Abstract
FIV is a significant pathogen in the cat and is, in addition, the smallest available natural model for the study of lentivirus infections. Although divergent at the amino acid level, the cat lentivirus has an abundance of structural and pathophysiological commonalities with HIV and thus serves well as a model for development of intervention strategies relevant to infection in both cats and man. The following review highlights both the strengths and shortcomings of the FIV/cat model, particular as regards development of antiviral drugs.
Introduction
FIV-induced AIDS in Cats
FIV causes an AIDS-like syndrome in the domestic cat, with many similarities to HIV-induced AIDS in man (1; Table 1). Like HIV, FIV can be transmitted via mucosal exposure, blood transfer, and vertically via prenatal and postnatal routes. FIV is tropic for T cells (1–6) macrophages (7–10), and central nervous system cells (CNS (11,12)). In vivo tissue tropism studies (11,12) have demonstrated viral RNA in T cells, macrophages, and CNS cells. FIV RNA has also been demonstrated in association with follicular dendritic cells (FDC) (13–15). Although CD4+ cell decline is a hallmark of FIV infection, FIV has a broader lymphocyte tropism than CD4+ T cells, with infection also evident in at least a subset of CD8+ T cells and B cells in vitro and in vivo (16–18). The course of the disease is also similar in cats and man, with a relatively short acute phase measure in weeks and denoted by increasing viral loads, febrile episodes, weight loss, lymphadenopathy, and neutropenia. During this time, CD4+ T cells decline as well as neutrophils and a percentage of cats will not recover and require humane euthanasia in experimental infections. However, most infected cats exhibit an increase in CD8+ T cells along with a strong humoral antibody response which allows them to weather this initial phase of the infection (19,20). The acute phase is followed by what is often referred to as an “asymptomatic”, or Latent phase denoted by relative quiescence of the infection in the face of strong antiviral immune responses, with lower viral titers and minimal clinical symptoms. As with HIV infections in man, this latent phase can be quite protracted in the cat, lasting from several months to several years. The rate of progression of the disease can dependant on the genotype of the infecting FIV and is also likely influenced by undefined genetic determinants of the particular cat. As with HIV-infected people, FIV-infected cats vary in the response to infection, with some animals remaining phenotypically normal throughout the course of the infection, while others suffer from assorted maladies including oral lesions, febrile episodes, bouts of diarrhea, and dehydration. The terminal phase is marked by further decline in the antiviral response, with resultant increase in plasma viral load and onset of clinical symptoms of immunodeficiency. Lymphoid tissue alterations are similar to those noted with primate lentivirus infections, including thymic depletion, lymphoid hyperplasia, plasmacytosis, and terminal lymphoid depletion (2,8–10,13,19,21). Neurological manifestations are often evident (12 22–25), noted by delays auditory evoked and visual evoked potential changes (22,23) and marked alterations in sleep patterns in experimentally infected animals (26). As with human AIDS, FIV-infected cats ultimately succumb to opportunistic infections.
Table 1.
COMPARATIVE PROPERTIES OF FIV AND HIV
| FIV | HIV | |
|---|---|---|
| Transmission | ||
| -blood contact | + | + |
| -mucosal contact | + | + |
| Target cell | ||
| CD4+ T cell | + | + |
| Macrophage | + | + |
| Dentritic cell, | + | + |
| Subset B cells | + | ? |
| Microglia | + | + |
| Disease symptoms: | ||
| Oral lesions | + | + |
| Lymphadenopathy | + | + |
| Neutropenia | + | + |
| CD4 T cell depletion | + | + |
| Hypergammaglobulinemia | + | + |
| Wasting, diahrrea | + | + |
| Secondary infections | + | + |
| CNS lesions | + | + |
| Viral genes encoded | ||
| Gag,Pol,Env,LTRs | + | + |
| Vif | + | + |
| Rev/RRE | + | + |
| Tat/TAR | − | + |
| Vpr | − | + |
| Vpu | − | + |
| OrfA | + | − |
| DU | + | − |
| Receptors utilized | ||
| -primary binding receptor | ||
| CD4 | − | + |
| CD134 | + | − |
| Heparans | + | + |
| -entry receptor | ||
| CXCR4 | + | + |
| CCR5 | − | + |
| -other chemokine receptors | ? | + |
Given the above parallels with HIV infection, both from the standpoint of target tissues and course of disease, study of FIV infection of cats can serve as a useful tool for identifying and understanding the immunological and cellular pressures that occur in the course of both lentivirus infections. As will be seen below, many facets of the virus life cycle are shared between the two lentiviruses, although distinct courses are taken in specific elements of the virus life cycle. Hopefully, the commonalities may lead to development of broad-based interventions that will block progression of the disease in both systems.
The Virus Genome
FIV and HIV share many features in their genomes, but also have important differences that influence the utility of comparative studies (27–29). The length of the FIV genome is around 9400 nucleotides, approximating that of HIV and other lentiviruses. The integrated provirus is bordered by long terminal repeats (LTRs) and possesses gag, pol, and env genes, common elements of all retroviruses. Also like other lentiviruses, FIV uses a tRNAlys primer-binding site to prime first strand synthesis by reverse transcriptase (RT). Transport of full-length and multiply spliced mRNAs is regulated by Rev, but the cis element acted on by Rev, the RRE, as well as second coding exon of Rev are located 3' of env instead of overlapping the TM coding region as in primate lentiviruses (30). FIV does not encode the vpr, vpu, or nef genes present in HIV and also lacks a Tat/Tar system for regulating viral gene transcription. FIV does encode a small gene product expressed along with Rev from a bicistronic mRNA, termed OrfA (or Orf2). Expression of OrfA is necessary for productive growth of FIV in T cells and it's expression results in a net increase in translation of gene products whose transcription is driven by the FIV LTR (31–33). However, OrfA does not act via a TAR element, as is the case with HIV-1 Tat and does not function in the manner of tradition gene activators (34). Gemeniano et al. (2003,2004) carried out studies that indicated that there is no wholesale increase in transcription in the presence of OrfA and thus, the increase in net translation may be the consequence of downstream action (35–36). They also showed that OrfA may have relatedness to Vpr and present indications of involvement in virus release from the cell and influence on cell cycle, similar to HIV Vpr (36). Thus, OrfA may be a multi-functional protein capable of substituting for more than one gene product lacking from FIV relative to HIV-1. Interestingly, OrfA shows the greatest sequence variability of all the viral genes, surpassing even the highly variable Env region when compared across viral clades. On one hand, if immunological pressure is driving this variability, it would be expected that the protein would reside on the surface of the cell or virion, which as far as we know is not the case. Alternatively. This variability might indicate a lack of any functional pressure to maintain sequence conservation. However, we know the protein plays an important role in virus replication in T cells and furthermore. Furthermore, a lack of function would likely result in even more variability and frequent appearance of stop codons and deletions, which is not the case. It remains to be determined the significance of this variability relative to the function of this interesting protein in the virus life cycle.
