Abstract
The endoderm of C. elegans arises entirely from a single progenitor cell, the E blastomere, whose identity is specified by GATA type transcription factors, including END-1. In response to an inductive interaction mediated through Wnt/MAP kinase signaling pathways, POP-1, a Lef/Tcf-type transcription factor (Lin et al., 1995), restricts end-1 transcription to the posterior daughter of the mesendoderm progenitor (EMS cell), resulting in activation of endoderm differentiation by the SKN-1 and MED-1/2 transcription factors. We purified a factor from semi-synchronized early embryos that binds to an end-1 cis regulatory region critical for its endoderm-specific expression. Mass spectrometry identified this protein, PLP-1, as a C. elegans orthologue of the vertebrate pur alpha transcription factor. Expression of end-1 is attenuated in embryos depleted for PLP-1. While removal of plp-1 activity alone does not prevent endoderm development, it strongly enhances the loss of endoderm in mutants defective for the Wnt pathway. In contrast, loss of PLP-1 function does not synergize with mutants in the endoderm-inducing MAPK pathway. Moreover, nuclear localization of PLP-1 during interphase requires components of the MAPK pathway, suggesting that PLP-1 is influenced by MAPK signaling. PLP-1 is transiently asymmetrically distributed during cell divisions, with higher levels in the chromatin of the future posterior daughter of EMS and other dividing cells shortly after mitosis compared to that in their sisters. These findings imply that PLP-1 acts as a transcriptional activator of end-1 expression that may be modulated by MAPK signaling to promote endoderm development.
Keywords: cell type specification, transcription, asymmetric cell division, germ layers, C. elegans
INTRODUCTION
Developmentally asymmetric cell division, in which the daughters of a single cell are instructed to adopt distinct fates, is a primary mechanism used to establish differences in cell identity during development (Gönczy, 2008). A striking example of an asymmetric cell division that endows daughter cells with dramatically different developmental properties is provided by the mesendodermal precursor, the EMS cell, in the four-cell C. elegans embryo (Maduro and Rothman, 2008; Maduro and Rothman, 2002a). EMS divides asymmetrically to generate two of the five embryonic somatic founder cells: E, which produces exclusively the entire endoderm of the animal, and MS, a mesodermal precursor (Deppe et al., 1978; Sulston et al., 1983). These two cells follow very different developmental trajectories as is first apparent in transcriptional differences shortly after their birth: while E transcribes genes encoding the END-1 and -3 GATA-type transcription factors (Zhu et al., 1997; Zhu et al., 1998; Maduro et al., 2005a), which are together necessary and individually sufficient for endoderm development, MS transcribes tbx-35, which encodes a T box transcription factor that activates mesodermal differentiation in this lineage (Broitman-Maduro et al., 2005; Broitman-Maduro et al., 2006).
The EMS cell generates an endoderm precursor cell in response to an inductive signal from its posterior neighbor, P2 (Goldstein, 1992, Goldstein 1993). In the absence of this induction, E adopts an MS fate. Induction of EMS by P2 activates Wnt and MAP kinase (MAPK) signaling pathways (Kaletta et al., 1997; Rocheleau et al., 1999; Shin et al., 1999; Rocheleau et al., 1997; Thorpe et al., 1997; Meneghini et al., 1999). These signaling pathways regulate the activity of a Lef-1/Tcf-4-type transcription factor called POP-1 (Lin et al., 1995). POP-1 performs a pivotal function not only in the asymmetric division of EMS, but in many, and perhaps most, asymmetric cell divisions throughout C. elegans development (Lin et al., 1998; Huang et al., 2007).
In response to the endoderm-inducing signals, the abundance of POP-1 is diminished in the nucleus and accumulates at a correspondingly higher level in the cytoplasm of the E cell (Lin et al., 1995; Maduro et al., 2002; Gay et al., 2003; Lo et al., 2004). This difference in nuclear concentration of POP-1 is observed during many asymmetric cell divisions throughout development of the animal: POP-1 is present at high levels in the nucleus of the anterior daughter of an asymmetrically dividing cell and low in its posterior sister (Maduro et al., 2002; Lin et al., 1995; Lin et al., 1998). POP-1, along with components of the Wnt and MAPK signaling pathways, is essential for the developmental asymmetry of these various cell divisions.
The MS cell does not receive the endoderm-inducing signal, and unsignaled POP-1 present in the MS lineage functions as a repressor of end-1 expression (Zhu et al., 1997; Calvo et al., 2001; Maduro et al., 2002). Endoderm induction in the E lineage causes POP-1 to switch from a repressor to an activator, allowing a maternally contributed transcription factor, SKN-1 (Bowerman et al., 1992; Bowerman et al., 1993), and its mesendoderm-determining targets, the MED-1 and –2 GATA factors (Maduro et al., 2001), to express end-1 in E (Zhu et al., 1997; Maduro et al., 2005b, Shetty et al., 2005). Thus, the different development fates of the EMS daughters are specified through integration of transcriptional activators and repressors whose activities are modulated by the endoderm-inducing signals. While endoderm is derepressed in MS in pop-1 mutants, mutants lacking both POP-1 and the SKN-1→MED-1/2 pathway generally lack endoderm (Maduro et al. 2001; Maduro et al., 2007). However, a minor fraction of such embryos continue to express end-1 and produce endoderm, suggesting that other factors provide a positive input into end-1 transcription.
In this study, we sought to identify additional factors that activate end-1 transcription by biochemically isolating proteins from early embryonic extracts that bind to essential transcriptional regulatory elements of end-1. Because fertilization cannot be synchronized in C. elegans, we developed methods to isolate nuclear factors from populations of semi-synchronized early embryos. We report the identification of a C. elegans protein, PLP-1, that is orthologous to the human transcription factor pur alpha, and that binds to, and activates expression of, end-1. PLP-1 is transiently asymmetrically distributed between the chromatin of E and MS immediately following division of the EMS blastomere and in other cell divisions throughout early embryogenesis, concentrating at higher levels in that of the future posterior nucleus. Elimination of PLP-1 function from early embryos, in combination with mutations in Wnt, but not MAPK components, results in a synergistic loss of endoderm, suggesting that PLP-1 may function in the MAPK pathway. Consistent with such a possibility, we found that MAPK components are required for PLP-1 nuclear localization. Thus, PLP-1 activates endoderm development through transcriptional regulation of end-1 expression and is regulated by MAPK signaling.
RESULTS
Detection of a factor from early C. elegans embryos that binds to the regulatory region of end-1
In an effort to identify transcriptional regulators that influence end-1 expression and that may have eluded detection in genetic screens, we devised a biochemical approach to isolate factors that bind end-1 regulatory DNA. We chose to identify such factors biochemically from embryonic extracts rather than, for example, a bacterial expression library, because their binding might require essential post-translational modifications not present in bacterially expressed proteins.
Many of the regulatory factors that function during early embryogenesis are active only transiently; thus, biochemical studies of such factors require isolation of large numbers of embryos at similar developmental stages. Previous biochemical approaches to identifying DNA binding factors and transcription factors present during early C. elegans embryogenesis either used methods that did not specifically involve isolation of early embryos (Lichtsteiner and Tjian, 1995) or used embryos that were allowed to accumulate at a synchronous stage of arrest by blocking development with fluorodeoxyuridine (Stroeher et al., 1994). The latter approach allowed the production of large quantities of embryos that, though containing the same number of cells, had been arrested for different lengths of time. Progress made in mass spectrometry has made it possible to identify a protein with as little as 100 attomoles of protein and, in conjunction with sequenced genomes, these factors can be unambiguously identified from extracts obtained from much smaller quantities of early embryos than was possible in the past. Indeed, we found that we can readily obtain sufficient quantities of semi-synchronized developing early embryos to allow identification of purified factors by mass spectrometry (see Materials and Methods).
