animals were sacrificed and mRNA extracted from liver specimens 6, 18, 24, or 48 hrs after treatment. Each RNA sample from a treated animal was compared with a pool of time-matched control mRNAs and analyzed in duplicate (dye reversal experiments) on Agilent rat oligonucleotide microarrays (Agilent Technologies, Palo Alto, CA). Acetaminophen exposure to the rat liver at 50 and 150 mg/kg is subtoxic. However, 1500 and 2000 mg/kg doses induce severe toxicity which peaks 24hrs after exposure but the rats show signs of recovery 48hrs after exposure. Scanning of the microarray chips, acquisition of data from scanned images and data analysis were as previously described. 2,6
2.2 Acetaminophen histopathology observations and clinical chemistry evaluations
Forty eight histopathological observations of the acetaminophen-treated rat liver specimen slides and ten clinical chemistry measurements on biosamples from the treated animals were collected as previously described. 5 Microscopic qualifiers were categorized as no, minimal, mild, moderate or marked. Elevated levels of Alanine Aminotransferase (ALT) and Aspartate aminotransferase (AST) correlate with liver injury. Missing values were imputed for rats #s 308 and 309 with either the group average or the overall mean value for each clinical chemistry evaluation.
2.3 Modified k (Modk) -prototypes objective function
The k-prototypes algorithm 7 which combines the k-means and the k-modes objective functions for clustering numeric data and categorical values respectively, was modified to include separate numeric components for the microarray and the clinical chemistry data resulting in the following Modk-prototypes 2 objective function:
| (1) |
where Xi is the ith sample, for i = 1 to N number of samples, Ql is the lth prototype, for l = 1 to K number of clusters, xij and qlj are values of the jth attribute for sample i and the prototype of cluster l respectively, mr is the number of microarray numeric attributes, ms is the number of clinical chemistry numeric attributes, mc is the number of histopathological categorical attributes, α, β and γ denote the weights (W) for the microarray, clinical chemistry and histopathology data domain dissimilarity measures, respectively. The weights for data domain d at the nth step (Wd[n]) are adapted (for controlling how much each data domain contributes to the clustering of the samples) as follows:
| (2) |
where tau (τ) is the exponential weighting update factor in the range [0,1] and avecorr (Xd, Qd) is the average correlation coefficient (Pearson for numeric data, Jaccard for categorical data) between the samples and the prototypes based on the feature values from domain d. The value of τ was set to 0.05 in order to adjust the weight of each domain by 5% at each iteration. The weights are non-negative and their sum is constrained to equal one. The weights can easily be constrained to always be above some lower bound, e.g. 0.05, or even fixed at proportions that are appropriate or reasonable to a domain expert. The data for the numeric features were scaled so that each yields a contribution between zero and one to the dissimilarity measure. For categorical feature values, the dissimilarity measure between and is defined by the total number of mismatches of the corresponding histopathologic features from the sample and the prototype and respectively such that
| (3) |
where
| (4) |
2.4 Simulated annealing of the modk (SA-Modk) -prototypes objective function
Simulated annealing is a simple and general algorithm for locating global minima. The process follows the cooling of a physical system whose possible energies correspond to the values of the objective function being minimized. The use of the Metropolis algorithm for Monte Carlo approximate numerical simulation in simulated annealing has been described. 8 The simulated annealing procedure involves a slow cooling process where a temperature starts out high and then is gradually lowered until a frozen state of the system is reached. At each temperature, the state of the system is perturbed many times to avoid initialization dependency. The process decreases the temperature along a predefined cooling schedule until the system reaches thermal equilibrium at a low temperature and on the basis of a decreasing energy objective function. For a given temperature T, the fraction of the time spent in any state w is proportional to exp(−E(w)/T). The states visited by the Metropolis algorithm have frequencies which follow the Boltzmann distribution with the probability of accepting a state of the system being
| (5) |
where K is Boltzmann’s constant and ΔE is the difference in the energy objective function between the trial and current states. In the simulated annealing Modk-prototypes algorithm (SA-Modk-prototypes) implementation N = 100 states are visited by the Metropolis algorithm, a state is the cluster assignment of the samples, the temperature of the system is decreased from an initial value of one to a final value of 0.01 and the system’s energy objective function is Eq. (1). If w0 and w1 denote current and trial cluster assignments respectively, with E(w0) and E(w1) denoting the energies of the objective function for the current and the trial states respectively, then w1 is accepted according to the acceptance probability
| (6) |
The rate that T decreases depends on a cooling schedule. The following cooling schedule is popular and was used in this study:
| (7) |
where T(k) is the temperature at the kth step and C is the cooling factor.
2.5 Determination of cluster number (k) and validation of cluster assignment
Determination of the number of clusters in the data set was by using the dynamic validity index (DVI) which is based on an intra/inter ratio validity index that also includes scaling of the intra-and the inter-cluster distances. 9
| (8) |
where
| (9) |
k is the number of clusters, N is the number of samples and intra is the average Euclidean distance between samples and the prototype Q of the cluster each sample is assigned to.
For mixed data with numeric and categorical values, DVI was modified to include a category utility (CU) measure10 that defines the probability of matching a categorical feature value given a cluster versus the probability of the categorical feature value given the entire data set
| (10) |
where P(Ai = Vij) is the unconditional probability of feature Ai taking on the value Vij, P(Ai = Vij | Ck) is the conditional probability of Ai = Vij given cluster Ck, and k is the cluster number from 1 to m. DVI modified with CU is DVI_CU = (DVI+1/CU).
Validation of the cluster assignment was carried out using the adjusted Rand index R’.12 Let nij be the number of samples that are in both class (designation of the level of centrilobular necrosis) ui and cluster vj of the U and V partitions. Let ni. and n.j be the number of samples in class ui and cluster vj respectively and n‥ = n, the total number of samples. Assuming a hypergeometric distribution as the model of the U and V partitions being picked at random such that the number of samples in the classes and clusters are fixed, the adjusted Rand index is
| (11) |
and ranges between 0 and 1. When two partitions agree totally, R’ is 1 and when the partitions are selected by chance, R’ is 0.
2.6 Extracting genes from the prototypes
Let the observed difference between the expression ratio of the gth gene (p in total) from the gene expression component of prototype q for the ith and jth (i not equal to j) clusters (k in total) be observed g = (q gi− q gi)and the expected change in expression be
| (12) |
Averaging over all genes gives an estimate of the expected difference between the expression ratio of a gene in the prototype of two clusters being compared. Assuming independence and an approximately normal distribution of differences, genes which have expression ratios which significantly distinguish between prototypes of clusters are evaluated and extracted using a standard chi-square (X2) goodness-of-fit test 2 where
| (13) |
and the null hypothesis is that the expression value of the gth gene does not distinguish between prototypes of a pair of clusters that are compared. The null hypothesis is rejected at a level of α (the probability of a type I error) if X2c is greater than or equal to X2 (1,α), the α-level critical value of a chi square distribution with 1 degree of freedom. An α of 0.05 gives reasonable results. Matlab code and a stand-alone executable program for the SA-Modk-prototypes algorithm are available at http://www.niehs.nih.gov/research/resources/software.
