Abstract
Objective
Evaluate KNAVE-II, a knowledge-based framework for visualization, interpretation, and exploration of longitudinal clinical data, clinical concepts and patterns. KNAVE-II mediates queries to a distributed temporal-abstraction architecture (IDAN), which uses a knowledge-based problem-solving method specializing in on-the-fly computation of clinical queries.
Methods
A two-phase, balanced cross-over study to compare efficiency and satisfaction of a group of clinicians when answering queries of variable complexity about time-oriented clinical data, typical for oncology protocols, using KNAVE-II, versus standard methods: Both paper charts and a popular electronic spreadsheet (ESS) in Phase I; an ESS in Phase II. The measurements included the time required to answer and the correctness of answer for each query and each complexity category, and for all queries, assessed versus a predetermined gold standard set by a domain expert. User satisfaction was assessed by the Standard Usability Score (SUS) tool-specific questionnaire and by a “Usability of Tool Comparison” comparative questionnaire developed for this study.
Results
In both evaluations, subjects answered higher-complexity queries significantly faster using KNAVE-II than when using paper charts or an ESS up to a mean of 255 seconds difference per query versus the ESS for hard queries (p=0.0003) in the second evaluation. Average correctness scores when using KNAVE-II versus paper charts, in the first phase, and the ESS, in the second phase, were significantly higher over all queries. In the second evaluation, 91.6% (110/120) of all of the questions asked within queries of all levels produced correct answers using KNAVE-II, opposed to only 57.5% (69/120) using the ESS (p<0.0001). User satisfaction with KNAVE-II was significantly superior compared to using either a paper chart or the ESS (p=0.006). Clinicians ranked KNAVE-II superior to both paper and the ESS.
Conclusions
An evaluation of the functionality and usability of KNAVE-II and its supporting knowledge-based temporal-mediation architecture has produced highly encouraging results regarding saving of physician time, enhancement of accuracy of clinical assessment, and user satisfaction.
Keywords: Medical Informatics, Clinical Decision-Support Systems, Human-Computer Interface, Information Visualization, Knowledge-Based Systems, Temporal Reasoning, Intelligent User Interfaces
1. INTRODUCTION: Knowledge-Based Visualization and Exploration of Time-Oriented Clinical Data and of Their Abstractions
Physicians are confronted on a daily basis with increasing amounts of time-oriented clinical data in patient records, especially of patients with chronic diseases. Electronic medical records rarely allow the visualization of more than one clinical parameter in a graphical view, much less of multiple clinically significant concepts and meaningful patterns derivable from parameters specific to the patient's clinical context. For example, as noted in a clinical survey, it would be extremely helpful to visualize graphically in the same timeline a hypertensive patient's blood pressure, anti-hypertensive medication doses, and interpretations of the blood pressure measurements (e.g., “above target”) [1]. This would allow for a quick analysis of the patient's response to anti-hypertensive therapy over time. A tool that assists in the visualization and exploration of clinical data and of their clinically significant interpretations, often interval-based and not only time-stamped, is highly desirable and would help physicians in many medical domains. This tool could have multiple implications for disease management, quality assessment and clinical research.
However, implied by the desiderata for such a tool are an underlying extensive, context-sensitive clinical knowledge base, and a comprehensive computational methodology for extraction of meaningful point- and interval-based temporal concepts and patterns from time oriented raw clinical data, given such a knowledge base. It is also our feeling that it is crucial to actually prove that such a tool is both functional and usable, and that it indeed reduces the interpretation time and enhances the accuracy of clinical interpretation.
In this paper, we describe an evaluation by clinicians--small in its scope, but rigorous, within its confinements, in its design--of KNAVE-II, an application for knowledge-based interactive visualization and exploration of clinical data regarding individual patients, accumulating over time from multiple sources, whose semantics and visual exploration operators we have previously described in detail elsewhere [2]. Underlying KNAVE-II is a distributed computational knowledge-based framework for computing abstractions from time-oriented clinical data (temporal abstractions), IDAN [3]. The combined IDAN/KNAVE-II architecture is based on our previous theoretical and practical research in the area of knowledge-based reasoning about longitudinal data in clinical domains and in the area of intelligent visualization of such data, and it extends our early work in the area of interactive exploration of time-oriented patient data [4,5] . This distributed Web-based architecture enables users to visually explore, for an individual patient or a small group of patients, both the time-stamped data and their context-sensitive interpretations, generated on the fly using domain-specific knowledge. (As we explain later, different tools are used to explore patient populations.)
The context-sensitive interpretations are computed from predefined abstractions of timestamped clinical data, using a methodology called knowledge-based temporal-abstraction [6]. Interpretation of longitudinal data in a clinical domain involves the integration of timestamped raw clinical parameters (e.g. hemoglobin values), external interventions (e.g. surgery or insulin administration) and a domain-specific knowledge base (KB) that contains knowledge regarding properties of time-oriented clinical data and basic or complex patterns that can be formed from them. The product of this temporal-abstraction process is a set of interval-based, context-specific parameters and/or patterns with their respective values (“a period of 3 weeks of myelotoxicity [bone-marrow toxicity] grade 3, as defined in the post-bone-marrow-transplantation context”). The computational process was originally implemented within the RÉSUMÉ system [7-9].
Previous approaches to visualization of longitudinal clinical data have usually not included the use of a domain specific KB or an abstraction process, and/or did not present a detailed evaluation of the usability of the interface. In an early research study, we implemented a visualization module specific for exploration of time-oriented data and abstractions called Knowledge-based Navigation of Abstractions for Visualization and Explanation, or KNAVE. Preliminary assessments of KNAVE in a medical domain [4,5] were encouraging and demonstrated the feasibility of knowledge-based exploration. In this study, we greatly enhanced the scope of both the architecture evaluated, and of the evaluation itself.
1.1 The IDAN Temporal-Abstraction Architecture
Based on our early research, we extended the scope of our architecture for support of temporal queries, into a fully distributed, web-based framework called IDAN [3]. IDAN is a mediator for goal-driven queries about time-oriented patient data and their abstraction, or a temporal-abstraction mediator, which uses data from one or more clinical data sources, knowledge from one or more medical KBs, and the ALMA temporal-abstraction computational service [10], to answer queries regarding time-oriented clinical data and their interpretations. These queries are generated by applications such as KNAVE-II. To access heterogeneous clinical data sources, the IDAN architecture assumes that each registered clinical database has been mapped to a set of standardized medical vocabularies, such as one of the common international coding systems for diagnoses or one of the North American or European standards for encoding laboratory tests. The leaves (raw data terms) of the derivation trees describing concepts within the temporal-abstraction medical KBs accessed by IDAN use only standard international codes, and thus, a KB can be mapped to any clinical database. Medical experts maintain each medical domain's temporal-abstraction knowledge within the KBs using specialized graphical temporal-abstraction knowledge-acquisition and maintenance tools [11].
An incremental, data-driven, continuous-abstraction version of the IDAN architecture, known as Momentum [12], which is highly useful for monitoring purposes and for storing abstractions in a persistent fashion, has also been implemented. A computational-complexity analysis demonstrated a log-linear (if data might arrive out of temporal order) and even linear (if data arrive only in temporal order) complexity of performing the basic incremental abstraction operations in Momentum [13], complementing the polynomial results for the method implemented by RÉSUMÉ and ALMA [6]. Abstractions of multiple patients can be visualized in population-specific exploration tools [13-16].
Artifacts in the displayed data and concepts are handled by the computational abstraction mechanisms, whose detailed description is out of the scope of this paper [3, 6-9]. In certain cases, the temporal interpolation mechanism or a probabilistic extension of it [17] can smooth the data during the abstraction process or complete missing data; in other cases, outliers might still show up in the display and can only be dismissed manually.
1.2 The KNAVE-II Intelligent Interactive Visualization and Exploration Client
KNAVE-II is an intelligent (i.e., knowledge-based) visualization and exploration client that supports formulation of temporal queries using the medical domain's ontology, receives the resulting answers (whether these are in the form of raw data or are concepts, possibly interval-based, that are derived from them) from the mediator, and enables the interactive and dynamic exploration of the results by the user, again exploiting the continuous access to the domain's ontology. For example, a user can graphically formulate all or a part of the query, “Show me the periods of myelotoxicity of Grade II or more, within the first 6 months following the bone-marrow transplantation (BMT) procedure, in the context of therapy by a Prednisone-Azathioprine (PAZ) protocol (a guideline for treatment of chronic graft-versus-host disease following bone-marrow transplantation).” The query defines the parameter (myelotoxicity), parameter value (Grade II), the context (Post-BMT and therapy by a PAZ protocol), and the time span (in this case, the timeline is a relative one: the time points following a particular, patient-specific reference point).
