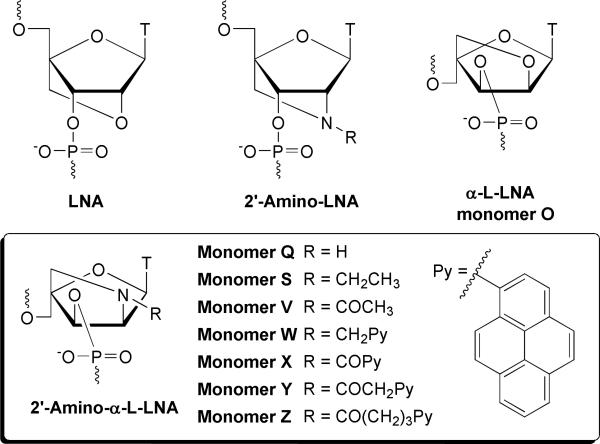
Abstract
Chemically modified oligonucleotides are increasingly applied in nucleic acid based therapeutics and diagnostics. LNA (Locked Nucleic Acid) and its diastereomer α-L-LNA are two promising examples hereof that exhibit increased thermal and enzymatic stability. Herein, the synthesis, biophysical characterization and molecular modeling of N2′-functionalized 2′-amino-α-L-LNA is described. Chemoselective N2′-functionalization of protected amino alcohol 1 followed by phosphitylation afforded a structurally varied set of target phosphoramidites, which were incorporated into oligodeoxyribonucleotides. Incorporation of pyrene-functionalized building blocks such as 2′-N-(pyren-1-yl)carbonyl-2′-amino-α-L-LNA (monomer X) led to extraordinary increases in thermal affinity of up to +19.5 °C per modification against DNA targets in particular. In contrast, incorporation of building blocks with small non-aromatic N2′-functionalities such as 2′-N-acetyl-2′-amino-α-L-LNA (monomer V) had detrimental effects on thermal affinity toward DNA/RNA complements with decreases of as much as −16.5 °C per modification. Extensive thermal DNA selectivity, favorable entropic contributions upon duplex formation, hybridization-induced bathochromic shifts of pyrene absorption maxima and increases of circular dichroism signals, and molecular modeling studies suggest that pyrene functionalized 2′-amino-α-L-LNA monomers W-Y having short linkers between the bicyclic skeleton and the pyrene moiety, allow high-affinity hybridization with DNA complements and precise positioning of intercalators in nucleic acid duplexes. This rigorous positional control has been utilized for the development probes for emerging therapeutic and diagnostic applications focusing on DNA-targeting.
Introduction
Oligonucleotides are widely used for modulation of gene expression (e.g., antigene/antisense/siRNA),1 for detection of nucleic acid targets,2 and as building blocks of novel self-assembling biomaterials.3 Chemical modification of oligonucleotides is often required to provide adequate protection from enzymatic degradation, to facilitate strong binding to complementary nucleic acid targets and to add functionality to oligonucleotides. Incorporation of conformationally restricted nucleotide monomers into oligonucleotides is a popular approach toward this end.4,5 Locked nucleic acid (LNA, β-d-ribo configuration, Fig. 1) is a very promising member of this class of compounds. LNA6-8 exhibits increases in thermal affinity toward DNA/RNA complements of up to +10 °C per modification along with markedly improved enzymatic stability relative to unmodified oligodeoxyribonucleotides (ONs).9,10 These properties render LNA with high therapeutic and diagnostic potential,11-14 which is underlined by ongoing Phase I/II clinical evaluations of LNA drug candidates against a variety of diseases. One of the diastereoisomers of LNA, i.e., α-L-LNA6,15 (α-l-ribo configuration, monomer O, Fig. 1) shares the beneficial properties of LNA and has been used as antisense ONs,16-18 triplex forming ONs,19 modified DNAzymes,20 and transcription factor decoy ONs.21
Figure 1.
Structures of LNA, 2′-Amino-LNA, α-L-LNA (monomer O), and 2′-Amino-α-L-LNA monomers Q-Z.
We have previously taken advantage of the known high-affinity hybridizations of 2′-amino-LNA6,22 (Fig. 1) to develop a series of N2′-functionalized 2′-amino-LNA, which precisely position functional entities in the minor groove of nucleic acid duplexes without compromising duplex stability.23 This has resulted in the development of tools for applications within therapeutics, diagnostics and material science including: a) probes yielding brightly fluorescent duplexes upon hybridization to complementary DNA/RNA with quantum yields approaching unity,23d,23k b) probes for single nucleotide polymorphism (SNP) detection,23b, 23h c) nucleic acid architectures auto signaling their self-assembly,23b,23h and d) artificial dinuclear ribonucleases.23j
Stimulated by these findings, we recently developed a synthetic route to 2′-amino-α-L-LNA (α-l-ribo configuration, monomer Q, Fig. 1),6,24 and N2′-functionalized analogs thereof (Fig. 1). Appended functional entities were anticipated to be positioned in the major groove of nucleic acid duplexes.24a However, initial studies with N2′-pyrene-functionalized 2′-amino-α-L-LNA suggest that the conjugated functional entity is directed toward the duplex core instead.25,26 This has already resulted in the development of promising tools for DNA-targeting,25a detection of single nucleotide polymorphisms25b and nucleic acid structural engineering.25d
Herein, full experimental details on the synthesis of a structurally varied set of N2′-functionalized 2′-amino-α-L-LNA phosphoramidites and their incorporation into ONs are presented (Fig. 1). Results from biophysical and computational studies are discussed together with the suggested binding mode of the appended functional entities.
Results and Discussion
Synthesis of N2′-functionalized 2′-amino-α-L-LNA
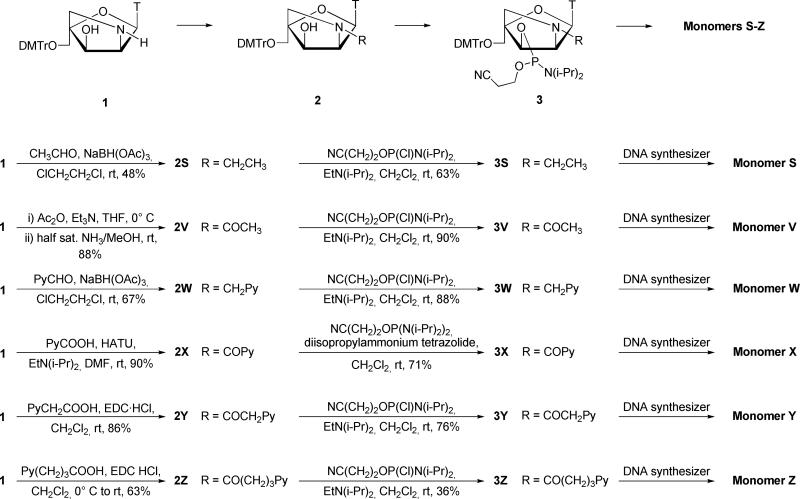
Known O5′-tritylated bicyclic nucleoside 124b, which is obtained from commercially available diacetone-α-D-glucose in 5% overall yield over seventeen steps involving eight chromatographic purification steps, was used as a suitable starting material for the synthesis of N2′-functionalized 2′-amino-α-L-LNA phosphoramidites 3Q-3Z (Scheme 1). The targets were selected to probe the available structural space in nucleic acid duplexes and fall into two groups based on the nature of the N2’-moiety, i.e., monomers with small non-aromatic units (monomers Q, S, and V) or with aromatic units (monomers W-Z). Sodium triacetoxyborohydride mediated reductive amination27 of secondary amine 1 with acetaldehyde or 1-pyrenecarbaldehyde furnished tertiary amines 2S and 2W25a in 48% and 67% yield, respectively. Chemoselective N-acylation of amino alcohol 1 was achieved using two different strategies. Treatment of nucleoside 1 with slight excess of acetic anhydride followed by selective O3′-deacylation using dilute methanolic ammonia furnished nucleoside 2V in excellent 88% yield over two steps. EDC-mediated coupling of amino alcohol 1 with 1-pyrenylcarboxylic acid, 1-pyrenylacetic acid or 4-(1-pyrenyl)butyric acid afforded nucleosides 2X, 2Y25b,25d and 2Z in 62%, 86% and 63% yield, respectively. A HATU-mediated (O-(7-azabenzotriazole-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate) coupling procedure successfully improved the yield of 2X to 90%. Disappearance of 1H NMR signals of the exchangeable 3′-OH protons upon D2O addition ascertained the N2′-functionalized constitution of nucleosides 2S-2Z, which subsequently were converted to the corresponding phosphoramidites 3S-3Z using 2-cyanoethyl N,N′-(diisopropyl)-phosphoramidochloridite and Hünig's base. While amidites 3S-3Y were obtained in good to excellent yields (60-90 %), 3Z was only obtained in 36% yield. The yield of 3X was improved using bis-(N,N-diisopropylamino)-2-cyanoethoxyphosphine in dichloromethane with diisopropylammonium tetrazolide28 as activator (71%). Synthesis of ONs was performed in 0.2 μmol scale using an automated DNA synthesizer. The corresponding phosphoramidites for incorporation of α-L-LNA thymine monomer O (obtained from commercial sources) and 2′-amino-α-L-LNA thymine monomer Q (synthesized by a previously described protocol)24b were incorporated into our preferred model system, i.e., a set of mixed sequence 9-mer ONs, as previously described.15b,24b Standard procedures were applied for incorporation of N2′-functionalized 2′-amino-α-L-LNA thymine monomers S-Z (Fig. 1) except for extended coupling times: 3S (10 min), 3V (10 min), 3W25a (30 min), 3Y25b (30 min), 3X (30 min) and 3Z (15 min)) using 1H-tetrazole as catalyst resulting in stepwise coupling yields of ~99% for monomers S, V, X, Y, Z and ~95% for monomer W (Fig. 1 for structures). Following standard workup and purification, the composition and purity (>80%) of all modified ONs was verified by MALDI-MS (Table S3)29 and ion-exchange HPLC, respectively. Please NOTE that the unmodified reference DNA and RNA strands are denoted D1/D2 and R1/R2, respectively, while ONs containing a single incorporation of a modified nucleotide in the 5′-GBG ATA TGC context are named O1, Q1, S1 etc. Similar conventions were used for ONs in B2-B7 series (Tables 1 and 2). In addition, the following descriptive nomenclature is used: α-L-amino-LNA (Q-series), Et α-L-amino-LNA (S-series), Ac-α-L-amino-LNA (V-series), PyMe-α-L-amino-LNA (W-series), PyCO-α-L-amino-LNA (X-series), PyAc-α-L-amino-LNA (Y-series) and PyBu-α-L-amino-LNA (Z-series).
Scheme 1.
Synthesis of N2′-functionalized 2′-amino-α-L-LNA phosphoramidites. DMTr = 4,4′-dimethoxytrityl, T = thymin-1-yl, Py = pyren-1-yl, EDC·HCl = 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride, HATU = O (7-Azabenzotriazole-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate.
Table 1.