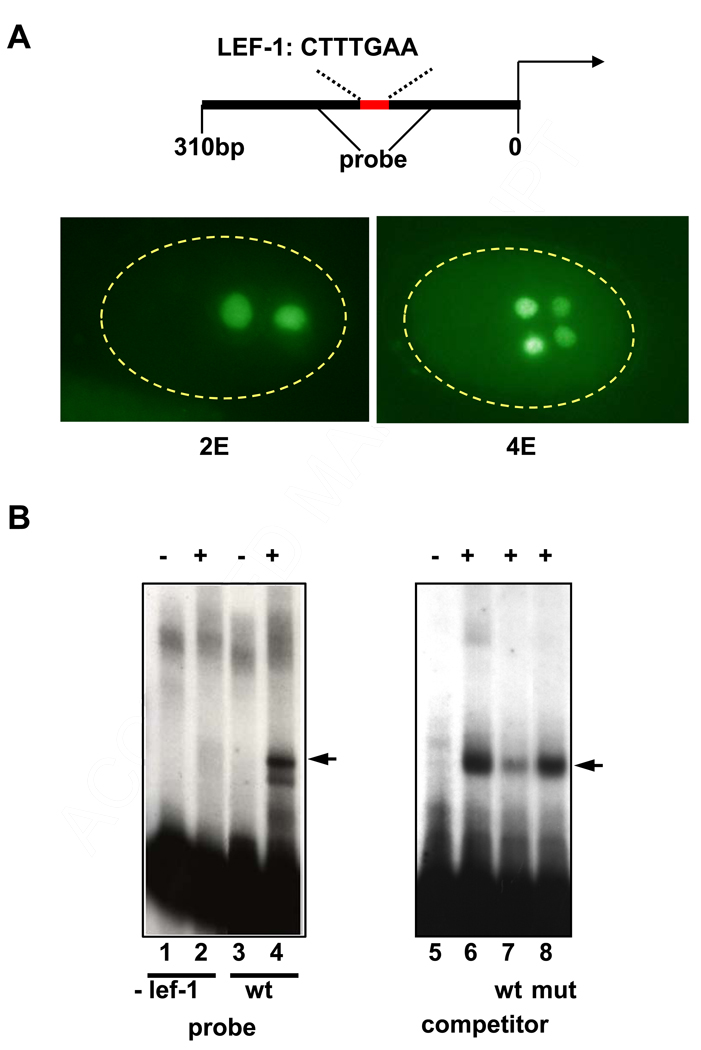
Factors that activate end-1 expression are expected to function at around the 8-cell stage, shortly before it is first expressed (Zhu et al., 1997). We prepared nuclear extracts from embryo populations in which a large fraction of the embryos were at that stage of development at the time of harvesting. We performed electrophoretic mobility shift assays with these extracts and a portion of the end-1 promoter. As the target DNA sequence, we focused on a 310 bp segment upstream of the end-1 coding sequence that is sufficient to direct E-specific expression of end-1 (Fig. 1A). Among the potential transcription factor binding sites in this region is one that matches the consensus for Lef-1, the orthologue of POP-1 (Love et al., 1995). Deletion of a 9 bp sequence containing this putative Lef-1 site from the 310 bp region abolishes expression of an end-1 reporter construct, demonstrating its essential requirement in activation of end-1 (Maduro et al, 2005b). We found that a factor in the early embryo extracts showed significant binding to a 100 bp segment containing this site (Fig. 1B). Moreover, the site that is essential for expression of end-1 is also required for normal binding of the factor: deletion of the site from the probe eliminated binding activity (Fig. 1B, lane 2). Competition experiments confirmed the specificity of the binding of this factor around the essential site: an oligonucleotide containing the consensus Lef-1 binding sequence CTTTGAA, which is present at the site, is able to compete the shift, while a mutant oligonucleotide containing two base changes in the sequence (CTATGAC) is unable to compete effectively (Fig. 1B lanes 7 and 8). (The relatively weak competition by this oligonucleotide may reflect that this small oligonucleotide is a less natural substrate for binding than a larger DNA fragment.) Thus, nuclear extracts of early embryos contain a factor that specifically recognizes end-1 regulatory DNA, including a site that is essential for activation of end-1 expression in early embryos.
Fig. 1. Detection of factors that bind the end-1 promoter.
(A) A 300 bp promoter region (schematized at top) containing the putative Lef-1 binding site CTTTGAA is sufficient to drive expression of a β-galactosidase reporter in the E lineage. Anti-β-galactosidase antibodies stain the daughters (2E) and granddaughters (4E) of the E blastomere in transgenic embryos expressing the reporter driven by this sequence.
(B) Electrophoretic mobility shift assays performed with extracts from developing early embryos detect a band (arrowhead, lane 4) that is eliminated when the putative Lef-1 site is deleted (lane 2). Small oligos containing the sequence CTTTGAA compete this shift (compare lanes 6 and 7; competing oligos were at ~500-fold excess). Mutant oligos with the sequence CTATGAC at the same concentration do not compete (compare lane 7 and 8). Presence (+) or absence (−) of extract is indicated at the top of each gel lane.
Mass spectrometric identification of the end-1-binding factor: a C. elegans orthologue of the mammalian transcription factor pur alpha
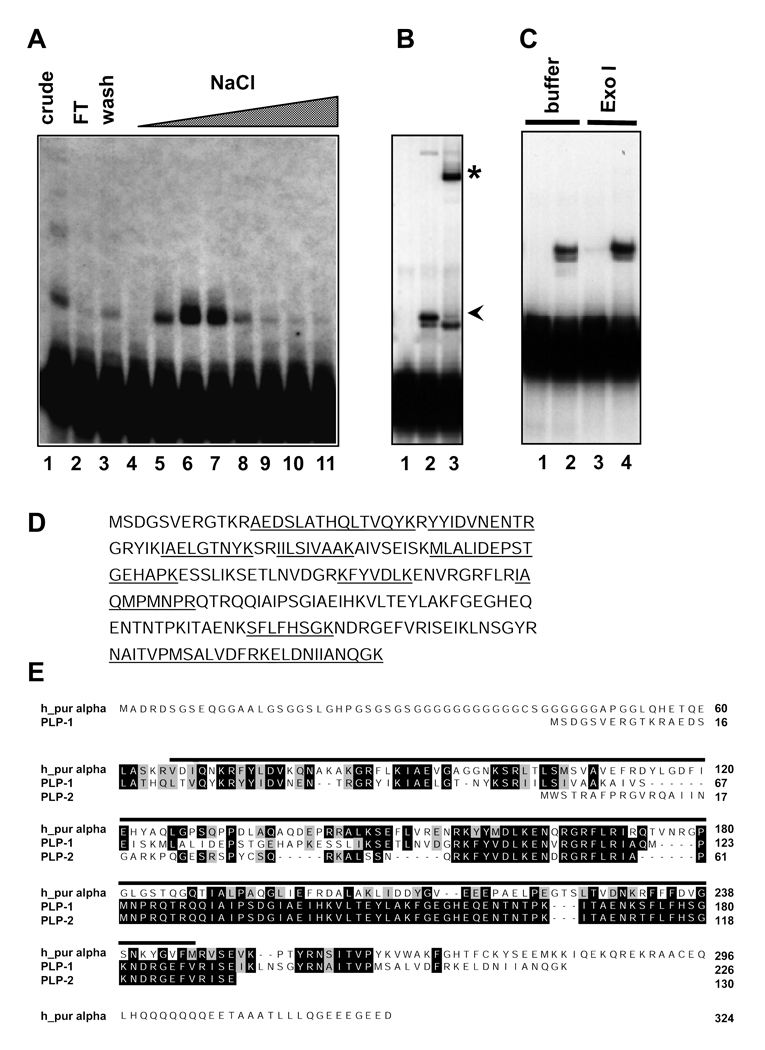
To identify the factor that binds to the essential element in the end-1 promoter, we purified it by affinity chromatography. A 50 bp fragment containing the essential element was labeled with biotin and incubated with nuclear extract. The reaction mixture was passed over a streptavidin column and bound proteins eluted off the column with a stepwise NaCl gradient. Each fraction from the column was assayed by the gel shift assay and significant enrichment of the DNA binding activity was observed in the 750 mM NaCl eluate (Fig. 2A, compare lane 6 to lane 1).
Fig. 2. Purification and identification of the C. elegans pur alpha transcription factor, PLP-1.
(A) Fractions from the DNA affinity column were assayed for binding activity to the end-1 probe. The peak DNA binding activity eluted from the column at ~750 mM NaCl (lanes 6 and 7).
(B) Antibodies against a PLP-1-specific peptide recognize the factor in the nuclear extract that binds the Lef-1 site, as evidenced by a supershift (asterisk). Lane 1, no extract, lane 2, extract plus preimmune serum, lane 3, extract plus PLP-1 antibody. The arrowhead indicates the position of the normal shift that is dependent on the critical end-1 regulatory sequences. The lower band that is not supershifted is seen only variably.
(C) The 100 bp region of the end-1 promoter that is bound by PLP-1 is resistant to the single-stranded exonuclease Exo I. Lanes 1 and 3, no extract, Lanes 2 and 4 extract included. Reactions contained either added Exo I buffer alone (Lanes 1 and 2) or buffer plus Exo I (Lanes 3 and 4).
(D) Digestion of the active fractions with trypsin followed by mass spectrometric analysis identified ten peptides that match the C. elegans orthologue (PLP-1) of the vertebrate transcription factor pur alpha (underlined on the PLP-1 sequence).
(E) Alignment of predicted C. elegans pur alpha orthologues (PLP-1 and PLP-2) with human pur alpha. Identical residues are shown in black boxes, similar residues in grey boxes. PLP-1 and PLP-2 are encoded by separate genes. The line above the sequences indicates the known pur alpha DNA binding domain.
While these fractions would be expected to contain a mixture of proteins, our mass spectrometric procedures are capable of resolving a large number of polypeptides in a mixture. Whole fractions were digested with trypsin and analyzed by electrospray mass spectrometry (see Materials and Methods). The resulting peptide maps were used to search the C. elegans sequence database to identify the corresponding genes. The purification and mass spectrometry were performed several times during optimization of the procedures, and as many as 19 proteins were identified in fractions containing binding activity, most of which were represented by peptides covering only 1–10% of the predicted protein sequence. Many of the identified proteins were obvious contaminants such as the vitellogenins, which are highly abundant in early embryos. The protein with the highest percentage of peptide coverage (10 peptides, 45% coverage) was a homologue of the vertebrate transcription factor pur alpha (Fig. 2 D and E). Since this protein was also found the most consistently in the peak activity fractions, it was the best candidate for the factor that specifically binds the end-1 regulatory region, as confirmed in subsequent experiments (see below). We named this protein PLP-1 (Pur Alpha Like protein-1).