3. Results
A workflow for the simulated annealing algorithm is shown in Fig. 1. At the start of the algorithm, parameters T0, Tf, C, N, α, β and γ are set for the initial temperature of the system, the final temperature of the system, the cooling factor, the number of steps for the Metropolis algorithm and the weights for the microarray, clinical chemistry and histopathology data respectively. The system is initialized by randomly selecting k samples as the initial prototypes of the k-clusters. Each sample is then assigned to a cluster based on Eq. (1). This assignment is set as the current state of the system (w0) and the energy of it E(w0) is determined by summing Eq. (1) for all samples and all clusters. Next, the system is perturbed using the Metropolis algorithm by repeated random assignment of a sample to a cluster. This assignment is set as the trial state of the system (w1) and the energy of it E(w1) is derived as was for E(w0). The change in the energy of the system [ΔE = E(w1)- E(w0) ] and the Boltzmann probability distribution (see Eq. (5)) are used to decide on whether or not to accept w1 (see Eq. (6)). Following that decision, the temperature is decreased according to the defined cooling schedule. Once the system is cooled to a final temperature, the algorithm ends by retuning the cluster assignment of the samples and the prototypes of the clusters.
Fig. 1.
Workflow for simulated annealing the Modk-prototype algorithm (SA-Modk-prototypes). N denotes no, Y denotes yes. Pr(w1) is obtained from a continuous uniform distribution on the [0,1] interval.
3.1 Estimating the number of k clusters
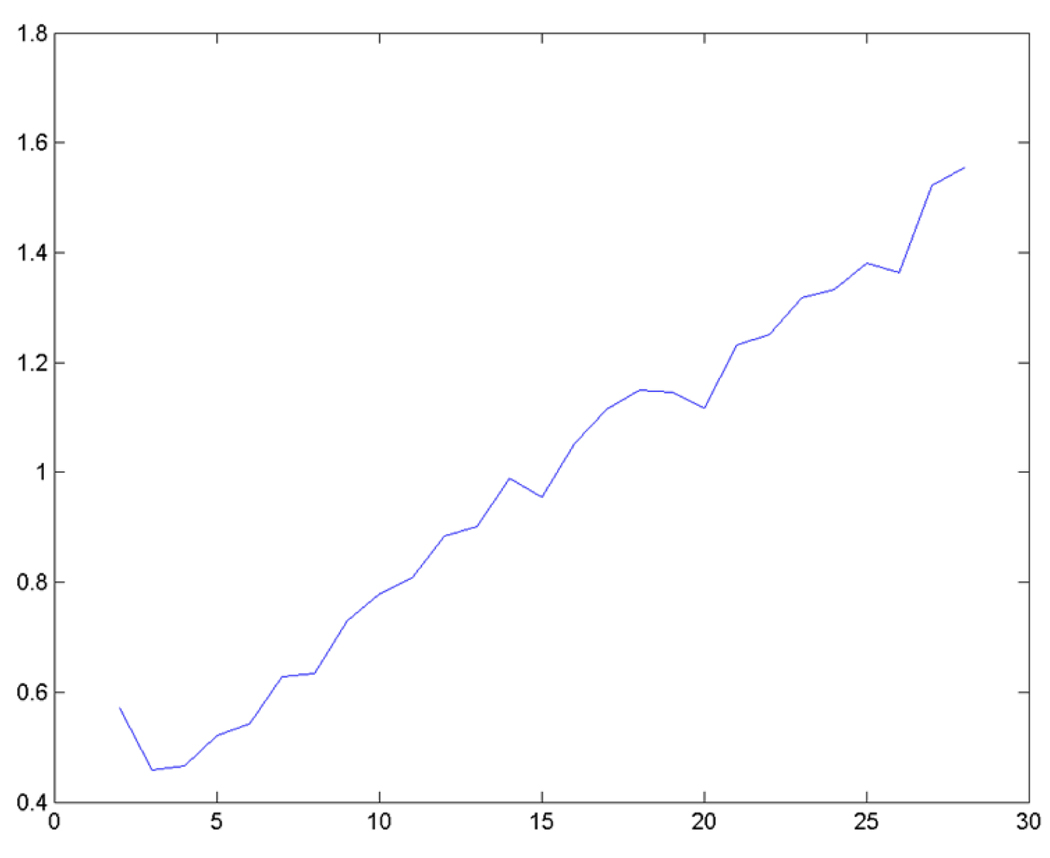
To estimate the number of clusters in the data set, the dynamic validity index 9 (DVI) was used for the numeric data and a category utility 10 (CU) measure of cluster “goodness” for categorical data (DVI_CU) to minimized over all k sets for each run of clustering mixed data using the Modk-prototypes algorithm with equal weighting of the domain data. An independent data set (cDNA microarray gene expression data with clinical chemistry and histopathology observations) with 436 differentially expressed genes acquired from the livers of rats exposed to 50, 150, 1500 or 2000 mg/kg of APAP for 6, 24 or 48 hours was used to empirically estimate the number of clusters. 11 As shown in Fig. 2, the plot of the DVI_CU validity measure minimizes at k = 3 but also very close to k = 4.
Fig. 2.
Determination of k clusters in an acetaminophen data set. The acetaminophen data was clustered using the Modk-prototypes algorithm with equal weighting at values of k increasing from 2. The dynamic validity index with category utility (DVI_CU) on the y axis was computed and plotted for the clustering of the data at each value of k on the x axis.
Having estimated the approximate number of clusters in a typical acetaminophen data set, the ability of the SA-Modk-prototypes algorithm to cluster the samples in the acetaminophen gene expression (oligonucleotide), clinical chemistry and histopathology data set according to the level of liver necrosis was assessed next. The indicator variable representing histopathological observations made by board-certified pathologists on the centrilobular region of the liver was removed from the data set prior to running the algorithm. This variable was then used as an external indicator to validate the assignment of samples to the clusters. This observation has four feature values for all the exposed samples denoting either no, minimal, mild, or moderate severity of necrosis of the centrilobular region of the liver. Using the SA-Modk-prototypes algorithm with k set at either three or four, equal weighting of the microarray, clinical chemistry and histopathology domain data and the cooling rate set at 0.1, 0.01 or 0.001, the adjusted Rand Index R’ 12 and the General Silhouette S 13 were computed to determine the agreement of the two partitions (known and assigned) and the stability of the clusters respectively. An R’ value of one denotes that the two partitions totally agree and zero when the agreement is by chance alone. A small or even negative value of S indicates the presence of one or more clusters with a sample at the outskirt of the group whereas a value close to one reflects clusters with samples more centrally located. As shown in Table 1, k = 3 and the two faster cooling rates (C set at 0.1 or 0.01) produce the higher values for R’ and S (0.67 and 0.65 respectively).
Table 1.
Validation of the clustering of the samples
| K | C | R' | S |
|---|---|---|---|
| 3 | 0.1 | 0.67 | 0.65 |
| 3 | 0.01 | 0.67 | 0.65 |
| 3 | 0.001 | ND | ND |
| 4 | 0.1 | 0.540 | 0.51 |
| 4 | 0.01 | 0.540 | 0.51 |
| 4 | 0.001 | 0.015 | −0.20 |
ND signifies not done. K, C, R’ and S denote the number of clusters, the cooling rate, the adjusted Rand Index and the General Silhouette respectively.