Thermal denaturation data for N2′-Functionalized 2′-amino-α-L-LNA and reference strands against DNA complements.a
|
Tm [ΔTm/mod] (°C) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ON | Duplex | B = | T | O | Q | S | V | W | X | Y | Z |
| B1 | 5′-GBG ATA TGC | 28.5 | 31.0 | 26.5 | 20.0 | 17.5 | 35.5 | 38.5 | 39.0 | 29.0 | |
|
D2
|
3′-CAC TAT ACG |
|
|
[+2.5] |
[–2.0] |
[–8.5] |
[–11.0] |
[+7.0] |
[+10.0] |
[+10.5] |
[+0.5] |
| B2 | 5′-GTG ABA TGC | 28.5 | 34.5 | 29.0 | 20.0 | 16.5 | 42.5 | 47.5 | 44.0 | 34.5 | |
|
D2
|
3′-CAC TAT ACG |
|
|
[+6.0] |
[+0.5] |
[–8.5] |
[–12.0] |
[+14.0] |
[+19.0] |
[+15.5] |
[+6.0] |
| B3 | 5′-GTG ATA BGC | 28.5 | 31.5 | 27.5 | 16.5 | 14.5 | 39.0 | 42.5 | 40.0 | ND | |
|
D2
|
3′-CAC TAT ACG |
|
|
[+3.0] |
[–1.0] |
[–12.0] |
[–14.0] |
[+10.5] |
[+14.0] |
[+11.5] |
|
| D1 | 5′-GTG ATA TGC | 28.5 | 32.0 | 28.0 | 17.0 | 12.0 | 35.0 | 39.0 | 38.5 | 29.0 | |
|
B4
|
3′-CAC BAT ACG |
|
|
[+3.5] |
[–0.5] |
[–11.5] |
[–16.5] |
[+6.5] |
[+10.5] |
[+10.0] |
[+0.5] |
| D1 | 5′-GTG ATA TGC | 28.5 | 36.5 | 31.0 | 22.5 | 19.0 | 44.0 | 48.0 | 45.0 | 35.0 | |
|
B5
|
3′-CAC TAB ACG |
|
|
[+8.0] |
[+2.5] |
[–6.0] |
[–9.5] |
[+15.5] |
[+19.5] |
[+16.5] |
[+6.5] |
| D1 | 5′-GTG ATA TGC | 28.5 | 36.0 | 27.5 | <10 | <10 | ND | 53.5 | 55.5 | ND | |
|
B6
|
3′-CAC BAB ACG |
|
|
[+3.8] |
[–0.5] |
|
|
|
[+12.5] |
[+13.5] |
|
| B7 | 5′-GBG ABA BGC | 28.5 | 36.0 | 27.0 | <10 | <10 | ND | ND | 69.0 | ND | |
| D2 | 3′-CAC TAT ACG | [+2.5] | [–0.5] | [+13.5] | |||||||
Thermal denaturation temperatures [Tm values/°C (ΔTm = change in Tm value calculated relative to D1:D2, D1:R2 and R1:D2 reference duplexes)] measured as the maximum of the first derivative of the melting curve (A260 vs temperature) recorded in medium salt buffer ([Na+] = 110 mM, [Cl–] = 100 mM, pH 7.0 (NaH2PO4/Na2HPO4)), using 1.0 μM concentrations of the two complementary strands. Tm values are averages of at least two measurements; A = adenin-9-yl DNA monomer, C = cytosin-1-yl DNA monomer, G = guanin-9-yl DNA monomer, T = thymin-1-yl DNA monomer, O = α-L-LNA thymin-1-yl monomer (Fig. 1). For structures of monomers Q-Z see Figure 1. ND = not determined.
Table 2.
Thermal denaturation data for N2′-functionalized 2′-amino-α-L-LNA and reference strands against RNA complements.a
|
Tm [ΔTm/mod] (°C) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ON | Duplex | B = | T | O | Q | S | V | W | X | Y | Z |
| B1 | 5′-GBG ATA TGC | 26.5 | 32.5 | 27.5 | 19.5 | 19.0 | 27.0 | 32.5 | 32.5 | 26.5 | |
|
R2
|
3′-CAC UAU ACG |
|
|
[+6.0] |
[+1.0] |
[–7.0] |
[–7.5] |
[+0.5] |
[+6.0] |
[+6.0] |
[±0] |
| B2 | 5′-GTG ABA TGC | 26.5 | 35.0 | 28.5 | 18.5 | 17.0 | 31.5 | 36.5 | 36.0 | 33.5 | |
|
R2
|
3′-CAC UAU ACG |
|
|
[+8.5] |
[+2.0] |
[–8.0] |
[–9.5] |
[+5.0] |
[+10.0] |
[+9.5] |
[+7.0] |
| B3 | 5′-GTG ATA BGC | 26.5 | 32.0 | 28.0 | 19.0 | 16.5 | 28.0 | 33.5 | 32.5 | ND | |
|
R2
|
3′-CAC UAU ACG |
|
|
[+5.5] |
[+1.5] |
[–7.5] |
[–10.0] |
[+1.5] |
[+7.0] |
[+6.0] |
|
| R1 | 5′-GUG AUA UGC | 24.5 | 31.0 | 27.5 | 19.5 | 17.5 | 23.5 | 29.0 | 30.5 | 19.5 | |
|
B4
|
3′-CAC BAT ACG |
|
|
[+6.5] |
[+3.0] |
[–5.0] |
[–7.0] |
[–1.0] |
[+4.5] |
[+6.0] |
[–5.0] |
| R1 | 5′-GUG AUA UGC | 24.5 | 34.5 | 29.0 | 21.5 | 20.5 | 32.0 | 36.0 | 36.5 | 31.0 | |
|
B5
|
3′-CAC TAB ACG |
|
|
[+10.0] |
[+4.5] |
[–3.0] |
[–4.0] |
[+7.5] |
[+11.5] |
[+12.0] |
[+6.5] |
| R1 | 5′-GUG AUA UGC | 24.5 | 35.5 | 28.5 | <10 | <10 | ND | 39.0 | 44.0 | ND | |
|
B6
|
3′-CAC BAB ACG |
|
|
[+5.5] |
[+2.0] |
|
|
|
[+7.3] |
[+9.8] |
|
| B7 | 5′-GBG ABA BGC | 26.5 | 44.0 | 31.0 | <10 | <10 | ND | ND | 52.5 | ND | |
| D2 | 3′-CAC UAU ACG | [+5.8] | [+1.5] | [+8.7] | |||||||
For conditions of thermal denaturation experiments see Table 1.
Thermal denaturation studies – experimental setup
The effect upon incorporation of one to three O-Z monomers (Fig. 1) into mixed sequence 9-mer ONs on thermal affinity toward DNA and RNA targets (Tables 1 and 2, respectively) was evaluated by UV thermal denaturation experiments using medium salt buffer ([Na+] = 110 mM), and compared to unmodified DNA. The UV thermal denaturation curves of all modified duplexes exhibited sigmoidal monophasic transitions with hyperchromicities (9-15%) which are comparable to the corresponding unmodified DNA:DNA or DNA:RNA duplexes (Fig. S1).29 All changes in thermal denaturation temperatures (Tm) of modified nucleic acid duplexes are discussed relative to Tm values of unmodified reference duplexes unless otherwise mentioned. In addition, the Watson-Crick specificity of ONs with a single central incorporation of monomer O-Z (B2-series) was evaluated by determining Tm values of the duplexes with DNA/RNA strands with central mismatches (Table 3).
Table 3.
Discrimination of mismatched DNA/RNA targets by N2′-functionalized 2′-amino-α-L-LNA.
|
Tm [ΔTm] (°C) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DNA: 3′-CAC TBT ACG |
RNA: 3′-CAC UBU ACG |
|||||||||
| ON | Sequence | B = | A | C | G | T | A | C | G | U |
| D1 | 5′-GTG ATA TGC | 28.5 | 12.0 [–16.5] | 19.0 [–9.5] | 11.5 [–17.0] | 26.5 | <10 [<–16.5] | 22.0 [–4.5] | <10 [<–16.5] | |
| O2 | 5′-GTG AOA TGC | 34.5 | 13.0 [–21.5] | 21.5 [–13.0] | 14.5 [–20.0] | 35.0 | 14.0 [–21.0] | 28.5 [–6.5] | 14.5 [–20.5] | |
| Q2 | 5′-GTG AQA TGC | 29.0 | <10 [<–19.0] | 17.5 [–11.5] | <10 [<–19.0] | 28.5 | <10 [<–18.5] | 22.5 [–6.0] | <10 [<–18.5] | |
| S2 b | 5′-GTG ASA TGC | 28.5 | <10 [<–18.5] | 17.0 [–11.5] | <10 [<–18.5] | 27.0 | <10 [<–17.0] | 25.5 [–1.5] | <10 [<–17.0] | |
| V2 b | 5′-GTG AVA TGC | 24.5 | <10 [<–14.5] | 14.5 [–10.0] | <10 [<–14.5] | 24.5 | <10 [<–14.5] | 28.0 [+3.5] | <10 [<–14.5] | |
| W2 | 5′-GTG AWA TGC | 42.5 | 30.0 [–12.5] | 37.0 [–5.5] | 39.0 [–3.5] | 31.5 | 19.5 [–12.0] | 30.5 [–1.0] | 27.0 [–4.5] | |
| X2 | 5′-GTG AXA TGC | 47.5 | 31.0 [–16.5] | 40.0 [–7.5] | 40.5 [–7.0] | 36.5 | 21.0 [–15.5] | 33.5 [–3.0] | 28.5 [–8.0] | |
| Y2 | 5′-GTG AYA TGC | 44.0 | 26.0 [–18.0] | 38.5 [–5.5] | 34.0 [–10.0] | 36.0 | 14.5 [–21.5] | 33.0 [–3.0] | 25.0 [–11.0] | |
| Z2 | 5′-GTG AZA TGC | 34.5 | 17.0 [–17.5] | 20.0 [–14.5] | 20.0 [–14.5] | 33.5 | <10 [<–22.5] | 17.5 [–16.0] | <10 [<–22.5] | |
For conditions of thermal denaturation experiments see Table 1. Tm values of fully matched duplexes are shown in bold. ΔTm = change in Tm value relative to fully matched DNA:DNA or DNA:RNA duplex.
Tm values for duplexes involving S2 and V2 are measured using high salt buffer, ([Na+] = 710 mM, [Cl–] = 100 mM, pH 7.0 (NaH2PO4/Na2HPO4)).
Thermal denaturation studies – α-L-LNA and 2′-amino-α-L-LNA – the reference ONs
α-L-LNA O1-O5 exhibit substantially increased thermal affinity toward DNA (ΔTm up to +8.0 °C, Table 1) and RNA complements (ΔTm = up to +10.0 °C, Table 2). The corresponding 2′-amino-α-L-LNA Q1-Q5 display notably smaller increases in thermal affinity (ΔTm up to +2.5 °C with DNA, Table 1; ΔTm up to +4.5 °C with RNA, Table 2). Similar ΔTm-values for duplexes between α-L-amino-LNA Q4 and DNA/RNA complements determined at different ionic strengths were observed (Table S4),29 suggesting that the 2-oxo-5-azabicyclo[2.2.1]heptane skeleton of monomer Q is not protonated at physiological pH. The increased binding affinity of α-L-LNA and 2′-amino-α-L-LNA was accompanied by improved discrimination of singly mismatched DNA and RNA targets relative to unmodified DNA D1 (e.g., ΔTm values for α-L-amino-LNA Q2 and D1 against DNA mismatches, Table 3).