As for other lentiviruses, the Gag polyprotein of FIV is expressed from the full-length viral mRNA and is comprised of a myristoylated matrix (MA) protein, a capsid (CA) protein, and a nucleocapsid (NC) protein that has two copies of a zinc finger motif. FIV lacks a p6 protein between Gag and Pol, but contains instead a p2 protein (37) that has a P(S/T)AP domain necessary for virus budding (38). Thus, it is likely that p2 serves the same purpose as HIV-1 p6. It is now recognized that the PTAP domain in HIV p6 recruits TSG 101, a cellular protein involved in the virus budding process (39–43) and a similar role is likely in the FIV life cycle.
The FIV Pol proteins are expressed from the full-length viral mRNA as a large Gag/Pol polyprotein generated via a −1 frameshift approximately once every fifteen translational events (44). FIV Pol is comprised of protease (PR), reverse transcriptase (RT), and integrase (IN) genes that are common with HIV-1, but also encodes a gene for deoxyuridine pyrophosphatase (DU) between RT and IN that is lacking from the human lentivirus (45,46). FIVs lacking DU are incapable of successful propagation in cells that are not undergoing division, such as primary macrophages, whereas wild type FIV will productively infect such cells (46). These findings are also true for equine infectious anemia virus (EIAV), which is also a DU+ lentivirus and primarily lives in macrophages (47,48). DU is not necessary for replication in rapidly dividing cells, due to high endogenous levels of DU in the replicating cell (46). The primary role of DU is to prevent mis-incorporation of uracil into DNA by limiting the concentration of dUTP through conversion to dUMP, a precursor for dTTP synthesis. If the virus is mutated to deactivate or remove DU, the mutant FIV shows a five- to eight-fold increase in G->A transition mutations compared to wild type FIV during replication in macrophages in vivo, consistent with mis-incorporation of uracil into viral DNA during reverse transcription. It is as yet unclear as to how HIV, which does not encode DU, avoids high-level mis-incorporation of uracil in viral DNA. However, Vpr may play a role via an association with uracil N-glycosylase (Ung), the enzyme responsible for excision of uracil mis-incorporated into DNA (49). Mutations of Vpr that knock out Ung association causes a phenotype remarkably similar to the DU− phenotype noted in FIV and EIAV (50).
In common with HIV, FIV encodes viral infectivity factor (Vif) immediately 3' of pol, as in other lentiviruses, with the exception of EIAV which lacks a vif gene. The primary role of Vif appears to be to reduce G->A transition mutations by preventing cytidine deamination by the cellular deaminase, APOBEC-3G (51,52). Vif interacts with APOBEC-3G and directs the enzyme to the proteosome for degradation, thus preventing its incorporation into the virus particle. FIV Vif shows substantial sequence divergence from HIV Vif, but is the same size and retains the consensus sequence, SLQ(Y/F)LA critical to Vif function and common to all Vifs. The role of Vif in the FIV life cycle is yet to be fully elucidated, but has been shown to be essential for virus propagation in available cat cell lines (53). Experiments indicated that FIV Vif cannot transcomplement Vif-Defective HIV (54; other publications by Malim and colleagues). Thus, the function of the protein is influenced by species divergent elements in spite of the common consensus sequence.
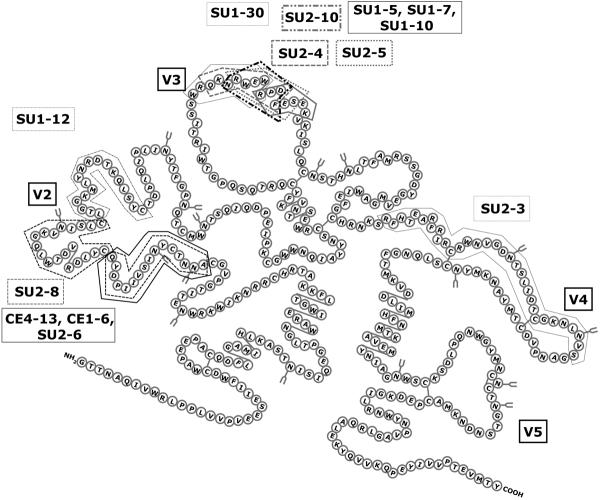
The env gene of FIV encodes heavily glycosylated SU and TM proteins, with 5–30% amino acid sequence divergence, similar to HIV Env. There are 5 consensus variable regions (V1–V5) in SU (29,55) (Figure 1). The mechanism of virus entry for FIV closely parallels SU/receptor interactions noted with HIV from the standpoint of employing both a primary binding and entry receptor for infection and sequence variability in Env. Four env subtypes (clades) plus numerous outliers have been defined via Env sequences (56). The intra- and inter-clade variation of FIV follows a pattern similar to that noted in HIV clades, as assessed by heteroduplex mobility shift assays (57). Assessments using synonymous/non-synonymous base change ratios and level of mutational saturation have been used to define distinctions between the most commonly found FIV env clades, A and B (58,59).
Figure 1.
Schematic diagram of the FIV SU, showing Variable (V) loops predicted from comparisons of multiple FIV Env sequences. Boxed areas in the structure indicate relative locations of the epitopes for a panel of monoclonal antibodies generated in mice using whole virions and mapped using nested synthetic peptides and deletion constructs (65,66; unpublished data)
In spite of the targeting of CD4+ T cells in vivo, FIV does not use CD4 as a primary binding receptor. However, all domestic cat FIVs we have examined to date share with certain T cell-tropic HIVs the utilization of the chemokine receptor, CXCR4 as an entry receptor as judged by the universal sensitivity of infection to the X4 antagonist, AMD3100 (60,61). Ex vivo studies have shown that FIV infection of certain cells may occur solely mediated by CXCR4 if expression of the chemokine receptor is sufficiently high (62,63). As to the initial binding receptor, Shimojima et al (64) demonstrated that this molecule is the activation marker, CD134, confirmed and extended in other studies (18,62,63). The demonstration that CD134 is up-regulated on activated CD4+ T cells (18) explains how FIV targets this cell population in vivo in spite of failure to bind CD4. Furthermore, soluble CD134 can interact with the virus to facilitate productive infection of CD134− CXCR4+ target cells (65), indicating that the binding receptor alters the conformation of SU to promote high affinity binding to CXCR4. This observation parallels findings with CD4 binding to HIV SU and indicates that although different primary binding receptors are utilized, both viruses use very similar mechanisms to infect target cells. Furthermore, neutralizing monoclonal antibodies have been identified that only neutralize the virus when soluble CD134 is present (65,66). These antibody epitopes, as well as the CXCR4 binding domain, reside on the V3 loop of FIV SU (65,66; Figure 1). The V2 loop also has many antibody binding epitopes, but none of these antibodies have been found to inhibit to date. If the epitope repertoire recognized in mice is any indicator, the V4 and V5 domains are relatively quiet from an immunological sense, likely reflecting a relatively low exposure to the environment. The findings of neutralizing epitopes exposed after virus binding parallel observations of such masked epitopes on HIV that become available upon interaction of SU with the CD4 binding receptor (67–71). The striking similarity and conservation of entry mechanisms between the two divergent lentiviruses likely the result of common immunological pressures in the two hosts. Thus, the feline lentivirus offers a valuable venue to study the mechanisms of lentivirus infection of T cells and for development of strategies to compromise the virus' ability to escape immune surveillance.