3.2 Weighting the domain data and clustering the samples
Perfect agreement of the clusters and the class labels derived from the observation of centrilobular necrosis of the liver (external indicator) was not attainable possibly due to the limitation in the grading of the necrotic regions of the rat liver (i.e. the amount of necrosis emphasized by the pathologists’ grading system is potentially affected by preferential sampling of necrotic and non-necrotic tissues). However, to test the influence of each domain data type on the clustering of the samples, various weighting schemes were used in the objective function (Eq. (1)) to control the proportion of the dissimilarity measure contributed by the microarray, clinical chemistry and histopathology measurements. As shown in Table 2, clustering the acetaminophen mixed data using the SA-Modk-prototypes algorithm with 49 times the weight applied to the microarray data relative to the clinical chemistry data and no weight given to the histopathology data (scheme 13) yielded the highest R’ measure of 0.81. The clustering result is better than applying all the weight to the microarray data (scheme 5, i.e., k-means clustering). Furthermore, k-means clustering of the microarray data or k-modes clustering of the histopathology data (scheme 7) is less valid than k-means clustering of the clinical data alone (scheme 6). Interestingly, when all (scheme 6) or some ([at least 0.02] schemes 1, 3, 4, 8–13, and 15) weight was given to the clinical chemistry data, clustering assignments were generated that yield an R’ of at least 0.67. The lowest R’ values were computed when all the weight was split between the microarray and histopathology data (scheme 2), applied to either data domain exclusively (schemes 5 and 7) or when 99% of the weight was given to the microarray data and only 1% to the clinical chemistry data (scheme 14). Note that the histopathology data had, essentially, a negligible effect on the validation of the clustering when combined with the other data (comparison of all the schemes).
Table 2.
Weighting the domain data during clustering of the samples
| Scheme | α | β | γ | R' |
|---|---|---|---|---|
| 1 | 0.333 | 0.333 | 0.333 | 0.67 |
| 2 | 0.5 | 0 | 0.5 | 0.27 |
| 3 | 0.5 | 0.5 | 0 | 0.67 |
| 4 | 0 | 0.5 | 0.5 | 0.67 |
| 5 | 1 | 0 | 0 | 0.29 |
| 6 | 0 | 1 | 0 | 0.67 |
| 7 | 0 | 0 | 1 | 0.23 |
| 8 | 0.60 | 0.20 | 0.20 | 0.67 |
| 9 | 0.90 | 0.10 | 0 | 0.70 |
| 10 | 0 | 0.10 | 0.90 | 0.67 |
| 11 | 0.90 | 0.05 | 0.05 | 0.70 |
| 12 | 0.95 | 0.05 | 0 | 0.70 |
| 13 | 0.98 | 0.02 | 0 | 0.81 |
| 14 | 0.99 | 0.01 | 0 | 0.27 |
| 15* | 0.26 | 0.39 | 0.35 | 0.67 |
Clustering performed with k=3 and C=0.01.
Adaptive weights.
3.3 Cluster assignment of the samples
The assignments of the samples from clustering the data with the weighting schemes that yielded the same R’ values were identical (data not shown). The assignment of the samples from the clustering with schemes 13 (R’ of 0.81) that produced the highest agreement with the original class labels of the samples differed from the assignment of the samples from the clustering with schemes 9, 11 and 12 (R’ of 0.70) in that the samples from rat #s 405, 423 and 518 treated with 1500 mg/kg for 18 hours, 1500 mg/kg for 48 hours and 2000 mg/kg for 18 hours of acetaminophen respectively, showing minimal centrilobular necrosis and having an average ALT and AST of 193 and 379 respectively, were placed in the cluster of samples that exhibited a mode of the score for the group on the centrilobular necrosis pathology observation equal to a mild severity (an average ALT and AST of 1558 and 2619, respectively).
In the other case, these samples were assigned to the cluster of samples where no centrilobular necrosis was observed and the samples had low ALT/AST enzyme measures.
The result of the clustering of the samples with the highest validity score (scheme 13) reveals that Cluster 1 contains all the low dosed samples along with high dosed samples exposed for 6 and 18 hrs except for the sample from rat #s 520 which was exposed to 2000 mg/kg of acetaminophen for 24 hrs. This exposure was expected to give at least a moderate hepatotoxic phenotype, but notably, the ALT and AST levels for this animal was far below the treatment group average for these enzymes. Cluster 3 consisted of exclusively high dosed samples that were exposed for 18 or 24 hrs and showed elevated levels of ALT and AST above 6,600 UI/L. Samples in Cluster 2 were essentially from animals which showed recovery from the toxic insult 48 hrs after exposure except for samples from rats #s 405, 407, 416, 420 and 518 which where exposed for only 18 or 24 hrs.
3.4 Phenotypic prototypes
The groups of samples from the SA-Modk-prototypes algorithm were analyzed for phenotypic prototypes by assessing the histopathologic feature value labels, clinical chemistry measurements, and genes from the prototypes of the clusters that (1) distinguish between pathologic outcomes and (2) best represent the underlying biology of the data. The clustering of the samples with k = 3 and α, β and γ set at 0.98, 0.02 and 0 respectively (scheme 13) allows the data from the microarray and clinical chemistry domains to contribute to the clustering of the data maximally and also permit the use of the histopathologic features as classifiers for discernment between the clusters of the samples by phenotype.
3.4.1 End-point components of the prototypes
Table 3 lists partial end-point prototypes of the resulting clusters. Samples in Clusters 2 and 3 were qualified by mild and moderate necrosis of the centrilobular region of the liver, respectively. By contrast, the majority of the samples in Cluster 1 had no centrilobular necrosis except for the two altered-responder rats (#s 520 and 423). The samples in Clusters 2 and 3 also showed moderately and markedly elevated ALT and AST enzyme levels respectively. The latter exhibited moderate congestion of the sinusoid region in the left medial lobe and minimal hypertrophy of the hepatocytes throughout. Furthermore, the samples from the rats in Cluster 2 were represented by a histopathologic prototype characterized by mild inflammatory cell infiltration in the centrilobular region and minimal regeneration of the hepatocytes, with the latter observed in the left lateral lobe region. Samples from rats #s 407, 416 and 420 were dosed with 1500 mg/kg acetaminophen for either 18 or 24 hrs durations, but had only moderately elevated levels of ALT and AST in the 490 to low 6,000 UI/L range. The rest of the histopathology feature values for the three clusters were not informative (all had no observed end-point) and therefore not included as representative features in the phenotypic prototypes.
Table 3.