Thermal denaturation studies – N2′-pyrene-functionalized 2′-amino-α-L-NA
Incorporation of a single PyMe/PyCO/PyAc-α-L-amino-LNA monomer W, X or Y, respectively, into ONs resulted in extraordinary increases in thermal affinity toward DNA complements (ΔTm from +6.5 °C to +19.5 °C, Table 1). Moderate increases were observed upon incorporation of PyBu-α-L-amino-LNA monomer Z (ΔTm up to +6.5 °C, Table 1). The observed trends in thermal affinity of singly modified strands toward DNA targets (X > Y > W >> Z) suggest that: a) alkanoyl linkers are thermally preferred over alkyl linkers of the same length (X > W), and b) shorter linkers are thermally preferred (X > Y >> Z). For a discussion on the sequence dependent variations of Tm-values observed for these ONs the reader is directed to the supporting information.29 Interestingly, additive increases in thermal affinity toward DNA targets are observed upon multiple incorporations of PyAc-α-L-amino-LNA Y monomers (e.g., compare Tm/mod values of Y6:D1, Y4:D1 and Y5:D1, Table 1), while subadditive increases are observed for the corresponding α-L-LNA O6, 2’-amino-α-L-LNA Q6 or PyMe-α-L-amino-LNA X6. Thus, PyAc-α-L-amino-LNA monomer Y lends itself as the building block of choice for applications necessitating densely functionalized ONs with maximal thermal affinity toward DNA targets.
PyCO/PyAc-α-L-amino-LNA (X1-X6 and Y1-Y7, respectively) exhibit prominent and additive increases in thermal affinity toward RNA complements (ΔTm from +4.5 °C to +12.0 °C, Table 2). In contrast, minor destabilizations to moderate increases were observed for PyMe/PyBu-α-L-amino-LNA W1-W5 and Z1-Z5, respectively, with the exception of Z4 which exhibited a very pronounced decrease in thermal affinity toward its RNA target (Table 2).
Accordingly, N2′-pyrene-functionalized 2′-amino-α-L-LNA exhibit a marked DNA selectivity, i.e., a positive ΔΔTm/mod (DNA-RNA) = ΔTm/mod (DNA) − ΔTm/mod (RNA). This is particularly noteworthy as the parent α-L-LNA and 2′-amino-α-L-LNA exhibit moderate RNA-selectivity (ΔΔTm/mod (DNA-RNA) = −3.5 to −1.7 °C, Tables 1 and 2 or, more conveniently, Table S529). PyMe/PyCO-α-L-amino-LNA exhibit the most pronounced DNA selectivity (ΔΔTm/mod (DNA-RNA) = +6.0 to +9.0 °C, Table S5), suggesting that short linkers between the pyrene and nucleoside moieties facilitate DNA-selectivity. While PyMe/PyCO-α-L-amino-LNA exhibit a similar degree of DNA selectivity as acyclic intercalating nucleic acids (INAs),30 2′-O-pyrenylmethyl uridines31 or pyrene-functionalized 4′-C-piperazinomethyl thymidines,32 they generally form stronger duplexes with DNA targets, which renders them as highly interesting probes for DNA-targeting applications.25a
Centrally modified PyMe-α-L-amino-LNA W2 exhibit less efficient discrimination of mismatched DNA/RNA targets than the corresponding reference strand D1 (Table 3). Interestingly, a change in linker chemistry from methylene to carbonyl (W→X) results in higher affinity toward DNA/RNA complements as well as significantly improved mismatch discrimination (Table 3). With the exception of T:T/U mismatches, PyCO-α-L-amino-LNA X2 displays mismatch discrimination comparable to reference strand D1. Increases in linker length result in progressively improved discrimination of DNA/RNA mismatches (compare ΔTm data for X2, Y2 and Z2, Table 3). Accordingly, PyBu-α-L-amino-LNA Z2 exhibits superior discrimination of RNA mismatches in general, and of the challenging T:rG mismatch in particular (ΔTm = −16.0 °C, data for Z2, Table 3), relative to the already highly discriminative α-L-LNA O2.
Thermal denaturation studies – N2′-ethyl/acetyl-modified 2′-amino-α-L-LNA
Et-α-L-amino-LNAs S1-S7 exhibit greatly decreased thermal affinities toward DNA/RNA targets in general and complementary DNA in particular (ΔTm/mod down to −12.0 °C, Table 1). These effects are even more pronounced with Ac-α-L-amino-LNA V1-V7 which exhibit decreases in Tm values down to −16.5 °C per modification (Table 1). Accordingly, no duplex transitions could be observed for ONs with two or three incorporations of S or V monomers and their DNA/RNA targets. Interestingly, the large decreases in thermal affinity toward DNA/RNA complements of Et/Ac-α-L-amino-LNA, generally did not compromise Watson-Crick specificity (see data for S2 and V2, Table 3), which suggests that base-pairing is preserved.
It is noteworthy that an exchange of a centrally positioned PyCO-α-L-amino-LNA monomer X with a corresponding Ac-α-L-amino-LNA monomer V (i.e., a formal change of pyrene to methyl), was accompanied by a decrease in thermal affinity toward complementary DNA of 31.0 °C (compare Tm values of X2:D2 and V2:D2, Table 1). This suggests very different binding mode for ONs modified with monomers S/V and W/Y, respectively, which was underlined upon additional biophysical characterization (vide infra).
Additional biophysical characterization of N2′-functionalized 2′-amino-α-L-LNA – experimental setup
To obtain additional insight into the highly divergent thermal affinities of N2′-functionalized 2′-amino-α-L-LNA the following biophysical studies were performed: a) determination of thermodynamic parameters for duplex formation, b) CD-spectra, c) UV-vis spectra (shifts of pyrene absorption maxima), and d) molecular modeling studies.
Thermodynamic parameters for duplex formation were determined by melting curve analysis assuming bimolecular reactions and two-state equilibrium hypothesis. Quality of the baseline permitting, thermodynamic parameters for two melting curves per investigated duplex were determined and an average value is listed. In full agreement with expectations, formation of all studied duplexes was favorable (ΔG298 < 0 kJ/mol), with favorable enthalpic (ΔH < 0 kJ/mol) and unfavorable entropic contributions (T298ΔS < 0 kJ/mol). The thermodynamic data rely on assumptions of two-state melting behavior and a heat capacity change ΔCp = 0 upon hybridization, which may not necessarily be fulfilled. However, apart from few exceptions (see footnote a in Table 4), the observed enthalpic/entropic contributions for hybridization of N2′-functionalized 2′-amino-α-L-LNA to DNA/RNA targets clearly followed monomer and sequence specific trends, which validates the utilized approach (data shown for the representative B2-series Tables 4 and 5; for data and full discussion of B1-B5 series see Table S629).
Table 4.
Energetics derived from thermal denaturation curves of duplexes between 2'-amino-α-L-LNA functionalized with non-aromatic moieties and DNA/RNA.a
|
Complementary DNA |
Complementary RNA |
||||||
|---|---|---|---|---|---|---|---|
| ON | Sequence | ΔG298 [ΔΔG298] (kJ/mol) | ΔH [ΔΔH] (kJ/mol) | T298ΔS [Δ(T298ΔS)] (kJ/mol) | ΔG298 [ΔΔG298] (kJ/mol) | ΔH [ΔΔH] (kJ/mol) | T298ΔS [Δ(T298ΔS)] (kJ/mol) |
| D1 | 5′-GTG ATA TGC | –41 | –327 | –286 | –38 | –275 | –237 |
| O2 | 5′-GTG AOA TGC | –46 [–5] | –354 [–27] | –308 [–22] | –46 [–8] | –361 [–86] | –315 [–78] |
| Q2 | 5′-GTG AQA TGC | –41 [0] | –312 [+15] | –271 [+15] | –40 [–2] | –302 [–27] | –261 [–24] |
| S2 | 5′-GTG ASA TGC | –33 [+8] | –349 [–22] | –316 [–30] | –31 [+7] | –352 [–77] | –321 [–84] |
| V2 | 5′-GTG AVA TGC | –29 [+12] | –295 [+32] | –266 [+20] | –30 [+8] | –303 [–28] | –273 [–36] |
Thermal denaturation curves were obtained as described in Tables 1 and 2. ΔΔG298, ΔΔH and Δ(T298ΔS) change in ΔG298, ΔH and Δ(T298ΔS) values, respectively, calculated relative to D1:D2 and D1:R2 reference duplexes). Values in italics indicate deviation from (or lack of) monomer trend. ND = not determined. See Table S6 for data from B1-B5 series.
Table 5.
Energetics derived from thermal denaturation curves of duplexes between N2'-pyrene-functionalized 2'-amino-α-L-LNA and DNA/RNA.a
|
Complementary DNA |
Complementary RNA |
||||||
|---|---|---|---|---|---|---|---|
| ON | Sequence | ΔG298 [ΔΔG298] (kJ/mol) | ΔH [ΔΔH] (kJ/mol) | T298ΔS [Δ(T298ΔS)] (kJ/mol) | ΔG298 [ΔΔG298] (kJ/mol) | ΔH [ΔΔH] (kJ/mol) | T298ΔS [Δ(T298ΔS)] (kJ/mol) |
| D1 | 5′-GTG ATA TGC | –41 | –327 | –286 | –38 | –275 | –237 |
| W2 | 5′-GTG AWA TGC | –53 [–12] | –307 [+20] | –254 [+32] | –44 [–6] | –328 [–53] | –284 [–47] |
| X2 | 5′-GTG AXA TGC | –59 [–18] | –348 [–21] | –289 [–3] | –49 [–11] | –343 [–68] | –294 [–57] |
| Y2 | 5′-GTG AYA TGC | –54 [–13] | –321 [+6] | –267 [+19] | –48 [–10] | –333 [–58] | –285 [–48] |
| Z2 | 5′-GTG AZA TGC | –46 [–5] | –336 [–9] | –290 [–4] | ND | ND | ND |
See Table 4 for conditions.
Duplexes between N2′-functionalized 2′-amino-α-L-LNA and DNA/RNA complements were studied by force field simulations. For this, DNA duplexes were built in silico and modified with an N2′-functionalized 2′-amino-α-L-LNA monomer. A starting B-type helix geometry was chosen as duplexes between α-L-LNA and DNA complements adopt helix geometries that are globally unperturbed relative to unmodified DNA:DNA duplexes.33 ONs with centrally positioned modifications (B2-series) were selected for the simulations to minimize the influence of fraying on the helix geometry near the modified nucleotides. The position of N2′-functionalities was explored using a truncated Monte Carlo search,29 and the partially constrained duplexes were subjected to stochastic dynamics simulations using the all-atom AMBER force field34 and GB/SA solvation model35 as implemented in the MacroModel V9.1 suite of programs.36
Integrated structural discussion - α-L-LNA and 2′-amino-α-L-LNA – the reference ONs
The markedly increased thermal affinity of α-L-LNA O1-O5 toward complementary DNA/RNA relative to unmodified ONs results from a more favorable enthalpic term that largely is counterbalanced by an unfavorable entropic term, i.e., ΔΔG298 (O2DNA) = ΔG298(O2:D2) − ΔG298(D1:D2) = −5 kJ/mol; ΔΔH (O2DNA) = ΔH(O2:D2) − ΔH(D1:D2) = −27 kJ/mol; Δ(T298ΔS) (O2DNA) = T298 ΔS (O2:D2) − T298ΔS (D1:D2) = −22 kJ/mol (Table 4). This suggests that high thermal affinity of the conformationally restricted α-L-LNA O1-O5 toward DNA/RNA complements is a result of a more favorable stacking/hydrogen bonding geometry and/or duplex solvation rather than preorganization of the single stranded probe.
