Abstract
Transcription factors regulate gene expression through their binding to DNA. In a living Escherichia coli cell, we directly observed specific binding of a lac repressor, labeled with a fluorescent protein, to a chromosomal lac operator. Using single-molecule detection techniques, we measured the kinetics of binding and dissociation of the repressor in response to metabolic signals. Furthermore, we characterized the nonspecific binding to DNA, one-dimensional (1D) diffusion along DNA segments, and 3D translocation among segments through cytoplasm at the single-molecule level. In searching for the operator, a lac repressor spends ~90% of time nonspecifically bound to and diffusing along DNA with a residence time of <5 milliseconds. The methods and findings can be generalized to other nucleic acid binding proteins.
In all kingdoms of life transcription factors (TFs) regulate gene expression by site-specific binding to chromosomal DNA, preventing or promoting the transcription by RNA polymerase. The lac operon of Escherichia coli, a model system for understanding TF-mediated transcriptional control (1), has been the subject of extensive biochemical (2–4), structural (5) and theoretical (6, 7) studies since the seminal work by Jacob and Monod (8). However, the in vivo kinetics of the lac repressor, and all other TFs, has only been studied indirectly by monitoring the regulated gene products. Traditionally, this was done on a population of cells (9), in which unsynchronized gene activity among cells masks the underlying dynamics. Recent experiments on single cells allow investigation of stochastic gene expression (10–15). However, direct observation of TF mediated gene regulation (16) remains difficult, because it often involves only a few copies of TF and their chromosomal binding sites. Here we report on a kinetics study of how fast a lac repressor binds its chromosomal operators and dissociates in response to a metabolic signal in a living E. coli cell.
Single molecule detection also makes it possible to investigate how a TF molecule searches for specific binding sites on DNA, a central question in molecular biology. Target location by TFs (and most nucleic acid binding proteins) is believed to be achieved by facilitated diffusion, in which a TF searches for specific binding sites through a combination of one-dimensional (1D) diffusion along a short DNA segment and 3D translocation among DNA segments through cytoplasm (17). However, real-time observation in living cells has not been available because of technical difficulties. Here we report on such an investigation, providing quantitative information of the search process.
The lac repressor (LacI) is a dimer of dimers. Under repressed conditions one dimer binds the major lac operator, O1, and the other dimer binds one of the weaker auxiliary operators, O2 or O3 (18) (Fig. 1A). LacI binding to O1 prevents RNA polymerase from transcribing the lac operon (lacZYA). Upon binding of allolactose, an intermediate metabolite in the lactose pathway, or a non-degradable analogue, such as IPTG (isopropyl β-d-1-thiogalactopyranoside), the repressor’s affinity for the operator is substantially reduced to a level comparable to that of nonspecific DNA interaction (19).
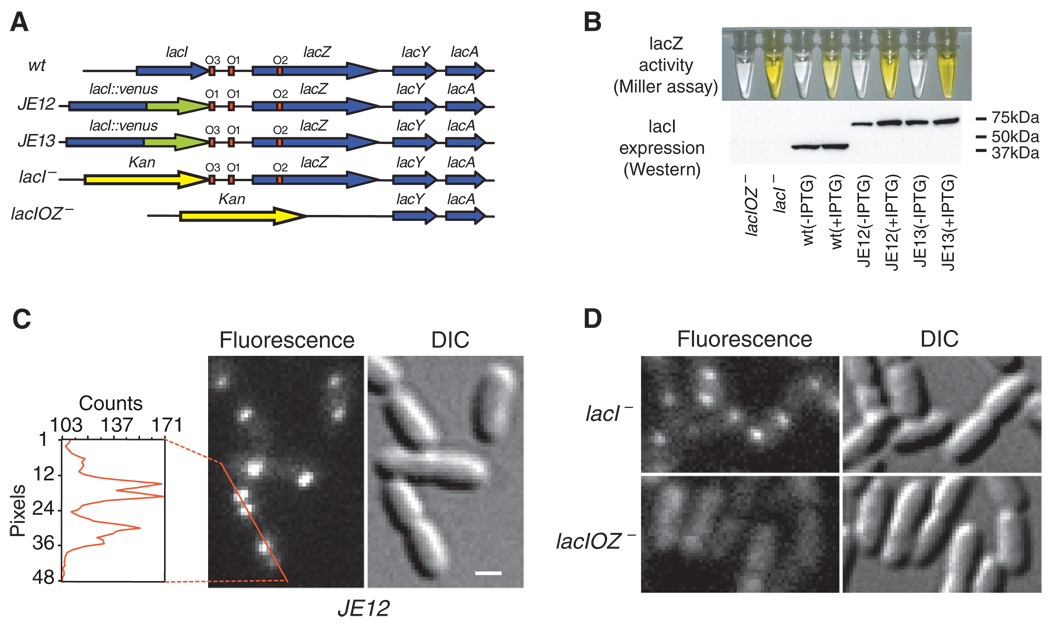
Fig. 1. Specific binding to lac operators.