To assess whether the factor that binds to the end-1 regulatory region is PLP-1, we prepared antibodies against PLP-1 and performed supershift experiments. We found that two independent PLP-1 antibodies made against two different peptides, but not preimmune serum, completely supershifted this band in the mobility shift assays (Fig. 2 B), confirming that PLP-1 is the relevant protein causing the shift.
PLP-1 shares overall 32% identity and 48% similarity with human pur alpha (Fig. 2 E). It contains a segment corresponding to the known DNA binding domain of the human protein, but lacks the glycine-rich region at the N-terminus (Fig. 2 E). A second pur alpha-encoding gene present in the C. elegans genome shares 88% identity and 93% similarity with PLP-1, but is 96 amino acids shorter than PLP-1 and lacks part of the DNA binding domain. None of the peptides we identified by mass spectrometry were specific to this second pur alpha homologue.
Mammalian pur alpha preferentially binds to purine-rich, single-stranded DNA (Bergemann and Johnson, 1992). We found that PLP-1 also binds strongly to a 25 nt purine-rich strand spanning the essential Lef-1 site but not to the complementary pyrimidine-rich single-stranded probe (not shown). The ability of PLP-1 to bind single-stranded DNA raised the possibility that the mobility shift originally observed with the double-stranded probe was caused by contaminating single-stranded DNA. To address this possibility, we repeated the experiments following incubation of the end-1 promoter probe with Exo I, a single-strand-specific exonuclease. We found that Exo I had no effect on the DNA binding activity of PLP-1 (Fig. 2 C), demonstrating that PLP-1 binds to double-stranded DNA from the end-1 promoter.
Much of the existing information on pur alpha has come from in vitro and cell culture systems. Pur alpha was first identified by its ability to bind to the promoter of the c-myc gene; it was also shown to increase transcription of the myelin basic protein gene both in vitro and in vivo (Bergemann et al., 1992; Haas et al., 1993; Haas et al., 1995). Stimulation of cultured cells with phorbol esters results in increased pur alpha protein and RNA levels and DNA binding activity (Shelley et al., 2002). Co-expression studies revealed that pur alpha causes a four-fold enhancement of CD11 gene expression and represses transcription of the beta integrin gene (Shelley et al., 2001). A mouse knockout revealed a role for pur alpha in postnatal brain development and cell proliferation (Khalili et al., 2003). Although these studies have implicated pur alpha in regulation of transcription, only limited information regarding its roles in regulating development has been obtained. For example, it has been shown to repress the gene encoding GATA2 in the zebrafish nervous system (Penberthy et al., 2004) and has been implicated in neurodegeneration in Drosophila (Jin et al., 2007). In this study, we have focused on the role of PLP-1 in end-1 expression and endoderm development. More detailed analysis of the interaction of PLP-1 with end-1 regulatory DNA will be described elsewhere (E. Witze and J. Rothman, in prep.).
plp-1 is required for normal end-1 expression levels
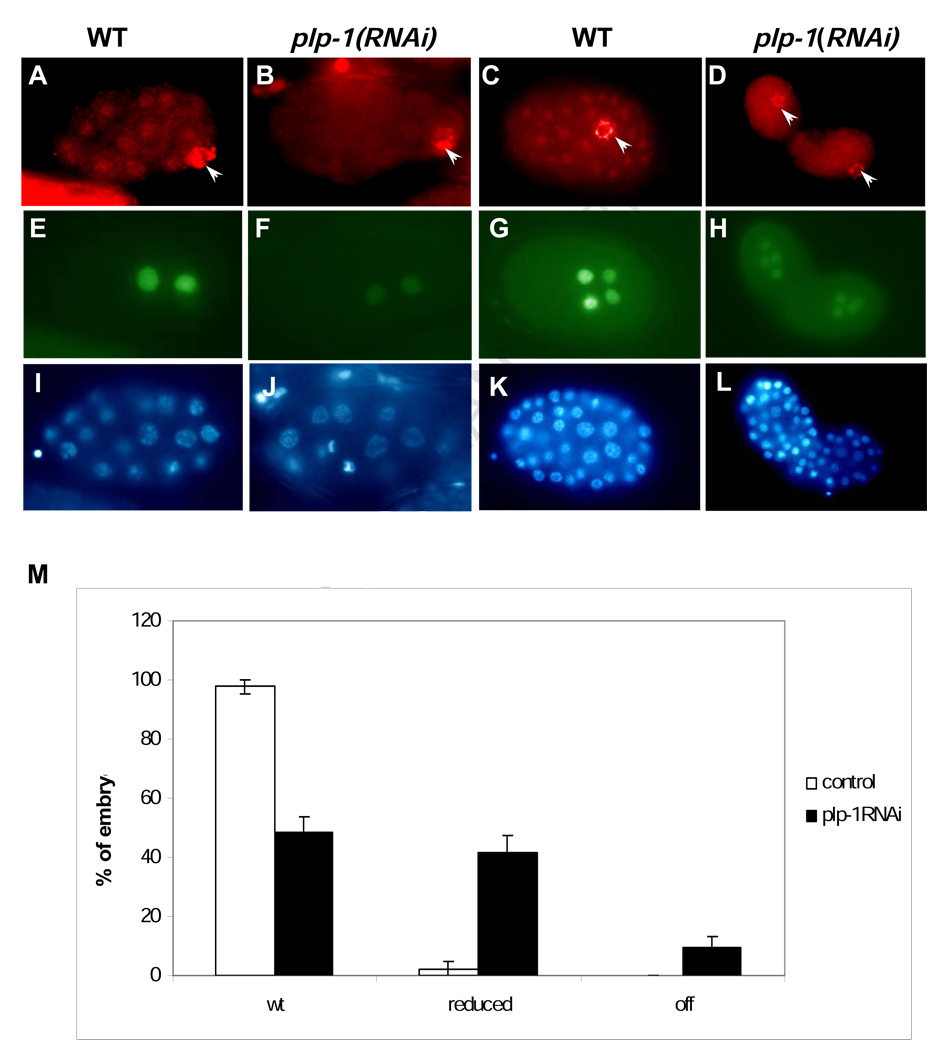
Given its ability to bind to end-1 regulatory sequences, we sought to determine whether PLP-1 might be important for end-1 expression. As there are two pur alpha homologues in C.elegans whose coding regions share very strong sequence identity, we designed a double-stranded RNA (dsRNA) that recognizes sequences present only in plp-1 (see Materials and Methods). As described below, immunoreactive PLP-1 localizes primarily to nuclei throughout embryos (Fig. 3); injection of this plp-1-specific dsRNA into adults effectively reduced or abolished immunoreactive PLP-1 from their progeny embryos (Fig. 3A–D).
Fig. 3. RNAi of plp-1 reduces end-1 expression.
(A–D) PLP-1 staining is diminished or eliminated by RNAi of plp-1 (B and D; compare with wild type, panels A and C). Antibody to PGL-1 (Kawasaki, et al., 1998), a component of the germline-specific P-granules, was used as a positive control for permeabilization (arrowheads).
(E–H) Anti-β-galactosidase antibody staining of the same embryos expressing an end-1 reporter. plp-1(RNAi) embryos show reduced expression of the reporter at the 2E stage (F) and 4E stage (H) compared to wild type (E and G).
(I–L) The same embryos as in (A–H) stained with DAPI to reveal nuclei.
(M) Distribution of embryos expressing end-1 at levels that appear normal (wt), strongly reduced, and undetectable (off). plp-1(RNAi) embryos that that were positive for P granule staining and that showed loss of PLP-1 immunostaining (to compensate for impenetrant reduction of plp-1) were scored for end-1 expression. The results were summed over eight experiments in total (143 wild-type control embryos and 138 plp-1(RNAi) embryos). Error bars indicate standard error of the mean.
Expression of end-1 was monitored using an end-1 reporter containing 1.5 kb of upstream DNA and the first exon of the gene. This reporter shows high levels of expression specifically in the E lineage (e.g., Fig. 3 E, G). We found that, while nearly all untreated embryos express this reporter at high levels, approximately half (49%; n = 73) of the plp-1(RNAi) embryos that were positive for a control marker (the immunoreactive germline P granules; Kawasaki et al., 1998) showed substantially reduced, or abolished expression of end-1 (e.g., Fig. 3 F, H; quantification in Fig. 3 M). In contrast, plp-1(RNAi) had no detectable effect on the expression of either med-1, which regulates, and is expressed immediately prior to, the onset of end-1 expression, or of the ectodermal transcription factor LIN-26, which is expressed much later in embryogenesis (not shown). Thus, PLP-1 does not appear to be generally required for efficient zygotic transcription. The ability of plp-1(RNAi) embryos to express end-1 (albeit at attenuated levels) is likely to be attributable to the continued presence of the MED-1/2 factors, which activate end-1 and are required for high levels of its expression (Maduro et al., 2001; Broitman-Maduro et al., 2005). We found that RNAi specifically directed against plp-2 showed no obvious effect and did not enhance the phenotypes we observed for plp-1RNAi); hence, PLP-2 does not appear to synergize with PLP-1 in regulating end-1 or endoderm formation (see below).