Similarly, the hybridization of 2′-amino-α-L-LNA Q1-Q5 to complementary RNA is also driven by favorable enthalpy that is partially counterbalanced by unfavorable entropy, although the individual contributions are less pronounced than for α-L-LNA O1-O5 (e.g., ΔΔH = −86 kJ/mol and −27 kJ/mol for O2RNA and Q2RNA, respectively, Table 4). The energetics for hybridization of Q1-Q5 to DNA complements are sequence dependent and could not be fitted to a clear pattern, Tables 4 and S629).
The applied molecular modeling protocol successfully reproduced expected global and local features of α-L-LNA duplex O2:D2 (Fig. S4) providing credibility to the applied computational protocol. These features of O2:D2 include a standard B-type global duplex geometry similar to D1:D2 (Fig. S3) and very characteristic local perturbations in the backbone needed to accommodate the inverted configurations at the C2′-, C3′- and C4′-positions of α-L-LNA monomer O.29,33 Interestingly, the global helix structures of O2:D2 and Q2:D2 (Fig. S5)29 are virtually identical, which is validated by very similar circular dichroism spectra of Q7:D2 and O7:D2 (Fig. S2). Thus, different solvation patterns rather than substantially altered helical geometries likely account for the diverging energetics observed for these closely related ONs upon hybridization with DNA/RNA complements.
Integrated structural discussion - N2′-ethyl/acetyl-modified 2′-amino-α-L-LNA
The dramatically destabilized duplexes between Et/Ac-α-L-amino-LNA and DNA/RNA targets (ΔΔG298 up to +12 kJ/mol) result from unfavorable entropic components that are only partially counterbalanced by favorable enthalpic components (e.g., Δ(T298ΔS) (S2RNA) = −84 kJ/mol and ΔΔH (S2RNA) = −77 kJ/mol, Table 4).
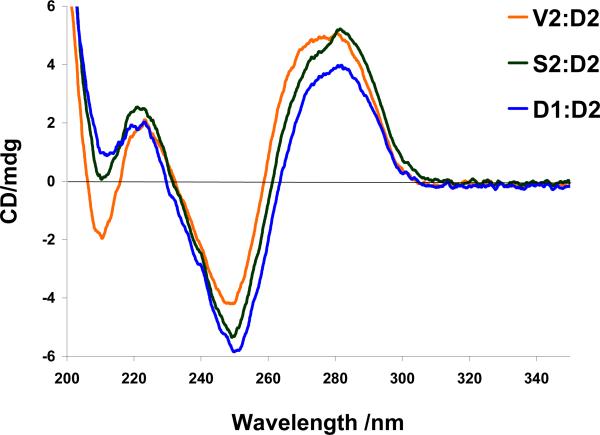
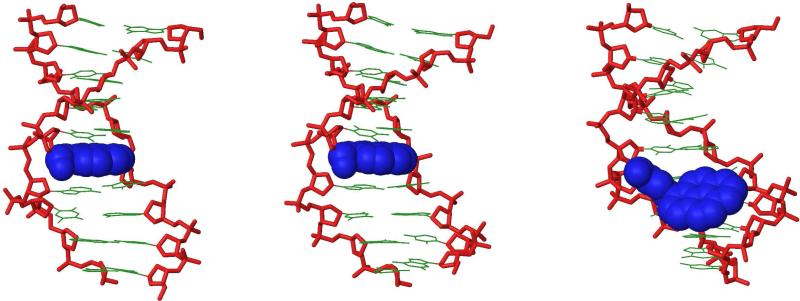
Intriguingly, very similar CD spectra are observed for S2:D2, V2:D2 and the reference duplex D1:D2 suggesting that incorporation of S and V monomers renders these duplexes globally unperturbed while dramatically lowering stability (Fig 2). In accordance with this, the lowest energy structures of Et-α-L-amino-LNA duplex S2:D2 (Figs. 3 and S6) and Ac-α-L-amino-LNA duplex V2:D2 (Figs. 3 and S7) globally resembled each other and α-L-amino-LNA duplex Q2:D2 (Fig. S5).29 In S2:D2 the ethyl group of S5 protrudes from the major groove valley to become involved in a steric clash with H5′ of A6 (for numbering scheme see Fig. 4). We speculate that unfavorable desolvation of the apolar ethyl moiety (whereby fewer water molecules are released) and interference with structural water along the sugar-phosphate backbone,37,38 in a similar manner as proposed for monomer Q,29 accounts for the unfavorable entropy observed upon hybridization of Et-α-L-amino-LNA S2 with DNA/RNA targets (Table 4). In a related manner, one part of the N2′-acetyl moiety of monomer V (i.e., either the -CO- or -CH3) in V2:D2 is directed toward the major groove where it can interfere with structural water while the other part simultaneously protrudes into the duplex core to disrupt π-π stacking (Figs. 3 and S7).29
Figure 2.
Circular dichroism spectra of D1:D2, S2:D2 and V2:D2.
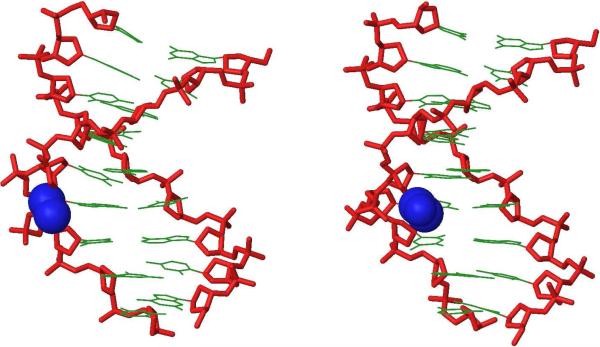
Figure 3.
Side view representations of the lowest energy structure of S2:D2 (left) and V2:D2 (right). For clarity, hydrogen atoms, sodium ions and bond orders have been omitted. Coloring scheme: nucleobases, green; sugar-phosphate backbone, red; ethyl moiety of monomer S and acetyl moiety of monomer V, blue.
Figure 4.
Nucleotide numbering for duplexes studied by molecular modeling; B = thymidine (D1) or monomers O-Z (O2-Z2).
To sum up, biophysical characterization and computer simulations jointly suggest that N2′-functionalization of 2′-amino-α-L-LNA, contrary to preliminary expectations,24a is not suitable to position small non-aromatic moieties in the major groove of duplexes with DNA or RNA complements.
Integrated structural discussion - N2′-pyrene-functionalized 2′-amino-α-L-LNA
The very pronounced stabilization of duplexes between PyMe-α-L-amino-LNA or PyAc-α-L-amino-LNA and complementary DNA (e.g. ΔΔG298 (W2DNA) = −12 kJ/mol) results from highly favorable entropy (e.g. ΔΔ(T298ΔS) (W2DNA) = +32 kJ/mol, Table 5). Stabilization of duplexes between PyCO-α-L-amino-LNA and DNA targets (e.g. ΔΔG298 (X2DNA) = −18 kJ/mol) is to a greater extent driven by favorable enthalpic factors. The moderate stabilization of duplexes between PyBu-α-L-amino-LNA and DNA complements (e.g. ΔΔG298 (Z2DNA) = −5 kJ/mol), originates from favorable enthalpy contributions that mostly were counterbalanced by unfavorable entropy components (ΔΔH (Z2DNA) = −9 kJ/mol, Δ(T298ΔS) (Z2DNA) = −4 kJ/mol, Table 5).
The observed stabilization of duplexes between PyMe/PyCO/PyAc-α-L-amino-LNA with RNA complements results from favorable enthalpy (ΔΔH = −53 kJ/mol, −68 kJ/mol and −58 kJ/mol for W2RNA, X2RNA and Y2RNA, respectively). However, comparison with 2′-amino-α-L-LNA reference strands instead of unmodified DNA suggests that entropic contributions also aid duplex formation with RNA targets (e.g. Δ(T298ΔS) = −17 kJ/mol, −28 kJ/mol, −17 kJ/mol and −51 kJ/mol for W3RNA, X3RNA, Y3RNA and Q3RNA, respectively, Table S6).29
Thus, energetics suggest that N2’-pyrene-functionalized 2’-amino-α-L-LNA exhibit binding modes that rely on preorganization unlike than 2’-amino-α-L-LNA modified with non-aromatic moieties.
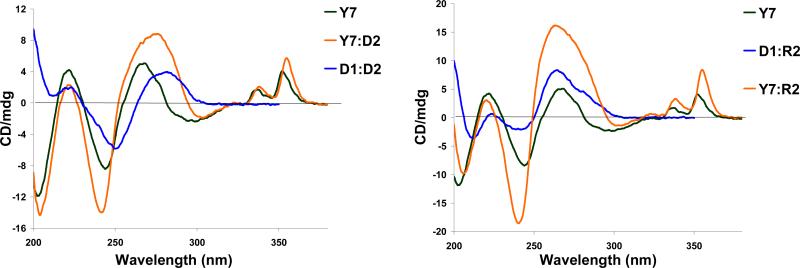
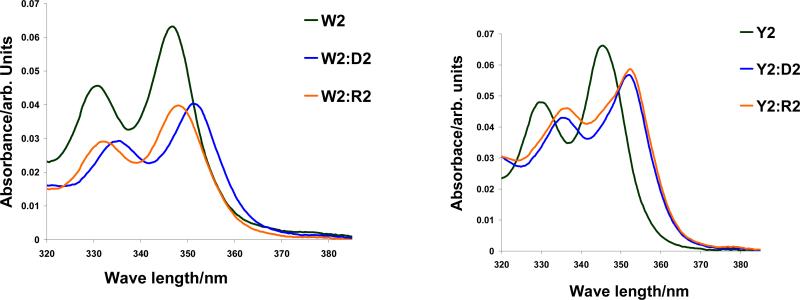
The pronounced DNA-selectivity of PyMe/PyCO/PyAc-α-L-amino-LNA suggests intercalation of the pyrene moieties as a likely binding mode.30-32,39 The CD spectra of PyAc-α-L-amino-LNA and duplexes with complementary DNA/RNA support this hypothesis as induced CD bands in the region of pyrene absorption (λ = 320-360 nm, Fig. 5), a feature indicative of intercalation,40 are observed upon hybridization. In addition, marked bathochromic shifts of pyrene absorption maxima of ONs containing monomers W-Y upon hybridization with DNA/RNA targets (Δλmax = 1-5 nm and 0-6 nm, respectively, Table 6) along with hypochromic shifts (illustrated for W2 and Y2, Fig. 6) suggest strong electronic interactions between the pyrene and nucleobase moieties in duplexes.40-42 A change in linker chemistry from alkyl to alkanoyl (PyMe-α-L-amino-LNA W → PyCO-α-L-amino-LNA X) resulted in small but consistently larger hybridization-induced bathochromic shifts, while further extension of the alkanoyl linker (PyCO-α-L-amino-LNA X → PyAc-α-L-amino-LNA Y → PyBu-α-L-amino-LNA Z) progressively reversed this trend. The very subtle hybridization-induced bathochromic shifts of pyrene absorption maxima observed with PyBu-α-L-amino-LNA indicate a non intercalating binding mode of the pyrene moiety of monomer Z (Table 6).
