Abstract
The goal of this work was to compare the differences between human immunodeficiency virus type 1 (HIV-1) of B and F1 subtypes in the acquisition of major and minor protease inhibitor (PI)-associated resistance mutations and of other polymorphisms at the protease (PR) gene, through a cross sectional study. PR sequences from subtypes B and F1 isolates matched according to PI exposure time from Brazilian patients were included in this study. Sequences were separated in four groups: 24 and 90 from children and 141 and 99 from adults infected with isolates of subtypes F1 and B, respectively. For comparison, 211 subtype B and 79 subtype F1 PR sequences from drug-naïve individuals were included. Demographic and clinical data were similar among B- and F1-infected patients. In untreated patients, mutations L10V, K20R, and M36I were more frequent in subtype F1, while L63P, A71T, and V77I were more prevalent in subtype B. In treated patients, K20M, D30N, G73S, I84V, and L90M, were more prevalent in subtype B, and K20T and N88S were more prevalent in subtype F1. A higher proportion of subtype F1 than of subtype B strains containing other polymorphisms was observed. V82L mutation was present with increased frequency in isolates from children compared to isolates from adults infected with both subtypes. We could observe a faster resistance emergence in children than in adults, during treatment with protease inhibitors. This data provided evidence that, although rates of overall drug resistance do not differ between subtypes B and F1, the former accumulates resistance at higher proportion in specific amino acid positions of protease when compared to the latter.
Keywords: Brazil, HIV-1, Subtype B, Subtype F1, Drug resistance mutations, Protease, Pediatric infection
1. Introduction
The genetic diversity of human immunodeficiency virus type 1 (HIV-1) allows its classification in several groups, subtypes, subsubtypes and circulating recombinant forms (CRF) (Robertson et al., 2000). To date, 9 known subtypes and at least 40 CRF are heterogeneously distributed around the world. While subtype B predominates in developed countries of Western Europe and in the U.S., other (non-B) subtypes or CRF account for the majority of infectious in the developing world (Hemelaar et al., 2006). The HIV-1 epidemic in Brazil is complex, even when compared to other South American countries, and is caused primarily by subtype B (approximately 70% of HIV infections). Appreciable numbers of HIV-1 infectious caused by clades F1 and B/F1 recombinants (approximately 10% each), and clades C and B/C recombinants (6% each) have been reported (Vicente et al., 2000; Brindeiro et al., 2003; E.A. Soares et al., 2003; M.A. Soares et al., 2003; Soares et al., 2005; Thomson et al., 2004; Rodrigues et al., 2005a,b; Sanabani et al., 2006; Santos et al., 2006; Lacerda et al., 2007). These different subtypes have been circulating among the same risk groups, but their prevalence may differ significantly across geographical regions, what account, in part, for the differences in estimates found in different studies. In the south region of Brazil, for example, the prevalence of subtype B equals that of subtype C (E.A. Soares et al., 2003; M.A. Soares et al., 2003; Soares et al., 2005). Recent studies suggest a discontinuous introduction of these different subtypes in the country. The onset of the HIV-1 subtype B Brazilian epidemic was estimated between mid 1960s and early 1970s, while the subtype F1 and C epidemics appears to date back to early 1980s and early 1990s, respectively (Salemi et al., 2005; Bello et al., 2006, 2007; Santos et al., 2007). Subtype F, the second most prevalent subtype in most part of Brazil, has also been reported in other countries like Argentina, Chile, Cameroon, France, Romania and Belgium (Apetrei et al., 1998; Masciotra et al., 2000; Hierholzer et al., 2002; Hemelaar et al., 2006; Rios et al., 2007).
An increasing body of experimental evidence suggested that different HIV-1 subtypes might exhibit disparate biological behaviors, and might respond differently to diagnostic, immunologic and therapeutic interventions (Jeeninga et al., 2000; Gonzalez et al., 2003; Renjifo et al., 2004). With respect to HIV antiretroviral (ARV) treatment, recent studies identified subtype-specific differences in viral susceptibility to specific drugs (Brenner et al., 2003; Carmona et al., 2005) and in signature mutations selected by treatment (Dumans et al., 2004; Gonzalez et al., 2004; Abecassis et al., 2005). An important question in this scenario is whether HIV-1 subtypes differ in the rate of fixation of mutations conferring drug resistance in individuals under ARV therapy, a point recently addressed in few reports (Carobene et al., 2004; Grossman et al., 2004; Montes et al., 2004; Soares et al., 2007). As Brazil exhibits a heterogeneous HIV-1 subtype distribution and a history of universal and free access to ARV therapy since 1996 (Teixeira et al., 2004), it represents an appropriate setting for retrospectively analyzing the rate of fixation of mutations conferring drug resistance under specific ARV class exposure, in different subtypes.
The main goal of this work was to compare the differences between B and F1 subtypes in patterns and time of acquisition of PI-related resistance mutations through a cross sectional study.
2. Materials and methods
2.1. Patient samples
This study was conducted on subtypes B and F1 HIV-1-infected and drug-experienced Brazilian patients with virological failure (detectable viral load after at least 3 months of PI treatment) and for whom treatment history and sequences of the protease genes were available. Initially, 24 and 90 children and 141 and 99 adults infected with isolates of subtypes F1 and B, respectively, were enrolled. Sequences from those individuals were isolated between 1998 and 2005. For comparison, 211 and 79 PR sequences isolated between 1989 and 2005 from drug-naïve Brazilian individuals infected with isolates of subtypes B and F1, respectively, were included. All HIV-1 subtype classification in this study refers solely to the PR region. Some nucleotide sequences used in the analyses were previously determined experimentally and had their subtype assigned by our group and others (Vicente et al., 2000; Caride et al., 2001; Brindeiro et al., 2002, 2003; E.A. Soares et al., 2003; M.A. Soares et al., 2003; Soares et al., 2005, 2007; Dumans et al., 2004; Machado et al., 2004; Pires et al., 2004; Thomson et al., 2004; Rodrigues et al., 2005a,b), whereas others were newly determined. New sequences were additionally determined and had their subtype assignment and absence of contamination assured by phylogenetic and bootscanning analyses as described previously (Soares et al., 2007). In both cases sequences were obtained after viral RNA extraction from plasma, RT-PCR and population sequencing (Soares et al., 2007). In case a mixture of nucleotides at a given position was observed (more than one peak in the sequence chromatogram), the predominant sequence in the virus quasispecies was kept. Irrespective of the subtype assignment in RT, PR all sequences were confirmed by phylogenetic and bootscanning analyses, and only pure (non-recombinant) subtype B and F1 PR sequences were kept for further analyses. Current exposure to ARV therapy at the time of sampling as well as previous treatment history were assessed through revision of the data at the Stanford University HIV Drug Resistance Database (http://hivdb.stanford.edu/; Rhee et al., 2003), from the respective previous studies mentioned above or directly from the medical records. For the report of RTV usage, that always referred to its use as a separate PI drug, either used as a single PI or in combination with other PI, but not as a boost to other PI. The only exception was in the case of LPV usage, as RTV was always present as a boost. Demographic data, CD4+ T-cell counts and plasma HIV viral load were also taken into account whenever possible. This study was approved by all Internal Review Boards from the centers involved.
2.2. Sequence analyses
To compare populations of patients infected with B and F1 HIV-1 variants, PR sequences obtained from both groups were matched by average exposure time to PI by excluding sequences with extreme values. After matching, demographic and clinical data from patients such as age, gender, time of HIV diagnosis, CD4 T-cell counts, HIV viral load, CDC immune staging, exposure times to individual PI as well as their usage frequency, the proportion of patients subjected to mono and/or dual therapy prior to highly active antiretroviral therapy (HAART), and the proportion of patients subjected to one or more HAART regimens were compiled to assure they did not vary between both groups. Finally, sequences were further grouped according to time in 12-month periods of PI exposure. Cumulative curves of proportions of mutant viruses over PI exposure time were plotted for each subtype. At each time point, the average number of mutations per mutant genome was also compared for both subtypes. Exposure times to individual PI were also compiled for each subtype group for evaluating differences with respect to specific drugs.