PLP-1 synergizes with the Wnt but not MAPK pathway to regulate endoderm specification
While RNAi of plp-1 results in diminished end-1 expression and embryonic lethality (50–100% in microinjection experiments), we found that plp-1(RNAi) embryos only rarely lack endoderm (Fig. 4 A and B; Table 1). Thus, while PLP-1 is required for normal end-1 expression, it presumably acts in concert with other regulators of end-1. Several independent regulatory systems, including Wnt and MAPK signaling, are known to be required for formation of the C. elegans endoderm (Rocheleau et al., 1999; Shin et al., 1999; Rocheleau et al., 1997; Thorpe et al., 1997; Meneghini et al., 1999). Elimination of any one of these regulatory inputs results in impenetrant loss of endoderm, while loss of multiple inputs leads to synergistic phenotypes.
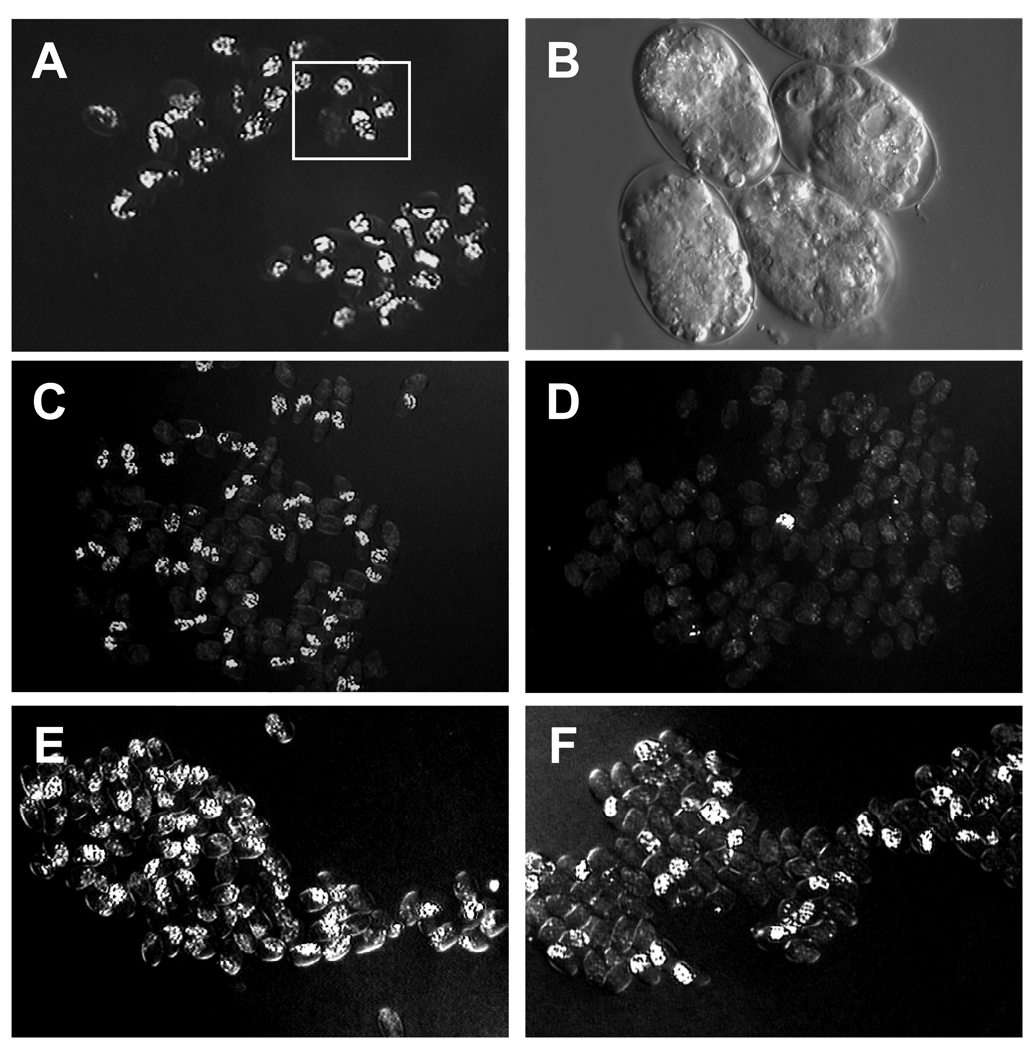
Fig. 4. Synergy of PLP-1 and Wnt signaling functions.
(A) plp-1(RNAi) embryos that lack immunoreactive PLP-1 (see Fig. 3) generally produce differentiated gut, detected by the presence of birefringent gut granules by polarized light, as seen in fields of embryos shown at low magnification.
(B) Higher magnification of field boxed in A under Nomarski optics.
(C) Arrested mom-2(or42) mutant embryos. ~40% produce differentiated gut.
(D) Arrested mom-2 (or42); plp-1(RNAi) double mutant embryos, only 6% of which produce differentiated gut.
(E) Arrested mom-2; pop-1(RNAi); unc-22(RNAi) embryos, nearly all of which produce gut. unc-22(RNAi) was used as a negative control for the plp-1(RNAi) experiments.
(F) Arrested mom-2; pop-1(RNAi); plp-1(RNAi) embryos. The synergistic loss of endoderm is independent of repression by POP-1.
Table 1.
Genetic interaction of PLP-1 with Wnt and MAPK mutants
| Genotype | % Lacking Endoderm (n) |
|---|---|
| plp-1(RNAi) | 3 (330) |
| mom-4(or39); unc-22(RNAi) | 41 (151) |
| mom-4(or39); plp-1(RNAi) | 42 (130) |
| mom-5(or57); unc-22(RNAi) | 3 (79) |
| mom-5(or57); plp-1(RNAi) | 35 (88) |
| mom-2(or42); unc-22(RNAi) | 66 (302) |
| mom-2(or42); plp-1(RNAi) | 96 (265) |
| mom-2(or42) ;plp-1(RNAi); pop-1(RNAi) | 42 (115) |
|
mom-2(or42); unc-22(RNAi); pop- 1(RNAi) |
0 (85) |
| pop-1(zu189); plp-1(RNAi) | 0 (286) |
Percentage of arrested embryos lacking endoderm was determined for each of the indicated genotypes. unc-22, which performs no relevant role in endoderm specification, was used as a negative control for the plp-1(RNAi) experiments. The data were taken from representative experiments repeated at least twice.
We investigated whether PLP-1 functions in concert with other regulatory processes by analyzing genetic interactions between them. Depletion of PLP-1 did not significantly change the penetrance of a mutant lacking MOM-4 (MAPKKK) function: 42% (n = 130) of mom-4(or39); plp-1(RNAi) embryos lacked endoderm, essentially indistinguishable from the mom-4(or39) single mutant embryos (40% without endoderm; n = 151) (Table 1). In contrast, when combined with mutants defective in the endoderm-inducing Wnt pathway, RNAi of plp-1 results in synergistic loss of endoderm (Fig. 4; Table 1). The magnitude of this synergy was particularly evident in double mutants with mom-5(or57), which carries a mutation in the MOM-5 Wnt receptor (Thorpe et al., 1997; Rocheleau et al., 1997): while, as with plp-1(RNAi) mutants, only a very small fraction (3%) of mom-5(or57) mutants lack endoderm, more than one-third of the mom-5 (or57); plp-1(RNAi) embryos do not make differentiated gut (Table 1). Synergy was also seen with the mom-2(or42) mutant, which carries a mutation in the MOM-2 Wnt ligand: mom-2(or42); plp-1(RNAi) embryos almost never make endoderm, whereas approximately one-third of mom-2 single mutants do (Table 1).
The loss of endoderm in Wnt pathway mutants results from the failure to convert POP-1 from its endoderm-repressive to endoderm-activating form (Maduro et al., 2005b; Shetty et al., 2005): in the absence of Wnt signaling, POP-1 acts as a repressor of the end genes in the E cell, just as it normally does in MS. Removal of POP-1 function restores endoderm in embryos mutant for the Wnt pathway (Thorpe et al., 1997; Rocheleau et al., 1997). Thus, the increased penetrance of the mom; plp-1(RNAi) double mutant condition could be attributable to an increase in POP-1 repressive activity rather than a direct positive role for PLP-1 in end-1 transcription. To test this possibility, we examined the effect of removing pop-1 activity in the mom-2(or42); plp-1(RNAi) double mutant. While this experiment resulted in some increase in the fraction of embryos making endoderm, as is as expected from derepression of endoderm in MS, 42% (n = 115) of the embryos still lacked endoderm (Fig. 4F and Table 1). This result demonstrates that the increased loss of endoderm in the mom-2; plp-1(RNAi) double mutant is largely independent of POP-1 repression, and implies that PLP-1 functions in parallel with the Wnt pathway to activate endoderm development.
The synergy seen between plp-1(RNAi) and the Wnt, but not MAPK, pathway mutants suggests that PLP-1 acts in parallel to the Wnt pathway and raises the possibility that it might function in the MAPK pathway.
PLP-1 is asymmetrically localized during cell division
The foregoing results suggest that PLP-1 modulates gene expression in response to signaling events that regulate a developmentally asymmetric cell division. We analyzed localization of PLP-1 protein to assess whether it might be influenced by events occurring during division of EMS. Immunoreactive PLP-1 localizes to the nuclei of all blastomeres beginning by the two-cell stage of embryogenesis (Figs. 3 and 5), implying that PLP-1 is a maternally encoded transcription factor. It is also present in the germline-specific P granules of early embryos (Fig. 5C, arrowhead). The same pattern was seen with two separate antibodies raised against distinct PLP-1 peptides, and both nuclear and P granule expression was largely eliminated in plp-1(RNAi) embryos (Fig. 3), confirming their specificity.