Figure 5.
Circular dichroism spectra of Y7 (5′-GYG AYA YGC) and its duplexes with DNA/RNA complements, and reference DNA:DNA (D1:D2) and DNA:RNA (D1:R2).
Table 6.
Pyrene absorption maxima for single stranded pyrene-functionalized 2′-amino-α-L-LNA and the corresponding duplexes with DNA/RNA complements.a
|
λmax (Δλmax)/ nm |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B = |
W |
X |
Y |
Z |
||||||||||
| ON | Sequence | ss | + DNA | + RNA | ss | + DNA | + RNA | ss | + DNA | + RNA | ss | + DNA | + RNA | |
| B1 | 5′-GBG ATA TGC | 348 | 350 [+2] | 349 [+1] | 349 | 351 [+2] | 351 [+2] | 350 | 351 [+1] | 352 [+2] | 346 | 348 [+2] | 348 [+2] | |
| B2 | 5′-GTG ABA TGC | 347 | 351 [+4] | 349 [+2] | 348 | 353 [+5] | 351 [+3] | 346 | 351 [+5] | 351 [+5] | 346 | 347 [+1] | 346 [±0] | |
| B3 | 5′-GTG ATA BGC | 348 | 351 [+3] | 350 [+2] | 350 | 354 [+4] | 351 [+1] | 348 | 351 [+3] | 351 [+3] | ND | ND | ND | |
| B4 | 3′-CAC BAT ACG | 348 | 350 [+2] | 348 [±0] | 348 | 351 [+3] | 349 [+1] | 350 | 351 [+1] | 351 [+1] | 346 | 347 [+1] | 346 [±0] | |
| B5 | 3′-CAC TAB ACG | 348 | 350 [+2] | 349 [+1] | 348 | 351 [+3] | 351 [+3] | 346 | 351 [+5] | 352 [+6] | 346 | 348 [+2] | 347 [+1] | |
Measurements were performed at room temperature on a spectrophotometer in the range 300-400 nm, using a quartz optical cell with a 1.0 cm path length and same conditions as for thermal denaturation experiments.
Figure 6.
Absorption spectra of W2 (left panel) and Y2 (right panel) and their duplexes with complementary DNA (D2) and RNA (R2) targets.
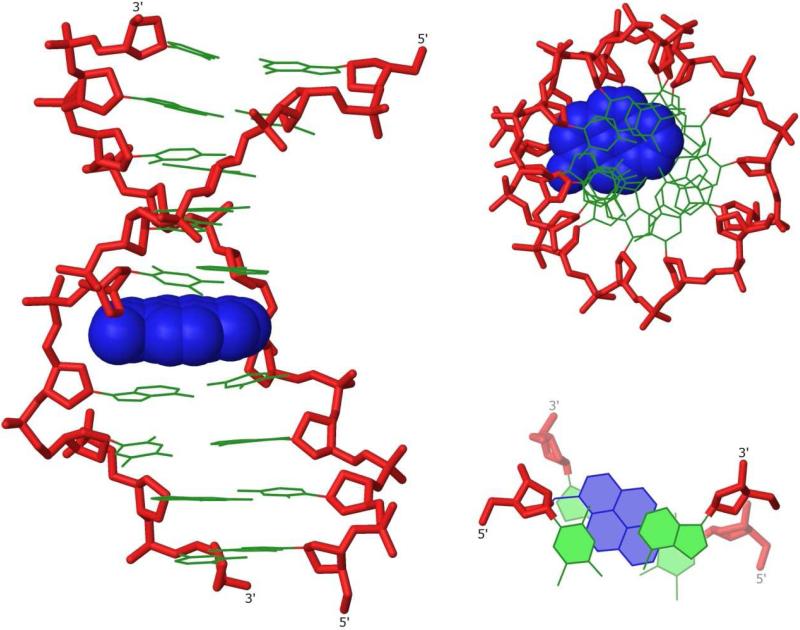
In full agreement with biophysical data, the lowest energy structure of the duplex between PyMe-α-L-amino-LNA W2 and complementary DNA D2 suggests precise intercalation of the pyrene moiety (Fig. 7). It is imperative to stress that the utilized simulation protocol did not initiate from a structure where the pyrene moiety was intercalated, i.e., the pyrene moiety moved from an extrahelical to an intercalated position during the simulation. As expected43 significant global unwinding, concomitant lengthening of the duplex and widening of the minor groove was observed upon intercalation. The pyrene moiety forms extensive π-π stacks with the nucleobase moieties of W5 and the 3′-flanking A6 and to a lesser extent with the nucleobase moieties of T13 and A14. The pseudorotational phase angle P and glycosidic torsion angle χ44 of PyMe-α-L-amino-LNA monomer W change little relative to the 2′-amino-α-L-LNA monomer in Q2:D2. However, P and χ of the adjacent A6 moiety increase markedly in response to intercalation (P from 97° to 127 °, and χ from −147 ° to −105 °, for Q2:D2 and W2:D2, respectively, Tables S7 and S8),29 to facilitate efficient π-π stacking between the pyrene and nucleobase moieties.
Figure 7.
Three representations of the lowest energy structure of W2:D2, side view (left), top view (upper right) and truncated top view showing W5A6:T13A14 (lower right). Coloring scheme as in Fig. 3 except that pyren-1-yl-methyl moiety of monomer W is in blue.
The lowest energy structures of duplexes between PyCO-α-L-amino-LNA X2 or PyAc-α-L-amino-LNA Y2 and complementary DNA D2 (Figs. 8, S8 and S9),29 exhibited similar key features as W2:D2, i.e., intercalation of the pyrene moiety and efficient π-π overlapping with flanking base pairs, similar sugar puckers and glycosidic torsion angles for X5/Y5 and A6, and unwinding of the duplex and widening of the grooves.29 Two minor structural differences observed with PyAc-α-L-amino-LNA duplex Y2:D2 (Figs. 8 and S9) relative to W2:D2 (Fig. 7) or X2:D2 (Figs. 8 and S8) included increased stacking interactions with T13 and A14 and an altered orientation of the pyrene moiety, i.e., H3py and H4py of monomer Y face the major groove while facing the minor groove in W2:D2 and X2:D2 (Fig. S10).29 Closer scrutiny of the molecular arrangement in PyMe/PyCO/PyAc-α-L-amino-LNA monomers W-Y reveals that the attachment points of the nucleobase and pyrene moieties (i.e., C1′ and N2′, respectively) are efficiently locked relative to each other (Fig. 9) as a consequence of the 2-oxo-5-azabicyclo[2.2.1]heptane skeleton. This, in concert with the short linker between the bicyclic skeleton and pyrene moiety and the strength of π-π stacking in aqueous environments, de facto directs the pyrene moiety of monomers W-Y into the duplex core to facilitate intercalation. This molecular arrangement leads to π-π stacking with the T5:A14 and A6:T13 base pairs and a reduction in buckle and propeller twist fluctuation in this nucleotide step (results not shown) to form a highly stablilized duplex segment. The observed thermodynamic data for PyMe/PyCO/PyAc-α-L-amino-LNA are in agreement with this preorganized binding mode of the pyrene as favorable entropic components were identified as important factors for duplex stabilization (Table 5). Desolvation upon intercalation of the highly apolar pyrene moiety is also likely to result in additional favorable entropic contributions upon duplex formation. Since the observed modeling structures of W2:D2, X2:D2 and Y2:D2 are very similar, it is likely that differential solvation of the single stranded probes or of their duplexes with DNA/RNA complements accounts for the observed differences in thermal affinity toward nucleic acid targets. For example, less pronounced desolvation of PyCO-α-L-amino-LNA X2 relative to PyMe-α-L-amino-LNA W2 upon hybridization to complementary DNA may account for less favorable entropy (fewer water molecules free upon hybridization) and more favorable enthalpy (formation of hydrogen bonds with surrounding water molecules).
Figure 8.
Side view representations of the lowest energy structures of X2:D2, Y2:D2 and Z2:D2 (non-intercalated binding mode), respectively. Coloring scheme as in Fig. 3 except that pyrene moieties are shown in blue.
Figure 9.
Illustration of directed positioning of pyrene moieties in duplex core by N2′-functionalized 2′-amino-α-L-LNA.
The binding mode of the pyrene moiety of PyBu-α-L-amino-LNA was expected to be more ambiguous since: a) Z1-Z5 exhibited lower increases in thermal affinity toward DNA complements in particular (Table 1), b) Z2 displayed markedly improved mismatch discrimination relative to PyMe/PyCO/PyAc-α-L-amino-LNA (Table 3), and c) more subtle hybridization-induced bathochromic shifts of pyrene absorption maxima were observed (Table 6). In full agreement with these biophysical observations, molecular modeling suggested at least two different binding modes. An intercalated binding mode was observed that exhibited the hallmarks described above for W2-Y2:D2 (Fig. S11). The model structure suggested that the long and relatively bulky butanoyl linker of PyBu-α-L-amino-LNA monomer Z: a) reduced π-π overlap between the pyrene and the nucleobase moieties of Z5 and A6 to a minimum, while increasing overlap with T13 and A14, b) was wedged into the duplex core in between Z5 and A6 to locally perturb the duplex and introduce a kink, and c) oriented the pyrene moiety with the H3pyr and H4pyr sides facing the major groove (Fig. S10).29
The second binding mode that is more in line with observations, the pyrene moiety is located at the floor of the major groove and is involved in non-specific contacts with the Hoogsteen faces of A6, C11, A12 and T13 (Figs. 8 and S12).29 Minor groove binders conjugated to ONs are well known to increase the strength and specificity of hybridization.45,46 By analogy, major groove binding of the pyrene moiety of monomer Z may explain the observed increased mismatch discrimination of Z2 (Table 3). Thus, biophysical characterization and computer simulations indicate that PyBu-α-L-LNA may stabilize duplexes with DNA/RNA complements by a wider variety of binding modes than PyMe/PyCO/PyAc-α-L-LNA exhibiting shorter linkers between the bicyclic skeleton and pyrene moiety.
Conclusion
Herein we demonstrate that the 2-oxo-5-azabicyclo[2.2.1]heptane skeleton of 2’-amino-α-L-LNA, in concert with short linkers, directs intercalators appended to the N2’-position very effectively to the nucleic acid cores. Consequently, dramatic increases in thermal affinity toward DNA complements of up to +19.5 °C per modification are observed. Directed positioning of intercalators inside nucleic acid duplex cores has many potential interesting applications within nucleic acid based diagnostics, therapeutics and nanotechnology,47 including detection of DNA/RNA complements and/or single nucleotide polymorphisms by fluorescence,25b,30a,48-50 study of charge transfer processes,51 formation of metal ion arrays within nucleic acid duplex cores,52,53 or development of artificial nucleases.54 Unlike previously reported building blocks, N2′-intercalator-functionalized 2′-amino-α-L-LNA effectively combines high-affinity hybridization with DNA complements and precise positioning of intercalators in nucleic acid duplexes. We propose N2’-intercalator-modified 2’-amino-α-L-LNA monomers as highly valuable monomers for established and emerging DNA-targeting applications.