We have also compared the differences in patterns and time of acquisition of PI resistance mutations between subtype B strains isolated from children and adults matched according to individual PI exposure times or to the PI ARV class, taking into account the proportion of patients subjected to mono and/or dual therapy prior to HAART and the proportion of patients subjected to one or more HAART regimens in both groups.
We considered mutations previously reported to be associated with PI resistance according to the International AIDS Society-USA consensus (Johnson et al., 2007) for each PI used. Mutations D30N (NFV), V32I (LPV), M46I/L (IDV/RTV), I47V/A (LPV/RTV), G48V (SQV), I50L/V (APV), V82A/F/T/S (LPV/IDV/RTV), I84V (APV/IDV/RTV) and L90M (SQV/NFV) were considered as major resistance mutations and were analyzed separately for each PI as well as quantitatively all together. Mutations L10F/I/R/V, K20M/R, L24I, L33F, M36I, F53L, I54V/L/A/M/T/S, L63P, A71V/T, G73C/S/T/A, V77I and N88D/S were considered as minor resistance mutations and were also analyzed separately and together as a group. In the quantitative analysis, L63P and M36I were not counted as minor mutations for subtypes B and F1, respectively, as they represent frequent polymorphisms found in those respective subtypes. All other amino acid differences between Brazilian subtypes B or F1 and the world B consensus (Los Alamos), including those in minor or major positions, were considered as other polymorphisms and were analyzed as a group. The exceptions were the signature positions 15, 35, 36, 41, 57, 61, 63 and 89 that were excluded from this quantitative analysis. Other polymorphisms at minor or major resistance positions were also analyzed individually. Identification of the cited mutations in PR genes was carried out following electronic submission to the Stanford database (Rhee et al., 2003).
2.3. Statistical analyses
Statistical analyses were performed with one-tailed Fisher's exact test for categorical variables and with one- and two-tailed Student t-test and two-tailed Mann–Whitney U-test for continuous variables as appropriate; p < 0.05 was considered significant. All analyses were conducted using the package XLSTAT, version 2008.4.01, with Microsoft Excel for Windows.
3. Results
Major demographic and clinical parameters for the four infected groups are summarized in Table 1. After matching time of exposure between groups and excluding 22 subtype B and 4 subtype F1 strains from adults with undetectable viral load, 69 and 90 strains isolated from subtype B-infected adults and children, and 76 and 126 strains isolated from subtype B- and F1-infected adults, were compared in Fig. 1A and B, respectively. In these subgroups similar proportions of adult patients were subjected to mono and/or dual therapy prior to HAART (46% of subtype B and 45% of subtype F1). However, 82% of subtype B-infected children were subjected to mono and/or dual therapy prior to HAART. A similar proportion of adult patients were subjected to multiple HAART regimens (58% of subtype B and 55% of subtype F1), but only 27% of subtype B-infected children were subjected to multiple HAART regimens. Viral load was higher in subtype B-infected children than in adults (median VL log of 4.5 in children versus 4.3 in adults, p = 0.01). Viral load was also higher in subtype F1-infected adults than in subtype B (median VL log of 4.7 in subtype F1 versus 4.2 in subtype B, p = 0.0002). Other demographic and clinical parameters depicted in Table 1 were checked again for these sub-groups of treated adults and no statistical differences were found.
Table 1.
Demographic and clinical characteristics of HIV-1-positive treated patient groups infected with B and F1 subtypes.
| Children | Adults | |||||
|---|---|---|---|---|---|---|
| Subtype B (n = 90) | Subtype F1 (n = 24) | p-value | Subtype B (n = 99) | Subtype F1 (n = 141) | p-value | |
| Age (yr/SD) | 6.7 (3.5) | 7.4 (3.8) | 0.44 | 40.6 (10.8) | 40.2 (10.0) | 0.83 |
| Gender (%)a | 0.40 | 0.26 | ||||
| Male | 33 (46.5) | 8 (36.4) | 59 (68.6) | 57 (61.0) | ||
| Average time of diagnosis (yr/SD) | 6.7 (3.5) | 7.4 (3.8) | 0.43 | 5.6 (3.8) | 6.3 (3.1) | 0.15 |
| CDC immune stage (%) | 0.91 | 0.62 | ||||
| 1 | 44 (54.0) | 12 (50.0) | 9 (10.7) | 9 (9.5) | ||
| 2 | 24 (29.0) | 7 (29.0) | 37 (44.1) | 36 (37.9) | ||
| 3 | 14 (17.0) | 5 (21.0) | 38 (45.2) | 50 (52.6) | ||
| Average CD4 T-cell counts at sample collection (SD) | 803 (1185) | 734 (721) | 0.79 | 263 (204) | 237 (174) | 0.37 |
| Median log 10 viral RNA (IQR) at sample collection | 4.5 (4.1–5.2) | 4.3 (3.9–4.8) | 0.14 | 3.1 (2.3–4.6) | 4.7 (4.1–5.2) | 0.0001b |
| Undetectable VL (%) (<80 copies/ml plasma) | 0 (0) | 0 (0) | 1 | 22 (26.5) | 4 (4.2) | 0.002 |
| Average PI treatment time (mo/SD) | 23.3 (12.3) | 22.6 (16.6) | 0.84 | 28.2 (20.7) | 36.2 (21.8) | 0.005 |
The total number of cases analyzed for each characteristic may vary, due to the lack of information in some medical charts.
Bold p-values are significant at the 0.05 level.
Fig. 1.
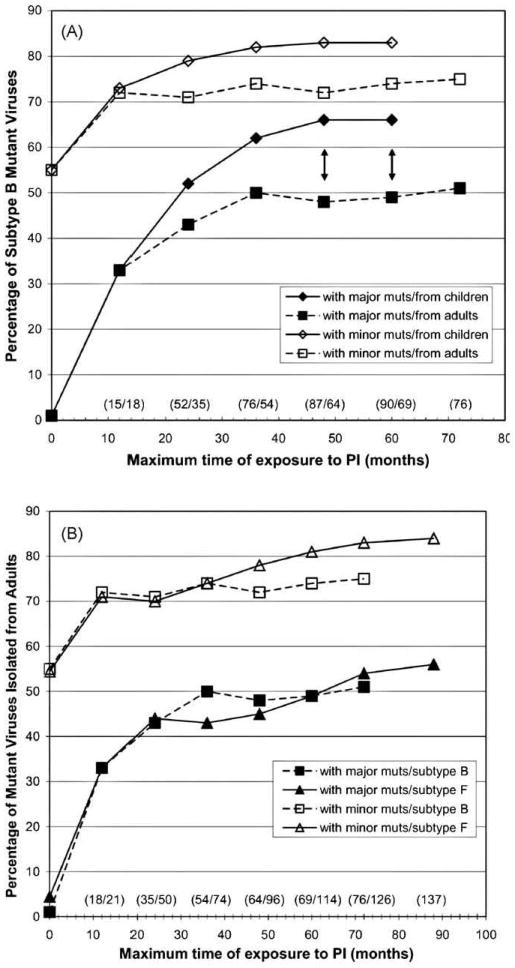
HIV-1 subtype B and F1 viruses with at least one major or minor resistance mutation. Different PI treatment exposure periods (in months) were plotted for each subtype and for each mutation class. (A) Comparison between subtype B isolated from infected adults and children; (B) comparison between subtypes B and F1 isolated from infected adults. Black double arrows denote significance in the difference of proportions between subtypes B isolated from children and adults at the 0.05 p-value level. The exposure time point 0 means the proportion of mutant strains isolated from untreated individuals. Numbers in parentheses above exposure time periods denote the number of sequences from children and adults (in panel A) and from subtypes B and F1 (in panel B), respectively.