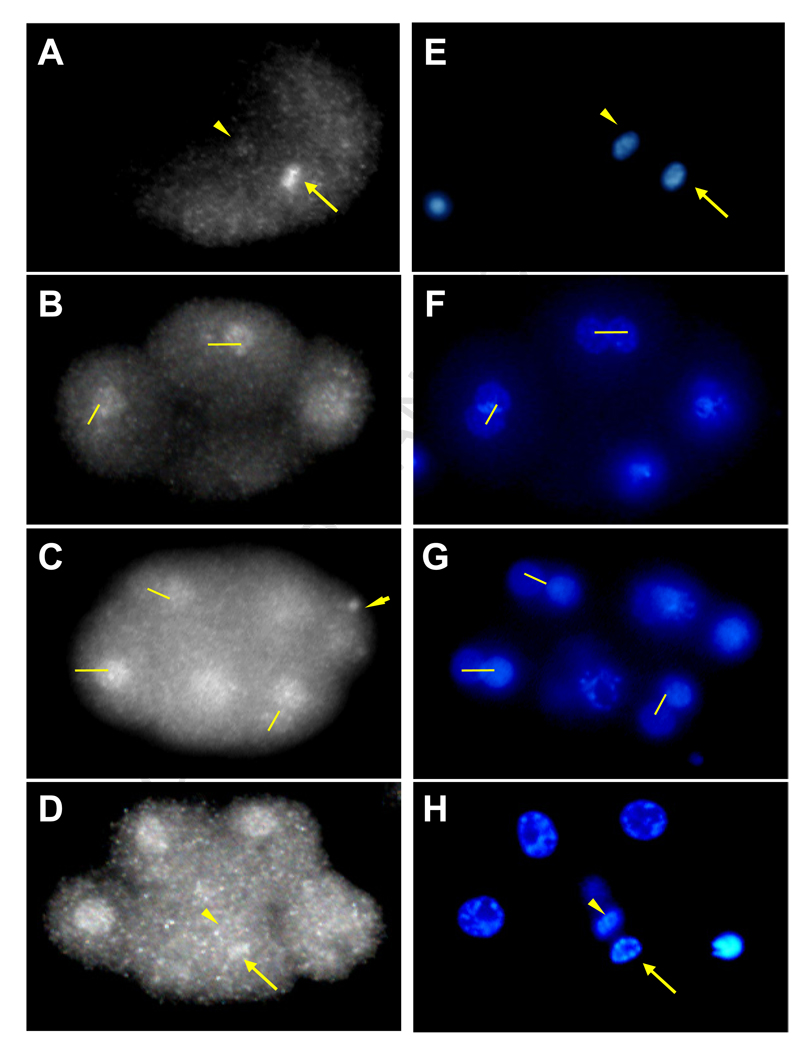
Fig. 5. Asymmetric distribution of PLP-1 protein in early embryos.
PLP-1 staining was analyzed in 10 separate experiments and asymmetries described observed in a total of 24 embryos (see text).
(A) PLP-1 is asymmetrically distributed during the first cell division with higher levels on what will become the nucleus of the posterior (P1) cell (arrow).
(B) At the 4 to 6 cell transition, immunoreactive PLP-1 is asymmetrically distributed in the left/right AB granddaughters (connected by lines).
(C) PLP-1 is asymmetric between the AB great granddaughters (connected by lines) with higher levels in posterior blastomeres and lower in the anterior. Germline specific P-granules also contain PLP-1 (arrowhead).
(D) During the following division PLP-1 is asymmetric in the dividing EMS blastomere, with higher levels in the chromatin of the future E (arrow) than in that of the future MS nucleus (arrowhead). Note that the AB granddaughters show symmetric PLP-1 staining later in the cell cycle.
(E–H) DAPI staining of the corresponding embryos shown in the left panels.
Consistent with a role in regulating gene expression in the E cell, we found that PLP-1 is transiently asymmetrically localized during telophase of the dividing EMS cell (observed in 12 embryos at the correct stage), with higher levels of the protein in the chromatin of the future E cell nucleus and low or undetectable levels in that of MS (Fig. 5). A similar transient asymmetry in PLP-1 levels was observed at many divisions throughout early development, starting at cleavage of the zygote, with higher levels seen in the cytoplasm and forming nucleus of the posterior daughter, P1 (observed in 5 embryos; e.g., Fig. 5A and B). The anteroposterior asymmetry in PLP-1 was also observed in the AB lineage during the division of the AB granddaughters (observed in 7 embryos): for example, PLP-1 is higher in the chromatin of the posterior daughter ABalp than that of its anterior sister ABala (Fig. 5C). In all cases, the asymmetry was observed only during telophase and at the time that nuclei were reassembling after cell division; the staining was symmetric at all other times. This asymmetry was also seen during the preceding division in the AB lineage (Fig. 5B). This was unexpected, as these are developmentally symmetric left/right divisions that produces initially equivalent daughter cells (Wood, 1991); however, these divisions do show a clear anteroposterior bias and, again, PLP-1 was always seen at higher levels in the forming nuclei of the posterior daughters.
Asymmetry of protein localization is a recurring theme during asymmetric cell division. For example, POP-1 is present at higher levels in anterior daughter nuclei of such divisions (Lin et al., 1995; Lin et al., 1998; Maduro et al., 2002) and a LIT-1::GFP fusion protein shows a reciprocal localization, being at higher levels in the posterior daughter cells than in their sisters (Lo et al., 2004). The asymmetric distribution of PLP-1 suggests that it may be influenced by the machinery that establishes anterior/posterior asymmetry during cell divisions.
Nuclear localization of PLP-1 requires MAPK components
The genetic evidence suggesting that PLP-1 might function in the MAPK pathway led us to examine whether its localization is affected by MAPK components. We found that, while PLP-1 is strongly localized to nuclei in interphase cells of wild-type embryos throughout early embryogenesis, it is at much lower levels in nuclei and higher in the cytoplasm in mutants defective in the LIT-1 MAPK (both lit-1(RNAi) and lit-1(t1512)) (Fig. 6). This effect on PLP-1 nuclear localization is dramatically enhanced in a double mutant that is also defective in the MOM-4 kinase, which functions upstream of LIT-1: PLP-1 accumulates in the cytoplasm and does not concentrate to appreciable levels in the nucleus of lit-1(RNAi); mom-4(or39) double mutants (Fig. 6D). This result does not reflect a general effect of MAPK components on localization of nuclear factors: for example, the nuclear MPM2 antigen (Fig. 6E–H) and POP-1 (Shin et al, 1999), are properly localized in lit-1; mom-2 double mutants. These findings suggest that MAPK components are required for efficient nuclear translocation or retention of PLP-1.
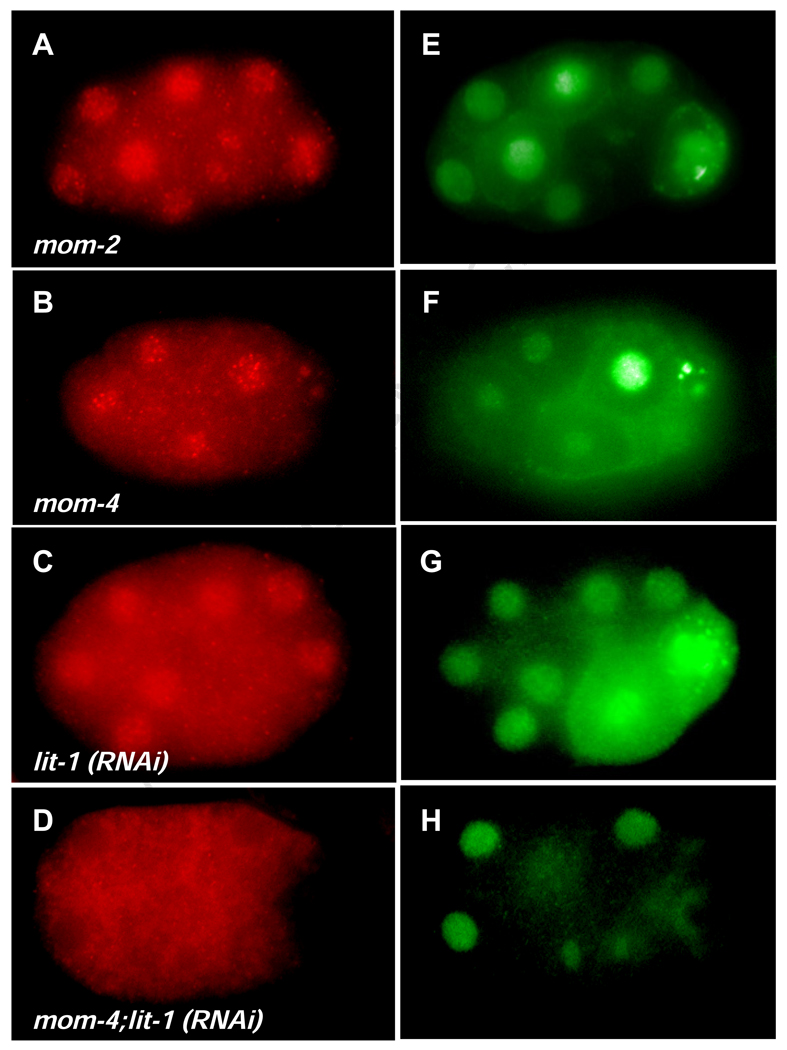
Fig. 6. PLP-1 nuclear localization requires lit-1 and mom-4 function.