Experimental Section
(1S,3R,4S,7R)-1-(4,4′-Dimethoxytrityloxymethyl)-5-ethyl-7-hydroxy-3-(thymin-1-yl)-2-oxa-5-azabicyclo[2.2.1]heptane 2S
Amino alcohol 1 (0.40 g, 0.70 mmol) was coevaporated with anhydrous 1,2-dichloroethane (2 × 8 mL) and dissolved in anhydrous 1,2-dichloroethane (8 mL). To this were added NaBH(OAc)3 (230 mg, 1.09 mmol) and CH3CHO (44 μL, 0.78 mmol) and after stirring the reaction mixture at rt for 40 h, it was diluted with EtOAc (35 mL) and washed with sat. aq. NaHCO3 (2 × 15 mL). The organic phase was evaporated to dryness and the resulting residue purified by silica gel column chromatography (0-5% i-PrOH in CH2Cl2, v/v) to afford nucleoside 2S (200 mg, 48%). Rf = 0.5 (10% MeOH in CH2Cl2, v/v); MALDI-HRMS m/z 622.2524 ([M + Na]+, C34H37N3O7Na+ Calcd 622.2522); 1H NMR (DMSO-d6)55 δ 11.26 (s, 1H, ex, NH), 7.49 (s, 1H, H-6), 7.21-7.44 (m, 9H, Ar), 6.87-6.92 (d, 4H, J = 8.8 Hz, Ar), 5.91 (d, 1H, J = 1.5 Hz, H-1′), 5.70 (d, 1H, ex, J = 3.5 Hz, 3′-OH), 4.31 (d, 1H, J = 3.5 Hz, H-3′), 3.74 (s, 6H, 2 × CH3O), 3.19-3.30 (m, 4H, H-2′, 2 × H-5′, H-5′′), 2.64-2.83 (m, 3H, CH2CH3, H-5′′), 1.83 (s, 3H, CH3Ar), 0.82 (t, 3H, J = 7.3 Hz, CH3). 13C NMR (DMSO-d6) δ 163.8, 158.0, 150.3, 144.8, 137.4, 135.4, 135.3, 129.7, 127.8, 127.6, 126.6, 113.1, 105.9, 90.0, 85.0, 74.4, 65.4, 60.8, 58.3, 54.9, 49.5, 14.8, 12.2; Anal. Calc. for C34H37N3O7: C, 68.10; H, 6.22; N, 7.01; Found: C, 67.96; H, 6.37; N, 6.54. Calcd with 1/8 i-PrOH: C, 68.00; H, 6.31; N, 6.92.
(1S,3R,4S,7R)-1-(4,4′-Dimethoxytrityloxymethyl)-7-hydroxy-5-(pyren-1-yl)carbonyl-3-(thymin-1-yl)-2-oxa-5-azabicyclo[2.2.1]heptane 2X
1-Pyrenylcarboxylic acid (162 mg, 0.65 mmol), O-(7-Azabenzotriazole-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HATU, 183 mg, 0.48 mmol) and N,N′-diisopropylethylamine (0.19 mL, 1.1 mmol) were dissolved in anhydrous DMF (4.2 mL) and the mixture was allowed to stir for 6 h at rt. To this was added a solution of nucleoside 1 (0.25 g, 0.44 mmol), which had been dried by coevaporation with anhydrous toulene (2 × 10 mL) ahead of time, dissolved in anhydrous DMF (4.2 mL). After stirring at rt for 12 h, the reaction mixture was diluted with CH2Cl2 (50 mL), washed with sat. aq. NaHCO3 (10 mL) and H2O (4 × 10 mL). The aqueous phase was back extracted with CH2Cl2 (2 × 30 mL), and the combined organic phase was evaporated to dryness, and resulting crude residue adsorbed on silica gel and purified by silica gel column chromatography (0-4% MeOH in CH2Cl2, v/v) to afford a rotameric mixture (~1:1.4 by 1H NMR) of nucleoside 2X as a white solid material (0.32 g, 90%). Rf = 0.2 (50% acetone in petroleum ether, v/v), MALDI-HRMS m/z 822.2786 ([M + Na]+, C49H41N3O8·Na+ Calcd 822.2746; Selected signals 1H NMR (DMSO-d6)56 δ 6.47 (d, 1H, ex, J = 3.7 Hz, 3′-OHB), 6.43 (d, 1.4H, ex, J = 3.8 Hz, 3′-OHA), 6.25 (s, 1H, H-1′B), 5.80 (s, 1.4H, H-1′A), 4.79 (d, 1H, J = 3.7 Hz, H-3′B), 4.56 (d, 1.4H, J = 3.8 Hz, H-3′A), 2.05 (s, 4.2H, CH3-A), 1.92 (s, 3H, CH3-A); 13C NMR (DMSO-d6) δ 169.6, 169.5, 164.2, 163.8, 158.3, 158.1, 150.3, 149.6, 144.8, 144.6, 135.4, 135.3, 135.2, 135.1, 134.6, 131.6, 131.4, 130.7, 130.5, 130.25, 130.18, 129.9, 129.8, 128.8, 128.53, 128.46, 128.1, 127.9, 127.8, 127.7, 127.3, 127.2, 126.92, 126.86, 126.8, 126.4, 126.2, 126.1, 125.9, 124.9, 124.5, 124.2, 123.8, 123.7, 123.6, 113.4, 113.2, 109.1, 108.5, 89.0, 88.8, 86.0, 85.6, 85.4, 72.4, 71.5, 64.9, 62.0, 60.3, 59.6, 55.1, 55.0, 53.9, 52.1, 12.6, 12.5; Anal. Calc. for C49H41N3O8·1 H2O: C, 71.96; H, 5.30; N, 5.14; Found: C, 71.63; H, 4.95; N, 4.77.
(1S,3R,4S,7R)-7-[2-Cyanoethoxy(diisopropylamino)phosphinoxy]-1-(4,4′-dimethoxytrityloxymethyl)-5-ethyl-3-(thymin-1-yl)-2-oxa-5-azabicyclo[2.2.1]heptane 3S
Nucleoside 2S (190 mg, 0.32 mmol) was coevaporated with anhydrous 1,2-dichloroethane (2 × 5 mL) and dissolved in a mixture of anhydrous EtN(i-Pr)2 in CH2Cl2 (2 mL, 20%, v/v). To this was added 2-cyanoethyl N,N′-(diisopropyl)phosphoramidochloridite (0.14 mL, 0.63 mmol) and the reaction mixture was stirred at rt for 1 h, whereupon it was diluted with CH2Cl2 (20 mL). The organic phase was washed with sat. aq. NaHCO3 (10 mL) and the aqueous phase back-extracted with CH2Cl2 (25 mL). The combined organic phase was evaporated to dryness and the resulting residue purified by silica gel column chromatography (0-50% EtOAc in petroleum ether, v/v) to afford amidite 3S (160 mg, 63%) as a white solid material. Rf = 0.5 (70% EtOAc in petroleum ether, v/v); MALDI-HRMS m/z 822.3636 ([M + Na]+, C43H54N5O8P·Na+ Calcd 822.3602); 31P NMR (CH3CN + DMSO-d6) δ 149.8, 147.9.
(1S,3R,4S,7R)-7-[2-Cyanoethoxy(diisopropylamino)phosphinoxy]-1-(4,4′-dimethoxytrityloxymethyl)-5-(pyren-1-yl)carbonyl-3-(thymin-1-yl)-2-oxa-5-azabicyclo[2.2.1]heptane 3X
Nucleoside 2X (0.31 g, 0.39 mmol) was dried by coevaporation with anhydrous 1,2-dichloroethane (2 × 5 mL) and dissolved in anhydrous CH2Cl2 (10 mL). To this was added N,N′-diisopropylammonium tetrazolide (112 mg, 0.66 mmol) and bis(N,N′-diisopropylamino)-2-cyanoethoxyphosphine (0.21 mL, 0.66 mmol) and the reaction mixture was stirred at rt for 12 h, whereupon it was diluted with CH2Cl2 (20 mL), washed with sat. aq. NaHCO3 (10 mL) and brine (10 mL). The aqueous phase was back extracted with CH2Cl2 (30 mL) and the combined organic phase was evaporated to dryness and the resulting residue purified by silica gel column chromatography (0-50% acetone in petroleum ether, v/v) to afford amidite 3X as a white solid material (276 mg, 71%). Rf = 0.5 (50% acetone in petroleum ether, v/v); MALDI-HRMS m/z 1022.3864 ([M + Na]+, C58H58N5O9PNa+ Calcd. 1022.3852; 31P NMR (CH3CN + DMSO-d6) δ 154.2, 153.8, 153.3, 151.9.
Protocol for synthesis of ONs
ONs containing 2′-amino-α-L-LNA monomers Q-Z (see Fig. 1 for structures) were synthesized on a 0.2 μmol scale using succinyl linked LCAA-CPG (long chain alkyl amine controlled pore glass) columns with a pore size of 500 Å on an automated DNA synthesizer. Synthesis of α-L-LNA and 2′-amino-α-L-LNA was performed as previously described.15b,24b For the incorporation of the N2′-functionalized 2′-amino-α-L-LNA monomers (S-Z), standard procedures were used, i.e., trichloroacetic acid in CH2Cl2 as a detrytilation reagent; 0.25 M 4,5-dicyanoimidazole (DCI) in CH3CN as activator; acetic anhydride in THF as cap A solution; 1-methylimidazole in THF as cap B solution, and 0.02 M iodine in H2O/pyridine/THF as the oxidizing solution. Extended coupling times were used for phosphoramidites 3S (10 min), 3V (10 min), 3W (30 min), 3Y (30 min), 3X (30 min), 3Z (15 min) using 1H-tetrazole as catalyst resulted in stepwise coupling yields of ~99% for monomers S, V, X, Y, Z and ~95% for monomer W. Coupling yields were determined by trityl monitoring. Removal of the nucleobase protecting groups of ONs and cleavage from solid support was accomplished using standard conditions (32% aq. ammonia for 12-16 h at 55 °C). Unmodified DNA and RNA strands were obtained from commercial suppliers and, if necessary, further purified as described below.
Purification of all modified ONs (till minimum 80% purity) was performed by two different methods: a) if overall yield > 90%: precipitation of crude ONs (DMT-OFF, abs. EtOH, −18 °C, 12-16 h, followed by washing with abs. EtOH (2 × 300 μL), b) purification of the ONs (DMT-ON) by RP-HPLC using the system described below, followed by detritylation (80% aq. AcOH, 20 min, rt) and precipitation/washing as outlined above. Purification of crude ONs (DMT-ONs) was performed using a HPLC system equipped with an Xterra MS C18 (10μm, 7.8×10mm) pre-column and an Xterra MS C18 (10μm, 7.8×150mm) column using the representative gradient protocol depicted in Table S1. The composition of all synthesized ONs were verified by MALDI-MS analysis (Table S3) recorded in negative ion mode using 3-hydroxypicolinic acid as a matrix, whereas the purity (>80%) was verified by ion-exchange HPLC system equipped with a Dionex PA100 column (4 × 250 mm) at pH 8 using the representative protocol shown in Table S2.