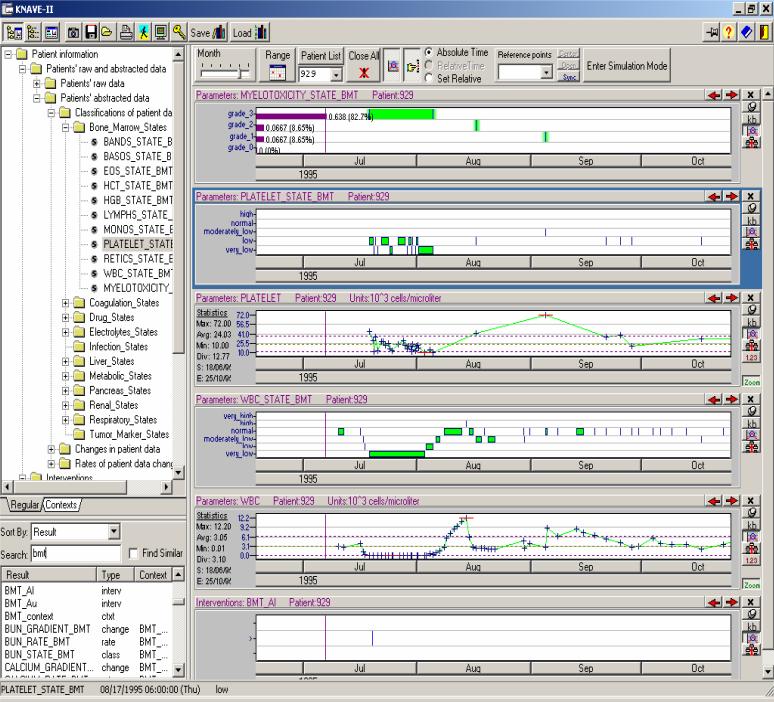
KNAVE-II includes a user-interface module that interacts with the user, and a computational module, which interacts with the temporal-abstraction mediator and processes information obtained from it. Figure 1 displays the KNAVE-II user-interface and explains the semantics of several typical operations performed when exploring patient data.
Figure 1.
The KNAVE-II visualization and exploration system's main interface. The knowledge browser, which shows in this case, the bone-marrow transplantation ontology, is on the left; panels containing raw clinical data and their abstractions are shown on the right. Panels are computed on the fly and displayed when a raw or abstract concept is selected within the left hand browser. Raw data, such as the allogenic bone-marrow transplantation (BMT-AL) event and the white blood cell (WBC) counts, are displayed in the bottom right-hand panels; abstracted concepts, specific to the BMT context, such as WBC States, Platelet-Count States, and Bone-Marrow Toxicity (Myelotoxicity) States, are typically displayed above the corresponding raw data. Selecting a time granule within a panel, such as July 1995, zooms all panels into the next detailed level (e.g., the 31 days of the July 1995). Icons on the right hand side of each panel, such as the “KB” icon, enable further exploration, such as examination of the knowledge defining the concept in that panel, exploration of the concepts it is derived from or that are derivable from it, display of additional statistics regarding relative durations of each value of the concept during a particular time span, etc.
A detailed exposition of the KNAVE-II interface is presented elsewhere [2] and is out of the scope of this study. For this study, it is sufficient to point out that a KNAVE-II user can dynamically view raw data or an abstract concept type by selection of a leaf node (concept) through the domain's hierarchical knowledge browser, or through the free-text search function (see Figure 1). A concept panel (and time line) then opens up to displays the corresponding raw contents of the database or the results of a knowledge-based temporal-abstraction process (see Figure 1). The temporal granularity of the panels (e.g., Days, Months) can be interactively changed using a variety of strategies, based on the temporal scope, the desired content on which the user desires to focus, and whether the relevant time line is absolute or relative to some reference point (e.g., “date of transplantation”) [2] (see Figure 1). Actual values can be read from a scale and also appear in a tool tip when moving over the data; statistics of raw and derived concepts are available as well, as are definitions of the abstracted concepts, supported by accessing the temporal-abstraction KB.
Note that the main interface shown in Figure 1 includes a “patient List” box at the top, whose value (a patient ID) can be changed before clicking on a concept name in the domain's ontology. Therefore, each panel in the main interface corresponds to a combination of a patient and a data type. Thus, multiple patients and multiple abstraction types can be displayed within a single instance of the KNAVE-II interface. However, the KNAVE-II interface is geared towards care providers who view patient data and their abstractions at the point of care. It is thus focused on efficient display of either multiple data types for the same patient, or a small number of raw or abstract data types (e.g., Hemoglobin, Anemia Levels) for a small number of patients (typically up to 5 or 10 at a time). To query and browse thousands of longitudinal patient records and their abstractions and aggregations concurrently, we use a combination of the Momentum system to generate a database of temporal abstractions, and a different interface, the VISITORS system [14-16].
2. BACKGROUND: EVALUATION OF INFORMATION-VISUALIZATION INTERFACES
Visualization and exploration of information in general, and of large amounts of time-oriented data in particular, is essential for effective decision making. Although deceptively intuitive, it is not that obvious, however, what precisely is the value of visualization, especially when combined with abstraction of the data and with interactive manipulation. Larkin and Simon [18] have demonstrated that the usefulness of visual representation is mainly due to (1) the reduction of logical computation through the use of direct perceptual inference, and (2) the reduction of necessary search for information through the use of efficient graphical representation.
Multiple methods have been developed for visualization of data in general, and of time-oriented data in particular, including data within medical domains (a brief overview of this area can be found in our previous study [2]). An excellent treatise on visualization of data is the series of books by Edward Tufte on methods to display information [19, 20]. However, the discussions typically ignore the aspects of (1) [knowledge-based] abstraction of the data, and (2) dynamic interaction, and focus mostly on display of static raw data. Assessment of the utility of the display is not a major focus either.
Several studies have been conducted on the usefulness of information visualization systems. The reports of usability studies and controlled experiments are helpful in understanding the potential and the limitations of the evaluated tools. The usability of software applications is a crucial element for user acceptance. Evaluating usability is important for the software development process, in which system functions and interfacing features are assessed from the user's point of view.
Nielsen [21] has defined five usability dimensions: (1) Learnability: how easy is it to learn the features of a system in order to start using it; (2) Efficiency: how efficient is the system to use; (3) Memorability: how easily can the user recall how to use a system after a period of time; (4) Error recuperation: what is the ability of a system to minimize the number of errors users make while using the system, and enhance their recovery speed after an error; and (5) Satisfaction: what is the subjective users’ satisfaction when using the system. According to Plaisant [22], the usability of information-visualization tools can be measured in a laboratory; however, to be convincing, utility needs to be demonstrated in a real setting, that is, a given application domain and set of users. Choosing and preparing convincing examples demonstrating realistic tasks and using real datasets with more than a few items, are very important. A literature survey by Komlodi et al. [23] confirms this fact by stating that generally accepted and used data sets, tasks, measures, and methods are needed, and that formal user-centered evaluation methods are necessary, if the true utility and effectiveness of information visualization tools are to be assessed. Another problem lies in the fact that comparative studies often report overall performance for a combined set of tasks, although reporting results per task is preferable, as noted by Plaisant et al. in another study [24]. Indeed, Chittaro et al. [25] had applied an integration of two-dimensional and three-dimensional interactive information visualization techniques, named the Interactive Parallel Bar Charts (IPBC) system for visual mining of temporal data, to the management of hemodialysis. The evaluation had led to changes in the system design: Several specific tasks achieved diverse levels of usability.
As Chen and Czerwinski point out in a special issue on empirical evaluations of information-visualization methods [26], the area of information visualization “ ...has began to reach a mature stage of its evolution. The insights and inspirations that one can draw from these studies will no doubt provide valuable forces to stimulate the further development of innovative information visualization techniques and a better understanding of the value of various information visualization techniques.”
A seminal special issue on information visualization in medicine has been edited by Chit-taro [27]. It included descriptions of systems such as the MedView project [28], whose goal is to develop models and tools to support clinicians in their daily diagnostic work, similar to the task for which KNAVE-II was designed, though with a very different underlying computational framework. Another approach described, though with a very different feel and look from KNAVE-II, whose goal is to display longitudinal abstractions, was Info-Zoom [29], which used a novel technique to display multiple data sets as a highly compressed table that always fits completely into the screen.
An approach closer in its spirit to KNAVE-II with respect to overall objectives and visual metaphores is the LifeLines project [30], which provides a general environment for visualization of personal histories. However, it includes no detailed, ontology-based domain-specific knowledge base, and no computational mechanisms that can use such a knowledge base, even if it existed.
Additional interfaces for display and exploration of temporal abstractions, though in different contexts and for somewhat different purposes, have been presented and discussed in detail. Examples include a study whose main objective was to combine temporal data abstraction with data mining to update domain knowledge with specific association rules [31], studies whose main goal is to support monitoring and clinical therapy in high-frequency domains, without using any predefined domain knowledge, but rather by transforming observed data curves (graphs) into a series of lines and bends [32, 33], or querying time-oriented databases based on an object-oriented data model and on the event-calculus [34]. Note that these studies focused on the temporal-abstraction methodology, rather than on interactive visualization and exploration models and on their usability.