Partial end-point components of the phenotypic prototypes
| Cluster | |||
|---|---|---|---|
| Features | 1 | 2 | 3 |
| Cong_Sinusoid* | None | None | Moderate |
| Necr_Cent | None | Mild | Moderate |
| Infl_Cent | None | Mild | None |
| Hypert_Hepa** | None | None | Minimal |
| Regen_Hepa** | None | Minimal | None |
| ALT (IU/L) | 104.2 | 1242.9 | 9649.4 |
| AST (IU/L) | 141.9 | 2102.1 | 20304 |
| BUN (mg/dL) | 15.2 | 18.5 | 23.6 |
| CHOLE (mg/dL) | 85.8 | 88.2 | 59.8 |
| TBA (umol/L) | 6.9 | 34.7 | 61.7 |
| SDH (IU/L) | 26.1 | 306.6 | 2.9 |
Observed in the left medial lobe only
Observed in the left lateral lobe only
Of the clinical chemistry measurements listed in Table 3 for each cluster prototype, ALT and AST levels clearly distinguish Cluster 3 samples labeled with the prototype feature as moderate necrosis of the centrilobular region of the liver from the two other clusters. In addition, elevated levels of TBA and decreased SDH levels (from about 9 to 100 fold) and blood cholesterol differentiate samples in Cluster 3 from samples in Clusters 1 and 2 reasonably well.
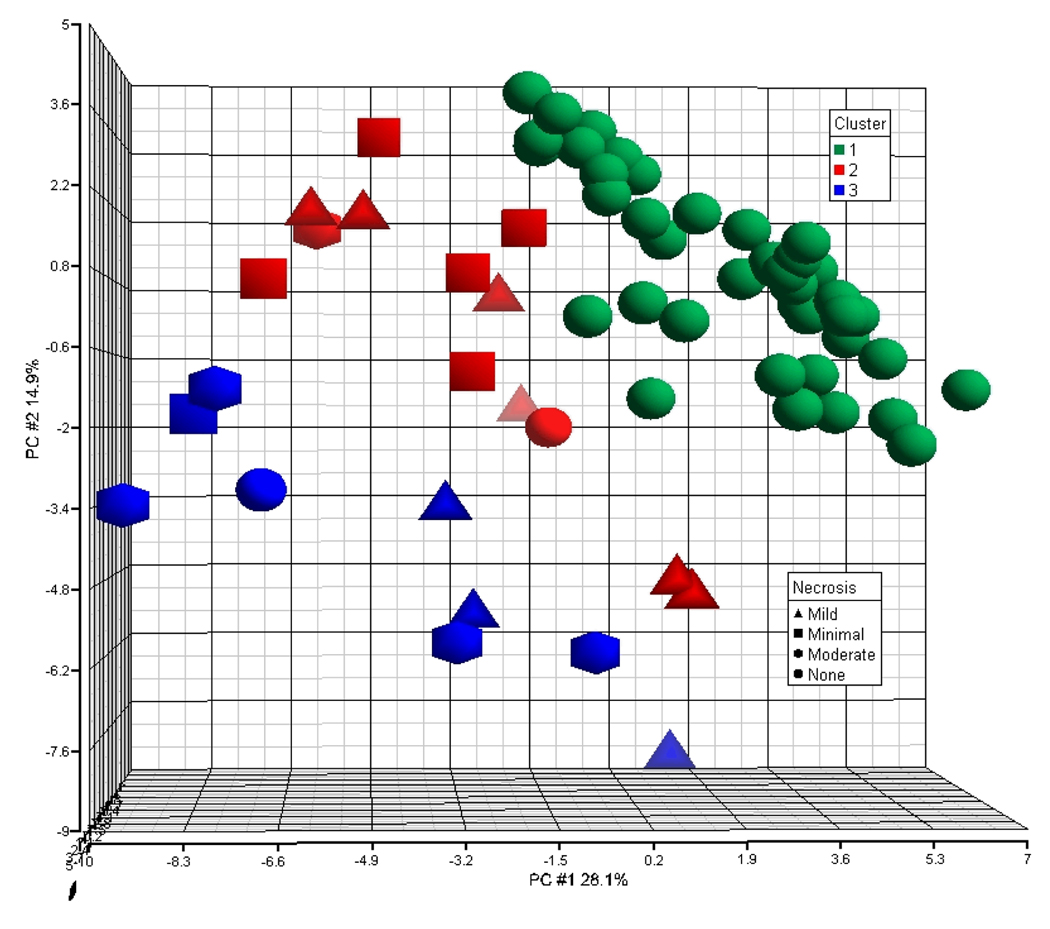
Fig. 3 illustrates the projection of the samples in 3-D space using the first 3 principal components (PCs) derived from the ~3100 differentially expressed genes. The samples are colored and shaped by their cluster assignment and severity of centrilobular necrosis respectively.
Fig. 3.
Principal component analysis of the gene expression data. The first 3 principal components (PCs) derived from the ~3100 differentially expressed genes were used to plot the samples into 3D space. The x axis is PC #1, the y axis PC #2 and the z axis PC #3. The samples are labeled by their respective cluster assignment (green-1, red-2, blue-3) and shaped by the severity of centrilobular necrosis observed.
The distribution of the samples from the top right corner of the graph to the lower left corner shifts from samples that were assigned to Clusters 1 to Cluster 3 denoting samples with cluster prototypes representing none, to mild, to moderate centrilobular necrosis. It is clear from the projection of the samples that PC1 and PC2 together contribute to the separation of the samples classified by the severity of centrilobular necrosis. However, a few samples in Cluster 1 (no centrilobular necrosis end-point prototype) are projected in the space occupied by Cluster 2 samples (mild centrilobular necrosis end-point prototype) and are therefore, difficult to separate into their respective class space.
3.4.2 Expression component of the prototypes
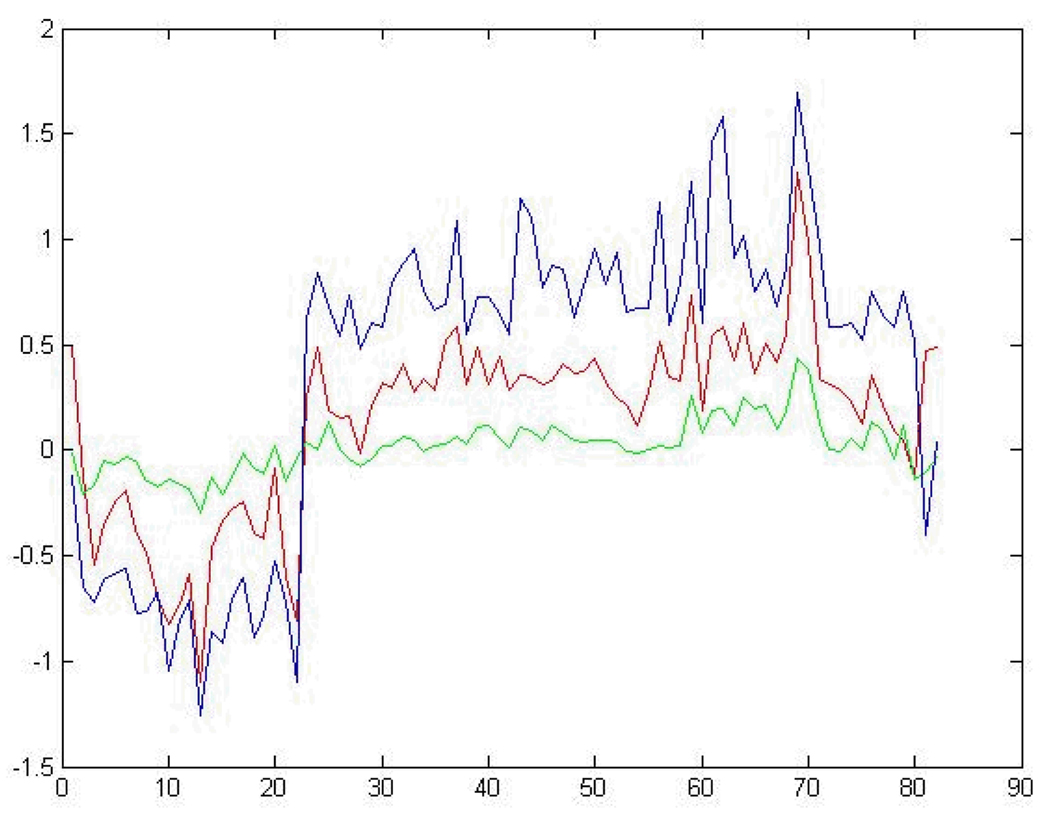
Differences in expression levels for each of the ~3100 genes between each cluster are shown in Fig. 4. The Cluster 1 prototype labeled with no necrosis of the centrilobular region had the least amount of differential gene expression of the samples in the group. Samples in Clusters 3 and 2 with moderate and mild necrosis of the centrilobular region as representative indicators respectively, had a few genes with over 2-fold differential expression. Pairwise comparisons of the expression component of the prototypes for the clusters were performed to extract genes that could statistically distinguish between levels of necrosis of the centrilobular region of the liver. A chi-square goodness-of-fit test 2 was employed using the observed difference in a gene’s expression ratios between two prototypes and the expected gene expression differences of all pairwise comparisons for all genes in the prototypes. With α set at 0.05, 86 genes, including two glucose metabolism genes (for glucose-6 phosphate [G6pc] and the glucose kinase regulatory protein [Gckr]) and two inflammatory response genes (calgranulin B [S100a9] and interleukin 1 beta [Il1b]), were identified as significant and unique in distinguishing contrasts between different levels of necrosis of the centrilobular region of the liver.