At 4–5-year exposure, 66% of subtype B viruses from children harbored major mutations, whereas only 49% of those isolated from adults carried them (p = 0.02) (Fig. 1A). Interestingly, subtype B isolated from adults showed higher numbers of average major mutations per genome from 2-year exposure, and this difference steadily increased up to 5-year exposure (1.3–1.6 major mutations per mutant genome in isolates from children versus 1.7–2.0 in isolates from adults; p < 0.05). Analysis of the number of average minor mutations per mutant viral genome in both groups failed to show significant differences in the same period.
Subtype B and F1 viruses from adults did not differ significantly with respect to the percentage of strains carrying major or minor mutations (Fig. 1B), but subtype B showed higher numbers of average major mutations per genome from 4- to 6-year exposure (1.6–1.7 major mutations per genome in subtype F1 versus 2.0 in subtype B; p = 0.05). However, the number of average minor mutations per mutant viral genome for the same PI exposure time did not differ significantly in both subtype groups.
The HIV-1 PR sequences from the treatment groups above were also compared with 211 and 79 sequences from drug-naïve Brazilian individuals infected with subtypes B and F1, respectively (time points “0” in both Fig. 1A and B). The percentage of strains carrying major mutations isolated from subtype B-and F1-infected untreated patients were 1% and 4%, respectively, and not significantly different (p = 0.20). At 12 months of PI exposure, some 33% of strains in all compared treatment groups already carried such mutations. The proportion of strains carrying minor mutations rose from 55% in subtype B- and F1-infected naïve patients to 71–73% in the three infected groups, adults infected by subtypes B and F1 and children infected by subtype B, by 12 months of PI exposure (Fig. 1A and B).
The number of subtype F1-infected children was very low and this group was not stratified in PI time exposure periods. All 24 subtype F1-infected children were compared with the 90 subtype B-infected infants, as there were no differences in the parameters depicted in Table 1 for these two infected groups. The percentages of subtype B and F1 viruses carrying major or minor mutations did not differ between both groups.
Additionally, we have compared the proportion of strains carrying other polymorphisms as well their average number per mutant viral genome in all patient groups. Protease sequences from the longest available comparable periods of Fig. 1A and B and those from drug-naïve patients were used in this analysis. Differently from what was found for major or minor mutations, the numbers of other polymorphisms did not always differ between viruses isolated from treated and naïve patients. This variability is already high in this last group of patients. Polymorphisms occurred significantly at higher levels in subtype F1 (98%) than in subtype B (92%) isolated from treated adults (p = 0.03) and than in subtype F1 isolated from naïve patients (87%; p = 0.003). The proportion of subtype B strains carrying other polymorphisms isolated from untreated (90%) patients did not differ significantly from those isolated from treated adults (92%), but differed from those isolated from treated children (97%; p = 0.05). Taking into account the global patient dataset, and after matching time of PI exposure between subtype groups, the number of average polymorphisms per genome were 3.7 for subtype F1 and 3.2 for subtype B strains (p = 0.02) versus 2.6 for both subtype groups isolated from naïve patients (p < 0.05).
In order to check whether specific PI-associated mutations could account for the differences observed between adults and children in Fig. 1A and if there were differences in qualitative patterns of drug resistance acquisition between subtype groups (Fig. 1B), the percentage of strains carrying specific major and minor mutations were calculated for the datasets compared. The results for the longest available periods of Fig. 1A and B (60 and 72 months, respectively) are shown in Tables 2 and 3, respectively. Significant differences in other polymorphisms at major and minor resistance positions are also shown.
Table 2.
Frequency of protease resistance mutations in sequences obtained from treated subtype B-infected children and adults compared with subtype B untreated patients.
| Mutation | Untreated patients (n = 211) | Treateda children (n = 90) | Treated adults (n = 69) | p-valueb |
|---|---|---|---|---|
| Major (%) | ||||
| D30N | 0 (0) | 14 (15.5) | 6 (8.7) | 0.15 |
| M46I | 0 (0) | 16 (17.8) | 9 (13.0) | 0.28 |
| M46L | 1 (0.5) | 7 (7.8) | 6 (8.7) | 0.53 |
| M46I/L | 1 (0.5) | 23 (25.6) | 15 (21.7) | 0.34 |
| G48V | 0 (0) | 0 (0) | 3 (4.4) | 0.08 |
| V82A | 1 (0.5) | 23 (25.5) | 13 (18.8) | 0.21 |
| V82F | 1 (0.5) | 0 (0) | 0 (0) | 1.0 |
| V82T | 0 (0) | 1 (1.1) | 2 (2.9) | 0.4 |
| V82S | 0 (0) | 2 (2.2) | 0 (0) | 0.32 |
| V82L | 0 (0) | 6 (6.7) | 0 (0) | 0.03 |
| V82A/F/T/S | 2 (0.9) | 26 (28.9) | 15 (21.7) | 0.2 |
| I84V | 0 (0) | 5 (5.6) | 9 (13.0) | 0.09 |
| L90M | 1 (0.5) | 24 (26.7) | 20 (29.0) | 0.44 |
| At least onec | 3 (1.4) | 59 (66.0) | 34 (49.0) | 0.03 |
| Minor (%) | ||||
| L10F | 3 (1.4) | 4 (4.4) | 2 (2.9) | 0.47 |
| L10I | 24 (11.4) | 24 (26.7) | 15 (21.7) | 0.30 |
| L10V | 6 (2.8) | 6 (6.7) | 9 (13.0) | 0.14 |
| L10F/I/R/V | 33 (15.6) | 34 (37.8) | 26 (37.7) | 0.56 |
| K20M | 2 (0.9) | 0 (0) | 3 (4.4) | 0.08 |
| K20R | 5 (2.4) | 14 (15.6) | 7 (10.2) | 0.22 |
| K20I | 0 (0) | 6 (6.7) | 1 (1.5) | 0.11 |
| K20T | 1 (0.5) | 2 (2.2) | 2 (2.9) | 0.58 |
| K20M/R | 7 (3.3) | 14 (15.6) | 10 (14.5) | 0.52 |
| L24I | 0 (0) | 5 (5.6) | 4 (5.8) | 0.61 |
| L33F | 0 (0) | 4 (4.4) | 3 (4.4) | 0.65 |
| M36I | 24 (11.4) | 29 (32.2) | 28 (40.6) | 0.18 |
| F53L | 0 (0) | 2 (2.2) | 3 (4.4) | 0.38 |
| I54V | 1 (0.5) | 26 (28.9) | 11 (15.9) | 0.04 |
| I54V/L/A/M/T/S | 2 (1.0) | 27 (30.0) | 12 (17.4) | 0.05 |
| L63P | 122 (57.8) | 66 (73.3) | 45 (65.2) | 0.18 |
| A71V | 4 (1.9) | 21 (23.3) | 11 (15.9) | 0.17 |
| A71T | 16 (7.6) | 7 (7.8) | 4 (5.8) | 0.45 |
| A71V/T | 20 (9.5) | 28 (31.1) | 15 (21.7) | 0.13 |
| G73S | 1 (0.5) | 0 (0) | 5 (7.2) | 0.01 |
| V77I | 54 (25.6) | 33 (36.7) | 12 (17.4) | 0.006 |
| N88D | 0 (0) | 7 (7.8) | 3 (4.4) | 0.29 |
| N88S | 0 (0) | 2 (2.2) | 1 (1.5) | 0.6 |
| N88D/S | 0 (0) | 9 (10.0) | 4 (5.8) | 0.25 |
| At least onec | 116 (55.0) | 75 (83.0) | 51 (74.0) | 0.10 |
Both treated datasets had detectable viral load and were matched according to PI exposure time and other criteria as described in the text.
p-values are related to the comparison between treated patients. Bold p-values are significant at the 0.05 level.