Of special note is the study by Chittaro and Combi [35] that evaluated the usability of three different metaphors (e.g., springs and weights) for display of temporal constraints as part of a methodology for specification of clinical temporal patterns. The focus of that study-was not on the display and exploration of the resulting longitudinal patterns. Similarly, Silva et al. [36] designed a formally based environment in which users can use various graphical widgets to define time scales and relationships for purposes of visual temporal queries.
However, both of these tools (and others similar to them) can be better compared to the temporal-abstraction knowledge-acquisition tool mentioned earlier [11], whose goal is to specify basic temporal abstractions and patterns, and even to the query-construction module in the VISITORS multiple-patient exploration system [16] rather than to the goal-driven, real-time-oriented IDAN/KNAVE-II system, whose goal is to exploit an existing domain-specific temporal-abstraction ontology to compute, visualize, and interactively explore a set of raw and derived point- and interval-based concepts, using various types of intuitive interactive temporal zoom-in and zoom-out operators, and navigating along the domain's semantic network (a part of its ontology) to quickly focus on the concepts and periods of interest.
Finally, it is interesting to note several graphical tools in recent years whose objective is to support interactive query and exploration of time-oriented data sets, such as the Time-Finder system, based on a direct-manipulation metaphore [37], and even an ontology-based approach that exploits a repository of meaningful terms or keywords to construct more meaningful queries regarding the data, a very different use of a domain ontology compared to the temporal-abstraction ontology underlying the KNAVE-II and IDAN systems.
In general, however, previously designed information-visualization methods typically either did not focus on visualization of domain-specific temporal abstractions, or did not focus on the issue of real-time interactive manipulation and exploration of the data and multiple levels of its abstractions, using domain-specific knowledge, or did not conduct a rigorous evaluation of the effect of using the tool. Typically, the temporal-abstraction capabilities have been omitted, because they require a formal, domain-independent representation of the domain-specific temporal-abstraction knowledge, considerable effort in modeling the visualized domain, availability of computational mechanisms for creation of the abstractions, and direct links among the display mechanisms, the computational mechanisms, and the domain knowledge base. Similarly, an evaluation of the functionality and usability of the tools by actual users, compared to a predefined gold standard and to existing tools is of course extremely time-consuming.
Such an underlying computational architecture and the necessary links amongst the domain ontology and the computational mechanisms do exist, however, within the combined IDAN/KNAVE-II architecture, and have enabled us to perform a detailed (though, indeed, rather time consuming) investigation of the usability and functionality of a knowledge-based, computationally driven interactive display of time-oriented clinical data and of their abstractions.
As we explain in more detail in the Methods section, our goal in the study was to evaluate the use of KNAVE-II versus the use of standard tools such as paper and an electronic spreadsheet with respect to answering in real time questions common in oncology protocols. Functionality, measured by time and accuracy, was the main yardstick, as well as usability.
Our main research questions were, broadly, therefore: (1) Will the participants answer queries faster when using KNAVE II than when using standard tools? (2) Will they answer questions more accurately? (3) Will they score KNAVE-II as usable at all, and in particular, higher than standard tools, using a standard usability scale? And (4) will they like KNAVE-II more than standard tools, when asked to directly compare them? .
3. METHODS
3.1 Summary of The Evaluation Methodology
We performed two consecutive user evaluations of the KNAVE-II interface, implicitly evaluating also the functionality of the combined IDAN/KNAVE-II distributed architecture: The clinical database and the computational framework (located at Ben Gurion University [BGU], Israel) were 7,400 miles apart from the users of the KNAVE-II interface (situated at the Veterans’ Administration's Palo Alto Health Care System [VAPAHCS], Palo Alto, California, USA).
The first evaluation focused on an assessment of two aspects of the KNAVE-II system's usability. We measured (1) the KNAVE-II system's effectiveness and (2) the users’ satisfaction, with respect to viewing and exploring context-sensitive clinical data to answer a set of typical time-oriented clinical queries extracted from oncology protocols, when referring to a set of data from a group of patients who had undergone bone-marrow transplantation (BMT) and who were monitored for up to four years.
The use of KNAVE-II to explore the data was compared to browsing the same (or comparable) data using two separate standard tools:
a popular electronic spreadsheet (ESS), (Excel™ by Microsoft Co.), which is a standard tool at the clinical environment in which our experiments were conducted, and with which all study participants were familiar; and
a paper chart (“Paper”) produced simply by a sorted printout of the ESS (the columns headed by clinical observation types, the rows headed by the dates, and the cells including actual values).
Both Paper and ESS included also all of the definitions of the knowledge needed to answer the questions (e.g., bone-marrow toxicity grade tables, given the hematological data values such as Platelet counts), organized as a list of tables containing the concept definitions, sorted by concept type (e.g., myelotoxicity) and context (e.g., post-BMT).
These tools, especially since we have organized the clinical data by type and time, and the knowledge by concept and context, actually represent typical or even better formats than the current standard for browsing of clinical data and knowledge in electronic medical records and paper charts.
However, of course, neither the paper nor the ESS provides any intelligent, context-sensitive temporal-abstraction capabilities. Our specific hypotheses regarding functionality and usability (listed at the end of this subsection) directly exploit the implications that we expected based on this observation.
Implicitly, as will be discussed, we had also assessed to some extent the aspects of learn-ability, commitment of errors, and memorability [21] for all tools.
Due to the small number of participants and the concerns of the VAPAHCS's Internal Review Board, we are not listing detailed demographic data. However, we have only included participants who had some level of clinical training (medical student, residents, or fellows), who said that they were very comfortable in using computers, who used computers daily, and who had moderate to high knowledge of using the specific ESS used in the study.
In all cases, we evaluated the users’ ability to view and explore raw data (e.g., hemoglobin levels) and abstractions of the data (e.g., anemia levels) after a short training period in using either KNAVE-II, the ESS, or Paper to answer such questions.
The second evaluation focused and enhanced the results of the first evaluation, while following its methodology. Since, as we will see in the Results section, the results of the first evaluation have indicated that using KNAVE led to significantly better results when answering medium to high complexity queries than when using either the ESS or a paper printout, the second evaluation focused explicitly on more complex queries (medium to high difficulty), using a different set of queries. Due to the consistently better results of users when using the ESS to answer questions about patients as compared to the use of Paper, we compared the use of the KNAVE-II interface in the second study only to the use of the ESS.
3.2 Specific Hypotheses
In both evaluations we tested the following four hypotheses:
Participants will answer queries at all difficulty levels (and, in particular, at the more difficult levels) faster when using KNAVE II rather than when using the ESS or Paper.
Participants will provide more correct answers to the queries of all difficulty levels when using KNAVE II than when using the ESS or Paper.
Participants will assign a higher Standard Usability Score (SUS) [21] to KNAVE II rather than to the ESS or the Paper, as a tool that helps them to answer clinical queries. (See details in the description of the first evaluation.)
Participants will explicitly rank KNAVE II first in the functionality and usability aspects of the “usability of tool comparison” (UTC) comparative questionnaire developed for this study. (See details in the description of the first evaluation.)
3.3 The Method Used in the First Evaluation
The first evaluation was a randomized two-period crossover study [39, 40] in which each participant used KNAVE-II, the ESS, and Paper to answer 10 clinical queries of increasing complexity.
The Efficiency of answering the queries (Hypothesis 1) was measured by the time in seconds required to answer each query.
The Correctness of the possible answers for each query (Hypothesis 2), with respect to the requested clinical value and/or requested time, was pre-determined on a scale of 0 (completely incorrect) to 1 (completely correct) by Dr. Kaizer, an oncology expert and a co-author of this paper.
User satisfaction was assessed by:
the well known and previously validated Standard Usability Score (SUS) questionnaire [41], which provides independent usability scores in the range 0 to 100 for each tool, and thus indirectly compares the tools’ separate scores (Hypothesis 3). The SUS is a Likert scale, i.e., one in which subjects are required to indicate a degree of agreement or disagreement with a set of questions. The SUS includes 10 questions, five of which typically elicit a strong agreement and five of which elicit a strong disagreement, in alternating order, thus forcing subjects to reflect on each item.
The UTC questionnaire, developed for this study, which directly compares all tools by requesting the users to rank the three tools with respect to overall preference for the tool, shortening of the time to find the answers using the tool, ease of finding the answers to clinical queries with the tool, and overall support for finding the answers using the tool (Hypothesis 4). The UTC questionnaire includes, for validation purposes, also a request to provide a relative ranking of the degree of familiarity with the tools.
We anticipated that KNAVE-II's major strength would be in answering queries of higher complexity (hard and hardest queries). A summary of the initial results was previously reported in a conference [42].
The primary outcome of this evaluation was the time to answer queries with the highest levels of difficulty: hard and hardest. Secondary analyses included: (1) the time to answer all queries; (2) the time to answer each query category; (3) the correctness of the answer for each query category; (4) the total correctness score for all queries; (5) the SUS questionnaire's tool-specific scores and (6) the relative rank of each tool in the UTC comparative questionnaire's questions.