Fig. 4.
Gene expression components of all the differentially expressed genes in the phenotypic prototypes. Plotting of the gene expression component of the prototypes from the clusters generated from clustering the acetaminophen data using all ~3100 genes detected as differentially expressed. The blue, red and green lines denote the gene expression prototype from Clusters 3 (moderate necrosis), 2 (mild necrosis) and 1 (no necrosis) respectively. The indices for the genes are denoted on the x axis and the log10 ratio values of the genes from the prototypes are signified on the y axis..
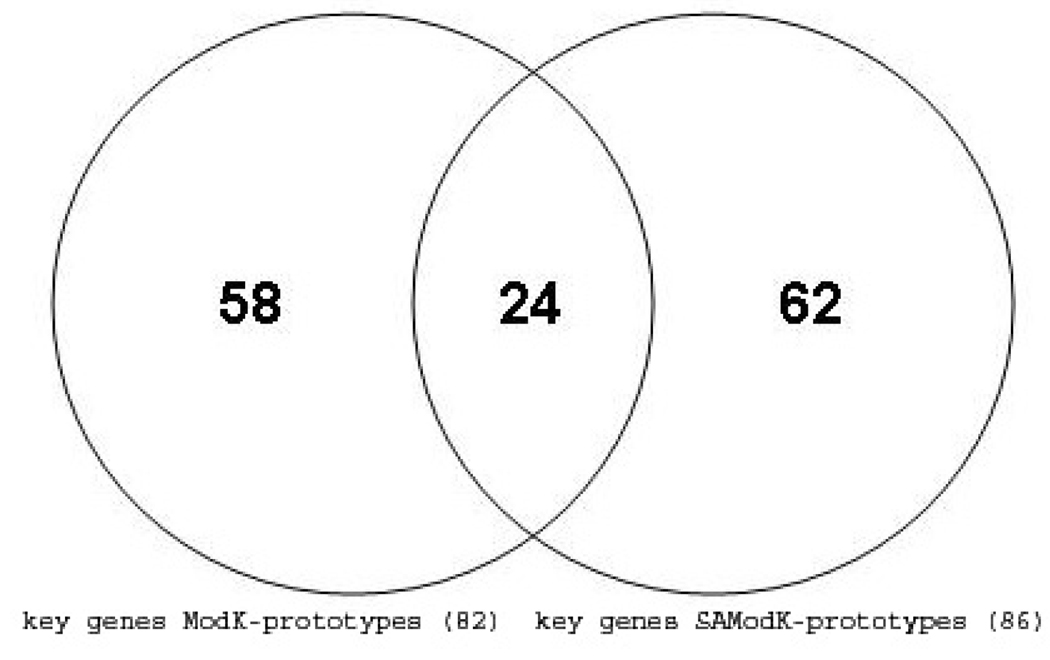
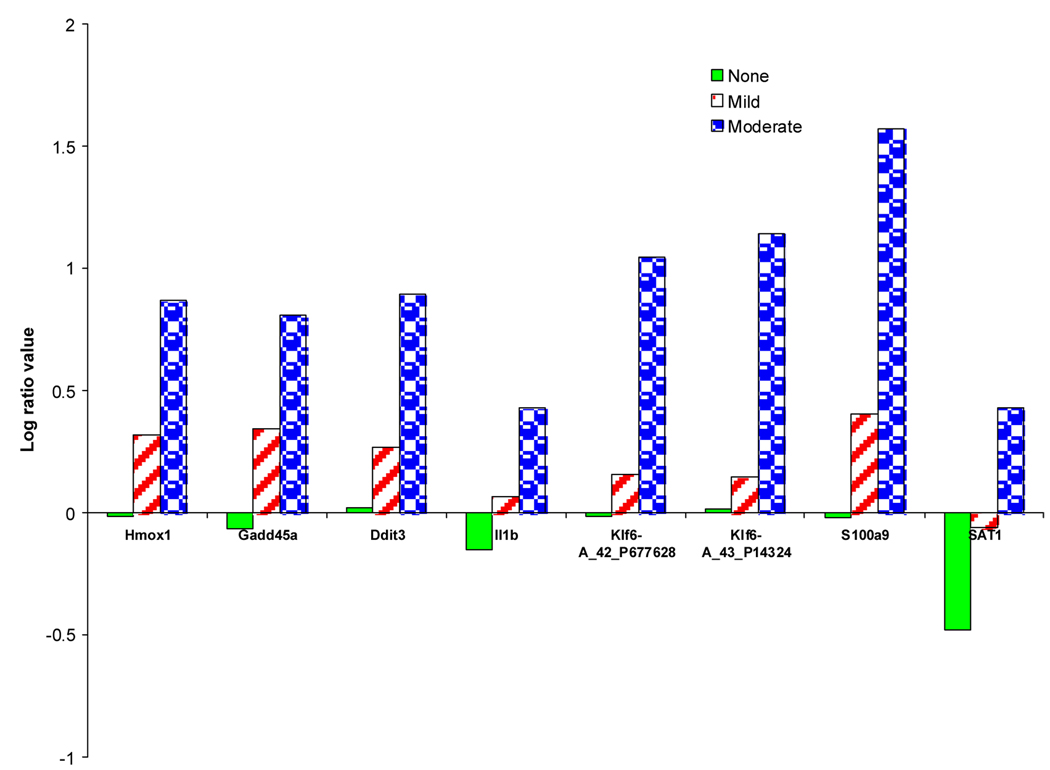
A more informative picture of the differences between the samples in the clusters labelled with either no, mild or moderate necrosis of the centrilobular region of the rat liver was obtained by comparisons of Clusters 1, 2 and 3 using just the expression values of the 86 genes extracted from the prototypes (Fig. 5). Most (about 75%) of the genes progressively increase or decrease in differential expression as the level of necrosis of the centrilobular region of the liver transitions from no, to mild, to moderate. A comparison of the 82 genes identified as significant in distinguishing contrasts between different levels of necrosis of the centrilobular region of the liver in the analysis of the data with the Modk-prototypes algorithm 2 and with the 86 genes from the analysis using the SA-Modk-prototypes algorithm in this paper reveals that 24 genes are in common between the two gene lists (Fig. 6) including a gene for heme oxygenase 1 (Hmox1). There are 62 unique genes from the analysis using the SA-Modk-prototypes approach. A subset of the genes represented in the Venn diagram (Fig. 6) is listed in Table 4.