Major and minor mutations refer to the definition used in Section 2.
Table 3.
Frequency of protease resistance mutations in sequences obtained from treated F1- and B-infected adult patients compared to sequences from untreated patients.
| Mutation | Untreated | p-valueb | Treateda | p-valueb | ||
|---|---|---|---|---|---|---|
| Subtype B (n = 211) | Subtype F (n = 79) | Subtype B (n = 76) | Subtype F (n= 126) | |||
| Major (%) | ||||||
| D30N | 0 (0) | 0 (0) | 1.0 | 6 (7.9) | 5 (4.0) | 0.19 |
| M46I | 0 (0) | 2 (2.5) | 0.07 | 10 (13.2) | 8 (6.4) | 0.05 |
| M46L | 1 (0.5) | 0 (0) | 0.73 | 6 (7.9) | 18 (14.3) | |
| M46I/L | 1 (0.5) | 2 (2.5) | 0.18 | 16 (21.1) | 26 (20.6) | 0.54 |
| G48V | 0 (0) | 0 (0) | 1.0 | 3 (4.0) | 6 (4.8) | 0.54 |
| I50V | 0 (0) | 0 (0) | 1.0 | 0 (0) | 4 (3.2) | 0.15 |
| V82A | 1 (0.5) | 2 (2.5) | 0.18 | 15 (19.7) | 35 (28.8) | 0.13 |
| V82F | 1 (0.5) | 0 (0) | 0.73 | 0 (0) | 0 (0) | 1.0 |
| V82T | 0 (0) | 0 (0) | 1.0 | 2 (2.6) | 3 (2.4) | 0.60 |
| V82S | 0 (0) | 0 (0) | 1.0 | 0 (0) | 1 (0.8) | 0.62 |
| V82L | 0 (0) | 0 (0) | 1.0 | 0 (0) | 1 (0.8) | 0.62 |
| V82A/F/T/S | 2 (0.9) | 2 (2.5) | 0.30 | 17 (22.4) | 39 (31.0) | 0.12 |
| I84V | 0 (0) | 0 (0) | 1.0 | 10 (13.2) | 4 (3.2) | 0.01 |
| L90M | 1 (0.5) | 0 (0) | 0.73 | 25 (32.9) | 30 (23.8) | 0.11 |
| At least onec | 3 (1.4) | 3 (3.8) | 0.20 | 39 (51) | 68 (54.0) | 0.41 |
| Minor (%) | ||||||
| L10F | 3 (1.4) | 0 (0) | 0.38 | 3 (4.0) | 1 (0.8) | 0.15 |
| L10I | 24 (11.4) | 5 (6.4) | 0.14 | 17 (22.4) | 38 (30.2) | 0.15 |
| L10V | 6 (2.8) | 15 (19.2) | <0.0001 | 10 (13.2) | 32 (25.4) | 0.03 |
| L10F/I/R/V | 33 (15.6) | 20 (25.3) | 0.04 | 30 (39.5) | 71 (56.4) | 0.01 |
| K20M | 2 (0.9) | 0 (0) | 0.53 | 6 (7.9) | 0 (0) | 0.002 |
| K20R | 5 (2.4) | 19 (24.1) | <0.0001 | 8 (10.5) | 61 (48.4) | <0.0001 |
| K20I | 0 (0) | 0 (0) | 1.0 | 1 (1.3) | 3 (2.4) | 0.52 |
| K20T | 1 (0.5) | 0 (0) | 0.73 | 3 (4.0) | 17 (13.5) | 0.02 |
| K20M/R | 7 (3.3) | 19 (24.1) | <0.0001 | 14 (18.4) | 61 (48.4) | <0.0001 |
| L24I | 0 (0) | 0 (0) | 1.0 | 4 (5.3) | 10 (7.9) | 0.34 |
| L33F | 0 (0) | 0 (0) | 1.0 | 3 (4.0) | 4 (3.2) | 0.53 |
| M36I | 24 (11.4) | 74 (93.7) | <0.0001 | 32 (42.1) | 123 (97.6) | <0.0001 |
| F53L | 0 (0) | 0 (0) | 1.0 | 4 (5.3) | 11 (8.7) | 0.27 |
| I54V | 1 (0.5) | 1 (1.3) | 0.47 | 14 (18.4) | 33 (26.2 | 0.14 |
| I54V/L/A/M/T/S | 2 (1.0) | 1 (1.3) | 0.62 | 15 (19.7) | 39 (30.9) | 0.06 |
| L63P | 122 (57.8) | 9 (11.4) | <0.0001 | 50 (65.8) | 38 (30.2) | <0.0001 |
| A71V | 4 (1.9) | 0 (0) | 0.28 | 14 (18.4) | 19 (15.1) | 0.33 |
| A71T | 16 (7.6) | 0 (0) | 0.005 | 6 (7.9) | 4 (3.2) | 0.12 |
| A71V/T | 20 (9.5) | 0 (0) | 0.001 | 20 (26.3) | 23 (18.3) | 0.12 |
| G73S | 1 (0.5) | 0 (0) | 0.73 | 7 (9.2) | 2 (1.6) | 0.02 |
| V77I | 54 (25.6) | 9 (11.4) | 0.005 | 14 (18.4) | 14 (11.1) | 0.11 |
| N88D | 0 (0) | 0 (0) | 1.0 | 3 (4.0) | 4 (3.2) | 0.53 |
| N88S | 0 (0) | 0 (0) | 1.0 | 1 (1.3) | 11 (8.7) | 0.02 |
| N88D/S | 0 (0) | 0 (0) | 1.0 | 4 (5.3) | 15 (11.9) | 0.09 |
| At least onec | 116 (55.0) | 41 (52.0) | 0.37 | 57 (75.0) | 105 (83.0) | 0.10 |
Both treated datasets had detectable viral load and were matched according to PI exposure time and other criteria as described in the text.
Bold p-values are significant at the 0.05 level.
Major and minor mutations refer to the definition used in Section 2.
Differences in the usage frequency of IDV, SQV and LPV between subtype B-infected children (5.5%, 6.7% and 0%, respectively) and adults (43.5%, 39.1% and 9.2%, respectively) were seen (p-values of n 0.001, n 0.001 and 0.006, respectively). Exposure times to all other PI, as well as their usage frequency, were not different for these two treatment groups. Only 1.5% and 3.3% of subtype B-infected adults and children were treated with APV, respectively. This means that mutations at positions 82 and 84 occurred predominantly in response to IDV and/or RTV in subtype Binfected adults and to RTV in children.
Among adults, we observed a longer RTV average exposure time in subtype B-infected patients (19.6 months versus 11.2 months in subtype F1; p < 0.01). Conversely, the average exposure time to IDV was longer in subtype F1-infected adults (22.8 months versus 16.8 months in subtype B; p < 0.01). The usage frequencies of all other PI analyzed, as well as their exposure times did not differ for these two subtype groups. For those viruses, the mutations at positions 82 and 84 occurred primarily in response to IDV and/or RTV as we have only 1.3% and 6.4% of subtype B- and F1-infected adults treated with APV, respectively, and 9.2% and 12.8% of subtype B- and F1-infected adults treated with LPV.
In the global dataset, D30N was only seen in isolates from patients treated with NFV and I84V in patients treated with IDV, RTV and/or APV. Ninety-seven percent of isolates with V82A/F/T/S mutations were from patients treated with IDV, RTV and/or LPV, and 89% of those with L90M were from patients treated with NFV and/or SQV. Thus, for those mutations, it is reasonable to compare different treatment groups after matching time of exposure under those specific PI or PI groups.