FIV as A Model for Drug Design
As outlined above, HIV and FIV have evolved along unique pathways that have led to development of alternative mechanisms to deal with certain aspects of replication, including transcriptional transactivation and uracil mis-incorporation. However, there are sufficient similarities to make the cat/FIV model a valuable tool for several lines of direct experimentation. The utilization of CXCR4 by FIV as one of the receptors used to enter target cells is an important similarity to a subset of HIV that can be explored in development of intervention strategies. In addition, commonalities in the target cell populations between FIV and HIV and most enzymes and structural components critical to virus replication present similar obstacles for perpetuation of species.
The enzymes encoded by pol, including PR, RT, and IN have common functions in the two viruses and in many cases, respond to the same inhibitors. In addition, defining the structural basis for failures to broadly inhibit the lentiviruses can provide essential information relevant to designing broad-based inhibitors. Use of the cat for development of broad-based protease inhibitors has been successful and has yielded an abundance of information regarding the regions that control both substrate and inhibitor sensitivities (72). Nucleoside analogs that interact with the active site of reverse transcriptase have been found efficacious against both FIV and HIV (73). It is likely that a similar will evolve for development of anti-integrase drugs. The structural proteins of Gag may also provide broad-based targets, since all lentiviruses share common morphological features. Elements of the virus core are likely to maintain commonalities in their mechanisms of action and orientations in the particles. The matrix, capsid, and nucleocapsid proteins may thus present effective targets for broad-based intervention strategies. As pointed out above, the P2 protein of FIV (37) is an apparent functional homologue to the P6 protein of HIV and shares late domain homologies (39,40). Both HIV and FIV encode Vif proteins, which may provide an additional target for intervention strategies to be used in both lentivirus systems.
Development of Inhibitors to FIV PR
The aspartic protease. PR, is responsible for viral Gag and Gag-Pol polyprotein processing into individual structural and enzymatic proteins during assembly and maturation (37,74–78). This processing step is highly specific, ordered, and essential to generate infectious retrovirus particles (77,79–81). Therefore, PR has been a very important target for antiviral therapies (82–85). Several approved protease inhibitors are available that are effective for treating HIV-1 infection (84–87) and combination drug therapies, termed highly active retroviral therapy (HAART), have been used successfully in suppressing HIV-1 replication to undetectable levels in patients (86–91). However, drug resistance development is a persistent problem (91–99). As many as 40% of the patients receiving HAART have a viral rebound within the first 3 years and this number is likely to be higher outside of controlled studies (100). In addition, transmission of resistant HIV has been observed and is likely to increase with more patients on combination therapy (100). Also, poor tolerance to current protease inhibitors by a significant number of patients may lead to increased non-compliance, which may be the leading reason for cases of failure of HAART therapy. Side effects resulting from long-term drug treatment have also been observed. Both of the latter problems might be allayed by development of drugs with better bioavailability and length of efficacy per dosage, which would reduce the drug regimen. Thus, there is a need to develop novel inhibitors with activities against drug-resistant isolates that exhibit delayed resistance development and show a high degree of specificity (95). Defining the determinants of substrate specificity of the lentiviral PRs is a logical first step in the development of such broad-based inhibitors.
FIV protease, like HIV-1 protease, is a homodimeric aspartic proteinase and the two enzymes are very similar at the crystallographic level, particularly within the substrate binding pocket (101). However, FIV is distinct in that each monomer is comprised of 116 amino acids, as opposed to 99 amino acids for HIV-1 protease, with only 27 conserved amino acids between FIV and HIV-1 PRs. Like HIV protease, FIV PR is responsible for processing Gag and Gag-Pol polyproteins (37). Similar to SIV and HIV-1 PRs, autoproteolysis of FIV protease is observed in vitro (102). Despite these similarities, FIV PR is specific to its respective substrates and inhibitors of HIV-1 protease currently employed in clinic do not inhibit FIV protease (103–105). FIV protease cleaves the FIV MA/CA cleavage junction efficiently. However, it does not appreciably cut the HIV-1 MA/CA cleavage junction, despite the presence of four identical residues in the P3-P3' position. HIV-1 protease prefers its own substrates as well, but can cleave FIV MA/CA cleavage junction to some degree. Important to the present discussion, there are at least 6 mutations found in HIV-1 proteases associated with drug resistance that are identical to structurally equivalent residues of wild type FIV protease (105). Two particularly interesting resistance mutations of HIV-1 protease, Val32→Ile (FIV Ile37) and Ile50→Val (FIV Val59), are located in the substrate binding pockets of the protease, which suggests they may play an important role in the inhibitor and substrate selectivity of retroviral protease. Studies (72,106) have shown that a major structural distinction between FIV and HIV-1 PRs is that the combined S1/S3 substrate binding pocket is restricted in size relative to the same site in HIV-1 PR. This finding offers a structural explanation for the failure of the current HIV-1 PR inhibitors, which possess bulky P3 groups, to inhibit FIV PR (72). Importantly, many drug-resistant HIV-1 PRs appear to have more restricted S1/S3 subsites as well (106), reducing inhibitor binding affinities in a manner similar to the feline enzyme. In addition, the nature of S2/S2' amino acids is particularly critical in directing PR substrate specificity as well as certain inhibitor efficacies. Thus, studies directed at understanding the structural basis for inhibitor and substrate specificity in the feline and human systems may lead to development of broad-based inhibitors with efficacy for a range of HIV variants.
Both FIV and HIV-1 PRs recognize, approximately, the P4-P4' residues of peptide substrates via a long cavity in the middle of the protease, as analyzed by biochemical experiments (78,107–111) and crystallographic analyses (112–114). Both homodimeric PRs utilize an acid-base hydrolysis mechanism in which aspartic acids 25 and 25' (of HIV-1 PR; 30 and 30' for FIV PR) activate a water to perform a nucleophilic attack on the amide carbonyl between the P1 and P1' positions in various peptide substrates (108). Like most aspartic proteases, optimal substrate cleavage occurs at approximately pH 4–5 (95,110,115).