We recruited eight subjects by convenience sampling among Stanford University's and VAPAHCS's MD/PhD students, residents, and fellows. Due to the self-contained nature of the queries, clinical expertise about BMT was not required from the subjects. Most had current clinical responsibilities, thus representative of clinical and research settings. As explained in the Summary of The Evaluation Methodology subsection, all subjects were very comfortable with computers and were familiar in the use of both paper charts and the ESS.
To counter-balance a potential sequence effect, in which the use of one tool might have an effect on the performance of the next one to be used (i.e., in the next period of the crossover study) by the same subject, a possible concern in a cross-over study design [39, 40], we randomized the order of the presentation of the tools to the subjects. Since the focus of this evaluation was on KNAVE-II, the Paper and ESS tools were not directly compared; therefore, we kept the order of these two tools fixed, as well as keeping them right after each other (always using Paper, then, following it, ESS) to maximize statistical power when analyzing a possible sequence effect. Thus, in half of the cases, KNAVE-II was used before Paper and ESS; in the other half, it was used after the Paper and ESS tools. The randomization of the use of the tools was unblinded for the participant at the start of each evaluation.
The BMT KB of the underlying IDAN framework, which was also available within the three tools to the users, was elicited in a previous study, using a graphical temporal-abstraction knowledge-acquisition tool [11], from Dr. Kaizer, an oncology expert and a co-author of this paper.
A sample case of a de-identified patient who had had a BMT was selected and then modified slightly to create two additional cases by shifting dates and changing data values. Thus, we had three similar clinical cases on which the same questions (with different values for answers) were asked, one case for each tool.
For each tool, subjects answered the same 10 clinical queries, which had four increasing levels of complexity: three easy, three moderate, three hard and one hardest. Each query might include one or more specific questions, such as both the time and value of a particular raw clinical parameter or derived (abstract) concept, for one or more patients. The type of data abstraction and the requirement to look at different abstractions simultaneously defined the complexity of the queries (Table 1). Easy queries inquired about a value of a single raw laboratory parameter. Moderate queries inquired about simple laboratory abstractions in a temporal relation to an intervention. Abstractions classify raw laboratory parameters into categories, for example high, normal, low. Hard queries inquired about more complex abstractions, based on two distinct laboratory parameters, such as myelotoxicity grade III, defined by the presence of “very low” white blood cell (WBC) count or “very low” platelet count. The hardest query required the clinicians to consider three distinct abstractions simultaneously, including a multiple parameter abstraction such as myelotoxicity.
Table 1.
Examples of the 10 clinical queries used in the first evaluation
| Complexity | Examples of queries |
|---|---|
| Easy | Find the highest value for this patient's serum creatinine between 07/20/95 and 12/31/95. |
| Moderate | During which period did this patient have a “very low” WBC count (defined in KB) post BMT? |
| Hard | What are the dates of the last period of grade 3 myelotoxicity (defined in KB) post BMT? |
| Hardest | Did this patient have a moderately high creatinine, moderately low hemoglobin and grade 2 liver toxicity (defined in KB) after BMT? On which dates? |
The study protocol included an initial training period in the use of the KNAVE-II interface of 10 to 20 minutes. The main features of KNAVE-II introduced during training included using the knowledge browser, the search function, zooming in/out, selecting a chart area, and scrolling the data. Subsequently subjects answered seven practice questions similar to the study queries and issues regarding use of KNAVE-II were clarified.
The evaluation consisted of three segments in which subjects answered 10 clinical queries using KNAVE-II, Paper, and the ESS with short breaks between segments. The queries required subjects to find laboratory data points relative to a single case. At the start of each segment subjects answered two practice questions to clarify issues regarding use of the tool. For Paper and ESS, we provided a printed (as well as electronic, in the case of the ESS tool) BMT KB with definitions of abstractions, such as myelotoxicity. A limit of five minutes was given to answer each query, and 30 minutes to answer all queries for each tool. A stopwatch was used to time the interval between completions of reading the query to completion of writing the answer on developed study forms.
Correctness scores between zero and one (correct) were determined for each query, based on predefined criteria. Where the five minute limit was reached without an answer, time was set to five minutes and correctness was set to zero for analysis. The correctness score was pre-determined by the oncology-domain expert for each answer. A score of zero indicated either an incorrect answer, or that the participant was unable to answer the query in the allotted time. A score of one indicated a correct answer. A score between 0 and 1 indicated that the answer was partially correct. The partial score depended on how close the answer was to the correct answer. Partial credit was determined prior to the analysis and without knowing which tool was being tested.
(Note: although only one expert determined the answers, the answer to all questions regarding the concept values (e.g., toxicity grade) was a simple application of a set of well-defined criteria taken from an oncology protocol, such as bone-marrow toxicity grades. Errors in the dates or durations were given predetermined penalties. There was therefore no need for additional experts other than the single senior expert who painstakingly looked at the data using all three tools, and who determined what the answers to the standard protocol-based queries should be. Furthermore, the expert's calculations were actually re-checked by several of the physicians in the research team, without any notable changes.)
Subjects completed SUS tool-specific questionnaires at the end of each segment and the UTC comparative questionnaire at the end of the three segments.
The data were analyzed using repeated measures analysis of variance, in which each subject acted as their own control, preceded by a test for a sequence effect. A sequence effect is an interaction between treatments and periods, and is often a concern when designing cross-over studies [39, 40]. When such an effect is present, the difference between the two treatments varies according to the order in which the two treatments are given.
Correctness scores and SUS questionnaires were analyzed using a paired t-test.
3.4 The Method Used in the Second Evaluation
A total of five physicians participated in the second evaluation, whose protocol and measures essentially followed the one used in the first evaluation. Four of the five participants had participated in the first study. The fifth was administered questions from the first evaluation so as to be to equally familiar with the new tool before participating in the second evaluation. As in the first evaluation, all participants had a medical background, and were proficient in the use of computers in general and in the ESS tool in particular.
Study participants were asked to answer six queries of increasing difficulty (Table 2) using both KNAVE-II and the ESS. The queries were at a moderate to a very hard level of difficulty. The levels were comparable in their semantics to those used in the first evaluation; thus, the two new, somewhat harder levels were referred to as “Hardest+1” and “Hardest+2”. Overall, the queries were considered by the domain expert (Dr. Herbert Kaizer, a co-author of this paper) to be more representative of queries that clinicians encounter and need assistance with in oncology protocols. The correctness scores were determined using a predetermined gold standard for concept values and dates, as in the first evaluation. Study participants were randomized to determine which tool they used first. Participants were given up to 30 minutes to answer all 6 queries when using KNAVE-II or the ESS. In the analysis, for those participants who ran out of time, the time was set to the average time that participant used to answer the previous queries using the ESS and the correctness score was set to zero (incorrect). This methodology was in fact highly conservative, as query six was the most difficult query and therefore expected to take much longer to answer than the other five queries.
Table 2.
The six queries used in the 2nd evaluation
| Query difficulty | Query | |
|---|---|---|
| 1 | Moderate | After the BMT, what was the longest period (give dates and number of days) that patient 911 had a “moderately low” WBC? |
| 2 | Hard | After the BMT, what was the longest period of grade 3 liver toxicity for patient 946? Give the number of days and dates. |
| 3 | Hard | Did patient 813 have a very high alkaline phosphatase and a high LDH on the same date(s) after the BMT? If so, how many times and on which dates? |
| 4 | Hardest +1 | For patients 813 and 946: After the BMT, did these patients have a pattern of “liver dysfunction”? If so, when was the last date? |
| 5 | Hardest +1 | Did patients 813, 946 and 929 recover from their myelotoxicity (recovery is defined as myelotoxicity grade 0)? If so, how long after BMT? Give the date and number of days from BMT to recovery. Which patient recovered in the shortest time after BMT? |
| 6 | Hardest +2 | Did patients 911, 929 and 946 develop simultaneous grade 3 mye-lotoxicity and grade 3 liver toxicity after their BMT? If yes, when? |
Analysis of Variance (ANOVA) was used to analyze differences in time required to answer the six queries using KNAVE-II versus the ESS. Prior to performing the analysis, a test for sequence effects was performed.
4. RESULTS
4.1 Results of the First evaluation
Subjects answered the practice questions without difficulty. Several subjects had problems using the search function and chart selection features due to the lack of familiarity with the full capabilities of KNAVE-II; however, due to the intentional redundancy inherent in the KNAVE-II interface, subjects were able to eventually use other features to adequately explore the data.
Prior to the main analysis, a standard test for sequence effects [40] was performed, as part of the multiple-variable Analysis of Variance (ANOVA) test, by examining the effect of the period variable (i.e., using the tool by the same subject within the first or second period in the cross-over study) to detect the presence of any sequence effect. There were no sequence effects when comparing KNAVE-II to Paper or to ESS.