The major protease mutations V32I and I47V/A and the minor mutations L10R, I54M, and G73C/T/A were not observed in subtype B and F1 viruses from treated or untreated patients in this study (Tables 2 and 3). Only V82L was present with increased frequency in subtype B (6.7%) and F1 (8.3%, not shown) isolates from children compared to subtype B (0%) and F1 (0.8%) isolates from adults (Table 2). Thus, albeit the proportion of strains carrying at least one major PI-related mutation is higher in subtype B isolated from children than from adults, no individual protease resistance sites could account for this result. This is in agreement with lower numbers of average major mutations per mutant viral genome in subtype B viruses isolated from children compared to those from adults.
As can be observed in Table 3, I84V was present with increased frequency in subtype B compared with F1. When checking the prevalence of this mutation at each time point, we observed its occurrence in subtype B already by 12 months of PI exposure, while for subtype F1 it appeared only by 36 months. Of note, M46I was more frequent than M46L in subtype B isolates, while M46L was more frequent than M46I in subtype F1 isolates (Table 3). The frequencies of specific major mutations in all patients under specific PI or PI groups were also examined. In agreement with previous analyses, the only significant difference (p = 0.008) was for the prevalence of I84V in subtype B (18.9%, 10/53 strains) and F1 (4.6%, 4/88) strains from patients matched for the same treatment time with IDV, RTV and/or APV.
In order to increase sample size, and considering that there were no differences in the frequencies of specific major mutations between isolates from adults and children infected with the same subtype (Table 2), we pooled both groups of each subtype to further examine the frequency of major mutations in both subtype groups matched for treatment time with specific PI or PI groups. Twenty-one percent (21/102) of subtype B-infected patients under NFV for an average time of 18.7 months presented D30N, while only 9.6% (10/104) of the subtype F1 isolates under 19.6 months of NFV exposure, showed that mutation (p = 0.02). When checking the prevalence of this mutation over treatment time with NFV as the unique PI, we observed higher levels in subtype B (16%) than in subtype F1 (6%) already by 36 months of NFV exposure (p < 0.05). To avoid confounding effects of the selection of L90M by NFV, SQV and other PI, it was possible to restrict this analysis to patients exclusively subjected to NFV as the unique PI over time. By 36 months, 16% of subtype B strains already carried L90M versus 5% of subtype F1 (p < 0.05). In agreement with previous analyses, 13.3% (15/113) of subtype B-infected patients under IDV, RTV and/or APV for an average time of 21.6 months presented I84V, while only 2.6% (3/117) of the subtype F1 isolates under 26.4 months of exposure to these PI, showed that mutation (p = 0.002).
Prevalence of minor resistance mutations in subtype B viruses from children and adults matched for PI exposure time is presented in Table 2. Mutation G73S was more prevalent in subtype Binfected adults. Conversely, mutations I54V and V77I were significantly more prevalent in subtype B-infected children.
Prevalence of minor resistance mutations in subtype B and F1 viruses from untreated and treated patients are presented in Table 3. In untreated patients, mutations L10V, K20R, and M36I were significantly more frequent in subtype F1 isolates, while mutations L63P, A71T, and V77I were more prevalent in subtype B isolates. Although mutations K20T, L63P, and N88S were present with increased frequency in subtype F1 from treated patients compared to untreated individuals (p = 0.0002, p = 0.001, and p = 0.004, respectively), the same differences were not observed when comparing subtype B from treated and untreated groups. Conversely, mutations L10V, K20M, L33F, M36I, G73S, and N88D were significantly more prevalent in subtype B viruses from treated patients compared to untreated individuals (p = 0.002, p = 0.005, p = 0.02, p < 0.0001, p = 0.0005, and p = 0.02, respectively), but this was not true for subtype F1.
4. Discussion
Some studies compared resistance mutations developed under treatment in patients infected with subtype B versus non-B HIV-1 variants (Caride et al., 2001; Carobene et al., 2004; Dumans et al., 2004; Grossman et al., 2004; Montes et al., 2004; Abecassis et al., 2005; Kantor et al., 2005; Rodrigues et al., 2005a,b; Deforche et al., 2007; Lacerda et al., 2007; Soares et al., 2007; Poveda et al., 2008). However, in part because of low numbers of available sequences from persons with well characterized treatment histories infected with specific non-B subtypes, these studies generally came from different human populations or the compared viruses were classified only by the classes of drugs to which they were exposed rather than by individual drugs or drug regimens. In our study, Brazilian patients infected with B and F1 HIV-1 variants were matched for similar treatment, being the study with the largest number of samples from subtype F1-infected patients under treatment ever analyzed in our country in search for drug resistance data.
In untreated patients, mutations L63P, A71T, and V77I were more prevalent in isolates of subtype B, while L10V, K20R, and M36I were significantly more frequent in subtype F1 isolates. Moreover, the proportion of L10I to L10V is inverted when comparing both groups of untreated patients. The positions more likely to be mutated in non-B than in B subtypes from untreated people can be considered subtype-specific polymorphisms (Kantor et al., 2005), and the ones found in subtype F1 at sites known to be associated with drug-resistance occurred at positions 10, 20, and 36 (Kantor et al., 2005). This is in agreement with our study, although Kantor et al. did not distinguish between different substitutions at the same position.
The mutations L10F/R, K20I, V32I, I47V/A, I50L/V, I54L/A/M/T/S, A71T, G73C/T/A, V77I, and V82F/T/S, either absent or observed in our subtype B and F1 viruses, were not statistically different in viruses isolated from treated or untreated patients. This suggests that these mutations were not positively selected by the drug regimens that were used by our cohort.
Kantor et al. (2005) pointed out some treatment-related positions for specific subtypes. As expected, these positions included 17 of 22 known PI resistance positions in subtype B and 14 of them were also observed in subtype F. Our results were similar. However, as we checked for different substitutions at the same position, we observed 19 mutations at 16 positions and 13 mutations at 12 positions more prevalent in treated than in untreated people infected with HIV-1 of subtypes B and F1, respectively. Moreover, despite the treatment history of B- and F1-infected adult patients was comparable, we could find subtype F1-specific treatment-related mutations, as K20T, L63P and N88S were more frequent only in treated than in untreated people infected with subtype F1, while L10V, K20M, D30N, L33F, M36I, M46I, G73S, N88D and I84V were more prevalent in treated than in untreated patients infected only with subtype B. In the case of L63P, we failed to detect differences between naïve and treated subtype B sequences because this polymorphism is high in Brazilian viruses of this subtype (E.A. Soares et al., 2003; M.A. Soares et al., 2003). Finally, the comparison of both subtype groups matched for treatment time exposure to the PI class or, when appropriated, to specific PI showed that mutations K20M, D30N (under NFV-containing regimens or under NFV as the unique PI), G73S, I84V, and L90M (under NFV as the unique PI) were more prevalent in subtype B. Conversely, K20T and N88S were specifically selected in subtype F1 as a consequence of PI treatment. Interestingly, Kantor et al. (2005) did not observe a higher prevalence of D30N, but rather a higher prevalence of A71V and V77I in subtype B compared with subtype F viruses isolated from treated patients. We may speculate the use of sequences isolated from people from different countries and the lack of detection of differences that depend on specific drugs in that work as reasons for those differences.
Although the PR mutations D30N and L90M both develop in non-B viruses during NFV therapy, they occur more commonly in subtype B (Carobene et al., 2004; Soares et al., 2007), what was confirmed in our comparison with subtype F1. We speculate that other polymorphisms in the protease, such as those at codon 89 (see below) may alternatively contribute to resistance to NFV in certain non-B subtypes, as previously suggested (Abecassis et al., 2005).