(A) Strains. The chromosomal lac region of the wild-type E. coli (BW25933) and various derivatives used in this report. (B) Bulk activity assay. The Miller assay (top) shows that the YFP fusion strains (JE12 and JE13) are active and respond to induction by IPTG (1 mM, 3 hours) by derepressing lacZ (yellow). The Western blot (bottom) for LacI shows that JE12 and JE13 express the full-length fusion protein (67kD) and that the expression in JE12, in the absence of IPTG, is strongly autorepressed as compared to the wild type and JE13. (C) Fluorescence (1-s exposure) and DIC images of JE12 grown in M9 glucose with amino acids. The YFP-labeled LacI binds persistently at one or two locations per cell depending on whether the operators have been replicated or not. The graph shows the fluorescence intensity along the red line. Scale bar, 1 µm. (D) DIC and fluorescence images (1-s exposure) of LacI-Venus expressed from plasmid in the lacI− and lacIOZ− strains, respectively. Plasmid expression is used to obtain similar expression levels in the two strains. No specific binding is observed in the absence of the lac operators.
To image the lac repressor, we expressed it from the native chromosomal lacI locus as a C-terminal fusion with the rapidly maturing (~7 min) yellow fluorescent protein (YFP) Venus (A206K) (15, 20) (Fig. 1A). The short maturation time prevents the lac operator sites from being occupied by immature fusion proteins. The C-terminal fusion avoids interference with the N-terminal DNA-binding domain (21). Our fusion protein forms a dimer, which like most other C-terminal fusions with LacI (22), does not tetramerize (fig. S1). The labeled dimer up-regulates the expression of lacZ ~100-fold in response to full induction by IPTG at 37°C (JE13 in Fig. 1B). This repression factor compares well with that of nonlabeled repressors (4), indicating that the fusion protein maintains regulatory activity.
The detection of specific binding of LacI to its operators is achieved through localization enhancement (15) [see Supporting Online Material (SOM)]. When E. coli cells are imaged with a wide-field fluorescence microscope and a charge-coupled device (CCD) camera with a long exposure time (1s), the fluorescence from TFs that are not specifically bound to DNA is collected from the entire area of the cell because of fast diffusion and is hence overwhelmed by strong cellular auto-fluorescence. However, a single TF specifically bound to the relatively stationary DNA emits from a highly localized region and can be detected above the auto-fluorescence background.
A necessary condition for detection through localization enhancement is that the copy number of TF must be low. LacI, like most other TFs in E. coli, is naturally expressed at a low level—about 20 monomers per lacI gene (23). This results from autorepression at the O3 operator which overlaps part of the lacI gene. We further reduced the expression level by replacing the O3 operator with an O1 operator sequence, thus enhancing auto-repression. This reduces the expression level by a factor of ~3 as compared to the wild type (Fig. 1B).