(Note: The significance level threshold was 0.05 throughout, but we tried to list an actual p value, when the result was significant, whenever it was reasonable to do so, as it allows the readers to better judge how significant each result was.)
No subject reached the 30 minute limit. Subjects answered the hard and hardest queries, our primary outcome, significantly faster when using KNAVE-II than when using either Paper or the ESS (p = 0.007 and p = 0.0006, respectively, when compared to ESS) (Table 3).
Table 3.
ANOVA results for KNAVE-II vs Paper and KNAVE-II vs the ESS for difference in mean time taken to answer queries in the first evaluation
| KNAVE-II vs. Paper | ||||
|---|---|---|---|---|
| Difference in mean times per query | 95% CI | F-value (df) | p-value | |
| Easy | 16 sec | 5, 27 | 3.43 (1) | 0.07 |
| Moderate | −21 sec | −31, −1 | 9.21 (1) | 0.004 |
| Hard | −69 sec | −97, −40 | 24.17 (1) | 0.00002 |
| Hardest | −93 sec | −145, −41 | 15.45 (1) | 0.008 |
| All queries | −31 sec | −45, −18 | 8.09 (1) | 0.005 |
| KNAVE-II vs. ESS | ||||
|---|---|---|---|---|
| Difference in Mean Times per Query | 95% CI | F-value (df) | p-value | |
| Easy | 21 sec | 8, 34 | 5.85 (1) | 0.02 |
| Moderate | 8 sec | −1, 18 | 2.06 (1) | 0.16 |
| Hard | −27 sec | −46, −8 | 8.21 (1) | 0.007 |
| Hardest | −49 sec | −66, −31 | 41.94 (1) | 0.0006 |
| All queries | −4 sec | −13, 5 | 0.15 (1) | 0.7 |
Pooling times for all queries, subjects answered queries faster when using KNAVE-II than when using Paper. There was no difference in time to answer all queries, when pooled together, when comparing KNAVE-II to the ESS.
Easy queries were answered faster in the ESS and had a slight trend of being answered faster in Paper when compared to KNAVE-II. Moderate queries were answered faster using KNAVE-II than Paper. There was no difference in time to answer moderate queries when comparing KNAVE-II to the ESS (Table 3)
Mean correctness scores for query categories answered with KNAVE-II, Paper, and ESS, were compared using paired t-test (Table 4). KNAVE-II had significantly higher correctness scores for all queries (p=0.01), and for hard queries (p=0.04) when compared to Paper. KNAVE II had higher correctness scores than the ESS for the hard and hardest queries and for all queries, but the results were not statistically significant.
Table 4.
Average correctness score per query category in the first evaluation
| Average Correctness Score | |||
|---|---|---|---|
| KNAVE-II | Paper | ESS | |
| Easy | 0.917 | 0.875 | 1 |
| Moderate | 0.994 | 0.997 | 0.958 |
| Hard | 0.917 | 0.792 | 0.885 |
| Hardest | 0.700 | 0.225 | 0.563 |
| All | 0.918 | 0.822 | 0.909 |
With respect to usability, the average SUS questionnaire score for KNAVE-II was 69.1, for Paper 46.3 and for the ESS 48.1. Based on a paired t-test of the difference in mean SUS scores, KNAVE-II's usability was significantly superior to both Paper (23, 95% CI = 9 to 37, p=0.006) and the ESS (21, 95% CI = 8 to 34, p=0.006). Note that these scores represent an average improvement in the score assigned to KNAVE-II of 50% over Paper (a difference of 23 points from a baseline of 46) and 43.75% over ESS (a difference of 21 points from a baseline of 48).
The UTC comparative ranking provided the following results: All subjects ranked KNAVE-II first in their preference; for overall support for finding answers. 7/8 (88%) subjects ranked KNAVE-II first in ease of finding answers to clinical queries. 6/8 (75%) subjects ranked KNAVE-II first in time to find answers to clinical queries. In terms of experience or familiarity with the tool, 63% (5/8) ranked Paper first and 37% (3/8) ranked the ESS first, thus providing evidence for the validity of the UTC scale and confirming that the users were indeed paying attention to each item in it (see the subsection The Method Used in the First Evaluation for the UTC scale description).
4.2 Results of the Second evaluation
Prior to performing the analysis, a test for sequence effect [40] was performed (again, by examining the period (1st, 2nd) variable as part of the ANOVA test). The test for sequence effects showed no sequence effect to be present. When performing the ANOVA, we noted that the overall period effect (i.e., whether the tool was used in the 1st or 2nd period) was close to significant (p=0.08), with subjects answering queries an average of 39 seconds faster in the second period (regardless of whether they used KNAVE first or the ESS first).
The five participants were able to answer all five queries within 30 minutes when using KNAVE-II. One study participant ran out of time on query 5 when using the ESS. Three of the four remaining participants ran out of time on query 6 when using the ESS. Analysis of Variance (ANOVA) was again used to analyze differences in time required to answer six queries using KNAVE-II versus the ESS (Table 5). Participants answered all queries significantly faster using KNAVE-II when compared to the ESS. There was no significant subject effect in the ANOVA; that is, none of the subjects using the tools was significantly different in their performance from the other subjects.
Table 5.
ANOVA results for KNAVE-II vs the ESS for difference in mean time taken to answer queries in the second evaluation
| KNAVE-II vs. ESS | ||||
|---|---|---|---|---|
| Types of Queries | Difference in mean times per query (sec) | 95% CI (sec) | F-value (df) | p-value |
| Moderate query (query 1) | −69 | −98, −40 | 42.50 (1) | 0. 007 |
| Hard (queries 2 & 3) | −150 | (−240, −60) | 13.96 (1) | 0.002 |
| Hardest+1 (queries 4-5) | −255 | (−368, −142) | 12.98 (1) | 0.003 |
| Hardest+2 (query 6) | −49 | (−422, 345) | 0.10 (1) | 0.77 |
| All queries | −155 | (−222, −88) | 14.76 (1) | 0.0003 |
When analyzing by query category, except for query 6 (Hardest +2), all categories were answered significantly faster using KNAVE-II (Table 5). There was no significant subject, nor period effect. The ANOVA results for the Hardest+2 Query (query 6) were not significant. However, four of the five participants ran out of time on query 6 when using the ESS; none ran out of time using KNAVE. The treatment coefficient (i.e., the difference in response time due to using KNAVE-II) was 49 seconds faster, although with a very broad 95% confidence interval of -422 to 345 seconds, for the hardest+2 query. Note that we used highly conservative, probably too conservative, estimates for the time needed to answer query 6 for the four subjects who ran out of time when answering that query (i.e., by replacing the time needed to answer query 6 by the average time to answer the significantly easier query 5). Nevertheless, the mean difference in time for answering queries was 155 seconds faster with KNAVE-II (p = 0.0003).
Answers to the Hardest+1 and Hardest+2 queries were significantly more correct when using KNAVE-II as compared to the ESS. The correctness counts are also statistically significant for all queries.
We also analyzed closely all of the questions and actual number of possible answers to them included within the various queries of the second evaluation. For example, query No. 4 actually asks two questions per patient for two different patients: Was there liver dysfunction, and if so, When exactly; in this case, the answer happened to be Yes to both questions and thus, a date is also expected for both; therefore, overall, 4 answers were expected to this particular query (If the answer happened to be No, we would not expect another answer regarding a date). Performing this analysis, altogether 91.6% (110/120) of the questions produced correct answers when using KNAVE-II, versus an accuracy of only 57.5% (69/120) when using the ESS (p<0.0001) (Table 6). The mean correctness scores for each type of query and 95% confidence intervals for the difference in correctness scores were higher when using KNAVE-II than when using the ESS for all categories except the moderate query, based on a t-test (not paired, since most of the participants did not finish answering all of the questions using both tools). The results were also statistically significant for all queries. The mean difference in the correctness score between queries answered using KNAVE-II and the ESS was 0 for the moderate difficulty query, 0.08 for the Hard queries, 0.34 for the Hardest+1 queries, and 0.8 for the Hardest+2 query. Overall, the mean difference in correctness was 0.36 (P<0.0001).
Table 6.