Mutations N88S and K20T are respectively reported by IAS consensus as major and minor mutations specifically associated with resistance to atazanavir, not used by our cohort, while N88D/S are considered minor mutations associated with resistance to NFV (Johnson et al., 2007). However, in agreement with our results, recent data identified selection of N88S and K20T by NFV treatment, which suggests that N88S might be more important than D30N and L90M in subtypes A1 and F1 and should be considered a major mutation, pointing out different roles for N88D and N88S in NFV resistance (Svicher et al., 2005; Deforche et al., 2007).
We cannot completely rule out the possibility that the difference in the average duration of exposure to RTV (longer in subtype B-infected adults) and IDV (longer in subtype F1-infected adults) observed in our cohort had an impact on the differences found, particularly at positions 84 and 46. However, it is unlikely that these differences could be attributed only to a differential exposure to these drugs. Indeed, we observed a higher prevalence of I84V in B versus F1 viruses also in a subset of isolates from patients matched for the same treatment time with IDV and/or RTV (_APV). According to the IAS consensus, RTV selects for mutations similar to those selected by IDV (Johnson et al., 2007). The higher prevalence of I84V in subtype B has been reported (Carobene et al., 2004; Soares et al., 2007).
We observed different proportions of M46I to M46L mutations between treated subtype groups (B and F1), and M46I behaved as a treatment-related mutation only for subtype B in our cohort. Hoffman et al. (2003) suggested that M46I and M46L lie along divergent evolutionary pathways, what has been supported by others (Svicher et al., 2005).
The data presented here showed that for positions 10, 20, 46, and 88 subtypes B and F1 showed different mutations at baseline and in response to the same treatment regimens. Taken together, our data also provided evidence that subtype B might accumulate resistance at higher proportion than subtype F1 at specific amino acid positions in PR. We cannot completely rule out the possibility that we have neglected potentially new, subtype F1-specific drug resistance mutations in positions not known to be associated with resistance in our analyses. As an example, mutation 89I, not considered by the IAS consensus, was found in 12.5% of subtype F1 viruses isolated from treated patients and in 1.3% of those from naïve persons. In fact, Abecassis et al. (2005) have associated mutations 89I/V to NFV resistance in certain non-B subtypes, including F1. Interestingly, the quantitative analysis showed that other polymorphisms occurred significantly at higher levels in subtype F1 than in subtype B isolated from treated patients and than in subtype F1 isolated from naïve patients.
Some authors recently reported rapid resistance emergence in treated children (Delaugerre et al., 2007; Vignoles et al., 2007) and that they were more likely to have virologic failure compared with adults (Kamya et al., 2007). Differences observed in this study between drug resistance acquisition in adults and children might be explained by the differences found in treatment regimens. Additionally, issues related to adherence (usually lower in children) and differences in viral replication rates may play a role in this finding. Interestingly, V82L reported by IAS consensus as a major mutation specifically associated with resistance to tipranavir, not used by our cohort, was observed in subtypes B and F1 isolated from treated children but not in adults.
The present study analyzes the emergence of certain drug resistance mutations in patients undergoing virologic failure, perhaps by acquisition of viral resistance. However, the genotypic/phenotypic correlates in non-B subtypes is not completely elucidated, and therefore the role of resistance mutations in subtype F1 needs to be further assessed by specific phenotyping assays.
In an HIV worldwide epidemic where an increasing number of non-B subtype infections accumulate, a better understanding of drug resistance acquisition and the role of specific ARV in resistance development is of pivotal importance. Our study provides a contribution to this understanding with respect to subtype F1, and further, larger subtype-specific studies should be conducted for this purpose.
Acknowledgments
We are indebted to the medical staff of all HIV/AIDS clinics who participated to this study for reviewing and compiling data from the patients' medical charts, and for sample collection. This study was supported by PN-DST/AIDS (Brazilian Ministry of Heath), Brazilian Ministry of Science and Technology (CNPq) grant no. 403589/2004-5 and Rio de Janeiro State Science Foundation (FAPERJ) grant no. E-26/171.337/2002.
References
- Abecassis AB, Deforche K, Snoeck J, Bacheler LT, McKenna P, Carvalho AP, Gomes P, Camacho RJ, Vandamme AM. Protease mutation M89I/V is linked to therapy failure in patients infected with the HIV-1 non-B subtypes C, F or G. AIDS. 2005;19:1799–1806. doi: 10.1097/01.aids.0000188422.95162.b7. [DOI] [PubMed] [Google Scholar]
- Apetrei C, Necula A, Holm-Hansen C, Loussert-Ajaka I, Pandrea I, Cozmei C, Streinu-Cercel A, Pascu FR, Negut E, Molnar G, Duca M, Pecec M, Brun-Vézinet F, Simons F. HIV-1 diversity in România. AIDS. 1998;12:1079–1085. [PubMed] [Google Scholar]
- Bello G, Eyer-Silva WA, Couto-Fernandez JC, Guimarães ML, Chequer-Fernandez SL, Teixeira SLM, Morgado MG. Demographic history of HIV-1 subtypes B and F in Brazil. Infect Genet Evol. 2007;7:263–270. doi: 10.1016/j.meegid.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Bello G, Guimarães ML, Morgado MG. Evolutionary history of HIV-1 subtype B and F infections in Brazil. AIDS. 2006;20:763–768. doi: 10.1097/01.aids.0000216377.84313.52. [DOI] [PubMed] [Google Scholar]
- Brenner B, Turner D, Oliveira M, Moisi D, Detorio M, Carobene M, Marlink RG, Schapiro J, Roger M, Wainberg MA. A V106 M mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to non-nucleoside reverse transcriptase inhibitors. AIDS. 2003;17:F1–F5. doi: 10.1097/00002030-200301030-00001. [DOI] [PubMed] [Google Scholar]
- Brindeiro PA, Brindeiro RM, Mortensen C, Hertogs K, De Vroey V, Rubini NPM, Sion FS, De Sá CAM, Machado DM, Succi RCM, Tanuri A. Testing genotypic and phenotypic resistance in human immunodeficiency vírus type 1 isolates of clade B and other clades from children failing antiretroviral therapy. J Clin Microbiol. 2002;40(12):4512–4519. doi: 10.1128/JCM.40.12.4512-4519.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindeiro RM, Diaz RS, Sabino EC, Morgado MG, Pires IL, Brígido L, Dantas MC, Barreira D, Teixeira PR, Tanuri A, Brazilian Network for Drug Resistance Surveillance Brazilian Network for HIV Drug Resistance Surveillance (HIV-BResNet): a survey of chronically infected individuals. AIDS. 2003;17:1063–1069. doi: 10.1097/00002030-200305020-00016. [DOI] [PubMed] [Google Scholar]