Fig. 1C shows a differential interference contrast (DIC) image and the corresponding fluorescence image with a 1s exposure time. In the image, specifically bound TFs appear as nearly diffraction-limited spots. Most cells have at least one spot per cell. Some have two, owing to replication of the operator region (24). We could not distinguish from the spot intensity whether one or two TF dimers were bound to the operator region containing two O1 operators (Fig. 1A and fig. S1). As a control, we showed that the specific LacI binding is dependent on the concentration of inducer (IPTG) (fig. S2). As another control, we proved that LacI binds specifically only to the lac operator. We expressed LacI-Venus from plasmid in two different strains, with and without specific chromosomal operators (lacI− and lacIOZ−, respectively) (Fig. 1A). The fluorescence images in Fig. 1D with 1s exposure demonstrate the lack of specific binding in lacIOZ− strain, proving that LacI binds persistently only at the lac operators.
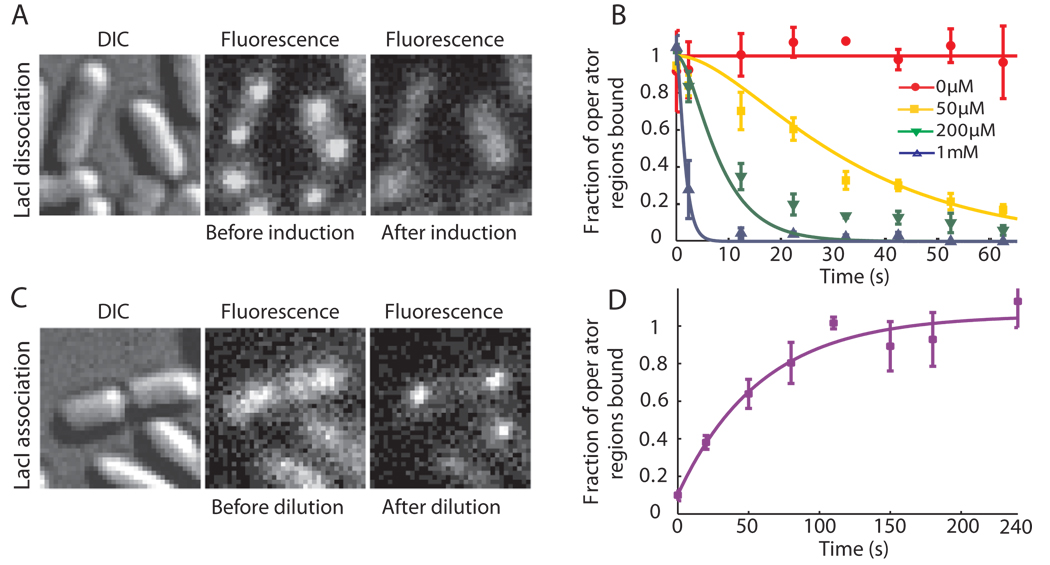
We next investigate the response time of TF-mediated induction. Figure 2A shows the same cells before and 40 s after addition of 1 mM IPTG to the growth media. During this 40 s time interval, the TFs dissociate from the operators in nearly all cells. As a control, we proved that the disappearance of localized fluorescence is not due to photobleaching (fig. S3). In Fig. 2B the fraction of operator regions with bound TF is plotted as a function of time after induction at different IPTG concentrations. The repressor dissociation kinetics was probed by imaging different groups of cells at different time points after induction (SOM). Because dissociation of the IPTG-bound LacI from the operator is significantly faster than 1 s−1 (2), we could attribute the kinetics in Fig. 2B to the binding of IPTG to the LacI-operator complex. However, fitting of Fig. 2B (SOM) yields an association rate constant of IPTG to the repressor-operator complex of ~851 M−1s−1. This value is one order of magnitude lower than the in vitro estimate (2), which indicates that IPTG's membrane permeability might be rate limiting for LacI binding.