There are three major structurally conserved regions that make up the substrate binding pockets of PR: 1) the active core region (residues 30–38 for FIV; 25–33 for HIV); 2) the flap (residues 54–60 for FIV; 45–51 for HIV); and 3) C-terminal “90s loop” region (residues 98–101 for FIV; “80s loop” for HIV, residues 80–84). Within these regions, there are 11 amino acids that differ between FIV and HIV-1 proteases. These residues have proved to be good candidate targets for mutational studies of substrate selectivity. The 11 different amino acid residues in the S4 - S4' subsites of FIV protease; Ile35, Ile37, Gln54, Asn55, Met56, Ile57, Val59, Ile98, Gln99, Pro100 and Leu101, most likely account for the specificity of the substrate/inhibitor binding. The corresponding residues in HIV-1 protease are Asp30, Val32, Lys45, Met46, Ile47, Gly48, Ile50, Pro81, Val82, Asn83 and Ile84, respectively. We have prepared a series of mutant FIV PRs in which HIV-1 amino acid residues have been substituted into the FIV PR background at equivalent positions. Confirmation of the involvement of several of these residues in both substrate and inhibitor specificities has been obtained. These wild type and mutant FIV PRs have, and will continue to serve, as a structural library for further defining substrate specificity and for inhibitor refinement in the proposed research.
Influence of Polyprotein Structure on Processing
An increasing body of evidence points to a pivotal role of the polyprotein folding/conformation in the temporal cleavage of the Gag and Gag-Pol proteins that is necessary for generation of infectious virus (77,79,116–118). The studies of Swanstrom and colleagues (77,79,80,119–121) pointed out that polyprotein cleavage occurs in a specific order and that alteration of the order by site-directed mutagenesis of certain sites resulted in production of non-infectious HIV. In particular, cleavage at the N-terminus of NC appeared to be the earliest cleavage event, at least in vitro, and subsequent studies have shown early cleavage on either side of NC (119). Differences in the rate of cleavage of synthetic substrates encompassing the cleavage sites suggested that the order of cleavage was in part, dictated by the relative cleavage efficacy of each junction. However, more recent studies have indicated that the availability of sites around NC, based on folding of the polyprotein relative to the “embedded” protease, is likely the critical trigger to the initiation of ordered processing. Interesting studies of Kaplan, Dunn, and colleagues have shown that subtle changes at the N-terminus of the embedded protease can markedly influence polyprotein processing in cis with no apparent influence on the ability of free protease to cleave the polyprotein in trans (119,121). This finding underscores the role of polyprotein conformation in processing and the importance of the temporal cleavage of Gag-Pol in the generation of infectious virus. Structural and biochemical analyses have been performed on FIV PR to define distinctions with HIV PR to aid in understanding of the basis for processing (122–124). The results show that similar events occur during processing of FIV Gag-Pol polyprotein and the critical nature of the proper temporal cleavage of Gag/Pol in generating infectious virus offers yet another important target for inhibitor development that can be pursued using FIV as a model system.
Acknowledgements
Work summarized in this review was supported from grants R01AI025825 and R01AI040882 of the National Institute of Allergy and Infectious Diseases of the National Institutes of Health.
References
- 1.Pedersen NC, Ho EW, Brown ML, Yamamoto JK. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science. 1987;235:790–3. doi: 10.1126/science.3643650. [DOI] [PubMed] [Google Scholar]
- 2.Beebe AM, Dua N, Faith TG, Moore PF, Pedersen NC, Dandekar S. Primary stage of feline immunodeficiency virus infection: viral dissemination and cellular targets. J. of Vir. 1994;68:3080–91. doi: 10.1128/jvi.68.5.3080-3091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Novotney C, English RV, Housman J, Davidson MG, Nasisse MP. Lymphocyte population changes in cats naturally infected with feline immunodeficiency virus. Aids. 1990;4:1213–8. doi: 10.1097/00002030-199012000-00005. [DOI] [PubMed] [Google Scholar]
- 4.O'Neil LL, Burkhard MJ, Hoover EA. Frequent perinatal transmission of feline immunodeficiency virus by chronically infected cats. J Vir. 1996;70:2894–901. doi: 10.1128/jvi.70.5.2894-2901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sparger EE, Luciw PA, Elder JH, Yamamoto JK, Lowenstine LJ. Feline immunodeficiency virus is a lentivirus associated with an AIDS-like disease in cats. Aids. 1989;3:S43–9. doi: 10.1097/00002030-198901001-00006. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto JK, Sparger E, Ho EW, Andersen PR, O'Connor TP, Mandell CP, Lowenstine L, Munn R, Pedersen NC. Pathogenesis of experimentally induced feline immunodeficiency virus infection in cats. Am J Vet Res. 1988;49:1246–58. [PubMed] [Google Scholar]
- 7.Brunner D, Pedersen NC. Infection of peritoneal macrophages in vitro and in vivo with feline immunodeficiency virus. J Virol. 1989;63:5483–8. doi: 10.1128/jvi.63.12.5483-5488.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callanan JJ, Racz P, Thompson H, Jarrett O. Morphologic characterization of the lymph node changes in feline immunodeficiency virus infection as an animal model of AIDS. S. Karger, Basel; Switzerland: 1993. [Google Scholar]
- 9.Parodi AL, Femenia F, Moraillon A, Crespeau F, Fontaine JJ. Histopathological changes in lymph nodes of cats experimentally infected with the feline immunodeficiency virus (FIV) J Comp Pathol. 1994;111:165–74. doi: 10.1016/s0021-9975(05)80048-9. [DOI] [PubMed] [Google Scholar]
- 10.Rideout BA, Lowensteine LJ, Hutson CA, Moore PF, Pedersen NC. Characterization of morphologic changes and lymphocyte subset distribution in lymph nodes from cats with naturally acquired feline immunodeficiency virus infection. Vet Pathol. 1992;29:391–9. doi: 10.1177/030098589202900504. [DOI] [PubMed] [Google Scholar]
- 11.Dow SW, Dritz MJ, Hoover EA. Feline immunodeficiency virus neurotropism: evidence that astrocytes and icroglia are the primary target cells. Vet Immunol. Immunopathol. 1992;35:23–35. doi: 10.1016/0165-2427(92)90118-a. [DOI] [PubMed] [Google Scholar]
- 12.Dow SW, Poss ML, et al. Feline immunodeficiency virus: a neurotropic lentivirus. J Acquir Immune Defic Syndr. 1992;3(7):658–68. 1990. [PubMed] [Google Scholar]