Protocol thermal denaturation studies
Concentrations of ONs were estimated using the following extinction coefficients for DNA (OD/μmol): G (12.01), A (15.20), T (8.40), C (7.05); for RNA (OD/μmol): G (13.70), A (15.40), U (10.00), C (9.00); and for pyrene (22.4). ONs (1.0 μM each strand) were thoroughly mixed, denatured by heating and subsequently cooled to the starting temperature of the experiment. Quartz optical cells with a pathlength of 1.0 cm were used. Thermal denaturation temperatures (Tm values/°C) were measured on a UV/VIS spectrometer equipped with a Peltier temperature programmer and determined as the maximum of the first derivative of the thermal denaturation curve (A260 vs. temperature) recorded in medium salt buffer (Tm-buffer; 100 mM NaCl, 0.1 mM EDTA and pH 7.0 adjusted with 10 mM NaH2PO4/5 mM Na2HPO4). For studies evaluating the dependence of Tm on ionic strength, Tm values were also determined in low and high salt buffers (composition as for medium salt buffer except that 0 mM and 700 mM NaCl were used, respectively). The temperature of the denaturation experiments ranged from at least 15 °C below Tm to 20 °C above Tm (although not below 1 °C). A temperature ramp of 1.0 °C/min was used in all experiments. Reported thermal denaturation temperatures are an average of two measurements within ±1.0 °C.
Protocol for determination of thermodynamic parameters
Thermodynamic parameters were obtained by analysis of the melting curves used to determine Tm values assuming bimolecular reactions and two state equilibrium hypothesis, using software accompanying the utilized UV/VIS spectrometer. The graphs of ln Ka (affinity constant) as a function of 1/T were approximated with straight lines facilitating parameter determination (ΔG, ΔH and ΔS, Tables 4 and 5). Quality of the baseline permitting, thermodynamic parameters for two melting curves per investigated duplex were determined, and an average value was listed. The changes in Gibbs free energy, ΔG, were determined at temperatures close to the Tm-value of the investigated duplexes to minimize errors (T = 298 K).
Supplementary Material
Acknowledgement
We greatly appreciate financial support from Idaho NSF EPSCoR, the BANTech Center at the University of Idaho, a University of Idaho Research Office and Research Council Seed Grant, NIH Grant Number P20 RR016448 from the COBRE Program of the National Center for Research Resources, and The Danish National Research Foundation. The Ph.D. school Nucleic Acid Based Drug Design (NAC DRUG) supported by the Danish Agency for Science Technology and Innovation is gratefully acknowledged. The Hrdlicka research team is thanked for proof-reading of the manuscript preparation.
Footnotes
Supporting Information Available: General experimental section; experimental section for synthesis of nucleosides 2 and 3 (V/W/Y/Z-series); copies of 1H NMR, 13C NMR, 31P NMR, 1H-1H COSY and/or 1H-13C HETCOR spectra for all novel nucleosides; protocols for RP-HPLC and ion exchange HPLC purification of ONs; MALDI-MS of synthesized ONs (Table S3); representative thermal denaturation curves (Fig. S1); Tm-values of N2′-functionalized 2′-amino-α-L-LNA at various ionic strengths (Table S4); discussion of sequence dependent variation of Tm-values; DNA-selectivity of N2′-functionalized 2′-amino-α-L-LNA (Table S5); thermodynamic data for B1-B5 (Table S6); protocol for acquisition and listing of additional CD-spectra (Fig. S2); protocol for molecular modeling studies; additional molecular modeling structures (Figs. S3-S9 and S11-S12), illustration of intercalation (Fig. S10); pseudorotational phase angle P and glycosidic torsion angle χ observed in simulated duplexes (Tables S7 and S8). This material is available free of charge via the Internet at http://pubs.acs.org.
References and Footnotes
∥A research center funded by the Danish National Research Foundation for studies on nucleic acid chemical biology.
- 1.a Duca M, Vekhoff P, Oussedik K, Halby L, Arimondo PB. Nucleic Acids Res. 2008;36:5123–5138. doi: 10.1093/nar/gkn493. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Simon P, Cannata F, Concordet J-P, Giovannangeli C. Biochimie. 2008;90:1109–1116. doi: 10.1016/j.biochi.2008.04.004. [DOI] [PubMed] [Google Scholar]; c Kurreck J. Eur. J. Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]; d Corey DR. J. Clin. Invest. 2007;117:3615–3622. doi: 10.1172/JCI33483. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Watts JK, Deleavey GF, Damha MJ. Drug Disc. Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 2.a Asseline U. Curr. Opin. Chem. 2006;10:491–518. [Google Scholar]; b Ranasinghe RT, Brown T. Chem. Commun. 2005:5487–5502. doi: 10.1039/b509522k. [DOI] [PubMed] [Google Scholar]
- 3.For representative reviews see: Wengel J. Org. Biomol. Chem. 2004;2:277–280. doi: 10.1039/b313986g., Gothelf KV, LaBean TH. Org. Biomol. Chem. 2005;3:4023–4037. doi: 10.1039/b510551j., Clever GH, Kaul C, Carell T. Angew. Chem. Int. Ed. 2007;46:6226–6236. doi: 10.1002/anie.200701185.
- 4.For reviews see Meldgaard M, Wengel J. J. Chem. Soc. Perkin Trans. 1. 2000:3539–3554., Leumann CJ. Bioorg. Med. Chem. 2002;10:841–854. doi: 10.1016/s0968-0896(01)00348-0.
- 5.For recent representative examples see Morita K, Takagi M, Hasegawa C, Kaneko M, Tsutsumi S, Sone J, Ishikawa T, Imanishi T, Koizumi M. Bioorg. Med. Chem. 2003;11:2211–2226. doi: 10.1016/s0968-0896(03)00115-9., Albæk N, Petersen M, Nielsen P. J. Org. Chem. 2006;71:7731–7740. doi: 10.1021/jo061225g., Honcharenko D, Vargese OP, Plashkevych O, Barman J, Chattopadhyaya J. J. Org. Chem. 2006;71:299–314. doi: 10.1021/jo052115x., Varghese OP, Barman J, Pathmasiri W, Plaskevych O, Honcharenko D, Chattopadhyaya J. J. Am . Chem. Soc. 2006;128:15173–15187. doi: 10.1021/ja0634977., Hari Y, Obika S, Ohnishi R, Eguchi K, Osaki T, Ohishi H, Imanishi T. Bioorg. Med. Chem. 2006;14:1029–1038. doi: 10.1016/j.bmc.2005.09.020., Plashkevych O, Chatterjee S, Honcharenko D, Pathmasiri W, Chattopadhyaya J. J. Org. Chem. 2007;72:4716–4726. doi: 10.1021/jo070356u., Srivastava P, Barman J, Pathmasiri W, Plaskevych O, Wenska M, Chattopadhyaya J. J. Am. Chem. Soc. 2007;129:8362–8379. doi: 10.1021/ja071106y., Sabatino D, Damha MJ. J. Am. Chem. Soc. 2007;129:8259–8270. doi: 10.1021/ja071336c., Abdur Rahman SM, Seki S, Obika S, Yoshikawa H, Miyashita K, Imanishi T. J. Am. Chem. Soc. 2008;130:4886–4896. doi: 10.1021/ja710342q., Enderlin G, Nielsen P. J. Org. Chem. 2008;73:6891–6894. doi: 10.1021/jo801081t.
- 6.We define LNA, α-L-LNA, 2′-amino-LNA and 2′-amino-α-L-LNA as a oligonucleotide containing one or more 2′-O,4′-C-methylene-β-d-ribofuranosyl monomer(s), 2′-O,4′-C-methylene-α-l-ribofuranosyl monomer(s), 2′-amino-2′-deoxy-2′-N,4′-C-methylene-β-d-ribofuranosyl monomer(s) or 2′-amino-2′-deoxy-2′-N,4′-C-methylene-α-l-ribofuranosyl monomer(s), respectively. Similar definitions are used for N2′-functionalized α-L-LNA derivatives.
- 7.a Singh SK, Nielsen P, Koshkin AA, Wengel J. Chem. Commun. 1998:455–456. [Google Scholar]; b Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. Tetrahedron. 1998;54:3607–3630. [Google Scholar]; c Wengel J. Acc. Chem. Res. 1999;32:301–310. [Google Scholar]
- 8.Obika S, Nanbu D, Hari Y, Andoh J, Morio K, Doi T, Imanishi T. Tetrahedron Lett. 1998;39:5401–5404. [Google Scholar]
- 9.Petersen M, Wengel J. Trends Biotechnol. 2003;21:74–81. doi: 10.1016/S0167-7799(02)00038-0. [DOI] [PubMed] [Google Scholar]
- 10.Kaur H, Babu BR, Maiti S. Chem. Rev. 2007;107:4672–4697. doi: 10.1021/cr050266u. [DOI] [PubMed] [Google Scholar]
- 11.Jepsen JS, Wengel J. Curr. Opin. Drug Discovery Dev. 2004;7:188–194. [PubMed] [Google Scholar]
- 12.Frieden M, Ørum H. IDrugs. 2006;9:706–711. [PubMed] [Google Scholar]
- 13.Grünweller A, Hartmann RK. Biodrugs. 2007;21:235–243. doi: 10.2165/00063030-200721040-00004. [DOI] [PubMed] [Google Scholar]
- 14.Stenvang J, Silahtaroglu AN, Lindow M, Elmen J, Kauppinen S. Sem. Cancer Biol. 2008;18:89–102. doi: 10.1016/j.semcancer.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 15.a Rajwanshi VK, Håkansson AE, Dahl BM, Wengel J. Chem. Commun. 1999:1395–1396. [Google Scholar]; b Sørensen MD, Kværnø L, Bryld T, Håkansson AE, Verbeure B, Gaubert G, Herdewijn P, Wengel J. J. Am. Chem. Soc. 2002;124:2164–2176. doi: 10.1021/ja0168763. [DOI] [PubMed] [Google Scholar]
- 16.Arzumanov A, Stetsenko DA, Malakhov AD, Techelt S, Sørensen MD, Babu BR, Wengel J, Gait MJ. Oligonucleotides. 2003;13:435–453. doi: 10.1089/154545703322860762. [DOI] [PubMed] [Google Scholar]
- 17.Frieden M, Christen SM, Mikkelsen ND, Rosenbohm C, Thrue CA, Westergaard M, Hansen HF, Ørum H, Koch T. Nucleic Acids Res. 2003;31:6365–6372. doi: 10.1093/nar/gkg820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fluiter K, Frieden M, Vreijling J, Rosenbohm C, De Wissel MB, Christensen SM, Koch T, Ørum H, Baas F. ChemBioChem. 2005;6:1104–1109. doi: 10.1002/cbic.200400419. [DOI] [PubMed] [Google Scholar]