Total number of correct, partially correct, incorrect, and interrupted answers to all questions for queries at all difficulty levels in the second evaluation, for the 5 subjects
| Query type (total no. of questions for query type) | Mean difference in correctness score between KNAVE-II and the ESS | 95% CI | Correctness level | Knave | ESS | Fisher's exact p-value |
|---|---|---|---|---|---|---|
| Total correct | 8 | 7 | ||||
| Moderate (2 [1+1]) | 0 | Total partial | 0 | 0 | p=1 | |
| (−0.40, 0.40) | Total wrong | 2 | 3 | |||
| |
|
|
Interrupted |
0 |
0 |
|
| Total correct | 23 | 22 | ||||
| Hard (5 [2+3]) | 0.08 | (−0.08, 0.24) | Total partial | 1 | 0 | p=0.61 |
| Total wrong | 1 | 3 | ||||
| |
|
|
Interrupted |
0 |
0 |
|
| Total correct | 57 | 38 | ||||
| Hardest+1 (12 [4+8]) | 0.34 | (0.20, 0.47) | Total partial | 0 | 0 | p<0.0001 |
| Total wrong | 3 | 14 | ||||
| |
|
|
Interrupted |
0 |
8 |
|
| Total correct | 22 | 2 | ||||
| Hardest+2 (5 [5]) | 0.80 | (0.63, 0.97) | Total partial | 0 | 1 | p<0.0001 |
| Total wrong | 3 | 2 | ||||
| |
|
|
Interrupted |
0 |
20 |
|
| Total correct | 110 | 69 | ||||
| Total | 0.36 | (0.27, 0.47) | Total partial | 1 | 1 | p<0.0001 |
| Total wrong | 9 | 22 | ||||
| Interrupted | 0 | 28 |
The average SUS questionnaire score for KNAVE-II was 64 (range 45-72.5, median 67.5), and for the ESS 45 (range 42.5-50, median 42.5). Based on a paired t-test of the difference in mean SUS scores, KNAVE-II's usability was significantly superior to the ESS (p = 0.011; 95% CI = 7 to 31) .
The UTC comparative ranking provided the following results: all subjects ranked KNAVE-II first with respect to overall preference, easiness to find answers to clinical queries, and time to find answers to clinical queries. 4/5 (80%) ranked KNAVE-II first in the overall support for finding answers. In terms of experience or familiarity with the tool, 80% (4/5) ranked the ESS first and 20% (1/5) ranked KNAVE-II first, validating again that the users paid attention to the various items of the UTC questionnaire.
5. DISCUSSION
5.1 Summary of the Results
To the best of our knowledge, this is one of the first cases in which an evaluation of an advanced medical decision-support system for intelligent interpretation and exploration of patient data, involving actual clinicians and patient records, and a full-fledged human-computer interaction (HCI) methodology, has been performed. Both functionality and usability, using separate, well defined measures, were assessed. We hope this study will be followed by additional ones, since a common issue raised in this area is the lack of sufficient evaluations and the lack of use of established HCI evaluation methodologies.
The study, focusing on an evaluation of the usability of intelligent (i.e., knowledge-based) visualization and exploration of longitudinal patient data, is different from previous studies focusing on the computation or detection of temporal abstractions, as well as from studies evaluating various creative metaphors for defining temporal constraints [35] or temporal queries [36]. As noted in the Background section, such studies might actually be compared to the temporal-abstraction knowledge-acquisition tool, through which the IDAN KB was elicited [11], rather than to the runtime environment of the KNAVE-II and IDAN frameworks.
Study participants were representative of clinicians with clinical responsibilities and good computer skills. Once we completed the first evaluation, we knew that KNAVE-II was more efficient, more accurate, and preferred by clinicians to explore clinical queries of higher complexity when compared to a printed chart. When compared to the ESS, KNAVE-II was also more efficient and was preferred by clinicians when answering clinical queries of higher complexity, without losing any accuracy, in spite of the shorter time used by KNAVE-II users for answering the queries, and in fact enhanced the accuracy when answering the more complex queries. The second evaluation therefore focused on increasing the complexity of the clinical queries, in particular, by requesting the users to identify specific clinical patterns defined by multiple abstractions (which are perhaps more representative of the needs of care providers using complex guidelines or protocols). In the second evaluation, the pattern noticed in the first evaluation was enhanced, demonstrating significantly faster speed and improved accuracy when using KNAVE-II versus the ESS.
In either evaluation phase, as the level of difficulty of the queries increased, the speed-up in response time for users answering the queries using KNAVE-II, relative to using either Paper or the ESS, kept increasing. The mean time for all difficulty levels was also significantly shorter.
In the first evaluation, there was actually no advantage to the use of KNAVE-II (or even a slight disadvantage) for answering queries at the easiest (in the case of Paper) or easiest and moderate (in the case of the ESS) levels of difficulty. This was probably due to the fact that, after only a few minutes of training with KNAVE-II, it was easier for the clinicians to simply look up, for example, the value of a single clinical parameter, or its maximal value within a sorted table column in the (electronic or printed) spreadsheet. However, applying the more demanding calculations needed for context-sensitive longitudinal clinical interpretations, which often appear within clinical guidelines and protocols for care of chronic patients, immediately brought to light the advantage of having an underlying temporal-abstraction framework and a time-oriented browsing interface. Furthermore, based on the experience of the physicians involved in the design of this study, we believe that once familiar with the KNAVE-II tool, even easy to moderate queries are answered faster by using the intuitive select-concept-and-zoom-within-timeline interface than by searching within a spreadsheet table.
The main advantage of using the new tool in the current study was apparent when users attempted to answer the more complex queries. Recall that for those participants who ran out of time on a query, which in the case of the second evaluation happened only when using the ESS, the time was set to the average time that participant used to answer the previous query using the ESS, and the correctness score was set to zero (incorrect). Although our goal here was to be conservative, this treatment was probably far too conservative, as query six was the most difficult query and therefore was expected to take much longer to answer than the other five queries.
Nevertheless, although we used this highly conservative methodology in the second evaluation to assign a time for users who ran out of time on the very hardest query, the mean difference in time for answering queries in the second evaluation was still 155 seconds faster when using KNAVE-II (p = 0.0003) than when using the ESS.
At the same time, 91.6% (110/120) of the total number of questions posed within queries of all levels of difficulty that were asked in the second evaluation, produced correct answers when using KNAVE-II, as opposed to only 57.5% (69/120) when using the ESS (p<0.0001). Obviously, time is not the only (or even most important) issue when answering complex clinical queries; users had significant trouble answering correctly common (though complex) clinical queries appearing in the context of oncology protocols.
As we noted in the Methods section, only one expert was needed to determine ahead of time what the answers should be, since they consisted of an application of well defined oncology protocols’ classification tables and predefine penalties for incorrect dates or durations. In fact, it is interesting to note that after the study there were actually no arguments regarding the “True” answers to all of the questions, once the participants were shown where they committed errors and had sufficient time to recompute their answers. Indeed, due to the cross-over nature of the study, all participants had an opportunity to be trained in and to use the KNAVE-II tool, and thus could much more easily verify these answers by comparing the raw data and abstract concepts panels within KNAVE-II; perhaps reaching a quick consensus is another advantage of using an integrated temporal-abstraction computational framework and an ontology-driven display.
Of notice is the fact that we did not train the subjects in the use of many of KNAVE-II features to facilitate tasks such as semantic navigation among concepts or answering of multiple questions in parallel, due to feasibility constraints such as keeping the duration of the time allocated to the training phase to a minimum. We also did not evaluate other major features of the KNAVE-II system that are useful for exploration of highly complex data abstractions [2]. Indeed, in the very short time allocated to the training, we focused mostly on the features enabling selection of the relevant clinical concept in the knowledge browser, and on the temporal zoom-in and zoom-out operators, which enable users to quickly focus on relevant time periods of points. Nevertheless, these features sufficed to enable the clinicians to answer all of the clinical queries in this study accurately (and quite fast), demonstrating the power of having a domain-specific knowledge base and a set of computational mechanisms underlying the exploration interface.
Although the studies included a small number of participants and one specific clinical domain, the results seem quite encouraging in light of the short time given the physicians for training. Furthermore, the functionality of the IDAN/KNAVE-II distributed architecture was clearly demonstrated: indeed, a 7,400 mile distance existed between the KNAVE II client interface and the computational modules. The response time to commands was in ‘real time’ and problems with ‘freezing’ or with a similar behavior were rare.
When assessing the significance of the results, it is important to note that clinicians with active clinical practice, none of whom specialized in the bone transplant domain, were able to answer clinical queries of higher complexity faster, more accurately and with higher user satisfaction when using KNAVE-II than when using paper charts or an ESS familiar to them. This result is important when considering the current trend towards empowerment of the general practitioner, who is often the only care provider with a clear overall view of the patient's consultation with multiple specialized experts.
5.2 Limitations of the Study
Although the IDAN/KNAVE-II framework was in fact applied, in real time, to a full clinical database, the evaluation, for methodological reasons, used only variations on one representative patient record. Since the main objective in this study was to assess the functionality and usability of the IDAN/KNAVE-II architecture for browsing individual patient data, and since indeed the same conditions held for paper charts and the ESS, we consider this choice reasonable.
One limitation of this study was the short training period. If the subjects had more experience using KNAVE-II, they would probably have been able to browse the data even more efficiently. As with any software, a power user will always be a more efficient user, both because of familiarity with the software interface and because of knowledge about features that enhance efficiency. (One potential remedy in the KNAVE-II interface, though, is that users can transfer knowledge and expertise from the use of other software programs that use similar metaphores, for example when using the search function.)