Fig. 2. lac repressor kinetics in living cells.
(A) JE12 bacteria before and 40 s after addition of IPTG to a final concentration of 1 mM. (B) Fraction of the lac operator regions that is TF-bound (±SEM, n~3) is plotted as a function of time after induction by various concentrations of IPTG. The data are globally fitted with a model in which IPTG binds independently to the two dimers in the operator region (SOM). (C) JE12 bacteria before and 1 min after dilution of IPTG from 100 to 2 µM with the addition of 1 mM ONPF (D) Fraction of the operator regions that is TF-bound (±SEM, n~3) as a function of time after rapid dilution of IPTG from 100 to 2 µM by addition of 1 mM ONPF. The data are fitted with an exponentially distributed binding time and yield a time constant of 59 s.
To investigate how fast the TFs find a specific operator site, we rapidly diluted IPTG from 100 to 2 µM by adding growth media with 1 mM ONPF (2-nitrophenyl-b-d-fucoside) (Fig. 2C). ONPF is an anti-inducer that competitively binds to LacI and therefore effectively prevents rebinding of IPTG after its dissociation (23). Kinetic analysis (Fig. 2D) yields a time constant of ~59 s for the exponential rise (SOM). Considering that IPTG dissociates from LacI in just a few seconds (2) and assuming that ONPF reaches sufficient intracellular concentration rapidly, we would expect the TF’s target searching to be rate limiting. Taking into account the possibility of a slower ONPF influx rate, we conclude that the upper bound of the time for the first TF to find one of the two unoccupied O1 operators to be 59 s. Because there are ~3 repressors per cell, it would take at most τ̣ ~ 59 s×2×3 = 354 s for a single lac repressor dimer in one cell to find a specific operator.
We next turn our attention to the search process. The conventional view is that the search is facilitated by a combination of 1D diffusion along short DNA segments separated by transfers between segments through cytoplasm. Recent in vitro experiments on DNA repair enzymes have directly demonstrated the 1D diffusion along nonspecific DNA (25). Similarly, we determined the 1D diffusion constant of the dimeric LacI-Venus to be D1 =0.046±0.01 µm2s−1 using single molecule tracking on flow-stretched DNA (SOM). Although the in vitro measurement was done at low salt concentration in order to obtain trajectories with long residence time, the diffusion constant, which is largely independent of ionic strength (25), is expected to be similar in a living cell. However, the residence time, which is dependent on salt concentration, is yet to be determined in vivo.