Fig. 5.
Gene expression components from selected genes in the phenotypic prototypes. Plotting of the gene expression component of the prototypes from the clusters generated from clustering the acetaminophen data using 86 genes selected as significant for distinguishing between levels of centrilobular necrosis. The lines and axes are as described in Fig. 4.
Fig. 6.
Venn diagram of genes identified as unique and significant.
Table 4.
Annotation of some of the unique and significant genes identified by SA-Modk-prototypes and Modk-prototypes clustering
| GenBank Acc. # | Description | Gene Symbol |
|---|---|---|
| NM_022229 | Heat shock protein 1 (chaperonin) | Hspd1 |
| NM_024127 | Growth arrest and DNA-damage-inducible 45 alpha | Gadd45a |
| NM_138863 | Leukotriene B4 12-hydroxydehydrogenase | Ltb4dh |
| NM_133578 | Dual specificity phosphatase 5 | Dusp5 |
| NM_013098 | Glucose-6-phosphatase, catalytic | G6pc |
| NM_024134 | DNA-damage inducible transcript 3 | Ddit3 |
| NM_053352 | Chemokine orphan receptor 1 | Cmkor1 |
| BI303289 | Growth arrest specific 5 | Gas5 |
| NM_031344 | Fatty acid desaturase 2 | Fads2 |
| NM_031642 | Kruppel-like factor 6 | Klf6 |
| NM_053453 | Regulator of G-protein signaling 2 | Rgs2 |
| NM_031971 | Heat shock 70kD protein 1A | Hspa1a |
| NM_024351 | Heat shock protein 8 | Hspa8 |
| NM_153312 | Cytochrome P450, family 3, subfamily a, polypeptide 2 | Cyp3a2 |
| BG378237 | Immediate early response 3 | Ier3 |
| NM_019376 | Tyrosine 3-monooxygenase/tryptophan 5- monooxygenase activation protein, gamma polypeptide |
Ywhag |
| NM_012580 | Heme oxygenase (decycling) 1 | Hmox1 |
| NM_138827 | Solute carrier family 2 (facilitated glucose transporter), member 1 |
Slc2a1 |
| NM_053319 | Dynein light chain LC8-type 1 | Dynll1 |
| BQ209232 | Heat shock 105kDa/110kDa protein 1 | Hsph1 |
| NM_031987 | Carnitine O-octanoyltransferase | Crot |
| NM_013048 | Tocopherol (alpha) transfer protein | Ttpa |
| NM_053587 | S100 calcium binding protein A9 (calgranulin B) | S100a9 |
| NM_013120 | Glucokinase regulatory protein | Gckr |
| NM_053516 | Nucleolar protein 3 (apoptosis repressor with CARD domain) |
Nol3 |
| NM_019186 | ADP-ribosylation factor-like 4A | Arl4a |
| NM_031512 | Interleukin 1 beta | Il1b |
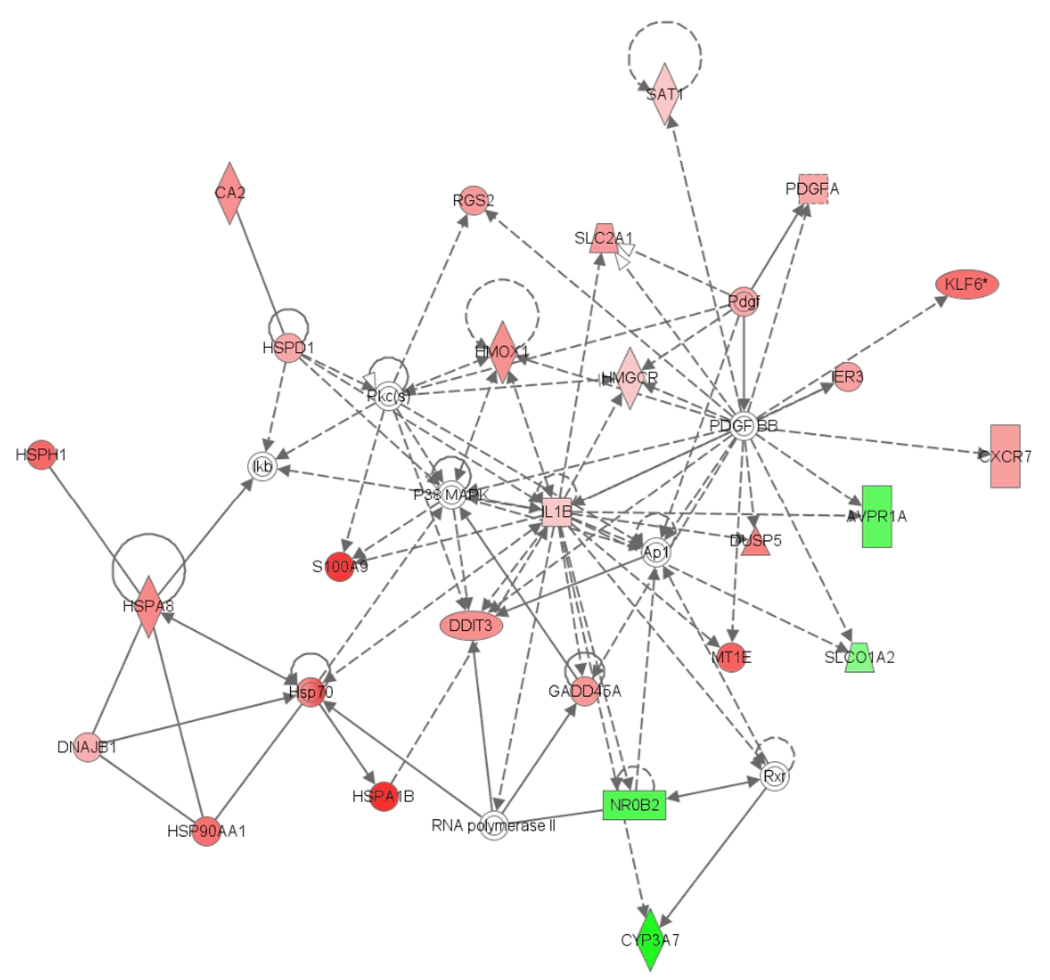
Gene Ontology and KEGG pathway analysis of the 144 genes by DAVID, the Database for Annotation, Visualization and Integrated Discovery 14, revealed that the Map kinase (MAPK) signaling pathway and the linoleic acid metabolism pathway are the significant biological processes that have genes which are influenced by the exposures of acetaminophen manifesting centrilobular necrosis. In addition, Ingenuity Pathway Analysis of the 86 genes from a sample exhibiting moderate centrilobular necrosis (Rat #508 exposed with 2000 mg/kg acetaminophen for 24 hrs) reveals a central regulatory roles of p38 MAP kinase (Mitogen-activated protein kinase) that regulates many cellular processes including inflammation, cell differentiation, cell growth and death and JNK, the c-Jun N-terminal kinase group of MAP kinases which contributes to inflammatory responses as well as cell death (Fig. 7). The complexes that interact with p38 MAP kinase and JNK (Gadd45a, IL-1β, Hmox1 and S100a9 for example) are induced substantially during stages of centrilobular necrosis in the rat liver while others are relatively unchanged in differential expression when compared to samples where no centrilobular necrosis is exhibited.