(A–D) PLP-1 staining in Wnt and MAPK mutants. PLP-1 is at higher levels in the nucleus than in the cytoplasm in wildtype (Fig. 3), mom-2, and mom-4 mutant embryos (A and B). Cytoplasmic levels of PLP-1 are increased in lit-1(RNAi) embryos (C). PLP-1 is largely excluded from the nucleus in lit-1(RNAi); mom-4 double mutants (D). (E–H) Staining with MPM2 control, showing very little or no effect on nuclear localization of the nuclear protein recognized by this antibody.
DISCUSSION
The simple lineage that generates the C. elegans endoderm is specified by a complex intersection of signaling systems and transcriptional cascades (Maduro and Rothman, 2008; Maduro and Rothman, 2002). While genetic approaches have proven highly effective at dissecting these regulatory events, components that modulate gene expression associated with, but not absolutely essential for, endoderm formation would be overlooked in screens for mutants with striking endoderm phenotypes, although elimination of such components might cause synergistic effects with loss of other regulatory inputs. We have taken a biochemical approach to identify a C. elegans orthologue of the vertebrate pur alpha factor, PLP-1, that is required for efficient expression of end-1 and for endoderm development when the endoderm-inducing Wnt signaling pathway is defective.
Biochemical purification methods, most notably with sea urchins from which massive quantities of synchronized embryos can be obtained (e.g., Calzone et al., 1991), have made it possible to identify many of the transcription factors that regulate cell identity in developing embryos (e.g., Davidson et al., 2002). A similar approach with C. elegans is more difficult, as it is not possible to fertilize large numbers of embryos synchronously and its embryos are vastly smaller. We have developed methods for isolating extracts from semi-synchronized embryos and have exploited more sensitive and accurate mass spectrometry methods to overcome the biochemical shortcomings of C. elegans. This approach allowed us to identify regulatory factors from small quantities of actively developing embryos. In addition, individual proteins can be identified in complex mixtures by these methods and we were able to resolve PLP-1 from other proteins in partially purified preparations. Subsequent studies with PLP-1 antibodies confirmed that it is the relevant end-1-binding protein.
It is conceivable that PLP-1 activity is modulated by endoderm-inducing signals in the E lineage to provide a positive input into end-1 expression. Several lines of evidence suggest that PLP-1 might be influenced by the endoderm-inducing MAPK pathway. First, consistent with its binding activity, PLP-1 is required to activate expression of end-1, which is also activated by the MAPK signaling pathway. Second, while PLP-1 functions synergistically with the Wnt pathway to control end-1 gene expression and endoderm formation, it does not synergize with MAPK pathway components, genetic evidence that it may operate in this pathway. Further, the MAPK components LIT-1 and MOM-4, are required for efficient nuclear localization of PLP-1. In the absence of MAPK signaling, PLP-1 accumulates in the cytoplasm, where it presumably would be unable to activate end-1 transcription. The activity of many transcription factors is regulated at the level of their nucleocytoplasmic localization (e.g. Maduro et al., 2002). PLP-1 might similarly be regulated by MAPK-dependent nuclear localization and/or activation as a transcription factor.
The MAPK pathway functions recursively with the Wnt pathway to control asymmetric cell division throughout C. elegans development (Kaletta et al., 1997; Rocheleau et al., 1999; Shin et al., 1999; Rocheleau et al., 1997; Thorpe et al., 1997; Meneghini et al., 1999). The reiterative use of these signaling systems to generate differences between daughter cells generated by anteroposterior divisions is associated with asymmetric nucleocytoplasmic distribution of their effector, POP-1. POP-1 is excluded from the nucleus after phosphorylation by LIT-1 kinase and becomes localized to the nucleus in response to acetylation by the p300/CBP acetyl transferase (Gay et al., 2003). Immunoreactive PLP-1 is strongly nuclear in early wild type embryos, but the protein is almost completely cytoplasmic in lit-1; mom-4 double mutants. The cytoplasmic localization of PLP-1 in the absence of MAPK signaling is reciprocal to that seen for POP-1, when it functions as a repressor of end-1 (e.g., Lo et al., 2004). Thus, MAPK signaling system might help to coordinate the differential activity of these two transcription factors during asymmetric cell divisions. We have found that POP-1 can compete with PLP-1 for binding to the fragment containing the Lef-1 site (not shown), though the significance of this competition in vivo is not clear. In vitro studies to assess physical interaction between PLP-1 and POP-1 in co-precipitation experiments did not reveal evidence for interaction between the two proteins (not shown).
PLP-1 is asymmetrically localized to the posterior chromatin during the division of EMS. It is conceivable that the asymmetry we observed in this and other cell divisions might reflect slight, reproducible differences in cell cycle timing of the daughters; however, no such difference in timing of formation of the nascent nuclei following cell division has been described. During early embryogenesis in the C. elegans embryo, at the time that E and MS are born, there are no G1 or G2 phases, implying that zygotic transcription must occur during S phase and/or mitosis (Edgar and McGhee 1988). The presence of PLP-1 in the newly forming E nucleus might allow end-1 to be poised for activation following the birth of the E cell. This very early and transient difference in PLP-1 levels may reveal an additional layer of transcriptional regulation that facilitates the rapid and restricted E-specific expression of the end genes and thereby the endoderm fate following asymmetric cell division.
Several observations suggest that the DNA binding activity of PLP-1 may be regulated by phosphorylation (E.W. and J.R., unpublished results). PLP-1 contains a putative proline-directed kinase site that is required for phosphorylation in vitro by a purified MAPK. In addition, we found that PLP-1 becomes phosphorylated when incubated with embryo extract, demonstrating the presence of a PLP-1 kinase in early embryos. Treatment of embryonic extracts with lambda phosphatase eliminates binding of PLP-1 to end-1. However, evidence that PLP-1 phosphorylation by the MAPK pathway might be responsible for regulating its transcriptional activity in vivo will require further investigation.
MATERIALS AND METHODS
Nuclear extract preparation
To obtain large numbers of early embryos for nuclear extract preparation, each of 30 150 mm peptone-enriched plates were seeded with ~80,000 starved L1 larvae, which were allowed to develop until shortly after they were visibly gravid. Hermaphrodites were harvested when they contained primarily embryos classified as “early” (>90% with 100 cells or less based on microscopic analysis). We found that this culturing method, involving growth on peptone-enriched solid medium rather than liquid culture, produced large quantities of early embryos specifically, as hermaphrodites tend to accumulate later embryos when grown in liquid. At the stages that they were harvested, nearly all of these embryos express end-1. Adults were washed off and allowed to settle on ice. The supernatant was removed and alkaline bleach was added to the worms, which were incubated on ice until no carcasses remained. The nuclear extract protocol was modified for C. elegans embryos from an existing protocol for cell culture (Ausubel et al., 2004). Embryos were washed three times in M9 buffer at 0°C and resuspended in 1 ml of hypotonic buffer (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 1 mM PMSF, 10 µM E64 (Sigma), 0.2mM EDTA), centrifuged, and the pellet resuspended in hypotonic buffer. Embryos were then homogenized with a Dounce homogenizer (pestle B) until they were all disrupted. The homogenate was immediately centrifuged for 10 min at 3,000 × g at 4°C. The supernatant was removed and the pelleted nuclei resuspended in one packed nuclear volume (pnv) of low salt buffer (20 mM HEPES, 25% glycerol, 0.2 M KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 1 mM PMSF, 10 µM E64). An equal volume of high salt buffer (same as the low salt buffer with 1.2 M KCl) was added slowly while stirring. The pellet was thoroughly resuspended and the nuclei were incubated on ice for 1 hour and then pelleted at 16,000 × g for 30 min. The supernatant (nuclear extract) was collected and centrifuged again for 30 min at 16,000 × g. The nuclear extract was transferred to dialysis tubing and dialyzed for 2 hours in 50 volumes of dialysis buffer (20 mM HEPES, 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 1 mM PMSF, 10 µM E64, 0.5 mM DTT). The extract was then centrifuged at 16,000 × g for 30 min, aliquotted, frozen in liquid nitrogen, and stored at −80°C.
Electrophoretic mobility shift assay
Typical gel shift reactions contained 50 fmoles of 32P-end labeled probe, 10 µg of crude nuclear extract, 1 µg of poly dIdC:dIdC non-specific competitor and 1 µl of 10× binding buffer (50% glycerol, 100 mM HEPES, pH 7.6, 10 mM EDTA, 600 mM KCl, and 10 mM DTT) in a final volume of 10 µl. Optimum amounts of probe, extract, and non-specific competitor were determined by preliminary titrations. Binding reactions were incubated for 15 minutes at room temperature, loaded onto a non-denaturing, 1.5 mm thick, 5% polyacrylamide gel with 1× TBE, and electrophoresed at 4°C. Gels were dried and exposed to film. For exonuclease treatment, probes were incubated for 2 hours at 37°C with 10 units of ExoI (New England Biolabs) in a 10 µl reaction. Activity was confirmed by digesting labeled single-stranded probe. For endonuclease treatment, probes were incubated for 2 hours in a 10 µl reaction containing 5 units of T7 endonuclease (New England Biolabs).