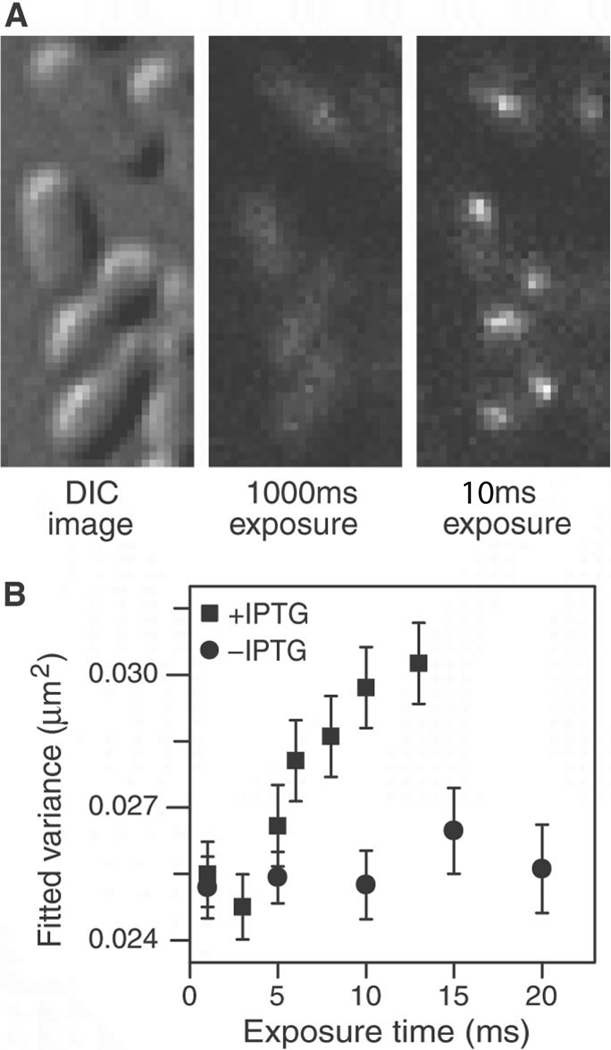
To determine the nonspecific residence time on DNA for IPTG-bound LacI-Venus in a living cell, we obtained fluorescence images at different exposure times. With 1-s exposure LacI cannot be imaged individually, as shown in Fig. 1D. With 10 ms exposure, however, LacI appear as nearly diffraction-limited spots (Fig. 3A). We observed two to four TFs in each cell, consistent with the expected LacI copy number. Fig. 3B (and fig. S4) shows that the spot sizes increase for exposure times longer than 5 ms. Because the 1D diffusion along a DNA segment on this time scale is much shorter than the diffraction-limit spot size, we attribute the increase in spot size to 3D translocation between nonspecific binding events. This result clearly indicates that the TF’s nonspecific residence time in cells, tR, was ≤5 ms.
Fig. 3. Imaging nonspecifically bound LacI.
(A) Two fluorescence images with different exposure times and the corresponding DIC images of IPTG-induced E. coli cells. At 1000 ms, individual LacI-Venus appear as diffuse fluorescence background. At 10 ms they are clearly visible as nearly diffraction-limited spots. (B) Fluorescence spot size as a function of exposure time. The size is represented as the average variance of a 2D Gaussian function fit to images of fluorescence spots (±SEM, n~100). The same total excitation energy is used for different exposure time. The spots are measured before (−IPTG, •) and after (+IPTG, ▪) induction. The size converges to the width of the point spread function (full width at half maximum = 370 nm) below 5 ms.
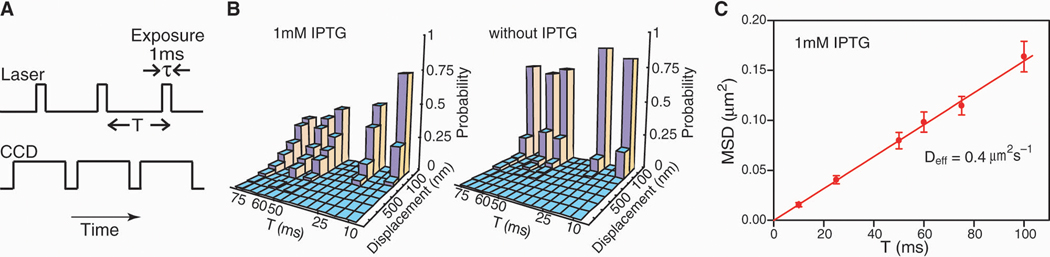
The ability to image nonspecifically bound TFs allowed us to track the movement of individual TFs using stroboscopic laser excitation. Laser pulses (1 ms) were synchronized to the frame rate of a fast EMCCD (electron-multiplying charge-coupled device) camera (Fig. 4A, SOM). Because individual molecules are bleached in three to four frames, we varied the frame rate (1/T) to construct net displacement histograms for different time intervals (T = 10 to 75 ms) (Fig. 4B). Without IPTG, both the specific and nonspecific binding events are observed, and the displacement histogram is strongly peaked at <100nm because most TFs are specifically bound. This shows that the lac operator region is confined to within 100 nm during 75 ms. With 1 mM IPTG, however, the displacement distribution broadens with increasing time intervals.
Fig. 4. Single molecule tracking with stroboscopic illumination.