- 13.Bach JM, Hurtrel M, Chakrabarti L, Ganiere JP, Montagnier L, Hurtrel B. (1994) Early stages of feline immunodeficiency virus infection in lymph nodes and spleen. AIDS Res Hum Retroviruses. 1994;10:1731–8. doi: 10.1089/aid.1994.10.1731. [DOI] [PubMed] [Google Scholar]
- 14.Hurtrel B, Chakrabarti L, Hurtrel M, Bach JM, Ganiere JP. Early events in lymph nodes during infection with SIV and FIV. Res Virol. 1994;145:221–7. doi: 10.1016/s0923-2516(07)80026-4. [DOI] [PubMed] [Google Scholar]
- 15.Toyosaki T, Miyazawa T, Furuya T, Tomonaga K, Shin YS. Localization of the viral antigen of feline immunodeficiency virus in the lymph nodes of cats at the early stage of infection. Arch Virol. 1993;131:335–47. doi: 10.1007/BF01378636. [DOI] [PubMed] [Google Scholar]
- 16.Brown WC, Bissey L, Logan KS, Pedersen NC, Elder JH, Collisson EW. Feline immunodeficiency virus infects both CD4+ and CD8+ T lymphocytes. J of Vir. 1991;65:3359–64. doi: 10.1128/jvi.65.6.3359-3364.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willett BJ, Hosie MJ, Callanan JJ, Neil JC, Jarrett O. Infection with feline immunodeficiency virus is followed by the rapid expansion of a CD8+ lymphocyte subset. Immunology. 1993;78:1–6. [PMC free article] [PubMed] [Google Scholar]
- 18.de Parseval A, Chatterji U, Sun P, Elder JH. Feline immunodeficiency virus targets activated CD4+T cells by using CD134 as a binding receptor. Proc Natl Acad Sci U S A. 2004;101:13044–9. doi: 10.1073/pnas.0404006101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.English RV, Nelson P, Johnson CM, Nasisse M, Tompkins WA. Development of clinical disease in cats experimentally infected with feline immunodeficiency virus. J Infect Dis. 1994;170:543–52. doi: 10.1093/infdis/170.3.543. [DOI] [PubMed] [Google Scholar]
- 20.de Rozieres S, Mathiason CK, Rolston MR, Chatterji U, Hoover EA. (2004) Characterization of a highly pathogenic molecular clone of feline immunodeficiency virus clade C. J Virol. 2004;78:8971–82. doi: 10.1128/JVI.78.17.8971-8982.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woo JC, Dean GA, Pedersen NC, Moore PF. Immunopathologic changes in the thymus during the acute stage of experimentally induced feline immunodeficiency virus infection in juvenile cats. J Virol. 1997;71:8632–41. doi: 10.1128/jvi.71.11.8632-8641.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lafrado L, Podell M, Krakowka S, Hayes K, Hanlon M, et al. FIV: a model for retrovirus-induced pathogenesis. AIDS Research Reviews. 1993;3:115–150. [Google Scholar]
- 23.Phillips TR, Prospero-Garcia O, Puaoi DL, Lerner DL, Fox HS. Neurological abnormalities associated with feline immunodeficiency virus infection. Journal of General Virology. 1994;75:979–87. doi: 10.1099/0022-1317-75-5-979. [DOI] [PubMed] [Google Scholar]
- 24.Phillips TR, Prospero-Garcia O, Wheeler DW, Wagaman PC, Lerner DL. (1996) Neurologic dysfunctions caused by a molecular clone of Feline Immundeficiency Virus, FIV-PPR. Journal of NeuroVirology. 1996;2:388–396. doi: 10.3109/13550289609146904. [DOI] [PubMed] [Google Scholar]
- 25.Prospero-Garcia O, Herold N, Phillips TR, Elder JH, Bloom FE. (1994) Sleep patterns are disturbed in cats infected with feline immunodeficiency virus. Pro Nat Acad Sci U S A. 1994;91:12947–51. doi: 10.1073/pnas.91.26.12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prospero-Garcia O, Herold N, Waters AK, Phillips TR, Elder JH. Intraventricular administration of a FIV-envelope protein induces sleep architecture changes in rats. Brain Research. 1994;659:254–8. doi: 10.1016/0006-8993(94)90888-5. [DOI] [PubMed] [Google Scholar]
- 27.Olmsted RA, Barnes AK, Yamamoto JK, Hirsch VM, Purcell RH. Molecular cloning of feline immunodeficiency virus. Proc Natl Acad Sci U S A. 1989;86:2448–52. doi: 10.1073/pnas.86.7.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Talbott RL, Sparger EE, Lovelace KM, Fitch WM, Pedersen NC. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc Natl Acad Sci U S A. 1989;86:5743–7. doi: 10.1073/pnas.86.15.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips TR, Lamont C, Konings DA, Shacklett BL, Hamson CA. Identification of the Rev transactivation and Rev-responsive elements of feline immunodeficiency virus. Journal of Virology. 1992;66:5464–71. doi: 10.1128/jvi.66.9.5464-5471.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phillips TR, Talbott RL, Lamont C, Muir S, Lovelace K. Comparison of two host cell range variants of feline immunodeficiency virus. J Vir. 1990;64:4605–13. doi: 10.1128/jvi.64.10.4605-4613.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Parseval A, Elder JH. (1999) Demonstration that orf2 encodes the feline immunodeficiency virus transactivating (Tat) protein and characterization of a unique gene product with partial rev activity. J Virol. 1991;73:608–617. doi: 10.1128/jvi.73.1.608-617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sparger EE, Shacklett BL, Renshaw-Gegg L, Barry PA, Pedersen NC. Regulation of gene expression directed by the long terminal repeat of the feline immunodeficiency virus. Virology. 1992;187:165–77. doi: 10.1016/0042-6822(92)90305-9. [DOI] [PubMed] [Google Scholar]
- 33.Waters AK, De Parseval AP, Lerner DL, Neil JC, Thompson FJ. (1996) Influence of ORF2 on host cell tropism of feline immunodeficiency virus. Virology. 1996;215:10–6. doi: 10.1006/viro.1996.0002. [DOI] [PubMed] [Google Scholar]
- 34.Chatterji U, de Parseval A, Elder JH. Feline immunodeficiency virus OrfA is distinct from other lentivirus transactivators. J Virol. 2002;76:9624–34. doi: 10.1128/JVI.76.19.9624-9634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gemeniano MC, Sawai ET, Sparger EE. Feline immunodeficiency virus Orf-A localizes to the nucleus and induces cell cycle arrest. Virology. 2004;325:167–74. doi: 10.1016/j.virol.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Gemeniano MC, Sawai ET, Leutenegger CM, Sparger EE. Feline immunodeficiency virus ORF-A is required for virus particle formation and virus infectivity. J Virol. 2003;77:8819–30. doi: 10.1128/JVI.77.16.8819-8830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elder JH, Schnolzer M, Hasselkus-Light CS, Henson M, Lerner DA, Phillips TR, Wagaman PC, Kent SB. Identification of proteolytic processing sites within the Gag and Pol polyproteins of feline immunodeficiency virus. J Virol. 1993;67:1869–76. doi: 10.1128/jvi.67.4.1869-1876.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manrique ML, Rauddi ML, Gonzalez SA, Affranchino JL. Functional domains in the feline immunodeficiency virus nucleocapsid protein. Virol. 