The fact that the domain concepts currently in the knowledge base did not include all possible synonyms forced several subjects to try the search function several times before going to the knowledge browser to find the concept, thus increasing their time to answer queries. Such particular limitations, however, are incidental and can be easily fixed in a future version of the oncology KB.
Another limitation was that although the evaluation was conducted within a hospital setting, it was rather close to a laboratory setting, since the pressure of a real patient encounter was not part of the conditions. Using KNAVE-II within actual clinical practice may prove even more beneficial in effectively detecting patterns that could lead to clinical interventions. Integrating KNAVE-II into a clinician's workflow requires further research, and is on our agenda.
Finally, due to the small number of participants in both studies, whose main objectives were to test the functionality and usability of the IDAN/KNAVE-II systems, it was difficult to perform any clustering of the results by subject features, such as gender and clinical training level, even if the anonymity constraints had been relaxed and we had been allowed to list them; we would not want to draw any definite conclusions in this limited study regarding any relationship, if at all, between any results and the features of the participants. Such a relationship might be included in a future study that will include many more participants.
5.3 Strengths of the Study
The evaluation of KNAVE-II's usability as a performance metric was comparable to standard interface evaluations performed in the HCI research area. The results were quite positive, providing objective evidence of the value of intelligent user interfaces to clinicians.
Furthermore, implicitly we had evaluated to some extent also the learnability, memorability and error recuperation capability components of usability, as described by Nielsen [15]. Subjects learned to navigate the KNAVE II interface effectively, at least sufficiently to answer the evaluation queries, in only 10 to 20 minutes; remembered how to use KNAVE II after starting the evaluation with Paper and the ESS (approximately a 60 minute gap); and were able to recuperate from mistakes when browsing KNAVE II because of the intentional redundancy built in the interface design. Although it is a client application interacting with the IDAN server, KNAVE-II is built with interface options similar to those used in modern web-based and other software applications. For example, the search tool for clinical concepts in the patient's medical record to be displayed enables a partial search for text and has an integrated synonym recognition feature as is common in other applications. Browsing the knowledge tree uses the familiar ‘+’ icon to indicate additional options under a class. Scroll bars, calendar and intuitive buttons help clinicians navigate the tool effectively. Of note is the fact that the co-author (Dr. S.B. Martins) who performed the two experiments with the physicians noticed that they had used many different patterns of navigation within the KNAVE-II interface to answer the same queries; apparently, the intentionally built in redundancy in the KNAVE-II interface design allowed for multiple ways to search for the same clinical data or its interpretations (abstractions).
In terms of efficiency, subjects using KNAVE-II answered clinical queries with a higher level of complexity faster and more accurately then when using Paper and faster when using the ESS, without loss of accuracy, in the first evaluation. In the second evaluation, this pattern was strengthened by increasing both speed and accuracy when comparing KNAVE-II to the ESS. Being able to find an answer for relevant complex clinical queries (such as occur within oncology and other protocols) quickly and accurately when browsing a patient's record has a significant potential to be highly beneficial to patient care and to patient safety.
User satisfaction was significantly superior for KNAVE-II, based on the SUS questionnaire results and of their ranking of overall preferences in the UTC comparative questionnaire.
Of note is that what we refer to in this study as “queries with a higher level of complexity” were actually quite simple relative to KNAVE-II's and the underlying IDAN system's capability for abstraction and exploration. The IDAN/KNAVE-II architecture has the capacity to detect and display abstractions with a much higher level of complexity, such as patterns of daily variability of glucose over time. These complex temporal patterns and abstractions represent clinically meaningful concepts defined in different domains, thus potentially supporting clinicians in disease management, quality assessment and research.
5.4 Conclusions
We consider this two-phase study, although including a relatively small number of clinicians, to be a useful milestone in the long path towards a routine use of effective, usable, clinically significant knowledge-based artificial intelligence systems in medical care.
Study participants using the combined IDAN/KNAVE-II knowledge-based architecture for visualization and exploration of large amounts of time-oriented clinical data, and of multiple levels of clinically meaningful abstractions derivable from these data, answered high-complexity clinical queries significantly faster and more accurately with KNAVE-II than with Paper or a popular ESS. Study participants found KNAVE-II a superior tool to use when answering the clinical queries as compared to the standard of care, either Paper or an ESS. This evaluation focused on use of simpler features and concepts of KNAVE-II.
5.5 Future Directions
One of our goals in this study was to prepare the way for a larger scale future clinical study of the IDAN and KNAVE-II systems, focusing on the task of patient treatment at the point of care, in particular when providing automated supported to guideline-based care within a more comprehensive framework. An example of such a framework is the DeGeL project's digital guideline library [43], and in particular, tools such as the Spock runtime guideline-application engine [44], which interacts with both the DeGeL library and the IDAN mediator to the patient's electronic medical record, to provide support to the care provider.
as explained in the Introduction section, KNAVE-II as well as its underlying IDAN temporal-abstraction mediator and ALMA temporal-abstraction computational tool are geared mostly towards the goal-directed, effective computation and display of temporal abstractions of individual patient records or of a small number of such records. To compute abstractions of multiple patients, or to support continuous, data-driven monitoring of such abstractions, the Momentum architecture [12, 13] might be the method of choice. Augmenting such a framework with the VISITORS multiple-record query and exploration system [14-16] would enable clinical researchers, administrators and quality assessors to continuously monitor large longitudinal patient populations. We have in fact initiated an evaluation of the VISITORS’ framework usability, assessing both graphical query construction and interactive exploration of abstractions and their time-oriented associations.
The current study, for the reasons explained earlier in this paper, did not examine the relationship between user features, such as gender, age, and levl of clinical training, and levels of performance. However, such a relationship might be included in a future study examining similar time and accuracy measures, but that will include many more subjects and their respective features.
As was noted, due to the very short time allocated to the training of the subjects, only a small (but important) subset of the KNAVE-II tool's features was introduced to the users. An assessment of the full potential of this application, using existing advanced features such as query profiles that are set by the user and dynamic sensitivity analysis, which are an essential part of the KNAVE-II tool [2], may prove in a future study even more meaningful for supporting disease management, quality management and clinical research.
Finally, given the trend for increasing time and accuracy gaps in favor of the KNAVE-II tool that was noted in the two evaluations reported here, a future evaluation of the full features of KNAVE II using queries of even higher complexity, such as those that arise in oncology protocols and in experimental clinical trials, might well result in even higher efficiency and user satisfaction improvement, when compared with standard tools.