Fig. 7.
Interaction network of core genes expressed during moderate centrilobular necrosis.
4. Discussion
Clustering microarray gene expression data has been useful for grouping genes that are co-expressed. Many clustering algorithms are suited to only cluster numeric or categorical data but not both. Clustering microarray gene expression data along with biological information about the samples has proven to be advantageous for grouping genes and samples that share biological relevance. For instance, a novel clustering approach called heritable clustering, incorporates epigenetic (genes monitored for hypermethylation according to a binary [0,1] status) and phenotypic data (clinical measurements encoded as ordinal categorical variables) to group tumor samples sufficiently well enough for discovery of informative pathways that adhere to strict heritability in breast cancer. 15 Other clustering methods have also accommodated either clinical data or histopathological observations about the samples in the grouping process by either linear models with regression coefficients representing strength of the association or by correlation with principal components of the microarray data. 16,17 In addition, a clustering method recently published partially integrates clinical measurements with microarray data through separate Bayesian networks that are joined by a single phenotype variable. 18 However, the extension of these types of clustering algorithms for full integration and optimized analysis of high dimensional gene expression data integrated with clinical data as continuous measurements and phenotypic data as categorical values simultaneously has not been investigated. The simulated annealing (SA)-Modk-prototypes algorithm presented in this paper is a continuation of the work of Bushel et al. 2 to permit grouping of biological samples based on microarray gene expression data and classes of known phenotypic variables in a more formal and optimized fashion. The method utilizes simulated annealing for optimization of an objective function comprised of the sum of the squared Euclidean distances for numeric microarray and clinical chemistry data and simple matching for histopathology categorical values in order to measure dissimilarity of samples. Separate weighting terms are used for microarray, clinical chemistry and histopathology measurements to control the influence of each data domain on the clustering of the samples.
A cluster’s prototype, formed from the mean of the values for numeric features and the mode of the categorical values of all the samples in the group, is representative of the phenotype of the cluster members. The advantage of SA-Modk-prototypes clustering is that the phenotypic prototypes are derived from optimally-formed clusters of the biological samples (Table 1 and Table 2).
From the clustering of samples based on gene expression, clinical and pathology data derived from rats exposed to acetaminophen, phenotypic prototypes were obtained from three clusters of the biological samples which showed signs of no, mild and moderate centrilobular necrosis of the rat liver. The clinical chemistry portion of the phenotypic prototype clearly indicates that the ALT and AST enzymes levels are elevated (indicative of liver injury) in the cluster of the samples with the moderate necrosis of the centrilobular region phenotype observation (Table 3). Furthermore, the genes in the phenotypic prototypes with expression ratio values that contribute the most to discerning the three clusters of samples partitioned by their manifestation of a given severity of centrilobular necrosis of the rat liver (Fig 5, Fig 7 and Fig 8), contain genes related to proliferation, hyperbilirubinemia, injury and hemorrhaging of the liver. Pathway analysis revealed that Map kinase (MAPK) signalling and the linoleic acid metabolism were significant biological processes that had genes which are influenced by the exposures of acetaminophen manifesting centrilobular necrosis (Table 5). Linoleic acid is a polyunsaturated fatty acid that the liver converts to arachidonic acid, a primary target for lipid peroxidation. The role of lipid peroxidation in acetaminophen-induced toxicity has been controversial for sometime. 19–21
Fig. 8.
Differential expression of network focus genes according to the centrilobular necrosis phenotype.
Only 24 genes were found to be in common between the lists of genes identified by Modk-prototypes 2 and SA-Modk-prototypes clustering as discerning between necrosis of the liver in rats from acetaminophen treatment. The difference in the number and the specific genes that statistically distinguish between the levels of centrilobular necrosis is likely due to the cluster assignment of the samples when grouped using the two approaches. The Modk-prototypes clustering algorithm searches for clusters formed closest to the global minima of the objective function but does not guarantee finding the optimal clustering solution. On the other hand, the SA-Modk-prototypes clustering algorithm uses simulated annealing of the objective function to escape local optima in search for the global optimum. This is advantageous for effectively linking the phenotype of samples to groups of genes. This process of phenotypic anchoring has been described previously and approached by ad-hoc methods to link cause of a disease or response with the effect observed. 22–25 Dugas et al. 26 approached phenotypic anchoring of gene expression data to characteristics of samples more formally by using multidimensional clustering (mdclust). However, the method can only match a single phenotype variable to a set of clustered samples. Li and Hong 27 proposed the use of the Rasch model (an item-response theory approach) for relating gene expression profiles to phenotypes described by latent factors. However, the method can suffer from the loss of information due to the discretization of the microarray data. The SA-Modk-prototypes method described in this paper goes further in that it allows a set of end-point measurements, either categorical or continuous, to be coupled to groups of samples that share gene expressions patterns and phenotypic characteristics without transforming the data to discrete values.
Acknowledgements
Many thanks to Gary Boorman and Rick Irwin of the NIEHS/National Toxicology Program (NTP) for the design of the acetaminophen study and for generation of the gene expression, clinical chemistry and the histopathology data. The data is publicly available at the Chemical Effects in Biological Systems (CEBS) database, accession number 002-00001-0011-000-5 and at the NTP Study Results & Research Projects for Toxicogenomics web site: [http://ntp.niehs.nih.gov/go/15716]. Appreciation goes to Judong Shen for the computation for the Dynamic Validity Index and Spencer Muse for the motivation to use simulated annealing in the clustering process. Thanks to Jennifer Fostel, Frank Chao and Hong Xu for their critical review of the manuscript. This research was supported in part by the Intramural Research Program of the NIH, and NIEHS.
Biography

Pierre R. Bushel received a M.S. in Molecular Biology from Long Island University, Brooklyn, New York in 1989 and a Ph.D. in Bioinformatics from North Carolina State University in 2005. In 1998, he joined the NIEHS Microarray Group where he was responsible for management of microarray database systems, data analysis and software development for computational biology. In 2006 he joined the Biostatistics Branch at NIEHS as a Staff Scientist and the Head of Microarray and Genome Informatics.