- 19.Kumar N, Nielsen KE, Maiti S, Petersen M. J. Am. Chem. Soc. 2006;128:14–15. doi: 10.1021/ja055483r. [DOI] [PubMed] [Google Scholar]
- 20.Vester B, Hansen LH, Lundberg LB, Babu BR, Sørensen MD, Wengel J, Doubtwaite S. BMC Mol. Biol. 2006;7:19. doi: 10.1186/1471-2199-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crinelli R, Bianchi M, Gentilini L, Palma L, Sørensen MD, Bryld T, Babu BR, Arar K, Wengel J, Magnani M. Nucleic Acids Res. 2004;32:1874–1885. doi: 10.1093/nar/gkh503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh SK, Kumar R, Wengel J. J. Org. Chem. 1998;63:10035–10039. doi: 10.1021/jo9806658. [DOI] [PubMed] [Google Scholar]
- 23.a Sørensen MD, Petersen M, Wengel J. Chem. Commun. 2003:2130–2131. doi: 10.1039/b307026c. [DOI] [PubMed] [Google Scholar]; b Hrdlicka PJ, Babu BR, Sørensen MD, Wengel J. Chem. Commun. 2004:1478–1479. doi: 10.1039/b404446k. [DOI] [PubMed] [Google Scholar]; c Babu BR, Hrdlicka PJ, McKenzie CJ, Wengel J. Chem. Commun. 2005:1705–1707. doi: 10.1039/b417101b. [DOI] [PubMed] [Google Scholar]; d Hrdlicka PJ, Babu BR, Sørensen MD, Harrit N, Wengel J. J. Am. Chem. Soc. 2005;127:13293–13299. doi: 10.1021/ja052887a. [DOI] [PubMed] [Google Scholar]; e Lindegaard D, Babu BR, Wengel J. Nucleosides Nucleotides Nucleic Acids. 2005;24:679–681. doi: 10.1081/ncn-200060247. [DOI] [PubMed] [Google Scholar]; f Bramsen JB, Laursen MB, Damgaard CK, Lena SW, Babu BR, Wengel J, Kjems J. Nucleic Acids Res. 2007;35:5886–5897. doi: 10.1093/nar/gkm548. [DOI] [PMC free article] [PubMed] [Google Scholar]; g Kalek M, Madsen AS, Wengel J. J. Am. Chem. Soc. 2007;129:9392–9400. doi: 10.1021/ja071076z. [DOI] [PubMed] [Google Scholar]; h Umemoto T, Hrdlicka PJ, Babu BR, Wengel J. ChemBioChem. 2007;8:2240–2248. doi: 10.1002/cbic.200700408. [DOI] [PubMed] [Google Scholar]; i Lindegaard D, Madsen AS, Astakhova IV, Malakhov AD, Babu BR, Korshun VA, Wengel J. Bioorg. Med. Chem. 2008;16:94–99. doi: 10.1016/j.bmc.2007.04.056. [DOI] [PubMed] [Google Scholar]; j Kalek M, Benediktson P, Vester B, Wengel J. Chem. Commun. 2008:762–764. doi: 10.1039/b712532a. [DOI] [PubMed] [Google Scholar]; k Østergaard ME, Maity J, Wengel J, Hrdlicka PJ. Abstracts of Papers, 235th ACS National Meeting; New Orleans. 2008; BIOL-032. [Google Scholar]
- 24.a Hrdlicka PJ, Kumar TS, Wengel J. Nucleosides Nucleotides Nucleic Acids. 2005;24:1101–1104. doi: 10.1080/15257770500276866. [DOI] [PubMed] [Google Scholar]; b Kumar TS, Madsen AS, Wengel J, Hrdlicka PJ. J. Org. Chem. 2006;71:4188–4201. doi: 10.1021/jo060331f. [DOI] [PubMed] [Google Scholar]
- 25.a Hrdlicka PJ, Kumar TS, Wengel J. Chem. Commun. 2005:4279–4281. doi: 10.1039/b506986f. [DOI] [PubMed] [Google Scholar]; b Kumar TS, Wengel J, Hrdlicka PJ. ChemBioChem. 2007;8:1122–1125. doi: 10.1002/cbic.200700144. [DOI] [PubMed] [Google Scholar]; c Andersen NK, Wengel J, Hrdlicka PJ. Nucleosides Nucleotides Nucleic Acids. 2007;26:1415–1417. doi: 10.1080/15257770701539153. [DOI] [PubMed] [Google Scholar]; d Kumar TS, Madsen AS, Østergaard ME, Wengel J, Hrdlicka PJ. J. Org. Chem. 2008;73:7060–7066. doi: 10.1021/jo800551j. [DOI] [PubMed] [Google Scholar]
- 26.Preliminary results have been briefly outlined in Kumar TS, Madsen AS, Wengel J, Hrdlicka PJ. Nucleosides Nucleotides Nucleic Acids. 2007;26:1403–1405. doi: 10.1080/15257770701538841.
- 27.Abdel-Magid AF, Carson KG, Harris BD, Maryanoff CA, Shah RD. J. Org. Chem. 1996;61:3849–3862. doi: 10.1021/jo960057x. [DOI] [PubMed] [Google Scholar]
- 28.Pedersen DS, Rosenbohm C, Koch T. Synthesis. 2002:802–808. [Google Scholar]
- 29.See Supporting information.
- 30.a Christensen UB, Pedersen EB. Nucleic Acids Res. 2002;30:4918–4925. doi: 10.1093/nar/gkf624. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Filichev VV, Hilmy KMH, Christensen UB, Pedersen UB. Tetrahedron Lett. 2004;45:4907–4910. [Google Scholar]
- 31.Yamana K, Iwase R, Furutani S, Tsuchida H, Zako H, Yamaoka T, Murakami A. Nucleic Acids Res. 1999;27:2387–2392. doi: 10.1093/nar/27.11.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bryld T, Højland T, Wengel J. Chem. Comm. 2004:1064–1065. doi: 10.1039/b402414a. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen KME, Petersen M, Håkansson AE, Wengel J, Jacobsen JP. Chem. Eur. J. 2002;8:3001–3009. doi: 10.1002/1521-3765(20020703)8:13<3001::AID-CHEM3001>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 34.a Weiner SJ, Kollman PA, Case DA, Singh UC, Ghio C, Alagona G, Profeta S, Weiner P. J. Am. Chem. Soc. 1984;106:765–784. [Google Scholar]; b Weiner SJ, Kollman PA, Nguyen DT, Case DA. J. Comp. Chem. 1986;7:230–252. doi: 10.1002/jcc.540070216. [DOI] [PubMed] [Google Scholar]
- 35.Still WC, Tempczyk A, Hawley RC, Hendrickson T. J. Am. Chem. Soc. 1990;112:6127–6129. [Google Scholar]
- 36.MacroModel version 9.1. S., LLC; New York, NY: 2005. [Google Scholar]
- 37.Egli M, Tereshko V, Teplova M, Minasov G, Joachimiak A, Sanishvili R, Weeks CM, Miller R, Maier MA, An H, Cook PD, Manoharan M. Biopolymers. 1998;48:234–252. doi: 10.1002/(SICI)1097-0282(1998)48:4<234::AID-BIP4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 38.Kielkopf CL, Ding S, Kuhn P, Rees DC. J. Mol. Biol. 2000;296:787–801. doi: 10.1006/jmbi.1999.3478. [DOI] [PubMed] [Google Scholar]
- 39.a Korshun VA, Stetsenko DA, Gait MJ. J. Chem. Soc. Perkin Trans. 1. 2002:1092–1104. [Google Scholar]; b Dioubankova NN, Malakhov AD, Stetsenko DA, Gait MJ, Volynsky PE, Efremov RG, Korshun VA. ChemBioChem. 2003;4:841–847. doi: 10.1002/cbic.200300678. [DOI] [PubMed] [Google Scholar]; c Kalra N, Babu BR, Parmar VS, Wengel J. Org. Biomol. Chem. 2004;2:2885–2887. doi: 10.1039/B411626G. [DOI] [PubMed] [Google Scholar]; d Donho C, Saito I. ChemBioChem. 2005;6:1075–1081. doi: 10.1002/cbic.200400325. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura M, Fukunaga Y, Sasa K, Ohtoshi Y, Kanaori K, Hayashi H, Nakano H, Yamana K. Nucleic Acids Res. 2005;33:5887–5895. doi: 10.1093/nar/gki889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dougherty G, Pilbrow JR. Int. J. Biochem. 1984;16:1179–1192. doi: 10.1016/0020-711x(84)90215-5. [DOI] [PubMed] [Google Scholar]
- 42.Okamoto A, Kanatani K, Saito I. J. Am. Chem. Soc. 2004;126:4820–4827. doi: 10.1021/ja039625y. [DOI] [PubMed] [Google Scholar]
- 43.a Wang AH-J, Ughetto G, Quigley GJ, Rich A. Biochemistry. 1987;26:1152–1163. doi: 10.1021/bi00378a025. [DOI] [PubMed] [Google Scholar]; b Spielmann HP, Wemmer DE, Jacobsen JP. Biochemistry. 1995;34:8542–8553. doi: 10.1021/bi00027a004. [DOI] [PubMed] [Google Scholar]; c Gallego J, Reid BR. Biochemistry. 1999;38:15104–15115. doi: 10.1021/bi9915869. [DOI] [PubMed] [Google Scholar]; d Mukherjee A, Lavery R, Bagchi B, Hynes JT. J. Am. Chem. Soc. 2008;130:9747–9755. doi: 10.1021/ja8001666. [DOI] [PubMed] [Google Scholar]
- 44.Following definitions of torsion angles are used: χ (O4′-C1′-N1-C2 or O4′-C1′-N9-C4 for pyrimidines or purines, respectively). The pseudorotation phase angle P is given as tan P = (ν4+ν1)-ν3+ν0) / [2ν2 (sin 36° + sin 72°)], where ν0 (C4′-O4′-C1′-C2′),ν1 (O4′-C1′-C2′-C3′), ν2 (C1′-C2′-C3′-C4′), ν3 (C2′-C3′-C4′-O4′) and ν4 (C3′-C4′-O4′-C1′). For further information see Saenger W. Principles of Nucleic Acid Structure. Springer-Verlag; Berlin: 1984.
- 45.Afonina I, Kutyavin I, Lukhtanov E, Meyer RB, Gamper H. Proc. Natl. Acad. Sci. USA. 1996;93:3199–3204. doi: 10.1073/pnas.93.8.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kutyavin IV, Afonina IA, Mills A, Gorn VV, Lukhtanov EA, Belousov ES, Singer MJ, Walburger DK, Lokhov SG, Gall AA, Dempcy R, Reed MW, Meyer RB, Hedgpeth J. Nucleic Acids Res. 2000;28:655–661. doi: 10.1093/nar/28.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Persil Ö, Hud NV. Trends Biotechnol. 2007;25:433–436. doi: 10.1016/j.tibtech.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 48.Wilson JN, Kool ET. Org. Biomol. Chem. 2006;4:4265–4274. doi: 10.1039/b612284c. [DOI] [PubMed] [Google Scholar]
- 49.Köhler O, Jarikote DV, Seitz O. ChemBioChem. 2005;6:69–77. doi: 10.1002/cbic.200400260. [DOI] [PubMed] [Google Scholar]
- 50.Benvin AL, Creeger Y, Fisher GW, Ballou B, Wagonner AS, Armitage BA. J. Am. Chem. Soc. 2007;129:2025–2034. doi: 10.1021/ja066354t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shao F, Augustyn K, Barton JK. J. Am. Chem. Soc. 2005;127:17445–17452. doi: 10.1021/ja0563399. [DOI] [PubMed] [Google Scholar]
- 52.Clever GH, Kaul C, Carell T. Angew. Chem. Int. Ed. 2007;46:6226–6236. doi: 10.1002/anie.200701185. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka K, Shionoya M. Coord. Chem. Rev. 2007;251:2732–2742. [Google Scholar]
- 54.Nakano S-I, Uotani Y, Uenishi K, Fujii M, Sugimoto N. J. Am. Chem. Soc. 2005;127:518–519. doi: 10.1021/ja045445s. [DOI] [PubMed] [Google Scholar]
- 55.Assignments of 1H NMR signals of H5′ and H5′′ (and of the corresponding 13C signals) may in principle be interchanged.
- 56.The integral of the H1′-signal of the least predominant rotamer (termed B) is set to 1.0.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.