2004;327:83–92. doi: 10.1016/j.virol.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 39.Demirov DG, Orenstein JM, Freed EO. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J Virol. 2002;76:105–17. doi: 10.1128/JVI.76.1.105-117.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freed EO. Viral late domains. J Virol. 2002;76:4679–87. doi: 10.1128/JVI.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 42.Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–9. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 43.VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc Natl Acad Sci U S A. 2001;98:7724–9. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morikawa S, Bishop DH. Identification and analysis of the gag-pol ribosomal frameshift site of feline immunodeficiency virus. Virol. 1992;186:389–97. doi: 10.1016/0042-6822(92)90004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elder JH, Lerner DL, Hasselkus-Light CS, Fontenot DJ, Hunter E. Distinct subsets of retroviruses encode dUTPase. J Virol. 1992;66:1791–4. doi: 10.1128/jvi.66.3.1791-1794.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lerner DL, Wagaman PC, Phillips TR, Prospero-Garcia O, Henriksen SJ. Increased mutation frequency of feline immunodeficiency virus lacking functional deoxyuridinetriphosphatase. Proc Natl Acad Sci U S A. 1995;92:7480–4. doi: 10.1073/pnas.92.16.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steagall WK, Robek MD, Perry ST, Fuller FJ, Payne SL. Incorporation of uracil into viral DNA correlates with reduced replication of EIAV in macrophages. Virol. 1995;210:302–13. doi: 10.1006/viro.1995.1347. [DOI] [PubMed] [Google Scholar]
- 48.Threadgill DS, Steagall WK, Flaherty MT, Fuller FJ, Perry ST. Characterization of equine infectious anemia virus dUTPase: growth properties of a dUTPase-deficient mutant. J Virol. 1993;67:2592–600. doi: 10.1128/jvi.67.5.2592-2600.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouhamdan M, Benichou S, Rey F, Navarro JM, Agostini I, Spire B, Camonis J, Slupphaug G, Vigne R, Benarous R, Spire J. Human immunodeficiency virus type 1 Vpr protein binds to the uracil DNA glycosylase DNA repair enzyme. J Virol. 1996;70:697–704. doi: 10.1128/jvi.70.2.697-704.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mansky LM, Preveral S, Selig L, Benarous R, Benichou S. The interaction of vpr with uracil DNA glycosylase modulates the human immunodeficiency virus type 1 In vivo mutation rate. J Virol. 2000;74:7039–47. doi: 10.1128/jvi.74.15.7039-7047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L. (2003) Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 52.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–8. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shacklett BL, Luciw PA. Analysis of the vif gene of feline immunodeficiency virus. Virology. 21994;04:860–7. doi: 10.1006/viro.1994.1609. [DOI] [PubMed] [Google Scholar]
- 54.Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by theproteasome in response to HIV-1 Vif. Nat Med. 2003;9:1404–7. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 55.Pancino G, Castelot S, Sonigo P. Differences in feline immunodeficiency virus host cell range correlate with envelope fusogenic properties. Virology. 1995;206:796–806. doi: 10.1006/viro.1995.1002. [DOI] [PubMed] [Google Scholar]
- 56.Bachmann MH, Mathiason-Dubard C, Learn GH, Rodrigo AG, Sodora DL, Mazzetti P, Hoover EA, Mullins JI. (1997) Genetic diversity of feline immunodeficiency virus: dual infection, recombination, and distinct evolutionary rates among envelope sequence clades. J Virol. 1997;71:4241–53. doi: 10.1128/jvi.71.6.4241-4253.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delwart EL, Gordon CJ. Tracking changes in HIV-1 envelope quasispecies using DNA heteroduplex analysis. Methods. 1997;12:348–54. doi: 10.1006/meth.1997.0489. [DOI] [PubMed] [Google Scholar]
- 58.Sodora DL, Courcelle J, Brojatsch J, Berson A, Wang YC. Analysis of a feline immunodeficiency virus provirus reveals patterns of gene sequence conservation distinct from human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1995;11:531–3. doi: 10.1089/aid.1995.11.531. [DOI] [PubMed] [Google Scholar]
- 59.Sodora DL, Shpaer EG, Kitchell BE, Dow SW, Hoover EA. Identification of three feline immunodeficiency virus (FIV) env gene subtypes and comparison of the FIV and human immunodeficiency virus type 1 evolutionary patterns. J Virol. 1994;68:2230–8. doi: 10.1128/jvi.68.4.2230-2238.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poeschla EM, Looney DJ. CXCR4 is required by a nonprimate lentivirus: heterologous expression of feline immunodeficiency virus in human, rodent, and feline cells. J Virol. 1998;72:6858–66. doi: 10.1128/jvi.72.8.6858-6866.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Willett BJ, Adema K, Heveker N, et al. The second extracellular loop of CXCR4 determines its function as a receptor for feline immunodeficiency virus. J Virol. 1998;72:6475–81. doi: 10.1128/jvi.72.8.6475-6481.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Parseval A, Elder JH. Binding of recombinant feline immunodeficiency virus surface glycoprotein to feline cells: role of CXCR4, cell-surface heparans, and an unidentified non-CXCR4 receptor. J Virol. 2001;75:4528–39. doi: 10.1128/JVI.75.10.4528-4539.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Parseval A, Chatterji U, Morris G, Sun P, Olson AJ. Structural mapping of CD134 residues critical for interaction with feline immunodeficiency virus. Nat Struct Mol Biol. 2005;12:60–6. doi: 10.1038/nsmb872. [DOI] [PubMed] [Google Scholar]
- 64.Shimojima M, Miyazawa T, Ikeda Y, McMonagle EL, Haining H. Use of CD134 as a primary receptor by the feline immunodeficiency virus. Science. 2004;303:1192–5. doi: 10.1126/science.1092124. [DOI] [PubMed] [Google Scholar]