Affinity purification of nuclear extract
100 pmoles of target DNA was biotinylated using terminal transferase (TdT) (New England Biolabs) to add biotin-ddUTP to the 3’ hydroxyl group of the DNA oligo. 100 pmole of oligo were incubated with 10 µl of 10× buffer, 20 µl of 2.5 mM CoCl2, 2 µl 1 nmol biotin-16-ddUTP (Boehringer Mannheim) and 40 units TdT, in a 100 µl volume for 1 hour at 37°C. The labeled oligo was precipitated, resuspended in water, and used immediately. 100 pmoles of biotin-labeled oligo was incubated with 250 µl of crude extract (10 mg/ml) for 1 hour at 4°C. The binding reaction was loaded onto a column of streptavidin beads (Sigma) and the flow-through passed over the column a second time. The column was washed with 3 bed volumes of 1× binding buffer and then eluted with 50 µl of elution buffer (5% glycerol, 20 mM Tris pH 8.5, 2 mM EDTA, 1 mM DTT with stepwise increases of NaCl from 90 mM to 1.5 M). 50ul fractions were collected and assayed by gel shift analysis.
In-solution Digestion of Proteins
Approximately 130 pmol of total protein from each DNA-affinity column fraction was equilibrated in 30 µL of 100 mM Tris buffer, pH 8.8. Modified trypsin (Promega, Madison, WI) was added in a ratio of 1:25 (enzyme:protein) and the digest was incubated at 37° C for 1 hour and then incubated at room temperature for approximately 18 hours. The enzymatic digestion was stopped by adding 2 µl of concentrated glacial acetic acid (HOAc, Sigma, St. Louis, MO, 99.99% pure). The sample was then stored at −36° C until mass spectrometric analysis was performed.
Electrospray ionization mass spectrometry
The entire sample volume from each proteolytically digested DNA-affinity column fraction was loaded onto a pre-column via helium bomb at ~500psi. After sample loading, the pre-column was washed with 0.1% HOAc in NANOpure (Barnstead, Dubuque, IA) water for approximately 10 min to desalt the sample. The pre-column was then Teflon-connected to the analytical column and interfaced to a modified LCQ quadrupole ion trap mass spectrometer (ThermoElectron Corporation, San Jose, CA).
The analytical column was connected to the HPLC line with ESI voltage (1.5–1.8 kV) being applied pre-column at a stainless steel union on the waste line. Tryptic peptides from the DNA-affinity column were eluted into the mass spectrometer using the following gradient: 0–15% B (0–4 min), 15–50%B (4–15 min), 50–100%B (25–28 min), 100% B (28–30 min), 100-0%B (30–32 min), 0% (32–44 min) where solvents A and B were 0.1 M HOAc in NANOpure water and 0.1 M HOAc in 70% acetonitrile (MeCN, Mallinkrodt, Paris, KY), respectively. The emitter tip was positioned about 1–2 mm in front of the orifice of the heated metal capillary prior to data acquisition. The flow rate of analyte solution out of the column was ~40 nl/min.
Data filtering and database searching
ProteoFarm (Hunt lab: Dr. Robert Settlage and Galvin Hsiu) was used as an interface between the raw mass spectrometric information and a database searching program, SEQUEST. Poor quality MS/MS spectra can be filtered out prior to database searching.
The RAW data file format (MS/MS spectra) created by the LCQ software was converted to DTA file format. All poor quality spectra were deleted based on the following parameters: minimum number of total fragment ions is 50, maximum of 10 ions present at above 30% abundance of the base peak (more than this is uncharacteristic of interpretable peptide MS/MS spectra), and a minimum total ion current of at least 15,000 ion counts. After deleting poor quality spectra, the charge states of peptides are assigned to the spectra.
All mass spectrometric data from the biotin-DNA affinity column fractions were searched against the C. elegans database that is maintained by the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov) using SEQUEST. The entire peptide mass in the database had to be within a 3.0 Da window of the monoisotopic experimental peptide mass to be considered a match and the fragment ions had to be within 0.35 Da to be considered a match with the average experimental fragment ions masses.
Antibody production
Synthetic peptides containing the sequences CLIDEPSTGEHAPKES (peptide #51), and CGSVERGTKRAEDSLA (peptide #52) were conjugated to maleimide-activated KLH (Pierce). The injection of rabbits, test bleeds, production bleeds, and exsanguination were performed by Cocalico Biologicals Inc. The conjugated peptides were injected separately into New Zealand White rabbits.
Antibody purification
GST::PLP-1 was isolated from 1 liter of culture and bound to a 1–2 ml bed volume of glutathione agarose beads. The bound protein was washed thoroughly in GST wash buffer (50 mM Tris pH 7.6, 150 mM NaCl) and 1 bed volume of 0.2% glutaraldehyde in PBS was added to the beads. The beads were incubated at room temperature for 10–20 minutes. The beads were centrifuged and washed three times in 10 ml volumes of PBS and incubated in PBS overnight at 4°C. 10% BSA in 1× PBS was added to the beads and incubated at room temperature for 0.5 hours. The beads were centrifuged and washed twice in 10 ml volumes of PBS. 2 ml of serum was added to the beads and incubated with gentle agitation at 4°C overnight. The beads were pelleted and washed twice in 10 ml of GST wash buffer. The beads were resuspended in one bed volume of Gentle Elution Buffer (Pierce) and incubated at room temperature for 10 min. The supernatant was removed and the antibodies concentrated with 30 kD MW cut-off ultrafilter (Millipore). The elution buffer was exchanged with GST wash buffer.
Immunostaining
Embryos were fixed for 15 min in 4% paraformaldehyde, 0.1% glutaraldehyde in PBS and immersed in dimethyl formamide at −20°C for 20 minutes. The slide was washed in PBST and blocked with 5% goat serum. It was then incubated with PLP-1 antibody diluted 1:50 in 5% goat serum in PBS overnight at 4°C, washed in PBST and goat anti-rabbit secondary antibody (1:500), and incubated at room temperature for 2 hours. The slide was then washed in PBST and mounted in Vectashield mounting media with DAPI (Vectashield Laboratories).
RNA interference
A 356 bp fragment of the plp-1 cDNA was PCR-amplified with the primers ATGTCGGACGGAAGTGTTGA and ATGGCGATTTGTTGGCGAGT tagged with the T7 promoter. Double stranded RNA was diluted 1:1 with nuclease-free water and injected into the intestine of adult hermaphrodites. The progeny were analyzed 8–12 hours after injection. For soaking experiments, the RNA was diluted 1:1 with 50% M9 buffer containing 3 mM spermidine hydrochloride (Sigma) and hermaphrodites were soaked for 12 hours in the RNA mixture.