- Caride E, Hertogs K, Larder B, Dehertogh P, Brindeiro R, Machado E, De Sá CAM, Eyer-Silva WA, Sion FS, Passioni LFC, Menezes JA, Calazans AR, Tanuri A. Genotypic and phenotypic evidence of different drug-resistance mutation patterns between B and non-B subtypes isolates of human immunodeficiency vírus type 1 found in Brazilian patients failing HAART. Virus Genes. 2001;23:193–202. doi: 10.1023/a:1011812810397. [DOI] [PubMed] [Google Scholar]
- Carmona R, Perez-Alvarez L, Munoz M, Casado G, Delgado E, Sierra M, Thomson M, Vega Y, Vázquez de Parga E, Contreras G, Medrano L, Nájera R. Natural resistance-associated mutations to enfivurtite (T20) and polymorphisms in the gp41 region of different HIV-1 genetic forms from T20 naïve patients. J Clin Virol. 2005;32:248–253. doi: 10.1016/j.jcv.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Carobene MG, Rubio AE, Carrillo MG, Maligne GE, Kijak GH, Quarleri JF, Salomón H. Differences in frequencies of drug resistance-associated mutations in the HIV-1 pol gene of B subtype and BF intersubtype recombinant samples. J Acquir Immune Defic Syndr. 2004;35:207–209. doi: 10.1097/00126334-200402010-00018. [DOI] [PubMed] [Google Scholar]
- Deforche K, Camacho R, Grossman Z, Silander T, Soares MA, Moreau Y, Shafer RW, Van Laethem K, Carvalho AP, Wynhoven B, Cane P, Snoeck J, Clarke J, Sirivichayakul S, Ariyoshi K, Holguin A, Rudich H, Rodrigues R, Bouzas MB, Cahn P, Brigido LF, Soriano V, Sugiura W, Phanuphak P, Morris L, Weber J, Pillay D, Tanuri A, Harrigan PR, Shapiro JM, Katzenstein DA, Kantor R, Vandamme AM. Bayesian network analysis of resistance pathways against protease inhibitors. Infect Genet Evol. 2007;7(3):382–390. doi: 10.1016/j.meegid.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Delaugerre C, Warszawski J, Chaix ML, Veber F, Macassa E, Buseyne F, Rouzioux C, Blanche S. Prevalence and risk factors associated with antiretroviral resistance in HIV-1-infected children. J Med Virol. 2007;79(9):1261–1269. doi: 10.1002/jmv.20940. [DOI] [PubMed] [Google Scholar]
- Dumans AT, Soares MA, Machado ES, Hue S, Brindeiro RM, Pillay D, Tanuri A. Synounymous genetic polymorphisms within Brazilian human immunodeficiency vírus type 1 subtypes may influence mutational routes to drug resistance. J Infect Dis. 2004;189:1232–1238. doi: 10.1086/382483. [DOI] [PubMed] [Google Scholar]
- Gonzalez LM, Brindeiro RM, Aguiar RS, Pereira HS, Abreu CM, Soares MA, Tanuri A. Impact of nelfinavir resistance mutations on in vitro phenotype, fitness and replication capacity of human immunodeficiency vírus type 1 with subtype B and C proteases. Antimicrob Agents Chemother. 2004;48:3552–3555. doi: 10.1128/AAC.48.9.3552-3555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez LMF, Brindeiro RM, Tarin M, Calazans A, Soares MA, Cassol S, Tanuri A. In Vitro hypersusceptibility of human immunodeficiency virus type 1 subtype C protease to lopinavir. Antimicrob Agents Chemother. 2003;47:2817–2822. doi: 10.1128/AAC.47.9.2817-2822.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman Z, Istomin V, Averbuch D, Lorber M, Risenberg K, Levi I, Chowers M, Burke M, Yaacov NB, Schapiro JM, Israel AIDS Multi-Center Study Group Genetic variation at NNRTI resistance-associated positions in patients infected with HIV-1 subtype C. AIDS. 2004;18:909–915. doi: 10.1097/00002030-200404090-00008. [DOI] [PubMed] [Google Scholar]
- Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global and regional distribution of HIV-1 genetics subtypes and recombinants in 2004. AIDS. 2006;20:W13–W23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- Hierholzer J, Montano S, Hoelscher M, Negrete M, Hierholzer M, Ávila MM, Carrillo MG, Russi JC, Vinoles J, Alava A, Acosta ME, Gianella A, Andrade R, Sanchez JL, Carrion G, Russell K, Robb M, Birx D, McCutchan F, Carr JK. Molecular epidemiology of HIV type I in Ecuador, Peru, Bolivia, Uruguay, and Argentina. AIDS Res Hum Retroviruses. 2002;18(18):1339–1350. doi: 10.1089/088922202320935410. [DOI] [PubMed] [Google Scholar]
- Hoffman NG, Schiffer CA, Swanstrom R. Covariation of amino acid positions in HIV-1 protease. Virology. 2003;314:536–548. doi: 10.1016/s0042-6822(03)00484-7. [DOI] [PubMed] [Google Scholar]
- Jeeninga RE, Hoogenkamp M, Armand-Ugon M, de Baar M, Verhoef K, Berkhout B. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J Virol. 2000;74:3740–3751. doi: 10.1128/jvi.74.8.3740-3751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson VA, Brun-Vézinet F, Clotet B, Gunthard HF, Kuritzkes DR, Pillay D, Schapiro JM, Richman DD. Update of the drug resistance mutations in HIV-1: 2007. Top HIV Med. 2007;15(4):119–125. [PubMed] [Google Scholar]
- Kamya MR, Mayanja-Kizza H, Kambugu A, Bakeera-Kitaka S, Semitala F, Mwebaze-Songa P, Castelnuovo B, Schaefer P, Spacek LA, Gasasira AF, Katabira E, Colebunders R, Quinn TC, Ronald A, Thomas DL, Kekitiinwa A, Academic Alliance for AIDS Care and Prevention in Africa Predictors of long-term viral failure among Ugandan children and adults treated with antiretroviral therapy. J Acquir Immune Defic Syndr. 2007;46:187–193. doi: 10.1097/QAI.0b013e31814278c0. [DOI] [PubMed] [Google Scholar]
- Kantor R, Katzenstein DA, Efron B, Carvalho AP, Wynhoven B, Cane P, Clarke J, Sirivichayakul S, Soares MA, Snoeck J, Pillay C, Rudich H, Rodrigues R, Holguin A, Ariyoshi K, Bouzas MB, Cahn P, Sugiura W, Soriano V, Brígido LF, Grossman Z, Morris L, Vandamme AM, Tanuri A, Phanuphak P, Weber JN, Pillay D, Harrigan PR, Camacho R, Schapiro JM, Shafer RW. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. Plos Med. 2005;2(4):325–337. doi: 10.1371/journal.pmed.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerda HR, Medeiros LB, Cavalcanti AMS, Ximenes RAA, Albuquerque MFPM. Comparison of the epidemiology, profile of mutations, and clinical response to antiretrovirals among subtypes B and F of the human immunodeficiency vírus type 1. Mem Inst Oswaldo Cruz. 2007;102(6):693–699. doi: 10.1590/s0074-02762007005000103. [DOI] [PubMed] [Google Scholar]
- Machado ES, Lambert JS, Watson DC, Afonso AO, da Cunha SM, Nogueira SA, Caride E, Oliveira RH, Sill AM, De Vico A, Tanuri A. Genotypic resistance and HIV-1 subtype in Brazilian children on dual and triple combination therapy. J Clin Virol. 2004;30(1):24–31. doi: 10.1016/j.jcv.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Masciotra S, Livellara B, Belloso W, Clara L, Tanuri A, Ramos AC, Baggs J, Lal R, Pieniazek D. Evidence of a high frequency of HIV-1 subtype F infections in a heterosexual population in Buenos Aires, Argentina. AIDS Res Hum Retroviruses. 2000;16(10):1007–1014. doi: 10.1089/08892220050058425. [DOI] [PubMed] [Google Scholar]