- 65.de Parseval A, Grant CK, Sastry KJ, Elder JH. Sequential CD134-CXCR4 interactions in feline immunodeficiency virus (FIV): soluble CD134 activates FIV Env for CXCR4-dependent entry and reveals a cryptic neutralization epitope. J Virol. 2006;80:3088–91. doi: 10.1128/JVI.80.6.3088-3091.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sundstrom M, et al. Mapping of the CXCR4 binding site within variable region 3 of the feline immunodeficiency virus surface glycoprotein. J Virol. 2008;82(18):9134–9142. doi: 10.1128/JVI.00394-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dey B, Del Castillo CS, Berger EA. neutralization of human immundeficiency virus type 1 by sCD4-17b, a single-chain chimeric protein based on sequential interaction of gp120 with CD4 and coreceptor. J Virol. 2003;77:2859–65. doi: 10.1128/JVI.77.5.2859-2865.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–82. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 69.Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol. 2003;77:10557–65. doi: 10.1128/JVI.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lusso P, Earl PL, Sironi F, Santoro F, Ripamonti C. Cryptic nature of a conserved, CD4-inducible V3 loop neutralization epitope in the native envelope glycoprotein oligomer of CCR5-restricted, but not CXCR4-using, primary human immunodeficiency virus type 1 strains. J Virol. 2005;79:6957–68. doi: 10.1128/JVI.79.11.6957-6968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moulard M, Phogat SK, Shu Y, Labrijn AF, Xiao X. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc Natl Acad Sci U S A. 2002;99:6913–8. doi: 10.1073/pnas.102562599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee T, Laco GS, Torbett BE, Fox HS, Lerner DL, Elder JH, Wong C-H. Analysis of the S3 and S3' subsite specificities of feline immunodeficiency virus (FIV) protease: Development of a broad-based protease inhibitor efficacious against FIV, SIV, and HIV in vitro and ex vivo. Proc Natl Acad Sci U S A. 1998;95:939–44. doi: 10.1073/pnas.95.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.North TW, North GL, Pedersen NC. Feline immunodeficiency virus, a model for reverse transcriptase-targeted chemotherapy for acquired immune deficiency syndrome. Antimicrob Agents Chemother. 1989;33:915–9. doi: 10.1128/aac.33.6.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dunn BM, Gustchina A, Wlodawer A, Kay J. Subsite Preferences of Retroviral Proteinases. Meth Enzymol. 1994;241:254–278. doi: 10.1016/0076-6879(94)41068-2. [DOI] [PubMed] [Google Scholar]
- 75.Katz RA, Skalka AM. The Retroviral Enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 76.Pettit SC, Simsic J, Loeb DD, Everitt L, Hutchison CA. d., Swanstrom R. Analysis of Retroviral Protease Cleavage Sites Reveals Two Types of Cleavage Sites and the Structural Requirements of the P1 Amino Acid. J Biol Chem. 1991;266(22):14539–14547. [PubMed] [Google Scholar]
- 77.Swanstrom R, Wills J. Synthesis, assembly, and processing of viral proteins. In: Coffin J, Hughes S, editors. Retroviruses. Cold Spring Harbor Laboratory Press; NY: 1997. [PubMed] [Google Scholar]
- 78.Tozser J, Bagossi P, Weber IT, et al. Studies on the Symmetry and Sequence Context Dependence of the HIV-1 Proteinase Specificity. J Biol Chem. 1997;272(27):16807–16814. doi: 10.1074/jbc.272.27.16807. [DOI] [PubMed] [Google Scholar]
- 79.Pettit SC, Moody MD, Wehbie RS, et al. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68(12):8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pettit SC, Sheng N, Tritch R, Erickson-Viitanen S, Swanstrom R. The regulation of sequential processing of HIV-1 Gag by the viral protease. Adv Exp Med Biol. 1998;436:15–25. doi: 10.1007/978-1-4615-5373-1_2. [DOI] [PubMed] [Google Scholar]
- 81.Tomasselli AG, Heinrikson RL. Specificity of retroviral proteases: an analysis of viral and nonviral substrates. Meth Enzymol. 1994;241:279–301. doi: 10.1016/0076-6879(94)41069-0. [DOI] [PubMed] [Google Scholar]
- 82.Flexner C. HIV drug development: the next 25 years. Nat Rev Drug Discov. 2007;6(2):959–66. doi: 10.1038/nrd2336. [DOI] [PubMed] [Google Scholar]
- 83.Schinazi RF, Larder BA, W. MJ. International Antiviral News. 1997;5(8):129–142. [Google Scholar]
- 84.McDonald D. The inside track on HIV. Nat Methods. 2006;3(10):782–783. doi: 10.1038/nmeth1006-782. [DOI] [PubMed] [Google Scholar]
- 85.Vacca JP, Condra JH. Clinically effective HIV-1 protease inhibitors. Drug Discovery. 1997;2(7):261–272. [Google Scholar]
- 86.Johnson VA, Brun-Vezinet F, Clotet B, et al. Update of the drug resistance mutations of HIV-1. Top HIV Med. 2005;13(4):125–131. [PubMed] [Google Scholar]
- 87.Richman DD, Morton SC, Wrin T, et al. The prevalence of antiretroviral drug resistance in the Unites States. Aids. 2004;18(10):1393–1401. doi: 10.1097/01.aids.0000131310.52526.c7. [DOI] [PubMed] [Google Scholar]
- 88.Collier AC, Coombs RW, Schoenfeld DA, Bassett RL, Timpone J, Baruch A, Jones M, Facey K, Whitacre C, McAuliffe VJ, Friedman HM, Merigan TC, Reichman RC, Hooper C, Corey L. N Engl J Med. 1996;334(16):1011–1017. doi: 10.1056/NEJM199604183341602. [DOI] [PubMed] [Google Scholar]
- 89.Gulick RM, Mellor JW, Havlir D, et al. Treatment with Indinavir, Zidovudine, and Lamivudine in Adults with Human Immunodeficiency Virus Infection and Prior Antiretroviral Therapy. N Engl J Med. 1997;337(11):734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 90.Kirk O, Katzenstein TL, Gerstoft, et al. Combination therapy containing ritonavir plus saquinavir has superior short-term antiretroviral efficacy: a randomized trial. Aids. 1999;13(1):F9–16. doi: 10.1097/00002030-199901140-00002. [DOI] [PubMed] [Google Scholar]
- 91.Palella FJ, Jr., Delaney KM, Moorman AC, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N Engl J Med. 1998;338(13):853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 92.Condra JH, Schleif WA, Blahy OM. In vivo emergence of HIV-1 variants resistance to multiple protease inhibitors. Nature. 1995;374(6522):569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 93.Jacobsen H, Hanggi M, Ott M, et al. Sensitivity to Saquinavir: An update on genotyping from phase I/II trials. J Infect Dis. 1996;173(6):1379–1387. [Google Scholar]
- 94.Kozal M. Cross-resistance patterns among HIV protease Inhibitors. AIDS Patient Care STDS. 2004;18(4):199–208. doi: 10.1089/108729104323038874. [DOI] [PubMed] [Google Scholar]
- 95.Kutilek VD, Sheeter DA, Elder JH, Torbett BE. Is resistance futile? Curr Drug Targets Infect Disord. 2004;3(4):295–309. doi: 10.2174/1568005033481079. [DOI] [PubMed] [Google Scholar]
- 96.Lawrence J, Schapiro J, Winters, et al. Clinical resistance patterns and responses to two sequential protease inhibitor regimens in Saquinavir and reverse transcriptase inhibitor- experienced persons. J Infect Dis. 1999;179(6):1356–1364. doi: 10.1086/314751. [DOI] [PubMed] [Google Scholar]
- 97.Molla A, Korneyeva M, Gao Q, et al. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2(7):760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 98.Young B, Johnson S, Bahktiari M, et al. Resistance mutations in protease and reverse transcriptase genes of human immunodeficiency virus type 1 isolates from patients with combination antiretroviral therapy failure. J Infect Dis. 1998;178(5):1497–1501. doi: 10.1086/314437. [DOI] [PubMed] [Google Scholar]