ACKNOWLEDGMENTS
We thank members of the Rothman lab for helpful discussions. We are grateful to A. Fire for pPD vectors. We thank the Worm Genome Consortium for providing sequences. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). This work was supported by grants from the NIH and the March of Dimes to J.H.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. John Wiley & Sons Publishers; 2004. Section 12.1. [Google Scholar]
- Broitman-Maduro G, Maduro MF, Rothman JH. The noncanonical binding site of the MED-1 GATA factor defines differentially regulated target genes in the C. elegans mesendoderm. Dev. Cell. 2005;8:427–433. doi: 10.1016/j.devcel.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Broitman-Maduro G, Lin KT, Hung WW, Maduro MF. Specification of the C. elegans MS blastomere by the T-box factor TBX-35. Development. 2006;133:3097–3106. doi: 10.1242/dev.02475. [DOI] [PubMed] [Google Scholar]
- Bergemann AW, Johnson EM. The HeLa pur factor binds single-stranded DNA at a specific element conserved in gene flanking regions and origins of DNA replication. Mol. Cell. Biol. 1992;12:1257–1265. doi: 10.1128/mcb.12.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B, Eaton AE, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryos. Cell. 1992;68:1061–1075. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- Bowerman B, Draper BW, Mello CC, Priess JR. The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell. 1993;74:443–452. doi: 10.1016/0092-8674(93)80046-h. [DOI] [PubMed] [Google Scholar]
- Calvo D, Victor M, Gay F, Sui G, Luke PS, Dufourcq P, Wen G, Maduro M, Rothman JH, Shi Y. A POP-1 repressor complex restricts inappropriate cell type-specific gene transcription during Caenorhabditis elegans embryogenesis. EMBO J. 2001;20:7197–7208. doi: 10.1093/emboj/20.24.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzone FJ, Hoog C, Teplow DB, Cutting AE, Zeller RW, Britten RJ, Davidson EH. Gene regulatory factors of the sea urchin embryo. I. Purification by affinity chromatography and cloning of P3A2, a novel DNA-binding protein. Development. 1991;112:335–350. doi: 10.1242/dev.112.1.335. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Rust AG, Pan Z, Schilstra MJ, Clarke PJ, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Deppe U, Schierenberg E, Cole T, Krieg C, Schmitt D, Yoder B, von Ehrenstein G. Cell lineages of the embryo of the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. 1978;75:376–780. doi: 10.1073/pnas.75.1.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar JD, McGhee JD. DNA synthesis and the control of embryonic gene expression in C. elegans. Cell. 1988;53:589–599. doi: 10.1016/0092-8674(88)90575-2. [DOI] [PubMed] [Google Scholar]
- Gay F, Calvo D, Lo M, Ceron J, Maduro M, Lin R, Shi Y. Acetylation regulates subcellular localization of the Wnt signaling nuclear effector POP-1. Genes Dev. 2003;17:717–722. doi: 10.1101/gad.1042403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B. Establishment of gut fate in the E lineage of C. elegans: the roles of lineage-dependent mechanisms and cell interactions. Development. 1993;118:1267–1277. doi: 10.1242/dev.118.4.1267. [DOI] [PubMed] [Google Scholar]
- Goldstein B. Induction of gut in Caenorhabditis elegans embryos. Nature. 1992;357:255–257. doi: 10.1038/357255a0. [DOI] [PubMed] [Google Scholar]
- Gönczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- Haas S, Gordon J, Khalili K. A developmentally regulated DNA-binding protein from mouse brain stimulates myelin basic protein gene expression. Mol Cell Biol. 1993;13:3103–3112. doi: 10.1128/mcb.13.5.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas S, Thatikunta P, Steplewski A, Johnson EM, Khalili K, Amini S. A 39-kD DNA-binding protein from mouse brain stimulates transcription of myelin basic protein gene in oligodendrocytic cells. J. Cell Biol. 1995;130:1171–1179. doi: 10.1083/jcb.130.5.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Shetty P, Robertson SM, Lin R. Binary cell fate specification during C. elegans embryogenesis driven by reiterated reciprocal asymmetry of TCF POP-1 and its coactivator beta-catenin SYS-1. Development. 2007;134:2685–2695. doi: 10.1242/dev.008268. [DOI] [PubMed] [Google Scholar]
- Jin P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC, Liu H, Feng Y, Warren ST. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletta T, Schnabel H, Schnabel R. Binary specification of the embryonic lineage in Caenorhabditis elegans. Nature. 1997;390:294–298. doi: 10.1038/36869. [DOI] [PubMed] [Google Scholar]
- Kawasaki I, Shim YH, Kirchner J, Kaminker J, Wood WB, Strome S. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell. 1998;94:635–645. doi: 10.1016/s0092-8674(00)81605-0. [DOI] [PubMed] [Google Scholar]
- Khalili K, Del Valle L, Muralidharan V, Gault WJ, Darbinian N, Otte J, Meier E, Johnson EM, Daniel DC, Kinoshita Y, Amini S, Gordon J. Puralpha is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse. Mol Cell Biol. 2003;23:6857–6875. doi: 10.1128/MCB.23.19.6857-6875.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtsteiner S, Tjian R. Synergistic activation of transcription by UNC-86 and MEC-3 in Caenorhabditis elegans embryo extracts. EMBO J. 1995;14:3937–3945. doi: 10.1002/j.1460-2075.1995.tb00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Thompson S, Priess JR. pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell. 1995;83:599–609. doi: 10.1016/0092-8674(95)90100-0. [DOI] [PubMed] [Google Scholar]
- Lin R, Hill RJ, Priess JR. POP-1 and anterior-posterior fate decisions in C. elegans embryos. Cell. 1998;92:229–239. doi: 10.1016/s0092-8674(00)80917-4. [DOI] [PubMed] [Google Scholar]
- Lo M-C, Gay F, Odom R, Shi Y, Lin R. Phosphorylation by the β-catenin/MAPK complex promotes 14-3-3-mediated nuclear export of TCF/POP-1 in signal-responsive cells in C. elegans. Cell. 2004;117:95–106. doi: 10.1016/s0092-8674(04)00203-x. [DOI] [PubMed] [Google Scholar]
- Love JJ, Li X, Case DA, Giese K, Grosschedl R, Wright PE. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Lin R, Rothman JH. Dynamics of a developmental switch: Recursive intracellular and intranuclear redistribution of Caenorhabditis elegans POP-1 parallels Wnt-inhibited transcriptional repression. Dev. Biol. 2002;248:128–142. doi: 10.1006/dbio.2002.0721. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Meneghini MD, Bowerman B, Broitman-Maduro G, Rothman JH. Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and GSK-3beta homolog is mediated by MED-1 and -2 in C. elegans. Mol. Cell. 2001;7:475–485. doi: 10.1016/s1097-2765(01)00195-2. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Rothman JH. Making worm guts: The gene regulatory network of the Caenorhabditis elegans endoderm. Dev. Biology. 2002;246:68–85. doi: 10.1006/dbio.2002.0655. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Hill RJ, Heid PJ, Newman-Smith ED, Zhu J, Priess JR, Rothman JH. Genetic redundancy in endoderm specification within the genus Caenorhabditis. Dev Biol. 2005a;284:509–522. doi: 10.1016/j.ydbio.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Kasmir JJ, Zhu J, Rothman JH. The Wnt effector POP-1 and the PAL-1/Caudal homeoprotein collaborate with SKN-1 to activate C. elegans endoderm development. Dev Biol. 2005b;285:510–523. doi: 10.1016/j.ydbio.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Broitman-Maduro G, Mengarelli I, Rothman JH. Maternal deployment of the embryonic SKN-1-->MED-1,2 cell specification pathway in C. elegans. Dev Biol. 2007;301:590–601. doi: 10.1016/j.ydbio.2006.08.029. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Rothman JH. In: Specification of the endoderm. The C. elegans Research Community, editor. WormBook; 2008. In press. [Google Scholar]
- Meneghini MD, Ishitani T, Clayton C, Hisamoto N, Ninomiya-Tsuji J, Thorpe CJ, Hamill DR, Matsumoto K, Bowerman B. MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature. 1999;399:793–797. doi: 10.1038/21666. [DOI] [PubMed] [Google Scholar]
- Penberthy WT, Zhao C, Zhang Y, Jessen JR, Yang Z, Bricaud O, Collazo A, Meng A, Lin S. Pur alpha and Sp8 as opposing regulators of neural gata2 expression. Dev Biol. 2004;275:225–234. doi: 10.1016/j.ydbio.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Rocheleau CE, Downs WD, Lin R, Whitman C, Bei Y, Cha YH, Ali M, Priess JR, Mello CC. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell. 1997;90:707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- Rocheleau CE, Yasuda J, Shin T, Ruyling L, Sawa H, Okano H, Priess JR, Davis RJ, Mello CC. WRM-1 activates the LIT-1 protein kinase to transduce anterior/posterior polarity signals in C. elegans. Cell. 1999;97:717–726. doi: 10.1016/s0092-8674(00)80784-9. [DOI] [PubMed] [Google Scholar]
- Shelley CS, Da Silva Nicolas, Teodoridis JM. During U937 monocytic differentiation repression of the CD43 gene promoter is mediated by the single-stranded DNA binding protein Pur alpha. Brit. J. Haematology. 2001;115:159–166. doi: 10.1046/j.1365-2141.2001.03066.x. [DOI] [PubMed] [Google Scholar]
- Shelley CS, Teodoridis JM, Park H, Farokhzad OC, Bottinger EP, Arnaout MA. During differentiation of monocytic cell line U937, Purα mediates induction of the CD11c β2 integrin gene promoter. J. Immunol. 2002;168:3887–3893. doi: 10.4049/jimmunol.168.8.3887. [DOI] [PubMed] [Google Scholar]
- Shetty P, Lo MC, Robertson SM, Lin R. C. elegans TCF protein, POP-1, converts from repressor to activator as a result of Wnt-induced lowering of nuclear levels. Dev. Biol. 2005;285:584–592. doi: 10.1016/j.ydbio.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Shin TH, Yasuda J, Rocheleau CE, Lin R, Soto M, Yanxia B, Davis RJ, Mello CC. MOM-1, a MAP kinase kinase kinase-related protein activates WRM-1/LIT-1 kinase to transduce anterior/posterior polarity signals in C. elegans. Mol. Cell. 1999;4:275–280. doi: 10.1016/s1097-2765(00)80375-5. [DOI] [PubMed] [Google Scholar]
- Stroeher VL, Kennedy BP, Millen KJ, Schroeder DF, Hawkins MG, Goszczynski B, McGhee JD. DNA-Protein interactions in the Caenorhabditis elegans embryo: oocyte and embryonic factors that bind to the promoter of the gut-specific esterase ges-1 gene. Dev. Biol. 1994;163:367–380. doi: 10.1006/dbio.1994.1155. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Thorpe CJ, Schlesinger A, Carter JC, Bowerman B. Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell. 1997;90:695–705. doi: 10.1016/s0092-8674(00)80530-9. [DOI] [PubMed] [Google Scholar]
- Wood WB. Evidence from reversal of handedness in C. elegans embryos for early cell interactions determining cell fates. Nature. 1991;349:536–538. doi: 10.1038/349536a0. [DOI] [PubMed] [Google Scholar]
- Zhu J, Fukushige T, McGhee D, Rothman JH. Reprogramming of early embryonic blastomeres into endodermal progenitors by a Caenorhabditis elegans GATA factor. Genes Dev. 1998;12:3809–3814. doi: 10.1101/gad.12.24.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Hill RJ, Heid PJ, Fukuyama M, Sugimoto A, Priess JR, Rothman JH. end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev. 1997;11:2883–2896. doi: 10.1101/gad.11.21.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]