- Montes B, Vergne L, Peeters M, Reynes J, Delaporte E, Segondy M. Comparison of drug resistance mutations and their interpretation in patients infected with non-B HIV-1 variants and matched patients infected with HIV-1 subtype B. J Acquir Immune Defic Syndr. 2004;35:329–336. doi: 10.1097/00126334-200404010-00001. [DOI] [PubMed] [Google Scholar]
- Pires IL, Soares MA, Speranza FAB, Ishii SK, Vieira MCG, Gouvêa MIFS, Guimarães MAAM, de Oliveira FE, Magnanini MMF, Brindeiro RM, Tanuri A. Prevalence of human immunodeficiency virus drug resistance mutations and subtypes in drug-naïve, infected individuals in the army health service of Rio de Janeiro, Brazil. J Clin Microbiol. 2004;42(1):426–430. doi: 10.1128/JCM.42.1.426-430.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poveda E, de Mendoza C, Parkin N, Choe S, Garcia-Gasco P, Corral A, Soriano V. Evidence for different susceptibility to tipranavir and darunavir in patients infected with distinct HIV-1 subtypes. AIDS. 2008;22:611–616. doi: 10.1097/QAD.0b013e3282f51eb9. [DOI] [PubMed] [Google Scholar]
- Renjifo B, Gilbert P, Chaplin B, Msamanga G, Mwakagile D, Fawzi W, Tanzanian Vitamin, HIV Study Group. Essex M. Preferential in-utero transmission of HIV-1 subtype C as compared to HIV-1 subtype A or D. AIDS. 2004;18:1629–1636. doi: 10.1097/01.aids.0000131392.68597.34. [DOI] [PubMed] [Google Scholar]
- Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31:298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios M, Delgado E, Pérez-Alvarez L, Fernandez J, Galvez P, de Parga EV, Yung V, Thomson MM, Nájera R. Antiretroviral drug resistance and phylogenetic diversity of HIV-1 in Chile. J Med Virol. 2007;79(6):647–656. doi: 10.1002/jmv.20881. [DOI] [PubMed] [Google Scholar]
- Robertson DL, Anderson JP, Bradac JA, Carr JK, Foley B, Funkhouser RK, Gao F, Hahn BH, Kalish ML, Kuiken C, Learn GH, Leitner T, McCutchan F, Osmanov S, Peeters M, Pieniazek D, Salminen M, Sharp PM, Wolinsky S, Korber B. HIV-1 nomenclature proposal. Science. 2000;288:55–56. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- Rodrigues R, Custódio RM, Bueno SM, Eira M, Ferreira JLP, Jamal L, Duarte AJS, Brígido LFM. Prevalence of ARV resistance mutations and impact of genotyping test in HIV patients with advanced disease in São Paulo, Brazil. J Clin Virol. 2005a;32(4):336–337. doi: 10.1016/j.jcv.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Rodrigues R, Vazquez CMP, Colares JK, Custódio RM, Bonásser-Filho F, Souza LR, Gianna MC, Marques CCA, Brigido LFM. Antiretroviral resistance mutations in human immunodeficiency virus type 1 infected patients enrolled in genotype testing at the Central Public Health Laboratory, São Paulo, Brazil: preliminary results. Mem Inst Oswaldo Cruz. 2005b;100(1):97–102. doi: 10.1590/s0074-02762005000100018. [DOI] [PubMed] [Google Scholar]
- Salemi M, de Oliveira T, Soares MA, Pybus O, Dumans AT, Vandamme AM, Tanuri A, Cassol S, Fitch WM. Different epidemic potentials of the HIV-1B and C subtypes. J Mol Evol. 2005;60:598–605. doi: 10.1007/s00239-004-0206-5. [DOI] [PubMed] [Google Scholar]
- Sanabani S, Neto WK, Kalmar EMN, Diaz RS, Janini LM, Sabino EC. Analysis of the near full length genomes of HIV-1 subtypes B, F, and BF recombinant from a cohort of 14 patients in São Paulo, Brazil. Infect Genet Evol. 2006;6:368–377. doi: 10.1016/j.meegid.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Santos AF, Schrago CG, Martinez AMB, Mendoza-Sassi R, Silveira J, Sousa TM, Lengruber RB, Soares EAJM, Sprinz E, Soares MA. Epidemiologic and evolutionary trends of HIV-1 CRF31_BC-related strains in southern Brazil. J Acquir Immune Defic Syndr. 2007;45(3):328–333. doi: 10.1097/QAI.0b013e3180690d6a. [DOI] [PubMed] [Google Scholar]
- Santos AF, Sousa TM, Soares EAJM, Sanabani S, Martinez AMB, Sprinz E, Silveira J, Sabino EC, Tanuri A, Soares MA. Characterization of a new circulating recombinant form comprising HIV-1 subtypes C and B in southern Brazil. AIDS. 2006;20(16):2011–2019. doi: 10.1097/01.aids.0000247573.95880.db. [DOI] [PubMed] [Google Scholar]
- Soares EA, Martinez AM, Sousa TM, Santos AF, Da Hora V, Silveira J, Bastos FI, Tanuri A, Soares MA. HIV-1 subtype C dissemination in southern Brazil. AIDS. 2005;19(Suppl. 4):S81–S86. doi: 10.1097/01.aids.0000191497.00928.e4. [DOI] [PubMed] [Google Scholar]
- Soares EA, Santos AF, Sousa TM, Sprinz E, Martinez AM, Silveira J, Tanuri A, Soares MA. Differential drug resistance acquisition in HIV-1 of subtypes B and C. PloS ONE. 2007;2(8):e730. doi: 10.1371/journal.pone.0000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares EA, Santos RP, Pellegrini JA, Sprinz E, Tanuri A, Soares MA. Epidemiologic and molecular characterization of human immunodeficiency vírus type 1 in southern Brazil. J Acquir Immune Defic Syndr. 2003;34:520–526. doi: 10.1097/00126334-200312150-00012. [DOI] [PubMed] [Google Scholar]
- Soares MA, De Oliveira T, Brindeiro RM, Diaz RS, Sabino EC, Brigido L, Pires IL, Morgado MG, Dantas MC, Barreira D, Teixeira PR, Cassol S, Tanuri A, Brazilian Network for Drug Resistance Surveillance A specific subtype C of human immunodeficiency virus type 1 circulates in Brazil. AIDS. 2003;17:11–21. doi: 10.1097/00002030-200301030-00004. [DOI] [PubMed] [Google Scholar]
- Svicher V, Ceccherini-Silberstein F, Erba F, Santoro M, Gori C, Bellocchi MC, Giannella S, Trotta MP, Monforte Ad'A, Antinori A, Perno CF. Novel human immunodeficiency virus type 1 protease mutations potentially involved in resistance to protease inhibitors. Antimicrob Agents Chemother. 2005;49(5):2015–2025. doi: 10.1128/AAC.49.5.2015-2025.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira PR, Vitória MA, Barcarolo J. Antiretroviral treatment in resourcepoor settings: the Brazilian experience. AIDS. 2004;18(Suppl. 3):S5–S7. doi: 10.1097/00002030-200406003-00002. [DOI] [PubMed] [Google Scholar]
- Thomson MM, Sierra M, Tanuri A, May S, Casado G, Manjón N, Nájera R. Analysis of near full-length genome sequences of HIV type1 BF intersubtype recombinant viruses from Brazil reveals their independent origins and their lack of relationship to CRF12_BF. AIDS Res Hum Retroviruses. 2004;20(10):1126–1133. doi: 10.1089/aid.2004.20.1126. [DOI] [PubMed] [Google Scholar]
- Vicente ACP, Otsuki K, Silva NB, Castilho MC, Barros FS, Pieniazek D, Hu D, Rayfield MA, Bretas G, Tanuri A. The HIV epidemic in the Amazon Basin is driven by prototypic and recombinant HIV-1 subtypes B and F. J Acquir Immune Defic Syndr. 2000;23:327–333. doi: 10.1097/00126334-200004010-00008. [DOI] [PubMed] [Google Scholar]
- Vignoles M, Barboni G, Agosti MR, Quarleri J, García MK, Giraudi V, Ayala SG, Salomón H. High frequency of primary mutations associated with antiretroviral drug resistance in recently diagnosed HIV-infected children. Antivir Ther. 2007;12:1133–1137. [PubMed] [Google Scholar]