- 99.Zhang YM, Imamichi H, Imamichi T, et al. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J Virol. 1997;71(9):6662–6670. doi: 10.1128/jvi.71.9.6662-6670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cohen OJ, Fauci AS. Perinatal Transmission of Human Granulocytic Ehrlichiosis. N Engl J Med. 1998;339(5):341–343. [PubMed] [Google Scholar]
- 101.Wlodawer A, Gustchina A, Reshetnikova L, et al. Structure of an inhibitor complex of the proteinase from feline immunodeficiency virus. Nat Struct Biol. 1995;2(6):480–488. doi: 10.1038/nsb0695-480. [DOI] [PubMed] [Google Scholar]
- 102.Laco GS, Schalk-Hihi C, Lubkowski J, Morris G, Zdanov A. Crystal structures of the inactive D30N mutant of feline immunodeficiency virus protease complexed with a substrate and an inhibitor. Biochemistry. 1997;36:10696–708. doi: 10.1021/bi9707436. [DOI] [PubMed] [Google Scholar]
- 103.Dunn BM, Pennington MW, Frase DC, Nash K. Comparison of inhibitors binding to feline and human immunodeficiency virus proteases: structure-based drug design and the resistance problem. Biopolymers. 1991;51(1):69–77. doi: 10.1002/(SICI)1097-0282(1999)51:1<69::AID-BIP8>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 104.Schnolzer M, Rackwitz HR, Gustchina A, Laco GS, Wlodawer A, Elder JH, Kent SB. Comparative properties of feline immunodeficiency virus (FIV) and human immunodeficiency virus type 1 (HIV-1) proteinases prepared by total chemica synthesis. Virology. 1996;224(1):268–275. doi: 10.1006/viro.1996.0528. [DOI] [PubMed] [Google Scholar]
- 105.Slee DH, Laslo KL, Elder JH, et al. Selectivity in the Inhibition of HIV and FIV Protease: Inhibitory and Mechanistic Studies of Pyrrolidine-Containing .alpha.-Keto Amide and Hydroxyethylamine Core Structures. J. Am. Chem. Soc. 1995;117:11867–11878. [Google Scholar]
- 106.Lee T, Le VD, Lim D, et al. Development of a New Type of Protease Inhibitors, Efficacious against FIV and HIV Variants. J Am Chem Soc. 1999;121:1145–1155. [Google Scholar]
- 107.Beck ZQ, Lin Y-C, Elder JH. Molecular Basis for the Relative Substrates Specificity of HIV-1 and FIV Proteases. J. Virol. 2001;75:9558–9469. doi: 10.1128/JVI.75.19.9458-9469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Silva AM, Cachau RE, Sham HL, Erickson JW. Inhibition and catalytic mechanism of HIV-1 aspartic protease. J Mol Biol. 1996;255(2):321–346. doi: 10.1006/jmbi.1996.0026. [DOI] [PubMed] [Google Scholar]
- 109.Tozser J, Gustchina A, Weber IT, et al. Studies on the role of the S4 substrate binding site of HIV proteinases. FEBS Lett. 1991;279(2):356–360. doi: 10.1016/0014-5793(91)80186-7. [DOI] [PubMed] [Google Scholar]
- 110.Tozser J, Weber IT, Gustchina A, et al. Kinetic and modeling studies of S3-S3' subsites of HIV proteinases. Biochemistry. 1992;31(20):4793–4800. doi: 10.1021/bi00135a008. [DOI] [PubMed] [Google Scholar]
- 111.Weber IT, Miller M, Jaskolski M, et al. Molecular modeling of the HIV-1 protease and its substrate binging site. Science. 1989;243(4893):928–931. doi: 10.1126/science.2537531. [DOI] [PubMed] [Google Scholar]
- 112.Erickson J, Neidhart DJ, VanDrie J, et al. Design, activity, and 2.8 A crystal structure of a C2 symmetric inhibitor complexed to HIV-1 protease. Science. 1990;249(4968):527–533. doi: 10.1126/science.2200122. [DOI] [PubMed] [Google Scholar]
- 113.Miller M, Schneider J, Sathyanarayana BK, et al. Structure of complex of synthetic HIB-1 protease with a substrate-based inhibitor at 2.3 A resolution. Science. 1989;246(4934):1149–1152. doi: 10.1126/science.2686029. [DOI] [PubMed] [Google Scholar]
- 114.Swain AL, Miller MM, Green J, et al. X-ray crystallographic structure of a complex between a synthetic protease of human immunodeficiency virus 1 and a substrate-based hydroxyethylamine inhibito. Proc Natl Acad Sci U S A. 1990;87(22):8805–8809. doi: 10.1073/pnas.87.22.8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Polgar L, Szeltner Z, Boros I. Substrate dependent mechanisms in the catalysis of human immunodeficiency virus protease. Biochemistry. 1994;33(31):9351–9357. doi: 10.1021/bi00197a040. [DOI] [PubMed] [Google Scholar]
- 116.Gross I, Hohenberg H, Wilk T, et al. A conformational switch controlling HIV-1 morphogenesis. Embo J. 2000;19(1):103–113. doi: 10.1093/emboj/19.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vogt V. Proteolytic processing and particle maturation. Curr. Top. Microbiol. Immunol. 1996;214:95–131. doi: 10.1007/978-3-642-80145-7_4. [DOI] [PubMed] [Google Scholar]
- 118.Wiegers K, Rutter G, Kottler H, et al. Sequential steps in himan immunodeficiency virus particle maturation revealed by alterations of individual gag polyprotein cleavage sites. J Virol. 1998;72(4):2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pettit SC, Lindquist JN, Kaplan AH, Swanstrom R. Processing sites in the human immunodeficiency virus tupe 1 (HIV-1) Gag-Pro-Pol precursor are cleaved by the viral protease at different rates. Retrovirology. 2005;2(1):66. doi: 10.1186/1742-4690-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pettit SC, Clemente JC, Jeung JA, Dunn BM, Kaplan AH. Ordered processing of the human immunodeficiency virus type 1 GagPol precursor is influenced by the context of the embedded viral protease. J Virol. 2005;79(16):10601–607. doi: 10.1128/JVI.79.16.10601-10607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pettit SC, Everitt LE, Choudhury S, Dunn BM, Kaplan AH. Initial cleavage of the human immunodeficiency virus type 1 gag/pol precursor by its activated protease occurs by an intramolecular mechanism. J Virol. 2004;78(16):8477–8485. doi: 10.1128/JVI.78.16.8477-8485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lin YC, Beck Z, Lee T, Le VD, Morris GM, Olson AJ, Wong CH, Elder JH. (2000) Alteration of Substrate and Inhibitor Specificity of Feline Immunodeficiency Virus Protease. J Virol. 2000;74(10):4710–4720. doi: 10.1128/jvi.74.10.4710-4720.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lin YC, Beck Z, Morris GM, Olson AJ, Elder JH. Structural Basis for Distinctions between Substrate and Inhibitor Specificities for Feline Immunodeficiency Virus and Human Immunodeficiency Virus Proteases. J Virol. 2003;77(12):6589–6600. doi: 10.1128/JVI.77.12.6589-6600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lin YC, Brik A, de Parseval A. Altered Gag Polyprotein Cleavage Specificity of Feline Immunodeficience Virus/Human Immunodeficiency Virus Mutant Proteases as Demonstrated in a Cell-Based Expression System. J Virol. 2006;80(16):7832–7843. doi: 10.1128/JVI.00374-06. [DOI] [PMC free article] [PubMed] [Google Scholar]