(A) Timing diagram for stroboscopic illumination. Each laser pulses (1 ms) is synchronized to the CCD frame time, which lasts time T. (B) Displacement histograms for different values of T. The absolute values of displacement along an arbitrary axis were calculated from 2D Gaussian fittings in two successive image frames. The displacement distribution of nonspecifically bound TFs broadens with time (left), whereas the distribution before induction (right) remains peaked at <100 nm. The contrast between them illustrates the change in TF mobility before and after induction. (C) Mean-square displacement for nonspecifically bound TFs at different time intervals. The red line shows a linear fit of the MSDs. Error bars are calculated as described in the SOM. The fitting agrees well with a normal diffusion in the imaging plane, <Δx2>=4Defft, with Deff=0.4 µm2s−1.
We next determine the apparent diffusion constant. The mean square displacement (MSD) of IPTG-bound LacI measured at various time intervals follows a linear dependence at the time scale >10 ms (Fig. 4C) and does not exhibit anomalous diffusion as was observed for mRNA in E. coli cytoplasm at longer time scales (26). The apparent diffusion constant, Deff =0.4±0.02 µm2s−1, is one order of magnitude higher than the 1D diffusion constant (D1) of LacI dimers on DNA. Therefore we attribute apparent diffusion to the contribution from 3D diffusion in between nonspecific bindings. Using fluorescence correlation spectroscopy (FCS) (27), we measured the in vivo diffusion constant of LacI-Venus without its DNA-binding domain to be D3 =3±0.3 µm2s−1 (SOM). This suggests that the LacI dimer spends ~87% of the time nonspecifically bound and diffusing along DNA (SOM). This fraction is consistent with the previous population averaged estimate (>90%) on the basis of the LacI tetramer concentration in minicells (28).
With the measurements of the diffusion constants and residence time (Table 1), we can give an estimate of the search time using a simple model for facilitated diffusion (7). Considering that the repressor spends most of its time bound to DNA, the search time is estimated as the nonspecific residence time (tR) multiplied by the average number of 1D diffusion events necessary to find the target (29). The latter is given by the accessible genome size (M ≈ 4.8×106 bp) divided by the number of base pairs visited per sliding event.
| (Eq. 1) |
Table 1.
Diffusion constants and characteristic times
| Diffusion Constant | Deff | D3 | D1 |
|---|---|---|---|
| Value | 0.4 ± 0.02 µm2/s | 3 ± 0.3 µm2/s | 0.046 ± 0.01 µm2/s |
| Method | in vivo SM tracking | in vivo FCS | in vitro SM tracking |
| Characteristic Time |
Search Time* (τ) | Residence Time on DNA (tR) |
|---|---|---|
| Value | 65†–360s | 0.3†–5ms |
| Method | Detection by immobilization upon IPTG dilution |
Spot size dependence on exposure time |
Defined as the time for a single TF in one cell to find a target.
Theoretical lower bound considering diffusion-limited association to nonspecific sites (29) (SOM)
This results in a search time of τ < 270 s for a single lac repressor in one cell to find one target. In the SOM, we further calculate a lower bound of the search time based on diffusion-limited association to nonspecific DNA (Table I). Hence, the measured and estimated search times are consistent. Despite the uncertainty of these numbers, our measurements provided quantitative information of the target search on DNA in vivo. This result has implications for other DNA binding proteins such as DNA-repair enzymes. Similar single-molecule experiments will advance our quantitative understanding of biochemistry and molecular biology in living cells.
Supplementary Material
Acknowledgments
We thank Paul Blainey, Peter Sims, Paul Choi, Otto Berg, and Martin Lovmar for helpful discussions. We are grateful to Paul Blainey, Peter Sims, and Jeremy Hearn for protein purification, Brian English for help with FCS, and Ji Yu for making pVS155 and pVS167. The plasmids pkO3, pKD30, and Venus gene were contributed by Profs. G. Church, B. Wanner and A. Miyawaki, respectively. This work was supported by NIH Director’s Pioneer Award Program, the Knut and Alice Wallenberg Foundation (J.E), and NSF Graduate Research Fellowship (G-W.L.).