References
- 1.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bushel PR, Wolfinger RD, Gibson G. Simultaneous clustering of gene expression data with clinical chemistry and pathological evaluations reveals phenotypic prototypes. BMC Syst Biol. 2007;1:15. doi: 10.1186/1752-0509-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou X, Wang X, Dougherty ER, Russ D, Suh E. Gene clustering based on clusterwide mutual information. J Comput Biol. 2004;11:147–161. doi: 10.1089/106652704773416939. [DOI] [PubMed] [Google Scholar]
- 4.Pan H, Zhu J, Han D. Genetic algorithms applied to multi-class clustering for gene expression data. Genomics Proteomics Bioinformatics. 2003;1:279–287. doi: 10.1016/S1672-0229(03)01033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irwin RD, Parker JS, Lobenhofer EK, Burka LT, Blackshear PE, Vallant MK, Lebetkin EH, Gerken DF, Boorman GA. Transcriptional profiling of the left and median liver lobes of male f344/n rats following exposure to acetaminophen. Toxicol Pathol. 2005;33:111–117. doi: 10.1080/01926230590522257. [DOI] [PubMed] [Google Scholar]
- 6.Heinloth AN, Irwin RD, Boorman GA, Nettesheim P, Fannin RD, Sieber SO, Snell ML, Tucker CJ, Li L, Travlos GS, Vansant G, Blackshear PE, Tennant RW, Cunningham ML, Paules RS. Gene expression profiling of rat livers reveals indicators of potential adverse effects. Toxicol Sci. 2004;80:193–202. doi: 10.1093/toxsci/kfh145. [DOI] [PubMed] [Google Scholar]
- 7.Huang Z. Clustering large data sets with mixed numeric and categorical values; Proceedings of the 14th International Joint Conference on Knowledge Discovery and Data Mining; 1997. [Google Scholar]
- 8.Kirkpatrick S, Gelatt CD, Jr, Vecchi MP. Optimization by Simulated Annealing. Science. 1983;220:671–680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- 9.Shen J, YD, Lee ES, Chang SI, Brown SJ. Determination of cluster number in clustering microarray data. Applied Math and Computation. 2005;169:1172–1185. [Google Scholar]
- 10.Gluck M, Corter J. Information, uncertainty, and the utility of categories; Proc 7th Ann Conf Cog Soc; 1985. pp. 283–287. [Google Scholar]
- 11.Bushel PR. Clustering of mixed data types with application to toxicogenomics. Ph.D. Dissertation. North Carolina State University; 2005. http://www.lib.ncsu.edu/theses/available/etd-03172005-091928/unrestricted/etd.pdf. [Google Scholar]
- 12.Yeung KY, Haynor DR, Ruzzo WL. Validating clustering for gene expression data. Bioinformatics. 2001;17:309–318. doi: 10.1093/bioinformatics/17.4.309. [DOI] [PubMed] [Google Scholar]
- 13.Famili AF, Liu G, Liu Z. Evaluation and optimization of clustering in gene expression data analysis. Bioinformatics. 2004;20:1535–1545. doi: 10.1093/bioinformatics/bth124. [DOI] [PubMed] [Google Scholar]
- 14.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 15.Wang Z, Yan P, Potter D, Eng C, Huang TH, Lin S. Heritable clustering and pathway discovery in breast cancer integrating epigenetic and phenotypic data. BMC Bioinformatics. 2007;8:38. doi: 10.1186/1471-2105-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selaru FM, Yin J, Olaru A, Mori Y, Xu Y, Epstein SH, Sato F, Deacu E, Wang S, Sterian A, Fulton A, Abraham JM, Shibata D, Baquet C, Stass SA, Meltzer SJ. An unsupervised approach to identify molecular phenotypic components influencing breast cancer features. Cancer Res. 2004;64:1584–1588. doi: 10.1158/0008-5472.can-03-3208. [DOI] [PubMed] [Google Scholar]
- 17.Jia Z, Xu S. Clustering expressed genes on the basis of their association with a quantitative phenotype. Genet Res. 2005;86:193–207. doi: 10.1017/S0016672305007822. [DOI] [PubMed] [Google Scholar]
- 18.Gevaert O, De Smet F, Timmerman D, Moreau Y, De Moor B. Predicting the prognosis of breast cancer by integrating clinical and microarray data with Bayesian networks. Bioinformatics. 2006;22:e184–e190. doi: 10.1093/bioinformatics/btl230. [DOI] [PubMed] [Google Scholar]
- 19.Wendel A, Feuerstein S, Konz KH. Acute paracetamol intoxication of starved mice leads to lipid peroxidation in vivo. Biochem Pharmacol. 1979;28:2051–2055. doi: 10.1016/0006-2952(79)90223-5. [DOI] [PubMed] [Google Scholar]
- 20.Knight TR, Fariss MW, Farhood A, Jaeschke H. Role of lipid peroxidation as a mechanism of liver injury after acetaminophen overdose in mice. Toxicol Sci. 2003;76:229–236. doi: 10.1093/toxsci/kfg220. [DOI] [PubMed] [Google Scholar]
- 21.Hinson JA, Bucci TJ, Irwin LK, Michael SL, Mayeux PR. Effect of inhibitors of nitric oxide synthase on acetaminophen-induced hepatotoxicity in mice. Nitric Oxide. 2002;6:160–167. doi: 10.1006/niox.2001.0404. [DOI] [PubMed] [Google Scholar]
- 22.Paules R. Phenotypic anchoring: linking cause and effect. Environ Health Perspect. 2003;111:A338–A339. doi: 10.1289/ehp.111-a338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moggs JG, Tinwell H, Spurway T, Chang HS, Pate I, Lim FL, Moore DJ, Soames A, Stuckey R, Currie R, Zhu T, Kimber I, Ashby J, Orphanides G. Phenotypic anchoring of gene expression changes during estrogen-induced uterine growth. Environ Health Perspect. 2004;112:1589–1606. doi: 10.1289/txg.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamadeh HK, Knight BL, Haugen AC, Sieber S, Amin RP, Bushel PR, Stoll R, Blanchard K, Jayadev S, Tennant RW, Cunningham ML, Afshari CA, Paules RS. Methapyrilene toxicity: anchorage of pathologic observations to gene expression alterations. Toxicol Pathol. 2002;30:470–482. doi: 10.1080/01926230290105712. [DOI] [PubMed] [Google Scholar]
- 25.Powell CL, Kosyk O, Ross PK, Schoonhoven R, Boysen G, Swenberg JA, Heinloth AN, Boorman GA, Cunningham ML, Paules RS, Rusyn I. Phenotypic anchoring of acetaminophen-induced oxidative stress with gene expression profiles in rat liver. Toxicol Sci. 2006;93:213–222. doi: 10.1093/toxsci/kfl030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dugas M, Merk S, Breit S, Dirschedl P. mdclust--exploratory microarray analysis by multidimensional clustering. Bioinformatics. 2004;20:931–936. doi: 10.1093/bioinformatics/bth009. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Hong F. Cluster-Rasch models for microarray gene expression data. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-8-research0031. RESEARCH0031. [DOI] [PMC free article] [PubMed] [Google Scholar]