Acknowledgements
This research was supported in part by NIH award No. LM-06806. We thank Drs. Aneel Advani, Eitan Lunenfeld, and Laszlo Vaszar for their participation in the studies and for their comments on the KNAVE-II interface, Dr. Meirav Taieb-Mimon for her comments regarding the analysis, and Mr. Denis Klimov for his comments on information visualization. The views expressed are those of the authors and not necessarily those of the Dept. Veterans Affairs or other affiliated organizations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goldstein M, Hoffman B. Graphical displays to improve guideline-based therapy of hypertension. In: Izzo JL Jr., Black HR, editors. Hypertension Primer. 3rd ed. Lippincot, Williams & Wilkins; Baltimore: 2003. [Google Scholar]
- 2.Shahar Y, Goren-Bar D, Boaz D, Tahan G. Distributed, intelligent, interactive visualization and exploration of time-oriented clinical data. Artificial Intelligence in Medicine. 2006;38(2):115–135. doi: 10.1016/j.artmed.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Boaz D, Shahar Y. A distributed temporal-abstraction mediation architecture for medical databases. Artificial Intelligence in Medicine. 2005;34(1):3–24. doi: 10.1016/j.artmed.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 4.Shahar Y, Cheng C. Intelligent visualization and exploration of time-oriented clinical data. Topics in Health Information Management. 1999;20(2):15–31. [PubMed] [Google Scholar]
- 5.Shahar Y, Cheng C. Model-Based Visualization of Temporal Abstractions. Computational Intelligence. 2000;16(2):279–306. [Google Scholar]
- 6.Shahar Y. A framework for knowledge-based temporal abstraction. Artificial Intelligence. 1997;90(1-2):79–133. doi: 10.1016/0933-3657(95)00036-4. [DOI] [PubMed] [Google Scholar]
- 7.Shahar Y, Musen M. Knowledge-based temporal abstraction in clinical domains. Artificial Intelligence in Medicine. 1996;8(3):267–298. doi: 10.1016/0933-3657(95)00036-4. [DOI] [PubMed] [Google Scholar]
- 8.Chakravarty S, Shahar Y. Specification and detection of periodicity in clinical data. Methods of Information in Medicine. 2001;40:410–420. [PubMed] [Google Scholar]
- 9.Chakravarty S, Shahar Y. CAPSUL: A Constraint-Based Specification of Repeating Patterns in Time-Oriented Data. Annals of Mathematics and Artificial Intelligence. 2000;30:3–22. [Google Scholar]
- 10.Balaban M, Boaz D, Shahar Y. Applying temporal abstraction in medical information systems. Annals of Mathematics, Computation, and Teleinformation. 2004;1(1):54–62. [Google Scholar]
- 11.Shahar Y, Chen H, Stites D, Basso L, Kaizer H, Wilson D, Musen M. Semiautomated acquisition of clinical temporal-abstraction knowledge. Journal of the American Medical Informatics Association. 1999;6(6):494–511. doi: 10.1136/jamia.1999.0060494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spokoiny A, Shahar Y. An active database architecture for knowledge-based incremental abstraction of complex concepts from continuously arriving time-oriented raw data. The Journal of Intelligent Information Systems. 2007;28(3):199–231. [Google Scholar]
- 13.Spokoiny A, Shahar Y. Incremental application of knowledge to continuously arriving time-oriented raw data. The Journal of Intelligent Information Systems. in press. [Google Scholar]
- 14.Klimov D, Shahar Y. A framework for intelligent visualization of multiple time-oriented medical records.. Proceedings of the American Medical Informatics Fall Symposium (AMIA-05); Hanley and Belfus, Philadelphia. 2005; pp. 405–9. [PMC free article] [PubMed] [Google Scholar]
- 15.Klimov D, Shahar Y. Intelligent visualization of temporal associations for multiple time-oriented patient records.. Notes of the 11th International Workshop on Intelligent Data Analysis in Medicine and Pharmacology (IDAMAP-2007); Amsterdam, The Netherlands. 2007. [Google Scholar]
- 16.Klimov D, Shahar Y. Intelligent querying and exploration of multiple time-oriented medical records.. Proceedings of the Twelfth World Congress on Medical Informatics (MEDINFO-07); Brisbane, Australia. IOS Press, Amsterdam. 2007; pp. 1314–8. [PubMed] [Google Scholar]
- 17.Ramati M, Shahar Y. Probabilistic abstraction of multiple longitudinal electronic medical records.. Proceedings of the 10th Conference on Artificial Intelligence in Medicine—Europe (AIME ‘05); Aberdeen, Scotland. Springer, Berlin. 2005; Lecture Notes in Artificial Intelligence 3581. [Google Scholar]
- 18.Larkin J, Simon H. Why a diagram is (sometimes) worth ten thousand words. Cognitive Science. 1987;11:65–99. [Google Scholar]
- 19.Tufte E. Envisioning Information. Graphics Press; Connecticut: 1990. [Google Scholar]
- 20.Tufte E. Visual Explanations. Graphics Press; Connecticut: 1997. [Google Scholar]
- 21.Nielsen J. Usability Engineering. 1st ed. Morgan Kaufmann; San Francisco: 1994. [Google Scholar]
- 22.Plaisant C. The Challenge of Information Visualization Evaluation.. Proceedings of The 7th International Working Conference on Advanced Visual Interfaces (AVI 2004); Gallipoli, Italy. ACM Press, New York. 2004. [Google Scholar]
- 23.Komlodi A, Sears A, Stanziola E. ISRC Technical Report UMBC-ISRC-2004-1. Dept. of Information Systems, University of Maryland; Baltimore County: 2004. Information visualization evaluation review. [Google Scholar]
- 24.Plaisant C, Grosjean J, Bederson BB. SpaceTree: Supporting exploration in large node-link tree: design evolution and empirical evaluation.. Proceedings of the IEEE Symposium on Information Visualization; IEEE Computer Society Press, Washington DC. 2002.pp. 57–64. [Google Scholar]
- 25.Chittaro L, Combi C, Trapasso G. Data mining on temporal data: a visual approach and its clinical application to hemodialysis. Journal of Visual Languages & Computing. 2003;14(6):591–620. [Google Scholar]
- 26.Chen C, Czerwinski M, editors. Introduction to the Special Issue on Empirical evaluation of information visualizations. International Journal of Human-Computer Studies. 2000;53(5):631–635. [Google Scholar]
- 27.Chittaro L. Chittaro L, editor. Information visualization and its application to medicine. Artificial Intelligence in Medicine. 2001;22(2):81–88. doi: 10.1016/s0933-3657(00)00101-9. Special issue on Information Visualization in Medicine. [DOI] [PubMed] [Google Scholar]
- 28.Falkman G. Chittaro L, editor. Information visualisation in clinical Odontology: multidimensional analysis and interactive data exploration. Artificial Intelligence in Medicine. 2001;22(2):133–158. doi: 10.1016/s0933-3657(00)00104-4. Special issue on Information Visualization in Medicine. [DOI] [PubMed] [Google Scholar]
- 29.Spenke M. Chittaro L, editor. Visualization and interactive analysis of blood parameters with Info-Zoom. Artificial Intelligence in Medicine. 2001;22(2):159–172. doi: 10.1016/s0933-3657(00)00105-6. Special issue on Information Visualization in Medicine. [DOI] [PubMed] [Google Scholar]
- 30.Plaisant C, Mushlin R, Snyder A, Li J, Heller D, Shneiderman B. LifeLines: Using Visualization to Enhance Navigation and Analysis of Patient Records. In: Bederson B, Shneiderman B, editors. The Craft of Information Visualization. Morgan Kaufmann; San Francisco: 2003. pp. 308–312. [Google Scholar]
- 31.Silvent A-S, Dojat M, Garbay C. Multi-level temporal abstraction for medical scenario construction. International Journal of Adaptive Control and Signal Processing. 2005;19(5):377–394. [Google Scholar]
- 32.Miksch S, Horn W, Popow C, Paky F. Utilizing temporal data abstraction for data validation and therapy planning for artificially ventilated newborn infants. Artificial Intelligence in Medicine. 1996;(8):543–576. doi: 10.1016/s0933-3657(96)00355-7. [DOI] [PubMed] [Google Scholar]
- 33.Miksch S, Seyfang A, Popow C. Abstraction and representation of repeated patterns in high-frequency data.. Proceedings of The Fifth Workshop on Intelligent Data Analysis in Medicine and Pharmacology (IDAMAP-2000); Workshop Notes of the 14th European Conference on Artificial Intelligence (ECAI-2000); Berlin, Germany. IOS Press, Amsterdam. 2000. [Google Scholar]
- 34.Combi C, Chittaro L. Abstraction on clinical data sequences: an object-oriented data model and a query language based on the event calculus. Artificial Intelligence in Medicine. 1999;17:271–301. doi: 10.1016/s0933-3657(99)00022-6. [DOI] [PubMed] [Google Scholar]
- 35.Chittaro L, Combi C. Visualizing Queries on Databases of Temporal Histories: New Metaphors and their Evaluation. Data and Knowledge Engineering. 2003;44(2):239–264. [Google Scholar]
- 36.Silva S, Catarci T, Schiel U. Formalizing visual interaction with historical databases. Information Systems. 2002;27(7):487–521. [Google Scholar]
- 37.Hochheiser H H, Shneiderman B. Dynamic query tools for time series data sets: Timebox widgets for interactive exploration. Information Visualization. 2004;3(1):1–18. [Google Scholar]
- 38.Catarci T, Mascio T, Franconi E, Santucci G, Tessaris S. An ontology based visual tool for query formulation support.. Proceedings of the European Conference on Artificial Intelligence (ECAI-04); IOS Press, Amsterdam. 2004. [Google Scholar]
- 39.Jones B, Donev AN. Modeling and design of cross-over trials. Statistics in Medicine. 1996;15:1435–1446. doi: 10.1002/(SICI)1097-0258(19960715)15:13<1435::AID-SIM278>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 40.Bate ST, Jones B. A review of uniform cross-over designs. Journal of statistical planning and inference. 2008;138:336–351. [Google Scholar]
- 41.Brooke J. SUS: A ‘quick and dirty’ usability scale. In: Jordan PW, Thomas B, Weerdmeester B, McClelland I, editors. Usability Evaluation in Industry. Taylor & Francis; London: 1996. pp. 189–194. [Google Scholar]
- 42.Martins SB, Shahar Y, Galperin M, Kaizer H, Goren-Bar D, McNaughton D, Basso LV, Goldstein MK. Evaluation of KNAVE-II: a tool for intelligent query and exploration of patient data.. Proceedings of the Eleventh World Congress on Medical Informatics (MEDINFO-04); San Francisco, CA. IOS Press, Amsterdam. 2004. [Google Scholar]
- 43.Shahar Y, Young O, Shalom E, Mayaffit A, Moskovitch R, Hessing A, Galperin M. A hybrid, multiple-ontology framework for specification and retrieval of clinical guidelines. The Journal of Biomedical Informatics. 2004;37(5):325–344. doi: 10.1016/j.jbi.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 44.Young O, Shahar Y, Lunenfeld E, Liel Y, Bar G, Martins SB, Vaszar LT, Marom T, Goldstein MK. Runtime application of hybrid-Asbru clinical guidelines. The Journal of BioMedical Informatics. 2007;40:507–526. doi: 10.1016/j.jbi.2006.12.004. [DOI] [PubMed] [Google Scholar]