References
- 1.Matthews KS, Nichols JC. Prog Nucleic Acid Res Mol Biol. 1998;58:127. doi: 10.1016/s0079-6603(08)60035-5. [DOI] [PubMed] [Google Scholar]
- 2.Dunaway M, et al. J Biol Chem. 1980;255:10115. [PubMed] [Google Scholar]
- 3.Winter RB, von Hippel PH. Biochemistry. 1981;20:6948. doi: 10.1021/bi00527a029. [DOI] [PubMed] [Google Scholar]
- 4.Oehler S, Amouyal M, Kolkhof P, von Wilcken-Bergmann B, Muller-Hill B. Embo J. 194;13:3348. doi: 10.1002/j.1460-2075.1994.tb06637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis M, et al. Science. 1996;271:1247. doi: 10.1126/science.271.5253.1247. [DOI] [PubMed] [Google Scholar]
- 6.Berg OG, Blomberg C. Biophys Chem. 1976;4:367. doi: 10.1016/0301-4622(76)80017-8. [DOI] [PubMed] [Google Scholar]
- 7.Berg OG, Winter RB, von Hippel PH. Biochemistry. 1981;20:6929. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 8.Jacob F, Monod J. J Mol Biol. 1961;3:318. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 9.Kepes A. Prog Biophys Mol Biol. 1969;19:199. doi: 10.1016/0079-6107(69)90006-6. [DOI] [PubMed] [Google Scholar]
- 10.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Science. 2002;297:1183. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 11.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Nat Genet. 2002;31:69. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 12.Ozbudak EM, Thattai M, Lim HN, Shraiman BI, Van Oudenaarden A. Nature. 2004;427:737. doi: 10.1038/nature02298. [DOI] [PubMed] [Google Scholar]
- 13.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Science. 2005;307:1962. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 14.Golding I, Paulsson J, Zawilski SM, Cox EC. Cell. 2005;123:1025. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 15.Yu J, Xiao J, Ren X, Lao K, Xie XS. Science. 2006;311:1600. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 16.Yao J, Munson KM, Webb WW, Lis JT. Nature. 2006;442:1050. doi: 10.1038/nature05025. [DOI] [PubMed] [Google Scholar]
- 17.von Hippel PH, Berg OG. J Biol Chem. 1989;264:675. [PubMed] [Google Scholar]
- 18.Oehler S, Eismann ER, Kramer H, Muller-Hill B. Embo J. 1990;9:973. doi: 10.1002/j.1460-2075.1990.tb08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang AC, Revzin A, Butler AP, von Hippel PH. Nucleic Acids Res. 1977;4:1579. doi: 10.1093/nar/4.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagai T, et al. Nat Biotechnol. 2002;20:87. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 21.Lewis M. C R Biol. 2005;328:521. doi: 10.1016/j.crvi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Fieck A, Wyborski DL, Short JM. Nucleic Acids Res. 1992;20:1785. doi: 10.1093/nar/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert W, Muller-Hill B. Proc Natl Acad Sci U S A. 1966;56:1891. doi: 10.1073/pnas.56.6.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bates D, Kleckner N. Cell. 2005;121:899. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. Proc Natl Acad Sci U S A. 2006;103:5752. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golding I, Cox EC. Phys Rev Lett. 2006;96:098102. doi: 10.1103/PhysRevLett.96.098102. [DOI] [PubMed] [Google Scholar]
- 27.Le TT, et al. Proc Natl Acad Sci U S A. 2005;102:9160. doi: 10.1073/pnas.0503311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kao-Huang Y, et al. Proc Natl Acad Sci U S A. 1977;74:4228. doi: 10.1073/pnas.74.10.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winter rb, Berg OG, von Hippel PH. Biochemistry. 1981;20:6961. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.